Abstract
Infiltration of monocyte-derived macrophages plays a crucial role in cardiac remodeling and dysfunction. The serum and glucocorticoid-inducible protein kinase 3 (SGK3) is a downstream factor of PI3K signaling, regulating various biological processes via an AKT-independent signaling pathway. SGK3 has been implicated in cardiac remodeling. However, the contribution of macrophagic SGK3 to hypertensive cardiac remodeling remains unclear. A cardiac remodeling model was established by angiotensin II (Ang II) infusion in SGK3-Lyz2-CRE (f/f, +) and wild-type mice to assess the function of macrophagic SGK3. Additionally, a co-culture system of SGK3-deficient or wild-type macrophages and neonatal rat cardiomyocytes (CMs) or neonatal rat fibroblasts (CFs) was established to evaluate the effects of SGK3 and the underlying mechanisms. SGK3 levels were significantly elevated in both peripheral blood mononuclear cells and serum from patients with heart failure. Macrophage SGK3 deficiency attenuated Ang II–induced macrophage infiltration, myocardial hypertrophy, myocardial fibrosis, and mitochondrial oxidative stress. RNA sequencing suggested Ndufa13 as the candidate gene in the effect of SGK3 on Ang II–induced cardiac remolding. Downregulation of Ndufa13 in CMs and CFs prevented the suppression of cardiac remodeling caused by SGK3 deficiency in macrophages. Mechanistically, the absence of SGK3 led to a reduction in IL-1β secretion by inhibiting the NLRP3/Caspase-1/IL-1β pathway in macrophages, consequently suppressing upregulated Ndufa13 expression and mitochondrial oxidative stress in CMs and CFs. This study provides new evidence that SGK3 is a potent contributor to the pathogenesis of hypertensive cardiac remodeling, and targeting SGK3 in macrophages may serve as a potential therapy for cardiac remodeling.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sustained hypertension is a significant risk factor for cardiac remodeling, which ultimately leads to heart failure (HF) and increased mortality. There is evidence that inflammation has a vital effect on the progression of cardiovascular diseases. Infiltration of monocyte-derived macrophages triggers the release of inflammatory cytokines, promoting the generation of mitochondrial reactive oxygen species (mtROS), myocardial hypertrophy, and collagen deposition in the heart [1,2,3]. However, the mechanisms of macrophage contribution to Ang II–induced cardiac remodeling are not fully understood.
The serum and glucocorticoid-inducible protein kinase (SGK) family, including SGK1, SGK2, and SGK3, is a downstream factor of PI3K signaling and shares similar structure and function with AKT. SGK3 has been reported to play a pivotal role in various biological processes through an AKT-independent signaling pathway, including cell proliferation and survival, ion and protein transport, and tumor malignant transformation [4,5,6]. A recent study has demonstrated that SGK3 overexpression in macrophages can enhance their chemotaxis and promote inflammatory responses [7]. However, the effect of SGK3 in macrophages on the regulation of myocardial remodeling has not been fully elucidated.
mtROS and mitochondrial dysfunction also have a vital influence on cardiovascular diseases [8]. Elevated mtROS production mediates cardiac fibrosis, hypertrophy, and cardiac dysfunction [9,10,11]. Nicotinamide adenine dinucleotide dehydrogenase (ubiquinone) 1 alpha subcomplex 13 (Ndufa13), also known as GRIM19, mainly exists on the inner membrane of mitochondria and is the basic subunit of mitochondrial respiratory chain complex I. Previous studies have implicated Ndufa13 in mitochondrial membrane potential, mitochondrial oxidative stress, apoptosis, and regulation of multiple signaling pathways in many diseases [12,13,14]. Nevertheless, the contribution of Ndufa13 to cardiac hypertrophy and fibrosis remains unknown.
In this research, we identified a previously unrecognized role of SGK3 in Ang II–induced cardiac remodeling. The specific deletion of SGK3 in macrophages prevented cardiac hypertrophy, fibrosis, and cardiac dysfunction in Ang II–treated mice. Transcriptome and bioinformatics analysis revealed Ndufa13 as a key molecule in the crosstalk between macrophages and cardiomyocytes (CMs) and cardiac fibroblasts (CFs). We further demonstrated that macrophage SGK3 regulated IL-1β release via the NLRP3 pathway to decrease Ndufa13 expression in CMs and CFs, thereby increasing mtROS-mediated myocardial hypertrophy and fibroblast activation after Ang II treatment. These findings establish a novel opinion that SGK3 is involved in macrophage-mediated cardiac hypertrophy and fibrosis and provide evidence that blockade of SGK3 may represent a potential therapeutic strategy for hypertensive cardiac remodeling and related diseases.
Materials and methods
Animal experiments
The macrophage-specific SGK3 deficiency mice (SGK3-Lyz2-CRE (f/f, +, SGK3 cKO) were constructed by Beijing Viewsolid Biotech Co., Ltd. SGK3 cKO and C57BL/6 wild-type (WT) mice were housed at 22 °C in a 12 h day/night lighting environment with free access to food and water. SGK3 cKO and WT male mice, 8 to 12-week-old, were anesthetized (tribromoethanol, Sigma-Aldrich, USA) and implanted and infused randomly with saline or Ang II (1000 ng/kg/min) with osmotic pumps (Alzet MICRO-OSMOTIC PUMP MODEL 1002, CA, USA) for 14 days. All procedures were performed in accordance with the Animal Care and Use Committee of Dalian Medical University and conformed to the NIH Guide for the Care and Use of Laboratory Animals.
For the injection of Recombinant Adeno-Associated Virus. Briefly, AAV9-sh-scramble and AAV9-sh-Ndufa13 (U6 promoter: GGACTATCATATGCTTACCG, intervention sequence: GCAATGCCAACTTCGGCTTCATTCAAGAGATGAAGCCGAAGTTGGCATTGCTTTTTT) were produced by Weizhen Bioscience Inc. (Shandong, China). 5 × 1011 vector genomes of each AAV9 in PBS solution were injected into the tail vein one day before saline or Ang II infusion, as described previously [15,16,17].
Echocardiography
The echocardiography measurements were performed with a 30-MHz transducer (Vevo2100, VisualSonics Inc., Canada) as previously described [17]. Mice were anaesthetized and heart rates were kept around 400 beats/min. M-mode images were obtained from the left parasternal long-axis view of the left ventricle at the level of the papillary muscle. Ejection fraction (EF) and fractional shortening (FS) were monitored on the M-mode tracings.
Isolation of BM-derived macrophages (BMDMs, Mφs)
SGK3 cKO and WT mice, 4–8 weeks old, were anesthetized with tribromoethanol. Mφs were isolated by flushing the tibias and femurs of mice with RPMI 1640. Then, Mφs were differentiated in RPMI 1640 supplemented with 10% FBS, 1% penicillin/streptomycin and 10 ng/ml macrophage colony-stimulating factor for 3 days [18].
Isolation of neonatal rat cardiomyocytes and fibroblasts
Neonatal rat cardiomyocytes (CMs) and neonatal rat cardiac fibroblasts (CFs) were cultured as previously described [15]. Briefly, heart samples of 1-2-day-old Sprague-Dawley rats were harvested, cut, and then digested in a solution of pancreatic enzymes and type II collagenase at 37 °C for 1–2 min per time. Cells from each digestion were suspended in DMEM/F-12 media (15% FBS, 1% penicillin/streptomycin). The single-cell suspensions were filtered using cell strainers (40 μm, Biosharp) and centrifuged at 1000 rpm for 5 min. Cells were collected and plated in culture dishes for 90 min. The supernatant containing CMs was resuspended and plated in 6 or 24-well cell culture dishes with DMEM/F-12 media (15% FBS, 0.1 mM 5-Brdu, 1% penicillin/streptomycin). CFs that adhered to the plates were cultured in DMEM media (10% FBS, 1% penicillin/streptomycin).
Adenoviral infection
Recombinant adenoviruses carrying Ndufa13 shRNA (Ad-sh-Ndufa13, intervention sequence: CTTTGGCTACTGGAGAATAAT), Ad-Ndufa13 (overexpression sequence: GCGATCGCCACCATGGCGGCGTCGAAGGTGAAGCAGGACATGCCCCCACCAGGGGGCTACGGCCCCATCGACTACAAGCGGAACCTGCCCCGCCGGGGACTGTCGGGATACAGCATGTTTGCTTTGGGCATCGGGGCCTTGATCTTTGGCTACTGGAGAATAATGAGGTGGAACCGGGAGCGTAGGCGCCTGCTGATCGAGGACTTGGAGGCCAGGATCGCCCTCATGCCACTCCTCCAGGCAGAGAAGGACCGCAGGACCCTGCAGATTCTTCGGGAGAACCTGGAGGAGGAGGCTATCATCATGAAAGATGTGCCCAACTGGAAGGTGGGGGAGTCTATGTTCCATACCACACGATGGGTGCCACCCCTCATCGGCGAGCTTTATGGGTTACGCACCAAGGAGGAGATGGAGAACGCCAACTTCGGCTTCACCGGGTACACTACGCGTACGCGGCCGCTCGAGGATTACAAAGATCACGACGGGGATTATAAGGACCATGACATCGACTATAAGGATGATGACGACAAGTAA) or control-scrambled shRNA (Ad-sh-scramble) were packaged from Weizhen Bioscience Inc. (Shandong, China). 24 h after plating, cells were infected with Ad-sh-scramble (50 MOI), Ad-sh-Ndufa13 (50 MOI) or Ad-Ndufa13 (50 MOI) for 24 h.
Measurement of IL-1β
CMs or CFs were infected by Ad-scramble or Ad-Ndufa13 for 24 h, followed by treatment with vehicle or recombinant rat IL-1β protein (10 ng/ml, 400-01B-1MG, Thermo Fisher, MA, USA) for 12 h.
Co-culture of Mφs and CMs or CFs
CMs or CFs were seeded in the lower chamber of a 24-well transwell device, and Mφs from SGK3 cKO (MφcKO) or WT mice (MφWT) were added to the upper chamber. Co-culture Mφs with CMs or CFs under the following conditions:
-
1.
CMs or CFs were co-cultured with MφWT and MφcKO and treated with PBS or Ang II (100 nmol/L).
-
2.
CMs or CFs were infected by Ad-sh-scramble or Ad-sh-Ndufa13 for 24 h, then co-cultured with MφWT and MφcKO and treated with Ang II.
-
3.
MφWT and MφcKO were treated with NLRP3 agonist (BMS-986299,1 µM, HY-139396, MedChemExpress, USA) for 24 h [19], then co-cultured with CMs or CFs and treated with Ang II.
Cells were co-cultured for 24 h, then CMs, CFs or Mφs were collected for follow-up experiments.
Histopathological analysis
Heart tissues were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. Blocks were sectioned to a thickness of 5 μm. Sections were stained with hematoxylin and eosin (H&E), Masson’s trichrome and wheat germ agglutinin (WGA) as previously described [17]. Immunohistochemical staining was conducted using anti-CD68 (1:200 dilution, 28058-1-AP, Proteintech, Chicago, USA). Digital images were captured under a microscope (DM6B Thunder, Laica, Wetzlar, Germany) and analyzed using Image J software.
Dihydroethidium (DHE) and mitoSOX staining
Heart tissues were quickly removed, frozen, embedded in OCT, and sectioned into 5 μm. Sections were stained with DHE (5 µM) or mitoSOX (5 µM) for 30 min at 37 °C. CMs or CFs, co-cultured with Mφs, were incubated with DHE (1 µM) or mitoSOX (1 µM) for 30 min at 37 °C. Images were captured under a microscope (DM6B Thunder, Laica, Wetzlar, Germany) and analyzed using Image J software.
Immunofluorescence
Cells were fixed with 4% formaldehyde for 15 min, permeabilized with 0.1% Triton X-100 for 10 min. CMs were incubated with anti-Actin, anti-Actinin, anti-Vimentin (23660-1-AP, 66895-1-Ig, 10366-1-AP, Proteintech, Cambridge, USA). CFs were incubated with anti-Actinin, anti-Vimentin. Mφs were incubated with anti-SGK3 and anti-STAT3 (CL488-60199, Proteintech, Cambridge, USA). Then cells were mounted with DAPI-containing Mounting Medium (S2110, Solarbio, Beijing, China). Images were acquired by a microscope (DM6B Thunder, Laica, Wetzlar, Germany) and analyzed by Image J analysis software.
Western blot
Total protein from heart tissues or cells was extracted with RIPA lysis buffer (Solarbio Science Technology Co.). Proteins (35–40 µg) were fractionated by 10–15% SDS-PAGE gels and transferred to PVDF membranes. Then, the membranes were incubated with antibodies and observed by the ECL Light Chemiluminescence Kit (Tanon, Shanghai, China). Antibodies against SGK3 (8156), Caspase-1 (E9R20), p-ERK1/2 (4370), IL-1β (12242), ERK1/2 (4695), phospho-Smad2/3 (8828), CaNA (12238) and TGF-β (3711) were obtained from Cell Signaling Technology (Danvers, MA, USA); NLRP3 (19771-1-AP), Ndufa13 (10986-1-AP), Smad2(12570-1-AP), Smad3(25494-1-AP), GAPDH (6004-1-Ig), Goat Anti-Rabbit IgG (SA00001-2) and Goat Anti-Mouse IgG (SA00001-1) were obtained from Proteintech (Chicago, USA). The protein intensities were analyzed using Image J software as previously described [16].
Quantitative PCR (qPCR)
Total RNA was extracted from heart tissues or cells using the RNAex Pro reagent (Accurate Biotechnology, Hunan, China). The isolated RNA (1 µg) was reverse-transcribed into complementary DNA with a reverse transcription kit (AG11711, Accurate Biotechnology, Hunan, China). qPCR amplification was performed with the RNAex Pro reagent (Accurate Biotechnology, Hunan, China) by a QuantStudio 5 system (Thermo Fisher, MA, USA). The PCR primers used in this study are shown in Table S1.
RNA sequencing
Total RNAs from the hearts of saline–treated WT mice, Ang II–treated WT mice, and Ang II–treated SGK3 cKO mice (n = 3) were subjected to transcriptome analysis. Genes with a P value lower than 0.05 and an absolute fold-change value higher than 1 were considered differentially expressed genes (DEGs). DEGs of the three groups (saline-treated WT mice vs. Ang II–treated WT mice, Ang II–treated WT mice vs. Ang II–treated SGK3 cKO mice) were identified with the DESeq2 R package (1.16.1). Heat map analysis was performed using the “heatmap” package of the R software. KEGG and GO pathway enrichment analysis was performed using the DAVID web service (https://david.ncifcrf.gov/) by entering DEGs. GO analysis consisted of cellular components (CC), biological processes (BP), and molecular functions (MF). DEGs were transferred to the online database STRING (https://string-db.org/) to analyze the protein–protein interaction (PPI) network of common DEGs, and the results were imported into Cytoscape software for visualization and correlation analysis. CytoHubba was used to identify important genes in this network as hub genes.
Mitochondrial complex I activity
Mitochondria from the Ad-sh-Ndufa13-treated CMs or CFs, co-cultured with MφWT and MφcKO, were isolated following the protocol of the detection kits. The activity of mitochondrial complex I was measured on an enzyme-labeled instrument at 340 nm using detection kits for complex I (RXWB0434-96, Ruixin Biotech, Fujian, China).
Caspase-1 activity
Mφs were treated with PBS or Ang II (100 nmol/L) along with vehicle or NLRP3 agonist (BMS-986299, 1µM) for 24 h. Then the Caspase-1 activity was measured on enzyme-labeled instrument at 405 nm using the Caspase-1 activity assay kit (C1102, Beyotime, Shanghai, China) according to the manufacturer’s instructions.
Patients
This study was approved by the Ethics Committees of the Second Affiliated Hospital of Dalian Medical University (No. 2023 − 239) and complied with the Declaration of Helsinki standards. Blood samples were obtained from 40 patients with heart failure with reduced ejection fraction (HFrEF), 19 patients with heart failure with preserved ejection fraction (HFpEF), and 40 healthy controls at the Second Affiliated Hospital of Dalian Medical University. Inclusion criteria: (1) diagnosis of HF was based on the 2021 ESC Guidelines for the diagnosis and treatment of acute and chronic HF [20]; (2) age > 18 years old; (3) patients with clinical symptoms, signs, and medical history of heart failure were diagnosed as chronic heart failure by two or more attending physicians; (4) heart function was above New York Heart Association functional class I; (5) evidence of cardiac dysfunction by echocardiography, left ventricular ejection fraction < 40% (HFrEF) or ≥ 50% (HFrEF). Healthy controls were defined as individuals with normal heart function and no cardiovascular risk factors, heart disease, or any family history of coronary artery disease, as previously described [15]. Patients and healthy controls underwent physical examination, cardiac ultrasound examination, and blood tests (Table S2). Written informed consent was signed by the participants before sample collection and data acquisition. Peripheral blood mononuclear cells (PBMCs) were isolated from 3 mL of whole blood collected in K2EDTA tubes. Ficoll-Paque Premium gradient centrifugation was performed for the diluted blood sample to isolate mononuclear cells.
ELISA analysis of SGK3 and IL-1β
The serum samples were obtained after centrifugation of whole blood collected in coagulant-containing tubes. PBMCs were lysed with 80 µL of PBS through repeated freeze-thaw cycles and centrifuged at 1000 x g for 10 min. The culture medium from Mφs was collected after treatments with saline or Ang II for 24 h. Then, 50 µL of serum, PBMC lysate or culture medium was added into a 96-well microtiter plate. The optical density was measured at a wavelength of 450 nm on a microplate reader. Levels of SGK3 (RX107069H, Ruixin Biotech, Fujian, China) in serum and PBMCs and levels of IL-1β (RX203063M, Ruixin Biotech, Fujian, China) in culture medium were analyzed according to the manufacturer’s protocols.
Statistical analysis
Statistical results were expressed with means ± SD for normally distributed quantitative variables, and median (25–75 IQ) for other quantitative variables. The distribution of data was assessed using the Shapiro-Wilk test. A two-tailed Student’s t-test was applied to compare the two groups if both groups exhibited normal distribution, whereas the rank-sum test was employed if at least one group did not follow a normal distribution. One-way ANOVA was utilized for the comparison of three or more groups. Statistical analysis was conducted using SPSS software version 19.0 (SPSS, Chicago, IL, USA). A P value of < 0.05 was deemed to be statistically significant.
Results
Macrophage-specific SGK3 deficiency attenuates Ang II–induced cardiac remodeling
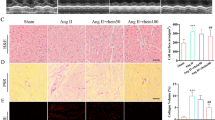
Initially, we examined the expression of SGK3 in patients with HF. Namely, PBMCs and serum were collected from 40 patients with HFrEF, 19 patients with HFpEF, and 40 healthy controls (Table S2). In comparison with the healthy control group, the concentrations of SGK3 in both PBMCs and serum were significantly increased in patients with HFrEF and HFpEF (Fig. S1a and S1b). Hence, SGK3 may play a role in cardiac remodeling. To investigate the contribution of macrophagic SGK3 to pathological cardiac remodeling and dysfunction, we generated macrophage-specific SGK3-deficient mice (SGK3 cKO) by crossing SGK3 flox/flox mice with Lyz2-CRE mice. Subsequently, both WT and SGK3 cKO mice were treated with saline or Ang II for 14 days. Echocardiography showed that Ang II infusion enhanced cardiac performance in WT mice, as indicated by increased EF% and FS%, whereas these were reduced in SGK3 cKO mice (Fig. 1a). WT mice exhibited characteristics of cardiac hypertrophy, as reflected by the augmented heart size, heart weight/body weight ratio (HW/BW), heart weight/tibia length (HW/TL) ratio, myocyte cross-sectional area, and the expression of hypertrophy-related genes (ANF and BNP), which were notably mitigated in SGK3 cKO mice after Ang II infusion (Fig. 1b and e). Moreover, SGK3 cKO substantially ameliorated Ang II–induced myocardial fibrosis, along with reduced expression levels of fibrosis-related genes (collagen I and collagen III) compared with those in WT mice (Fig. 1f and g). Accordingly, Ang II–induced upregulation of p-ERK1/2, calcineurin A (CaNA), TGF-β, and p-Smad2/3 protein levels in the WT group was suppressed in the SGK3 cKO group (Fig. 1h and i). Notably, there was no significant difference in cardiac function, cardiac remodeling, or signaling pathways between the WT and SGK3 cKO groups after saline infusion (Fig. 1a and i). Taken together, these findings affirm the crucial role of macrophagic SGK3 in the regulation of Ang II–induced cardiac remodeling.
SGK3 cKO attenuates Ang II–induced cardiac hypertrophy and fibrosis. WT and SGK3 cKO mice were treated with saline or Ang II for 14 days. (a) Representative M-mode echocardiography of left ventricular chamber in mice (left). Measurement of EF% and FS% (n = 6, right). (b) Representative gross images of whole hearts (scale bar 5 mm). (c) The ratios of HW/BW and HW/TL (n = 12). (d) WGA staining of heart sections (left), the quantification of the relative myocyte cross-sectional area (n = 6, scale bar 100 μm, right). (e) qPCR analysis of ANF and BNP levels in heart tissues (n = 4). (f) Masson staining of heart sections (left), the quantification of the fibrotic area (n = 6, scale bar 100 μm, right). (g) qPCR analysis of COL I and COL III levels in heart tissues (n = 4). (h) Western blot analysis of p-ERK1/2, ERK1/2, and CaNA in heart tissues, and quantification of the relative protein levels (n = 4). (i) Western blot analysis of TGF-β, p-Smad2/3, Smad2, and Smad3 in heart tissues, and quantification of the relative protein levels (n = 4). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001, ****P < 0.0001. HW/BW: heart weight to body weight; HW/TL: heart weight to tibia length; EF%: ejection fraction; FS%: fractional shortening; COL: collagen
Ablation of SGK3 in macrophages suppresses Ang II–induced inflammation and mitochondrial oxidative stress
Subsequently, we tested the effect of SGK3 on the accumulation of proinflammatory cells (especially macrophages) and mitochondrial oxidative stress levels in heart tissues. H&E and immunostaining of heart tissues illustrated that Ang II–induced elevation of proinflammatory cells infiltration (including CD68-positive macrophages) and mRNA levels of proinflammatory cytokines (IL-1β and IL-6) were markedly lower in SGK3 cKO mice than in WT mice (Fig. 2a and b). Furthermore, DHE and mitoSOX staining suggested that SGK3 cKO significantly mitigated the Ang II–induced increase in total superoxide and mtROS levels (Fig. 2c). In addition, the upregulated expression of NLRP3, Caspase-1, and mature IL-1β in the Ang II–treated WT group was suppressed in the SGK3 cKO group (Fig. 2d). Notably, there was no significant difference in inflammation response, total superoxide and mtROS levels, or signaling pathways of heart tissues between the WT and SGK3 cKO groups after saline infusion (Fig. 2a and d). These results suggest that SGK3 knockout in macrophages contributes to both cardiac inflammation and mitochondrial oxidative stress.
SGK3 cKO protect against Ang II–caused inflammation and mtROS. WT and SGK3 cKO mice were treated with saline or Ang II for 14 days. (a) Representative H&E staining (upper) and CD68 immunohistochemistry (lower) in the heart tissues. Quantification of CD68-positive area (n = 6, right). (b) qPCR analysis of the mRNA expression of IL-1β and IL-6 in heart tissues (n = 4). (c) DHE (upper) and mitoSOX (lower) staining of heart sections and quantitative results (n = 6, right). (d) Western blot analysis of NLRP3, Caspase-1, and IL-1β in heart tissues, and quantification of the relative protein levels (n = 4). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, **P < 0.01, ***P < 0.001, ****P < 0.0001. Scale bar: 100 μm. DHE: Dihydroethidium
Loss of SGK3 in macrophages impedes cardiomyocyte hypertrophy and fibroblast activation in vitro
To further evaluate whether BM-derived SGK3-deficient macrophages ameliorate Ang II–caused cardiomyocyte hypertrophy, fibroblast activation, and mtROS accumulation, we isolated and identified CMs and CFs with staining of α-actinin and vimentin (Fig. S2) and then co-cultured CMs or CFs with MφWT or MφcKO in a transwell co-culture system (Fig. 3a). The results showed that CMs co-cultured with MφcKO exhibited reduced myocyte size, levels of hypertrophy-related genes (ANF and BNP), and protein levels of p-ERK1/2 and CaNA compared with those co-cultured with MφWT with Ang II treatment (Fig. 3b and e). MφcKO also significantly decreased Ang II–induced upregulation of TGF-β and p-Smad2/3 protein expression as well as collagen I and collagen III mRNA expression in CFs compared with MφWT (Fig. 3f and g). Additionally, total superoxide and mtROS levels in CMs and CFs co-cultured with MφWT were increased after Ang II treatment but were significantly alleviated in those co-cultured with MφcKO (Fig. 3h and i).
SGK3-deficient macrophages reduced cardiomyocyte hypertrophy, fibroblast activation and mtROS levels. (a) The structure of co-culture system. CMs or CFs were seeded in the lower chamber of a 24-well transwell device, and MφWT and MφcKO were added to the upper chamber, then treated with Ang II or PBS for 24 h. (b) CMs were stained with α-actin (red), DAPI (blue) was used as a nuclear marker. (c) Quantification of myocyte size (n = 5, right). (d) qPCR analysis of ANF and BNP mRNA levels in CMs (n = 3). (e) Western blot analysis of p-ERK1/2, ERK1/2, and CaNA in CMs, and quantification of the relative protein levels (n = 3). (f) Western blot analysis of TGF-β, p-Smad2/3, Smad2 and Smad3 in CFs, and quantification of the relative protein levels (n = 3). (g) qPCR analysis of COL I and COL III mRNA levels in CFs (n = 3). (h) DHE (upper) and mitoSOX (lower) staining of CMs and quantitative results (n = 5). (i) DHE (upper) and mitoSOX (lower) staining of CFs and quantitative results (n = 5). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm. DHE: Dihydroethidium; COL: collagen
To test the role of SGK3 in proinflammatory response in macrophages, MφWT and MφcKO were treated with Ang II or PBS. Ang II infusion upregulated mRNA levels of IL-1β and IL-6 and increased the concentration of IL-1β in the culture medium of MφWT, which were reversed in MφcKO (Fig. S3a and S3b). Accordingly, the increased protein levels of NLRP3, Caspase-1, and IL-1β in MφWT were markedly attenuated in Ang II–treated MφcKO compared with Ang II–treated MφWT (Fig. S3c). Then, we found that SGK3 deficiency reduced the Caspase-1 activity compared with MφWT after Ang II treatment (Fig. S3d). Furthermore, silencing of SGK3 reduced Ang II–induced nuclear STAT3 localization in Mφ (Fig. S3e), which acts as a vital transcriptional regulator of NLRP3. These data suggest that SGK3 deficiency inhibits IL-1β release by the STAT3/NLRP3 pathway in macrophages.
Identification of Ndufa13 as a key factor in SGK3-based regulation of cardiac remodeling
To further confirm the underlying mechanism of SGK3’s involvement in cardiac remodeling, we analyzed the transcriptomes of saline-treated WT, Ang II–treated WT, and Ang II–treated SGK3 cKO mice (n = 3). Based on the threshold of |log2FoldChange| > 1 and P value < 0.05, we screened 383 downregulated genes in the Ang II–treated WT group vs. the saline-treated WT group. Then, 415 upregulated genes were found in the Ang II–treated SGK3 cKO group vs. the Ang II–treated WT group. The Venn diagram (Fig. 4a) showed 185 DEGs that were downregulated in the Ang II–treated WT group compared with the saline-treated WT group while upregulated in the Ang II–treated SGK3 cKO group. A heat map (Fig. 4b) was created to show the expression patterns of these 185 DEGs. To evaluate the biological functions of these DEGs, we performed GO and KEGG enrichment analyses. The results revealed significant enrichment in terms related to energy metabolism, particularly linked to mitochondria and ROS production (Fig. 4c and f).
Identification of differentially expressed genes and enrichment analysis. (a) Bulk RNA-seq analysis of the transcriptome of the hearts of saline-treated WT mice, Ang II-treated WT mice, and Ang II-treated SGK3 cKO mice (n = 3). Venn plot showing the overlapped genes among differentially expressed genes (DEGs) which a total of 185 DEGs were downregulated in the Ang II-treated WT group vs. saline-treated WT group, while upregulated in the Ang II-treated SGK3 cKO group vs. Ang II-treated WT group (|log2 FC| > 1, p < 0.05). (b) The heatmap shows the expression of the 185 DEGs. (c) KEGG pathway enrichment results. (d) GO cell component (CC) enrichment results. (e) GO molecular function (MF) enrichment results. (f) GO biological process (BP) enrichment results. KEGG: Kyoto Encyclopedia of Genes and Genomes; GO: Gene ontology
A DEG-encoded protein interaction network was constructed using STRING and visualized using Cytoscape. Next, the cytoHubba plugin was used to identify the hub genes. By intersecting the results of the five cytoHubba algorithms (MCC, MNC, DMNC, Degree, and Closeness), we identified five hub genes, namely Atp5k, Cox5b, Ndufa13, Uqcrq, and Atp5e. These genes were the most important biomarkers in the PPI network and may play a pivotal role in the pathogenesis of Ang II–induced cardiac remodeling (Fig. S4a). Furthermore, we performed qPCR of heart tissue to validate the genes selected from RNA sequencing and found that downregulated gene expression of Ndufa13 in the Ang II–treated WT group was ameliorated in the Ang II–treated SGK3 cKO group, which was confirmed by immunoblotting CMs and CFs in the in vitro studies (Fig. S4b and S4c). Taken together, Ndufa13 was singled out as a contributor to macrophagic SGK3–mediated effects in Ang II–induced cardiac remodeling.
SGK3, via Ndufa13, regulates Ang II–induced cardiac remodeling
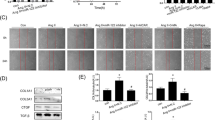
To determine whether SGK3 plays a crucial role in cardiac remodeling via Ndufa13-mediated mtROS, we administered tail vein injections of AAV9-sh-scramble or AAV9-sh-Ndufa13 to the WT or SGK3 cKO group, followed by Ang II infusion for 2 weeks. We found that EF%, FS%, heart size, HW/BW, HW/TL, myocyte cross-sectional area, myocardial fibrosis, and mRNA levels of ANF, BNP, collagen I, and collagen III were significantly exacerbated in the AAV9-sh-Ndufa13-WT group compared with the AAV9-sh-scramble-WT group, and these effects were blocked by macrophage SGK3 deficiency (Fig. 5a and f). Furthermore, relative to the AAV9-sh-scramble-WT group, the protein levels of p-ERK1/2, CaNA, TGF-β, and p-Smad2/3 were further activated in the AAV9-sh-Ndufa13-WT group but were inhibited in the AAV9-sh-Ndufa13-SGK3 cKO group (Fig. 5g and h). Additionally, the reduction of Ndufa13 significantly enhanced the infiltration of proinflammatory cells (including CD68-positive macrophages), mRNA expression of IL-1β and IL-6, total superoxide and mtROS levels, and protein levels of NLRP3, Caspase-1, and IL-1β, whereas SGK3 cKO ameliorated these outcomes (Fig. 6a and d).
SGK3 via Ndufa13 regulates cardiac hypertrophy and fibrosis in vivo. (a) Representative M-mode echocardiography of left ventricular chamber in mice (left). Measurement of EF% and FS% in each group (n = 8, right). (b) Representative gross images of whole hearts (scale bar 5 mm, left). The ratios of HW/BW and HW/TL (n = 9, right). (c) WGA staining of heart sections (n = 6, left), the quantification of the relative myocyte cross-sectional area (n = 6, right). (d) qPCR analysis of ANF and BNP levels in heart tissues (n = 4). (e) Masson staining of heart sections (n = 6, left), the quantification of the fibrotic area (n = 6, right). (f) qPCR analysis of COL I and COL III levels in heart tissues (n = 4). (g) Western blot analysis of Ndufa13, p-ERK1/2, ERK1/2, and CaNA in heart tissues, and quantification of the relative protein levels (n = 4). (h) Western blot analysis of TGF-β, p-Smad2/3, Smad2, and Smad3 in heart tissues, and quantification of the relative protein levels (n = 4). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. HW/BW: heart weight to body weight; HW/TL: heart weight to tibia length; EF%: ejection fraction; FS%: fractional shortening; COL: collagen
SGK3 via Ndufa13 regulates inflammation and mtROS in vivo. (a) Representative H&E staining (upper) and CD68 immunohistochemistry (lower) in the heart tissues. Quantification of CD68-positive area (n = 6, right). (b) qPCR analysis of the mRNA expression of IL-1β and IL-6 in heart tissues (n = 4). (c) DHE (upper) and mitoSOX (lower) staining of heart sections and quantitative results (n = 6, right). (d) Western blot analysis of NLRP3, Caspase-1, and IL-1β in heart tissues, and quantification of the relative protein levels (n = 4). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm. DHE: Dihydroethidium
To further investigate the role of Ndufa13 in the SGK3 regulation of cardiomyocyte hypertrophy and fibroblast activation, we further infected CMs or CFs with Ad-shRNAs to reduce the levels of Ndufa13 and then co-cultured them with MφWT and MφcKO under Ang II treatment (Fig. 7a). We found that knockdown of Ndufa13 further enhanced Ang II–caused myocyte hypertrophy, mRNA levels of ANF and BNP, and protein levels of p-ERK1/2 and CaNA in CMs co-cultured with MφWT. However, these effects were suppressed in Ad-sh-Ndufa13-CMs co-cultured with MφcKO (Fig. 7b and d). Furthermore, Ad-sh-Ndufa13-CFs co-cultured with MφWT exhibited an additional increase in protein expression of TGF-β and p-Smad2/3 and mRNA expression of collagen I and collagen III compared with Ad-sh-Scramble-CFs co-cultured with MφWT. However, these effects were hindered when co-culturing with MφcKO (Fig. 7f and g). In addition, we measured the mitochondrial complex I activity of CMs and CFs in the co-culture system and found that Ad-sh-Ndufa13-CMs or CFs showed an increased mitochondrial activity, which was reduced by co-culturing with MφcKO(Fig. 7h). Moreover, compared with the Ad-sh-scramble groups, we found that the deficiency of Ndufa13 further magnified Ang II–caused total superoxide and mtROS levels in CMs and CFs co-cultured with MφWT, whereas these effects were mitigated through co-culturing with MφcKO (Fig. 7i and j). Hence, these in vitro data suggest that deficiency of SGK3 in macrophages regulates cardiomyocyte hypertrophy and fibroblast activation by Ndufa13-mediated mtROS in CMs and CFs.
SGK3-deficient macrophages suppress Ndufa13-silence-induced cardiomyocyte hypertrophy and fibroblast activation in vitro. (a) The structure of co-culture system. Ad-sh-Scramble or Ad-sh-Ndufa13 treated CMs or CFs were seeded in the lower chamber, and MφWT and MφcKO were added to the upper chamber, then treated with Ang II. (b) CMs were stained with α-actin (red), DAPI (blue) was used as a nuclear marker. (c) Quantification of myocyte size (n = 5,). (d) qPCR analysis of ANF and BNP mRNA levels in CMs (n = 3). (e) Western blot analysis of Ndufa13, p-ERK1/2, ERK1/2, and CaNA in CMs, and quantification of the relative protein levels (n = 3). (f) Western blot analysis of TGF-β, p-Smad2/3, Smad2, and Smad3 in CFs, and quantification of the relative protein levels (n = 3). (g) qPCR analysis of COL I and COL III mRNA levels in CFs (n = 3). (h) The Complex I activity of CMs and CFs (n = 6). (i) DHE (upper) and mitoSOX (lower) staining of CMs and quantitative results (n = 5). (j) DHE (upper) and mitoSOX (lower) staining of CFs and quantitative results (n = 5). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm. DHE: Dihydroethidium; COL: collagen
SGK3-deficient macrophages suppress mtROS in CMs and CFs by inhibiting IL-1β release
We further confirmed that deletion of SGK3 suppressed mtROS in CMs and CFs by inhibiting IL-1β secretion via the NLRP3/Caspase-1/IL-1β pathway. We first treated MφWT and MφcKO with the NLRP3 agonist (BMS-986299, 1 µM) or vehicle for 24 h before co-cultivating with CMs or CFs (Fig. 8a). CMs co-cultured with BMS-986299-MφWT showed increased myocyte size, as well as higher mRNA levels of ANF and BNP compared with those co-cultured with vehicle-MφWT, which were decreased in CMs co-cultured with BMS-986299-MφcKO (Fig. 8b and d). Similarly, BMS-986299-MφWT significantly increased mRNA levels of collagen I and collagen III in CFs, whereas this effect was attenuated in CFs co-cultured with BMS-986299-MφcKO (Fig. 8e). Moreover, MφWT treated with BMS-986299 amplified Ang II–induced total superoxide and mtROS levels in CMs and CFs compared with the control group, but co-culture with MφcKO attenuated this effect (Fig. 8f and g). Mechanically, BMS-986299 further increased the concentration of IL-1β in the culture medium of MφWT, whereas this increase was attenuated in the culture medium of BMS-986299-MφcKO (Fig. S5a). In addition, mRNA levels of IL-1β and IL-6; protein expression levels of NLRP3, Caspase-1, and IL-1β; and Caspase-1 activity were further upregulated in MφWT treated with BMS-986299, but this effect was suppressed in MφcKO (Fig. S5b and S5d). Therefore, these findings suggest that SGK3 regulates IL-1β secretion via the NLRP3/Caspase-1/IL-1β pathway in macrophages, thereby contributing to cardiac remodeling.
SGK3-deficient macrophages aggravate hypertrophy and fibrosis through NLRP3. (a) The structure of co-culture system. CMs or CFs were seeded in the lower chamber of a 24-well Transwell device, and BMS-986299- or vehicle-treated MφWT and MφcKO were added to the upper chamber, then treated with Ang II. (b) CMs were stained with α-actin (red), DAPI (blue) was used as a nuclear marker. (c) Quantification of myocyte size (n = 5). (d) qPCR analysis of ANF and BNP mRNA levels in CMs (n = 3). (e) qPCR analysis of COL I and COL III mRNA levels in CFs (n = 3). (f) DHE (upper) and mitoSOX (lower) staining of CMs and quantitative results (n = 5). (g) DHE (upper) and mitoSOX (lower) staining of CFs and quantitative results (n = 5). Data are expressed as mean ± SD, one-way ANOVA for multiple comparisons, *P < 0.05, **P < 0.01, ***P < 0.001. Scale bar: 100 μm. DHE: Dihydroethidium; COL: collagen
To investigate the involvement of Ndufa13 in IL-1β–mediated mtROS in CMs and CFs, we infected CMs or CFs with Ad-Ndufa13 to increase Ndufa13 protein levels, followed by treatment with recombinant rat IL-1β protein. Our findings revealed that the protein expression of Ndufa13 was reduced in CMs and CFs treated with IL-1β compared with the control group (Fig. S6a and S6b). Furthermore, IL-1β significantly increased total superoxide and mtROS levels in both CMs and CFs, which was attenuated in CMs and CFs treated with Ad-Ndufa13 (Fig. S6c and S6d). Taken together, our study demonstrated that IL-1β modulated mtROS in CMs and CFs via the involvement of Ndufa13.
Discussion
In this study, we identified a previously unrecognized role of macrophagic SGK3 in Ang II–induced cardiac remodeling. The concentrations of SGK3 were elevated in both PBMCs and serum from patients with HF. Macrophage-specific SGK3 deficiency attenuated inflammatory response, thereby ameliorating cardiac hypertrophy, fibrosis, mtROS, and dysfunction in Ang II–infused heart tissue in vivo. MφcKO suppressed hypertrophy of CMs and activation of CFs in a co-culture system in vitro. Moreover, the absence of SGK3 led to a decreased IL-1β release by inhibiting the STAT3/NLRP3/Caspase-1/IL-1β pathway in macrophages, which increased Ndufa13 level and suppressed Ndufa13-mediated mtROS in CMs and CFs. Consequently, these results indicate SGK3 as a potent contributor to the progression of cardiac remodeling. A schematic is shown in Fig. 9.
The involvement of the SGK family in cardiac diseases has been analyzed in previous studies [21,22,23,24]. It has been shown that SGK1 inhibitors can improve heart function and reduce heart injury and ROS in ischemia/reperfusion (I/R) injury rats [21]. Moreover, the inhibition of SGK1 prevents TGF-β1–induced fibroblast activation via the MAPK/ERK1/2/p38/JNK pathway through the regulation of FoxO3a and ROS [22]. Conversely, SGK3 overexpression in cardiomyocytes modulates myocardial repair after I/R injury through the regulation of the GSK-3β pathway [23]. Another study has indicated that activation of SGK3 and its downstream GSK-3β/β-catenin/cyclin D1 pathways facilitates cardiomyocyte proliferation and myocardial regeneration after myocardial infarction [24]. Nevertheless, the role of macrophagic SGK3 in Ang II–induced cardiac remodeling has not been elucidated. Contrary to the effects of SGK3 on I/R injury, we revealed that macrophage-specific deletion of SGK3 attenuated cardiac hypertrophy, fibrosis, and inflammation by suppressing mtROS.
ROS play a crucial role in the pathogenesis of HF, diabetic cardiomyopathy, I/R injury, and Ang II–induced pathological cardiac hypertrophy [11, 12, 25, 26]. There is increasing evidence that the upregulation of ROS production in cardiomyocytes leads to a cascade of adverse effects, including further ROS production, mitochondrial DNA damage, cardiac hypertrophy, fibrosis, inflammation, and cardiac dysfunction. mtROS, the major cellular source of ROS, are implicated in many diseases. Decreased mtROS generation and suppressed NLRP3 inflammasome activation are involved in obesity-induced cardiac remodeling and dysfunction [27]. In addition, the inhibition of mitochondrial dysfunction and mtROS production has been demonstrated to suppress Ang II–induced cardiac hypertrophy [28].
According to a recent study, Ndufa13, a gene associated with the mitochondrial oxidative respiratory chain, can affect mitochondrial membrane potential, the respiratory chain, and intracellular ROS levels [12, 13]. Ndufa13 knockout results in complex I assembly disruption and embryonic lethality [29]. It has been shown that partial loss of Ndufa13 causes an electron leak within mitochondrial complex I, leading to an increase in hydrogen peroxide, STAT3 dimerization, and Bcl-2 upregulation, which significantly suppresses the infarct size during the I/R process [12]. Moreover, knockdown of Ndufa13 decreases complex I activity and increases ROS production [30]. However, the role of Ndufa13 in Ang II–induced cardiac remodeling is not yet known, nor is the relationship with SGK3. Consistent with the results from RNA sequencing, our study showed that Ndufa13 expression was significantly downregulated in Ang II–treated heart tissues and cells both in vitro and in vivo. Knockdown of Ndufa13 resulted in decreased mitochondrial complex I activity and additional mtROS activation in CMs and CFs, which were suppressed by SGK3 deficiency.
Recent studies have revealed that crosstalk between macrophages and CMs or CFs plays an important role in the development of cardiac disease in co-culture systems. Many cytokines (IL-1β, TNF-α, IL-12, IFN-β, and CXCL10/IP10) or exosomes released from macrophages act directly on CMs to decrease ROS, cardiomyocyte apoptosis, cardiomyocyte pyroptosis, or cardiomyocyte hypertrophy [31,32,33,34,35]. Moreover, activated macrophages increase the secretion of inflammatory factors (GRN, IL-1β, TNF-α, IL-6, CCL5, CXCL3, IFN-β, and CXCL10/IP10) and promote myofibroblast activation and collagen synthesis by binding or antagonizing the receptors on CFs [35,36,37,38]. In our study, reduced IL-1β secretion via inhibiting the activation of NLRP3 in MφcKO increased Ndufa13 expression in CMs and CFs in Ang II–treated co-coculture system, which effectively inhibited mtROS-mediated myocardial hypertrophy through the ERK1/2/CaNA pathways and myocardial fibroblasts activation by TGF-β/Smad2/3 pathways. We further investigated the interaction between SGK3 and NLRP3, and no chain contacts were found between SGK3 and NLRP3 using the ZDOCK method [39]. Then, we determined the ZDOCK score values of SGK3 and STAT3 (a transcription factor of NLRP3) was 1401.25. Our study demonstrated that SGK3 deficiency reduced the Ang II–induced nuclear localization of STAT3 in macrophages, leading to the activation of NLRP3 and secretion of IL-1β.
Our study has some limitations. First, other mechanisms may also be involved in the crosstalk between SGK3-deficient macrophages and CMs or CFs in Ang II–induced cardiac remodeling, such as ncRNA or exosomes [33, 40]. Second, we did not demonstrate the mechanism by which IL-1β upregulates Ndufa13 expression in CMs and CFs. Third, we were unable to detect the level of SGK3 in monocytes of patients with HF. Finally, we did not elucidate the exact mechanisms by which SGK3 interacts with STAT3. Hence, further studies are needed to confirm our findings.
Conclusion
This study demonstrates that SGK3 promotes a proinflammatory process in macrophages via the NLRP3 pathway, thereby aggravating myocardial hypertrophy and fibrosis by Ndufa13-mediated mtROS. Therefore, SGK3 may be a potential therapeutic target for Ang II–induced cardiac remodeling.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Mezzaroma E, Toldo S, Farkas D, Seropian IM, Van Tassell BW, Salloum FN, Kannan HR, Menna AC, Voelkel NF, Abbate A (2011) The inflammasome promotes adverse cardiac remodeling following acute myocardial infarction in the mouse. Proc Natl Acad Sci U S A 108(49):19725–19730
Longevity O (2023) Retracted: Alleviation of Inflammation and Oxidative Stress in Pressure Overload-Induced Cardiac Remodeling and Heart Failure via IL-6/STAT3 Inhibition by Raloxifene. Oxid Med Cell Longev 2023:9815047
Wang S, Li J, Bai J, Li JM, Che YL, Lin QY, Zhang YL, Li HH (2018) The immunoproteasome subunit LMP10 mediates angiotensin II-induced retinopathy in mice. Redox Biol 16:129–138
Wang H, Huang F, Zhang Z, Wang P, Luo Y, Li H, Li N, Wang J, Zhou J, Wang Y, Li S (2019) Feedback activation of SGK3 and AKT contributes to Rapamycin Resistance by reactivating mTORC1/4EBP1 Axis via TSC2 in breast Cancer. Int J Biol Sci 15(5):929–941
Bhandaru M, Kempe DS, Rotte A, Capuano P, Pathare G, Sopjani M, Alesutan I, Tyan L, Huang DY, Siraskar B, Judenhofer MS, Stange G, Pichler BJ, Biber J, Quintanilla-Martinez L, Wagner CA, Pearce D, Foller M, Lang F (2011) Decreased bone density and increased phosphaturia in gene-targeted mice lacking functional serum- and glucocorticoid-inducible kinase 3. Kidney Int 80(1):61–67
Peng LQ, Zhao H, Liu S, Yuan YP, Yuan CY, Mwamunyi MJ, Pearce D, Yao LJ (2018) Lack of serum- and glucocorticoid-inducible kinase 3 leads to podocyte dysfunction. FASEB J 32(2):576–587
Wang C, Zheng Y, Fan Q, Li Z, Qi X, Chen F, Xu L, Zhou S, Chen X, Li Y, Zhu J, Su C (2024) Mir-155 promotes Acute Organ Injury in lps-Induced Endotoxemic mice by enhancing Ccl-2 expression in macrophages. Shock 61(4):611–619
Peoples JN, Saraf A, Ghazal N, Pham TT, Kwong JQ (2019) Mitochondrial dysfunction and oxidative stress in heart disease. Exp Mol Med 51(12):1–13
Zhang B, Yang J, Li X, Zhu H, Sun J, Jiang L, Xue C, Zhang L, Xu C, Xing S, Jin Z, Liu J, Yu S, Duan W (2023) Tetrahydrocurcumin ameliorates postinfarction cardiac dysfunction and remodeling by inhibiting oxidative stress and preserving mitochondrial function via SIRT3 signaling pathway. Phytomedicine 121:155127
Souza-Neto FV, Islas F, Jimenez-Gonzalez S, Luaces M, Ramchandani B, Romero-Miranda A, Delgado-Valero B, Roldan-Molina E, Saiz-Pardo M, Ceron-Nieto MA, Ortega-Medina L, Martinez-Martinez E, Cachofeiro V (2022) Mitochondrial Oxidative Stress Promotes Cardiac Remodeling in Myocardial Infarction through the Activation of Endoplasmic Reticulum Stress. Antioxidants (Basel) 11(7)
Luo YX, Tang X, An XZ, Xie XM, Chen XF, Zhao X, Hao DL, Chen HZ, Liu DP (2017) SIRT4 accelerates Ang II-induced pathological cardiac hypertrophy by inhibiting manganese superoxide dismutase activity. Eur Heart J 38(18):1389–1398
Hu H, Nan J, Sun Y, Zhu D, Xiao C, Wang Y, Zhu L, Wu Y, Zhao J, Wu R, Chen J, Yu H, Hu X, Zhu W, Wang J (2017) Electron leak from NDUFA13 within mitochondrial complex I attenuates ischemia-reperfusion injury via dimerized STAT3. Proc Natl Acad Sci U S A 114(45):11908–11913
Chen Y, Lu H, Liu Q, Huang G, Lim CP, Zhang L, Hao A, Cao X (2012) Function of GRIM-19, a mitochondrial respiratory chain complex I protein, in innate immunity. J Biol Chem 287(32):27227–27235
Yang Y, Cheng L, Wang Y, Han Y, Liu J, Deng X, Chao L (2017) Expression of NDUFA13 in asthenozoospermia and possible pathogenesis. Reprod Biomed Online 34(1):66–74
Li J, Wang S, Zhang YL, Bai J, Lin QY, Liu RS, Yu XH, Li HH (2019) Immunoproteasome Subunit beta5i promotes Ang II (angiotensin II)-Induced Atrial Fibrillation by Targeting ATRAP (Ang II type I receptor-Associated protein) degradation in mice. Hypertension 73(1):92–101
Li J, Wang S, Bai J, Yang XL, Zhang YL, Che YL, Li HH, Yang YZ (2018) Novel role for the Immunoproteasome Subunit PSMB10 in Angiotensin II-Induced Atrial Fibrillation in mice. Hypertension 71(5):866–876
Wang S, Bai J, Che Y, Qu W, Li J (2023) Fucoidan inhibits apoptosis and improves cardiac remodeling by inhibiting p53 transcriptional activation through USP22/Sirt 1. Front Pharmacol 14:1164333
Zhang YL, Bai J, Yu WJ, Lin QY, Li HH (2024) CD11b mediates hypertensive cardiac remodeling by regulating macrophage infiltration and polarization. J Adv Res 55:17–31
Gao G, Fu L, Xu Y, Tao L, Guo T, Fang G, Zhang G, Wang S, Qin T, Luo P, Shen X (2022) Cyclovirobuxine D ameliorates Experimental Diabetic Cardiomyopathy by inhibiting Cardiomyocyte pyroptosis via NLRP3 in vivo and in vitro. Front Pharmacol 13:906548
McDonagh TA, Metra M, Adamo M, Gardner RS, Baumbach A, Bohm M, Burri H, Butler J, Celutkiene J, Chioncel O, Cleland JGF, Crespo-Leiro MG, Farmakis D, Gilard M, Heymans S, Hoes AW, Jaarsma T, Jankowska EA, Lainscak M, Lam CSP, Lyon AR, McMurray JJV, Mebazaa A, Mindham R, Muneretto C, Francesco Piepoli M, Price S, Rosano GMC, Ruschitzka F, Skibelund AK, Group ESCSD (2023) 2023 focused update of the 2021 ESC guidelines for the diagnosis and treatment of acute and chronic heart failure. Eur Heart J 44(37):3627–3639
Souri F, Badavi M, Dianat M, Mard SA, Sarkaki A (2023) Protective effects of gallic acid and SGK1 inhibitor on oxidative stress and cardiac damage in an isolated heart model of ischemia/reperfusion injury in rats. Iran J Basic Med Sci 26(3):308–315
Gonzalez-Herrera F, Catalan M, Anfossi R, Maya JD, Pedrozo Z, Diaz-Araya G, Vivar R (2023) SGK1 is necessary to FoxO3a negative regulation, oxidative stress and cardiac fibroblast activation induced by TGF-beta1. Cell Signal 109:110778
Li YF, Wei TW, Fan Y, Shan TK, Sun JT, Chen BR, Wang ZM, Gu LF, Yang TT, Liu L, Du C, Ma Y, Wang H, Sun R, Wei YY, Chen F, Guo XJ, Kong XQ, Wang LS (2021) Serine/Threonine-Protein kinase 3 facilitates myocardial repair after Cardiac Injury possibly through the glycogen synthase Kinase-3beta/beta-Catenin pathway. J Am Heart Assoc 10(22):e022802
Sun J, Yang T, Wei T, Zhou L, Shan T, Chen J, Gu L, Chen B, Liu L, Jiang Q, Du C, Ma Y, Wang H, Chen F, Guo X, Ji Y, Wang L (2022) CDK9 binds and activates SGK3 to promote cardiac repair after injury via the GSK-3beta/beta-catenin pathway. Front Cardiovasc Med 9:970745
Peng ML, Fu Y, Wu CW, Zhang Y, Ren H, Zhou SS (2022) Signaling pathways related to oxidative stress in Diabetic Cardiomyopathy. Front Endocrinol (Lausanne) 13:907757
Wu C, Zhang Z, Zhang W, Liu X (2022) Mitochondrial dysfunction and mitochondrial therapies in heart failure. Pharmacol Res 175:106038
Xie Q, Wei T, Huang C, Liu P, Sun M, Shen W, Gao P (2016) Nebivolol ameliorates Cardiac NLRP3 Inflammasome activation in a juvenile-adolescent animal model of Diet-Induced obesity. Sci Rep 6:34326
Lu Y, Zhang J, Han B, Yu Y, Zhao W, Wu T, Mao Y, Zhang F (2023) Extracellular vesicles DJ-1 derived from hypoxia-conditioned hMSCs alleviate cardiac hypertrophy by suppressing mitochondria dysfunction and preventing ATRAP degradation. Pharmacol Res 187:106607
Higashikuni Y, Liu W, Numata G, Tanaka K, Fukuda D, Tanaka Y, Hirata Y, Imamura T, Takimoto E, Komuro I, Sata M (2023) NLRP3 inflammasome activation through Heart-Brain Interaction initiates cardiac inflammation and hypertrophy during pressure overload. Circulation 147(4):338–355
He X, Zhou A, Lu H, Chen Y, Huang G, Yue X, Zhao P, Wu Y (2013) Suppression of mitochondrial complex I influences cell metastatic properties. PLoS ONE 8(4):e61677
Zhou T, Qian H, Zheng N, Lu Q, Han Y (2022) GYY4137 ameliorates sepsis-induced cardiomyopathy via NLRP3 pathway. Biochim Biophys Acta Mol Basis Dis 1868(12):166497
Al-Qazazi R, Lima PDA, Prisco SZ, Potus F, Dasgupta A, Chen KH, Tian L, Bentley RET, Mewburn J, Martin AY, Wu D, Jones O, Maurice DH, Bonnet S, Provencher S, Prins KW, Archer SL (2022) Macrophage-NLRP3 activation promotes right ventricle failure in pulmonary arterial hypertension. Am J Respir Crit Care Med 206(5):608–624
Heymans S, Corsten MF, Verhesen W, Carai P, van Leeuwen RE, Custers K, Peters T, Hazebroek M, Stoger L, Wijnands E, Janssen BJ, Creemers EE, Pinto YM, Grimm D, Schurmann N, Vigorito E, Thum T, Stassen F, Yin X, Mayr M, de Windt LJ, Lutgens E, Wouters K, de Winther MP, Zacchigna S, Giacca M, van Bilsen M, Papageorgiou AP, Schroen B (2013) Macrophage microRNA-155 promotes cardiac hypertrophy and failure. Circulation 128(13):1420–1432
Wang Y, Qiu Z, Yuan J, Li C, Zhao R, Liu W, Deng W, Gu N, Zhang W, Hu S, Bai Z, Shi B (2021) Hypoxia-reoxygenation induces macrophage polarization and causes the release of exosomal miR-29a to mediate cardiomyocyte pyroptosis. Vitro Cell Dev Biol Anim 57(1):30–41
Hu S, Gao Y, Gao R, Wang Y, Qu Y, Yang J, Wei X, Zhang F, Ge J (2022) The selective STING inhibitor H-151 preserves myocardial function and ameliorates cardiac fibrosis in murine myocardial infarction. Int Immunopharmacol 107:108658
Xu Y, Jiang K, Su F, Deng R, Cheng Z, Wang D, Yu Y, Xiang Y (2023) A transient wave of Bhlhe41(+) resident macrophages enables remodeling of the developing infarcted myocardium. Cell Rep 42(10):113174
Zhang N, Ma Q, You Y, Xia X, Xie C, Huang Y, Wang Z, Ye F, Yu Z, Xie X (2022) CXCR4-dependent macrophage-to-fibroblast signaling contributes to cardiac diastolic dysfunction in heart failure with preserved ejection fraction. Int J Biol Sci 18(3):1271–1287
Zhao J, Chen Y, Chen Q, Hong T, Zhong Z, He J, Ni C (2021) Curcumin ameliorates Cardiac Fibrosis by regulating macrophage-fibroblast crosstalk via IL18-P-SMAD2/3 signaling pathway inhibition. Front Pharmacol 12:784041
Li DD, Fan HX, Yang R, Li YY, Zhang F, Shi JS (2022) Dendrobium Nobile Lindl. Alkaloid suppresses NLRP3-Mediated pyroptosis to alleviate LPS-Induced neurotoxicity. Front Pharmacol 13:846541
Glezeva N, Horgan S, Baugh JA (2015) Monocyte and macrophage subsets along the continuum to heart failure: misguided heroes or targetable villains? J Mol Cell Cardiol 89(Pt B):136–145
Acknowledgements
The authors thank Prof. Jie Bai (Dalian Medical University) and Prof. Yun-Peng Xie (First Affiliated Hospital of Dalian Medical University) for their constructive discussion.
Funding
This work was supported by grants from National Natural Science Foundation of China, Grant/Award Number: 81900223 (Jing Li), 91739119, 81670406 (Peng Qu), 82103667 (Yilin Che); Natural Foundation of Liaoning Province, Grant/Award Number: 2021-BS-208 (JL).
Author information
Authors and Affiliations
Contributions
Jiayu Ren, Heyu Li, Haijun Gao, Yue Wang, Ying Wang, Hongtong Su: methodology, data curation, validation, formal analysis; Zhihan Li: software; Jing Li, Yilin Che: visualization, original draft; Jing Li, Peng Qu: conceptualization, project administration, supervision, resources, writing, review and editing, funding acquisition.
Corresponding authors
Ethics declarations
Ethical approval
This study was approved by the Ethics Committees of the Second Affiliated Hospital of Dalian Medical University (No. 2023 − 239) and complied with the Declaration of Helsinki standards. Animal experiments were approved and supervised by the Animal Ethics Committee of Dalian Medical University. All methods were carried out in accordance with relevant guidelines and regulations.
Consent to publish
Not applicable.
Conflict of interest
The authors have declared that no confict of interest exists.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ren, J., Che, Y., Li, H. et al. SGK3 deficiency in macrophages suppresses angiotensin II–induced cardiac remodeling via regulating Ndufa13–mediated mitochondrial oxidative stress. Cell. Mol. Life Sci. 81, 359 (2024). https://doi.org/10.1007/s00018-024-05395-w
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00018-024-05395-w