Abstract
Humans can be exposed to per- and polyfluoroalkyl substances (PFASs) via many exposure routes, including diet, which may lead to several adverse health effects. So far, little is known about PFAS transport across the human intestinal barrier. In the current study, we aimed to assess the transport of 5 PFASs (PFOS, PFOA, PFNA, PFHxS and HFPO-DA) in a human induced pluripotent stem cell (hiPSC)-derived intestinal epithelial cell (IEC) model. This model was extensively characterized and compared with the widely applied human colonic adenocarcinoma cell line Caco-2 and a human primary IEC-based model, described to most closely resemble in vivo tissue. The hiPSC-derived IEC layers demonstrated polarized monolayers with tight junctions and a mucus layer. The monolayers consisted of enterocytes, stem cells, goblet cells, enteroendocrine cells, and Paneth cells that are also present in native tissue. Transcriptomics analysis revealed distinct differences in gene expression profiles, where the hiPSC-derived IECs showed the highest expression of intestinal tissue-specific genes relative to the primary IEC-based model and the Caco-2 cells clustered closer to the primary IEC-based model than the hiPSC-derived IECs. The order of PFAS transport was largely similar between the models and the apparent permeability (Papp) values of PFAS in apical to basolateral direction in the hiPSC-derived IEC model were in the following order: PFHxS > PFOA > HFPO-DA > PFNA > PFOS. In conclusion, the hiPSC-derived IEC model highly resembles human intestinal physiology and is therefore a promising novel in vitro model to study transport of chemicals across the intestinal barrier for risk assessment of chemicals.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Introduction
Per- and polyfluoroalkyl substances (PFASs) are a group of more than 4700 man-made organic compounds that are used in a wide variety of industrial sectors and consumer products (OECD 2018; Gluge et al. 2020). PFASs consist of a partly or fully fluorinated carbon chain with different chain lengths and attached chemical groups, giving them the desired properties. Due to the strong fluorine–carbon bond, PFASs have an extremely high thermal stability, which, in combination with their hydrophobic and oleophobic properties, renders them desirable for the use in many applications, including firefighting foams, food contact materials, textiles, household products, construction materials and electronics (Gluge et al. 2020; Panieri et al. 2022). However, due to these properties, PFASs are also highly persistent causing them to accumulate in the environment and pose health concerns to all living organisms (Fenton et al. 2021). To that end, perfluorooctane sulfonate (PFOS) was identified as a persistent organic pollutant and included in the Stockholm Convention in 2009, followed by perfluorooctanoic acid (PFOA) in 2019 and by perfluorohexane sulfonate (PFHxS) in 2022, to restrict their production and use (EU 2019, 2020; UNEP 2020, 2022).
PFASs are omnipresent and can be found in the environment including soil, surface water and groundwater, but are also detected in living organisms such as plants, wildlife and humans (Sedlak et al. 2017; Jian et al. 2018; Brusseau et al. 2020; Death et al. 2021; Panieri et al. 2022). Humans can be exposed to PFASs via several routes including ingestion of contaminated food and drinking water, inhalation of air and dust, and dermal absorption (Jian et al. 2018; Kotlarz et al. 2020; Poothong et al. 2020), of which oral exposure is shown to be the predominant route (Vestergren and Cousins 2009; Haug et al. 2011; Poothong et al. 2020). Once ingested, PFASs are absorbed into the bloodstream, where they primarily bind to albumin and are distributed throughout the body with a high affinity for tissues containing high protein levels, mainly blood, liver and kidneys (Pizzurro et al. 2019; ATSDR 2021). PFASs have highly variable serum elimination half-lives ranging from several days for short-chain PFASs to several years for longer-chain PFASs (Olsen et al. 2007; Li et al. 2018; Xu et al. 2020).
Epidemiological studies have demonstrated associations between human exposure to PFASs and hepatoxicity, immunotoxicity, developmental toxicity, elevated cholesterol levels and decreased thyroid hormone levels (EFSA CONTAM Panel 2018, 2020). Although several of these effects have been corroborated by animals studies, there are also inconsistencies. The same holds true for the bioavailability data of PFASs. While most in vivo data show that PFOS and PFOA are readily absorbed in rodents and humans, little information is available for other PFASs and there are inconsistencies in the literature (EFSA CONTAM Panel 2020). Bioavailability appears to depend on the type of PFAS, the absorption rate of PFASs across the human intestinal barrier and the sex, but variations in the study setup hamper comparability of studies. Furthermore, translation from rodents to humans is known to be challenging due to differences in toxicokinetic parameters between humans and rodents. To that end, alternative human cell-based in vitro methods to study absorption of various PFASs under comparable conditions could greatly add to the evaluation of the toxicokinetics of PFASs in humans.
Well-established intestinal barrier models resembling the intestinal physiology are lacking. Currently, the most widely applied and accepted in vitro model for transport studies is an intestinal barrier model based on the human intestinal epithelial cell (IEC) line Caco-2. The model consists of Caco-2 cells that are cultured in a monolayer on Transwell inserts and differentiated into IECs that display characteristics of in vivo human intestinal epithelial cells. However, the Caco-2 cell-based intestinal barrier model has several limitations when comparing it with the in vivo human intestinal epithelium, like high transepithelial electrical resistance values, presence of only enterocytes and the lack of a mucus layer (Sun et al. 2008). Human induced pluripotent stem cells (hiPSCs)-derived IEC models are promising novel in vitro models, which better reflect human intestinal tissue make-up and functionality than Caco-2 cells. For example, whereas Caco-2 cells have no or limited expression of the most important intestinal CYP enzyme CYP3A4 (Prueksaritanont et al. 1996; Ozawa et al. 2015; Küblbeck et al. 2016), hiPSCs-derived intestinal organoids demonstrate CYP3A4 mRNA expression and possess CYP3A4 activity (Onozato et al. 2018; Janssen et al. 2021). Moreover, hiPSCs-derived intestinal organoids contain multiple cell types including enterocytes, stem cells, goblet cells, Paneth cells, enteroendocrine cells and mesenchymal cells (Spence et al. 2011; Tamminen et al. 2015; Takayama et al. 2019; Janssen et al. 2021; Grouls et al. 2022). However, the spheric configuration of intestinal organoids poses a practical limitation, as their microarchitecture limits easy access to the apical epithelium to facilitate transport studies. Alternatively, hiPSCs can also be differentiated into IEC layers (Kabeya et al. 2018, 2020; Grouls et al. 2022). This layer model has been shown to also contain multiple cell types, including enterocytes and goblet cells, possesses CYP activity, as well as transporter activity (Kabeya et al. 2018, 2020).
In the current study, we studied the transport of PFASs in hiPSC-derived IEC layers as a model for the human intestinal barrier and compared the data with transport data obtained with the Caco-2 cell model and a commercially available human primary IEC-based model (EpiIntestinal). The EpiIntestinal model is a small intestinal microtissue based on human primary cells, which has been described to appropriately predict human intestinal absorption (Ayehunie et al. 2018; Cui et al. 2020). The hiPSC-derived IEC layers were obtained based on a differentiation method described by Kabeya et al. (2020). Before studying PFAS transport, we extensively characterized the hiPSC-derived IEC layers by evaluating their differentiation during various differentiation stages, followed by evaluation of their cellular make-up, gene expression profiles and transport functionality. Characterization data of hiPSC-derived IEC layers was compared with data obtained from Caco-2 cells and the EpiIntestinal model where applicable. Four PFASs, namely perfluorooctanoic acid (PFOA), perfluorooctane sulfonate (PFOS), perfluorononanoic acid (PFNA) and perfluorohexanesulfonate (PFHxS), were selected for bidirectional transport studies. These substances have been described to currently contribute most to human exposure and in 2020 EFSA established a health based guidance value of 4.4 ng/kg bw for the sum of these four PFASs (EFSA CONTAM Panel 2020). In addition, also hexafluoropropylene oxide-dimer acid (HFPO-DA) was included in this study because it is increasingly used as a replacement for PFOA and other PFASs and is also associated with several health concerns (EPA 2021).
Materials and methods
Chemicals
For characterization of transport functionality of the intestinal cell layers, the following model chemicals were used: warfarin, propranolol, quinidine, atenolol, enalapril and acebutolol. The tested PFASs were: perfluorooctanoic acid (PFOA), perfluorooctane sulfonate (PFOS), perfluorononanoic acid (PFNA), perfluorohexane sulfonate (PFHxS) and hexafluoropropylene oxide dimer acid (HFPO-DA). All stocks were prepared in 100% dimethyl sulfoxide (DMSO HybriMax, Sigma, St. Louis, MO). More information about suppliers, purity, catalog numbers and CAS numbers is presented in Supplementary Table 1.
Caco-2 cell culture
Human colorectal adenocarcinoma cells (Caco-2, HTB-37) were obtained from American Type Culture Collection (ATCC, Manassas, VA). Cells were cultured in a humidified incubator (37°C, 5% CO2) in Dulbecco’s modified Eagle’s medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (Gibco, Thermo Fisher Scientific, Waltham, MA), 1% non-essential amino acids (Gibco) and 1% penicillin/streptomycin (Sigma). For experiments, Caco-2 cells below passage number 50 were used within 10 passages. They were harvested at 80% confluence using Trypsin/EDTA (0.25%/0.05%; PAN Biotech, Aidenbach, Germany) and seeded at a density of 4 × 105 cells/cm2 into inserts with a polycarbonate membrane (0.4 µm pore, Corning, New York, NY). After 21 days of differentiation (using 500 µl cell culture medium in the apical and 1500 µl in the basolateral compartment), monolayer integrity was determined by measuring transepithelial electrical resistance (TEER) with a Millicell ERS Volt-Ohm meter (Millipore, Bedford, MA). Only monolayers exhibiting TEER values exceeding 400 Ω · cm2 were used for experiments.
EpiIntestinal culture
The human EpiIntestinal small intestinal microtissues, consisting of epithelial cells and an underlying lamina propria containing intestinal fibroblasts, were obtained from MatTek Life Sciences (Ashland, USA). Cell culture inserts (8.8 mm ID) were placed in 12-well plates and cells were maintained in 100 µl maintenance medium (MatTek Life Sciences) in the apical compartment and 5 ml maintenance medium in the basolateral compartment upon arrival. The transport studies were performed 1 day after arrival.
hiPSC culture
The hiPSC cell line (CS83iCTR-33n1) was provided by the Cedars-Sinai Medical Center’s David and Janet Polak Foundation Stem Cell Core Laboratory. These cells had been generated through episomal reprogramming of fibroblasts of a 31-year-old healthy female. The cell line was fully characterized and no karyotype abnormalities were found. hiPSCs were cultured on human embryonic stem cell qualified Matrigel-coated (Corning #354277) cell culture plates in mTeSR1 medium (Stem Cell Technologies, Vancouver, Canada) and passaged using gentle cell dissociation reagent (Stem Cell Technologies).
hiPSC differentiation into IEC layers and intestinal organoids
For differentiation, hiPSC were dissociated into single cells using Accutase (Stem Cell Technologies) and cultured on human embryonic stem cell qualified Matrigel-coated 24-well plates in mTeSR1 supplemented with 10 µM Y-27632 (Stem Cell Technologies) for 1 day. Human iPSCs were subsequently differentiated into definitive endoderm (DE) by incubation in RPMI1640 medium containing 1% non-essential amino acids (Gibco) and 100 ng/ml Activin A (Cell Guidance Systems, Cambridge, UK), with increasing concentrations of fetal bovine serum (0%, 0.2% and 2% on day 1, 2 and 3, respectively). 50 ng/ml BMP4 (R&D Systems, Minneapolis, MN) was also added during the first day of definitive endoderm formation.
For differentiation into IECs, definitive endoderm cells were subsequently differentiated into intestinal stem cells by changing medium into DMEM/F12 containing 2% fetal bovine serum, 1% GlutaMAX (Gibco) and 250 ng/ml FGF2 (R&D Systems). After 4 days, cells were dissociated using Accutase and seeded onto polyester Transwell inserts (0.4 µm, Corning) coated with Growth Factor Reduced Matrigel #356231. Cells were cultured for 7 days in intestinal differentiation medium (DMEM/F-12 containing 2% fetal bovine serum, 1% non-essential amino acids (Gibco), 1 × B27 (Gibco), 1 × N2 (Gibco), 1% penicillin/streptomycin (Gibco) and 2 mM L-glutamine (Gibco)) supplemented with 20 ng/ml EGF (R&D Systems) and 30 µM Forskolin (Sigma). ROCK inhibitor, y-27632 (final concentration 10 µM; Stem Cell Technologies), was added during the initial 24 h after seeding to reduce cell death. The cells were subsequently cultured for 12 days in intestinal differentiation medium supplemented with 20 ng/ml EGF, 30 µM Forskolin, 5 µM 5-aza-2′-deoxycytidine (Sigma), 20 µM PD98059 (Stem Cell Technologies) and 0.5 µM A-83–01 (Sigma). The medium was refreshed every 2–3 days. Barrier integrity was determined by measuring apical to basolateral translocation of Lucifer yellow (see Supplementary Data for a detailed description).
Definitive endoderm cells were also differentiated into human intestinal organoids (HIOs), as previously described (Janssen et al. 2021). Briefly, definitive endoderm cells were differentiated into hindgut endoderm using FGF4 (R&D Systems) and Chiron99021 (Stemgent). After 4 days, free-floating spheroids were collected and embedded into domes of Matrigel (Corning #354234) and further differentiated using 50 ng/ml EGF (R&D Systems), 100 ng/ml Noggin (R&D Systems) and 500 ng/ml R-spondin-1 (R&D Systems) for 38 days. The medium was refreshed every 2–3 days and HIOs were passaged every 10–14 days. To promote intestinal differentiation, 0.5 µM A-83-01 (Sigma), 20 µM PD98059 (Stem Cell Technologies), 5 µM 5-aza-2’-deoxycytidine (Sigma) and 5 µM DAPT (Sigma) were added to the medium the last 7 days.
RNA isolation and qPCR
Total RNA was extracted from the hiPSCs-derived IEC layers using the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). Subsequently, 500 ng RNA was used to synthesize cDNA using the iScript cDNA synthesis kit (Bio-Rad Laboratories, Veenendaal, The Netherlands). Changes in gene expression were determined by real-time PCR on a CFX384 real-time PCR detection system (Bio-Rad Laboratories) using SensiMix (Bioline; GC Biotech, Alphen aan den Rijn, The Netherlands). A standard curve of pooled cDNA samples was used to calculate amplification efficiency, which was for each primer pair within the acceptable range between 90 and 110% (Rogers-Broadway and Karteris 2015). The expression of the gene of interest, calculated using this standard curve, was subsequently normalized to the expression of the housekeeping gene RPL27. PCR conditions are described in the Supplementary Data and the used primers are listed in Supplementary Table 2.
Immunofluorescence staining
hiPSC-derived IEC layers were fixed by gently replacing the medium in the apical compartment with 200 µl room temperature paraformaldehyde (1% v/v in PBS; Thermo Fisher Scientific). For immunocytochemistry, cells were fixed with Carnoy’s fixative (60% ethanol, 30% chloroform, 10% glacial acetic acid and 1 g ferric chloride; all from Sigma) and processed as previously described (van der Hee et al. 2018, 2020). The Transwell inserts were subsequently transferred to wells containing 1 ml of their corresponding fixative and incubated overnight at 4 °C. After fixation, the Transwells were gently cut using a scalpel and processed in 1.5 ml tubes (Eppendorf). For whole mount staining, the Transwells were washed three times in TBS-triton buffer (8.8 g/l NaCl, 6.06 g/l Tris, 0.02% triton X-100, pH 7.4; all from Sigma) and blocked using normal goat serum (5% v/v in TBS-t, #10000C, Thermo-Fisher) for 30 min at RT. The Transwells were then incubated with primary antibodies overnight at 4 °C. After fixation and after each incubation with antibodies or other staining solution, Transwell membranes were washed three times with TBS-t (5 min incubation per wash). After incubation with the first antibody, Transwells were incubated with a secondary antibody (1:400 in TBS-t) for 1 h at RT. Nuclei were stained with 4′6-diamidino-2-phenylindole (DAPI 1 µg/ml in PBS; Sigma-Aldrich) for 10 min at RT. The primary and secondary antibodies are listed in Supplementary Table 3. Transwell membranes were mounted in Diamond anti-fade mountant (Thermo Fisher) on a poly-l-lysine-coated glass slide and a coverslip. All slides were imaged on a DM6 microscope fitted with DFC365 FX camera (Leica), or on a Stellaris 8 confocal point scanning system at 40 × magnification (Leica) and images were processed using Leica Application Suite X software (LAS X, version 3.7.4.23463, Leica).
RNA isolation for RNA seq and data analysis
To compare whole genome expression of the various intestinal models, RNA was extracted from Caco-2 cells, hiPSC-derived IEC layers or EpiIntestinal microtissues using the RNeasy Mini Kit (Qiagen, Venlo, The Netherlands). Total RNA was extracted from hiPSC-derived organoids using the NucleoSpin RNA isolation kit (Macherey-Nagel, Düren, Germany).
Total RNA was quantified using a Qubit fluorometer (Invitrogen, Life Technologies) and RNA integrity was analyzed using a total RNA Pico chip in an Agilent 2100 Bioanalyzer (Agilent Technologies). Subsequently, RNA Library preparations and RNA sequencing were performed at the Genomics Facility of Wageningen University and Research, Business Unit Bioscience. A detailed description of the RNA Library preparations and RNA seq, and the processing of RNA seq reads is given in the Supplementary Data. RNAseq data were deposited in the NCBI Gene Expression Omnibus under accession number GSE268603.
Multidimensional scaling analysis was performed to visualize multidimensional variation of the differentially expressed genes between the various models and samples. Limma (version 3.50.3) was used to create a multidimensional scaling (MDS) plot. Differential gene expression in the various models was evaluated by comparison with the EpiIntestinal model, a p value < 0.01 was considered statistically significantly different based on empirical Bayes moderate t-statistic. Venn diagrams, showing differentially expressed genes in the various models were made using the open access online tool Venny (version 2.1.0; https://bioinfogp.cnb.csic.es/tools/venny/). Additionally, a heatmap was made using Morpheus (https://software.broadinstitute.org/morpheus/) to visualize the differentially expressed genes [showing the respective genes’ counts per million (cpm)] that were specific for one or two models only. Therefore, differentially expressed genes that were shared by all three models (the center of the Venn diagram) were excluded. Gene expression, relative to the EpiIntestinal microtissues, was hierarchically clustered based on a Euclidean distance algorithm.
Gene set enrichment analysis (GSEA; version 4.3.2; Broad Institute, Cambridge, MA) was used to identify enriched or suppressed gene sets (Subramanian et al. 2005). Gene sets were derived from Biocarta, KEGG and WikiPathways pathway databases and ranked based on the normalized enrichments score (NES) (cutoff nominal p value < 0.05 and FDR q value < 0.25). Only gene sets comprising more than 15 and fewer than 500 genes were taken into account. Statistical significance of GSEA results was determined using 1000 permutations.
Transport studies
Transport studies were performed in Caco-2 cells, hiPSC-derived IECs and EpiIntestinal microtissues. To that end, Caco-2 cell layers were differentiated for 21 days and hiPSCs were differentiated into IEC layers for 26 days, as described above. EpiIntestinal microtissues were subjected to transport studies 1 day after arrival. Transport studies with the model compounds (warfarin, propranolol, quinidine, atenolol, enalapril and acebutolol) were performed in apical to basolateral, and basolateral to apical direction in HBSS containing 10 µM of each model compound, except for the EpiIntestinal microtissues where only transport of Warfarin and Atenolol was assessed in apical to basolateral direction. For all models, PFAS transport was assessed in apical to basolateral, and basolateral to apical direction using HBSS containing a mixture of PFASs containing 1 µM of each PFAS. See Supplementary Data for a detailed description of the method used for the transport studies of the model compounds and PFASs.
Quantification of model compounds using LC–MS/MS
For the transport studies of the model compounds, samples of the donor and receiver compartment and cell lysates were analyzed on a Waters Acquity I UPLC (Milford, USA) system. The system was equipped with a Waters Acquity UPLC BEH C18 (100 × 2.1 mm, 1.7 μm) column. The column oven was kept at 60 °C and the temperature of the autosampler was kept at 10 °C. The mobile phase consisted of water (mobile phase A) and 90% methanol (mobile phase B) both containing 1 mM ammonium formate and 0.1% formic acid. The gradient started at 0% B, was kept at 0% B for 1 min, was then linearly increased to 50% B in 2 min and was kept at 50% B for 1 min. Then, the gradient linearly increased to 100% B in 2 min and was kept at 100% B for 2 min. The gradient returned to 0% B within 0.5 min and was kept at this condition for 2.5 min before the next injection. The injection volume used was 5 μl.
Mass spectrometric detection was performed using a Micromass Quattro Ultima mass spectrometer (Waters, Milford, USA) equipped with an electrospray ionization interface (ESI). A capillary voltage of 2.50 kV, a source temperature of 120 °C, a desolvation temperature of 350 °C, a cone gas flow of 194 L/h and a desolvation gas flow of 568 l/h was used. The collision-induced dissociation gas used was argon. Cone voltage and collision energy were optimized by direct infusion for each compound. Parameters for multiple reaction monitoring (MRM) such as ion mode, mass charge (m/z) transitions, cone voltage and collision energy for each compound are described in Supplementary Table 4.
Quantification of PFASs from transport experiment using LC–MS/MS
Samples of culture medium from the transport experiments were prepared for PFAS measurement by adding 50 μl of the culture medium to 850 μl methanol (Actuall Chemicals, Oss, The Netherlands) containing internal standards [13C4-PFOA, 13C5-PFNA, 18O2-PFHxS, 13C4-PFOS and 13C3-HFPO-DA (Wellington Laboratories, Canada)]. After vortexing, the samples were centrifugated at maximum speed (20,817×g) for 10 min at 4 °C. PFAS concentrations were determined in the supernatant using LC–MS/MS analysis. LC separation was performed using either a Shimadzu UHPLC system containing: two pumps (LC 20AD xr); a column oven (Shimadzu CTO-20AC); a pump switch (Shimadzu FCV-11AL); a degasser (Shimadzu DGU-20A3); and a sample tray holder (Shimadzu SIL-20 AC XR model) (Shimadzu Corporation, Kyoto, Japan) or a Sciex UHPLC system containing: two pumps (ExionLC AD); column oven (ExionLC AC); controller (ExionLC); degasser (ExionLC); and sample tray holder (Exion AD) (Sciex Framingham, MA, USA). Optimized PFAS measurement protocols for each system were used. To separate the PFASs, an Acquity BEH-C18 analytical column (50 × 2.1 mm i.d., 1.7 μm, Waters, Milford, MA, USA) (Shimadzu system) or Luna Omega PS C18 analytical column (100 Å, 100 × 2.1 mm i.d., 1.6 µm, Phenomenex, Torrance, CA, USA) (Sciex system) was used at a column temperature of 35 °C (Shimadzu system) or 40 °C (Sciex system). Additionally, a symmetry C18 analytical column (50 × 2.1 mm i.d., 5 µm, Waters, Milford, MA, USA) (Shimadzu system) or Gemini C18 analytical column (110 Å, 50 × 3 mm i.d., 3 µm, Phenomenex, Torrance, CA, USA) (Sciex system) was used as an isolator column, which was placed between the pump and the injector valve to isolate and delay interferences out of the LC system. The mobile phase consisted of 2 mM (Shimadzu system) or 20 mM (Sciex system) ammonium acetate (Merck Millipore, Darmstadt, Germany) in Milli-Q water (prepared using a Milli-Q system with a resistivity of at least 18.2 M Ω cm−1 (Merck Millipore)) (Shimadzu system) or Ultra LC/MS grade water (Actu-All Chemicals, Oss, The Netherlands) (Sciex system) (mobile phase A) and Acetonitrile ULC/MS grade (Actu-All Chemicals, Oss, The Netherlands) (mobile phase B). The injection volume used was 20 μl. The chromatographic gradient was operated at a flow rate of 0.3 ml/min (Shimadzu system) or 0.8 ml/min (Sciex system) starting from 25% (Shimadzu system) or 15% (Sciex system) mobile phase B in the first 0.1 (Shimadzu system) or 1.0 (Sciex system) minute followed by a linear increase to 100% (Shimadzu system) or 98% (Sciex system) B in 6 min which was held for 2.5 (Shimadzu system) 0.5 (Sciex system) minutes. Then, the gradient returned to 25% (Shimadzu system) or 15% (Sciex system) B within 0.1 min and was held for 3.9 (Shimadzu system) or 0.7 (Sciex system) minutes to equilibrate before the next injection, resulting in a total run time of 12.5 (Shimadzu system) or 8.3 (Sciex system) minutes. Mass spectrometric detection was carried out by MS/MS using a Sciex QTRAP 5500 (Shimadzu system) or Sciex QTRAP 7500 (Sciex system) system (Sciex, Framingham, MA, USA) in negative electrospray ionization (ESI-) mode. The following conditions were used: an ion spray voltage (IS) of − 4500 V (Shimadzu system) or − 1500 V (Sciex system); curtain gas (CUR) of 30 l/h (Shimadzu system) or 45 psi (Sciex system); a source temperature (TEM) of 350 °C (Shimadzu system) or 400 °C (Sciex system); gas 1 (GS1) of 55 l/h (Shimadzu system) or 40 psi (Sciex system); gas 2 (GS2) of 60 l/h (Shimadzu system) or 80 psi (Sciex system); and collision gas (CAD) high (Shimadzu system) or 9 (Sciex system). The PFASs were fragmented using collision induced dissociation (CID) using argon as target gas. The analyses were performed in multiple reaction monitoring (MRM) mode, using two mass transitions per component selected based on the abundance of the signal and the selectivity of the transition. In Supplementary Table 5 and 6 information on the MRM transitions, declustering potential (DP), entrance potential (EP), collision energy (CE) and cell exit potential (CXP) are presented for the Shimadzu and Sciex system. Data were acquired using Analyst software (Shimadzu system) or SciexOS software (Sciex system) and processed using MultiQuantTM software (Sciex, Framingham, MA, USA).
Statistical analysis
Data are presented as mean ± SEM. A one-way ANOVA, followed by Bonferroni’s post hoc multiple comparison test was used for comparisons of the Papp values of model compound and PFAS transport between the three different models. An unpaired Studen’s t test was used for comparison of the Papp values of basolateral to apical model compound transport between hiPSC-derived IECs and Caco-2 cells. p < 0.05 was considered statistically significant. Prism software (version 9.2.0; GraphPad, San Diego, CA) was used for statistical analysis.
Results
Differentiation and characterization of hiPSC-derived intestinal epithelial cell layers
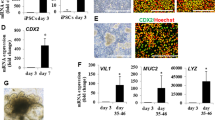
The differentiation procedure of the human hiPSC line CS83iCTR-33n1 into definitive endoderm was based on a previous report (Janssen et al. 2021). Definitive endoderm was subsequently differentiated into IEC layers based on methods described by Kabeya et al. (2018, 2020) (Fig. 1A). To assess the differentiation procedure, terminally differentiated hiPSC-derived IEC layers were characterized using gene expression measurements and immunohistochemical evaluation. The expression levels of the differentiation markers were normalized to the expression of the housekeeping gene RPL27, which remained stable throughout all phases of differentiation phases in both independent experiments (Supplementary Fig. 1A). The intestinal differentiation markers CDX2 (intestinal transcription factor), IFABP2 (cellular protein specific for mature enterocytes), VIL1 (major component of brush border cytoskeleton specific for enterocytes) and SI (a glucosidase enzyme located on the brush border of the small intestine) were well expressed in the IEC phase (Fig. 1B; Supplementary Table 7). Villin1 expression could also readily be detected as confirmed by immunofluorescence (Fig. 1D). The intestinal stem cell marker LGR5 gradually increased over time with the highest expression in the ISC phase (average Ct value of 27.1; Fig. 1B). Despite the highest expression of LGR5 in the ISC phase, some expression of LGR5 was still present in the IEC phase and could also still be visualized in this phase (Fig. 1D; Supplementary Table 7). In addition to enterocytes and stem cells, the IEC layer also contained Paneth cells, goblet cells and enteroendocrine cells as evidenced by an upregulation of LYZ, MUC2 and CHGA in the IEC phase, respectively (Fig. 1C; Supplementary Table 7). These findings were corroborated by immunofluorescence analysis using GLP1 to visualize enteroendocrine cells, the lectin Ulex Europaeus Agglutinin I (UEA-1), which is known to stain Paneth cells and goblet cells including secreted mucins, and Trefoil Factor 3 (TFF3), staining glycosylated mucus proteins present in mature goblet cells (Fig. 1D). The IECs grow in a confluent layer, which was confirmed to be a monolayer by evaluation of a cross section of the IEC layer (Fig. 1D). The expression of the tight junction protein occludin and the adherens junctions protein E-cadherin gradually increased over time during differentiation with the highest expression in the IEC phase (Fig. 1E; Supplementary Table 7). The tight junction protein ZO-1 was highly expressed throughout all phases of differentiation (Fig. 1E, Supplementary Table 7). The presence of ZO-1 was also visualized using immunofluorescence staining (Fig. 1F). In addition, ZO-1 staining (Fig. 1F), VILLIN staining and UEA-1 staining (Fig. 1D) were mainly located at the apical surface, indicating polarization of the IEC layer with formation of tight junctions, a brush border and a mucus layer on the luminal side. Gene expression data from each independent experiment are presented in Supplementary Fig. 1B. While most differentiation markers were well expressed in the IEC phase (Supplementary Table 7), there was some variability in the expression of LGR5, CHGA, ZO-1 throughout the differentiation process. The TEER remained constant from day 19 of the differentiation protocol onwards, amounting to ~ 360 Ω cm2, and there was no quantifiable apical to basolateral transport of lucifer yellow in terminally differentiated IECs, indicating a proper barrier integrity (data not shown).
Differentiation of hiPSCs into intestinal epithelial cell (IEC) layers. A Schematic procedure for the differentiation of hiPSCs into the IEC layers. B Relative expression of intestinal differentiation and stem cell markers during the course of the differentiation from hiPSCs up to IEC layers. The highest expression level of each gene was set at one. Data are presented as mean values ± SEM from two independent experiments with triplicate wells. C Relative expression of markers of Paneth cells (LYZ), goblet cells (MUC2) and enteroendocrine cells (CHGA) during the course of the differentiation from hiPSCs up to IEC layers. The highest expression level of each gene was set at one. Data are presented as mean values ± SEM from two independent experiments with triplicate wells. D Representative immunofluorescent pictures of IEC layers stained for various intestinal differentiation markers. Nuclei were counterstained with 4′6-diamidino-2-phenylindole (DAPI). LGR5: leucine-rich repeat-containing G-protein-coupled receptor 5, TFF3: trefoil factor 3, GLP-1: glucagon-like peptide 1, UEA-1: Ulex europaeus agglutinin-1. The VIILIN and UEA-1 images show a cross section (CS) and a maximum intensity projection of all imaged layers. E Relative mRNA expression of the tight junctional proteins zonula occludens 1 (ZO-1) and occludin, and the adherens junction protein E-cadherin during the course of the differentiation from hiPSCs up to the IEC layers. The highest expression level of each gene was set at one. Data are presented as mean values ± SEM from two independent experiments with triplicate wells. F Representative immunofluorescent pictures of IEC layers stained for ZO-1. Both images show maximum intensity projection of all imaged layers. Nuclei were counterstained with DAPI
Transcriptomics of four intestinal models
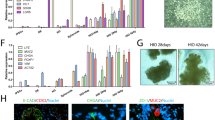
Transcriptomics analysis was performed to assess the base level gene expression profiles of the following four intestinal models: hiPSC-derived IEC layers, Caco-2 cell layers, hiPSC-derived HIOs, and a human primary model for the intestine (EpiIntestinal). After normalization and filtering of lowly expressed genes, multidimensional scaling (MDS) analysis was employed to compare global gene expression patterns of the four intestinal models. The MDS plot shows that the replicate samples of each intestinal model cluster together indicating low variability within the sample groups (Fig. 2A). The biggest source of variation was explained by dimension 1 (56%), which showed a clear separation between the hiPSC-derived IECs and HIOs, and the Caco-2 cell layers and EpiIntestinal model. Subsequently, differentially expressed genes were selected using the human primary model for the intestine (EpiIntestinal) for benchmarking. When applying a statistical cutoff of p < 0.01 (empirical Bayes moderated t-statistic), 8,820 genes were differentially expressed in the Caco-2 cells, 12,646 genes in the hiPSC-derived HIOs and 13,611 genes in the hiPSC-derived IECs, of the total of 19,619 genes (Fig. 2B). A Venn diagram of these differentially expressed genes shows that there was an overlap of 5,038 genes that were differentially expressed in all three intestinal models, when compared to the EpiIntestinal model. The number of genes that were statistically differentially expressed in only one model compared to the EpiIntestinal model was higher for the hiPSC-derived models (2560 genes in hiPSC-derived IECs and 2162 genes in HIOs) than for the Caco-2 cell layers (793 genes) (Fig. 2C). Next, the 12,739 genes that were differentially expressed in either one or two models, as compared to the EpiIntestinal model, were hierarchically clustered and presented in a heatmap (Fig. 2D). Like in the MDS plot, the heatmap also shows little variation in the gene expression patterns of the technical replicates suggesting robust differentiation methods of Caco-2 cell layers and hiPSC-derived HIOs and IEC layers. Furthermore, the heatmap clearly shows the differential expression between the Caco-2 cell layers, hiPSC-derived IEC layers and the hiPSC-derived HIOs versus the EpiIntestinal model, but it also shows apparent differences between the hiPSC-derived models and the Caco-2 cell layers. In these models, several differentially expressed genes showed an opposing gene expression when compared to the EpiIntestinal model. Subsequently, a list of 698 intestine-specific genes was obtained from the Human Protein Atlas that have been annotated to be enriched or enhanced in the intestine (Uhlén et al. 2015). 492 (70,5%) of these genes were also expressed in our data set, each having an expressing level greater than approximately ten counts in at least one of the triplicates. These genes were hierarchically clustered and presented in a heatmap again using the EpiIntestinal model as a benchmark (Fig. 3). When comparing the number of differentially expressed intestine-specific genes between the Caco-2 cell layers, hiPSC-derived IEC layers and the hiPSC-derived HIOs versus the EpiIntestinal model, it was found that the hiPSC-derived IEC model has the largest number of differentially expressed intestine-specific genes with a log twofold change > 1 and the smallest number of differentially expressed intestine-specific genes with a log twofold change > − 1 (Table 1). This suggests that of the four intestinal models, the hiPSC-derived IEC layers have the highest expression of intestine-specific genes.
Transcriptomics analysis of the EpiIntestinal model, hiPSC-derived IEC layers, Caco-2 cell layers and hiPSC-derived HIOs. A Multidimensional scaling plot of transcriptome-wide gene expression in the four intestinal models. Numbers in parentheses represent the percentage of total variance explained by the first and second dimension. B Volcano plots showing relative changes in gene expression (expressed as signal log2 ratio, x-axis) plotted against statistical significance (expressed as − log10 p value of empirical Bayes moderated t statistic p value, y-axis) of the hiPSC-derived IEC layers vs. EpiIntestinal model, Caco-2 cell layers vs. EpiIntestinal model and hiPSC-derived HIOs vs. EpiIntestinal model. Dotted line represents cutoff of p < 0.01. C Venn diagrams showing the number of differentially expressed genes (p < 0.01) in the hiPSC-derived IEC layers, Caco-2 cell layers and hiPSC-derived HIOs compared to the EpiIntestinal model. D Heatmap obtained upon hierarchical clustering of genes that were differentially expressed in either one or two models, as compared to the EpiIntestinal model. Expression is normalized against average expression of the EpiIntestinal model (arbitrarily set at zero), showing Log2 ratio of expression in the hiPSC-derived IEC layers, Caco-2 cell layers and hiPSC-derived HIOs versus expression in the EpiIntestinal model
Heatmap obtained upon hierarchical clustering of the gene expression data of 492 intestine-specific genes in the EpiIntestinal model, hiPSC-derived IEC layers, Caco-2 cell layers and hiPSC-derived HIOs. Expression is normalized against average expression of the EpiIntestinal model (arbitrarily set at zero), showing Log2 ratio of expression in the hiPSC-derived IEC layers, Caco-2 cell layers and hiPSC-derived HIOs versus expression in the EpiIntestinal model
To obtain insight into the biological function of the differentially expressed genes in the three intestinal models in comparison with the EpiIntestinal model, gene set enrichment analysis (GSEA) was performed (Fig. 4). Various pathways related to cytokine receptor interaction were enriched in both hiPSC-derived IEC and HIO models, and the IL6 and IL2RB pathways were enriched in the Caco-2 model, indicating higher expression of genes related to immune function and signaling in all three intestinal models as compared with the EpiIntestinal model. In addition, all three models had elevated expression of gene sets related to cell–matrix adhesion. When comparing pathways with a lower expression, the three models shared lower expression of gene sets related to DNA replication and DNA repair pathways. In addition, in the hiPSC-derived IEC layers and Caco-2 cell layers, there was lower expression of gene sets related to ribosome and RNA processing, whereas in the hiPSC-derived HIOs more pathways related to the cell cycle had a lower expression as compared to the EpiIntestinal model.
In the current study, the main aim was to study the transport of PFASs across the intestinal barrier in the three intestinal epithelial cell layer models. To that end, gene expression analysis specifically focused on comparison of gene expression levels of the most important intestinal transporters in these three models, as visualized in a heatmap (Fig. 5). The heatmap illustrates significant variation between the three different intestinal epithelial cell layer models for the selected genes. For instance, the gene expression of SLC1A5 and SLC7A6 was higher in the Caco-2 cell layers, but it was significantly lower in the hiPSC-derived IEC layers as compared to the EpiIntestinal model. In addition, mRNA levels of SLC51B and SLC7A5 were elevated in hiPSC-derived IEC layers, but reduced in the Caco-2 cell layers in comparison with the EpiIntestinal model. Interestingly, out of the 37 selected intestinal transporter genes, 8 genes (ABCG2, SLC19A3, SLC22A2, SLC22A3, SLC28A1, SLC28A3, SLC51A and SLC5A1) were highler expressed and 6 genes (ABCC4, SLC10A2, SLC19A2, SLC29A2, SLC5A11 and SLCO1A2) were found to be lower expressed in the Caco-2 and hiPSC-derived IEC models as compared to the EpiIntestinal model. Taken together, these data suggest that the models could have different active transport properties based on the gene expression levels of the transporters.
Comparative heatmap analysis on gene expression of intestinal transporters in the EpiIntestinal model, hiPSC-derived IEC layers and Caco-2 cell layers. Expression is normalized against average expression of the EpiIntestinal model (arbitrarily set at zero), showing Log2 ratio of expression in the hiPSC-derived IEC layers and Caco-2 cell layers versus expression in the EpiIntestinal model
Transport of model compounds
Transport studies were performed in the hiPSC-derived IEC layers, in the primary model for the intestine (EpiIntestinal), and in the Caco-2 cell layers. Transport in hiPSC-derived HIOs was not evaluated due to their microarchitecture that limits easy access to the apical epithelium. To characterize the transport properties of the hiPSC-derived IEC layers, the transport of six model compounds was assessed. Model compounds were selected based on known human absorption data and on the availability of permeability data in the EpiIntestinal model. Based on the biopharmaceutics classification system (BCS), which classifies drugs based on their solubility and permeability, three model compounds with a high permeability (warfarin, propranolol and quinidine) were selected and three model compounds with a low permeability (atenolol, enalapril and acebutolol). The EpiIntestinal model has been described to appropriately predict human intestinal absorption (Ayehunie et al. 2018). Transport studies were conducted in the EpiIntestinal model for warfarin and atenolol in apical to basolateral (A–B) direction (Fig. 6A, B; Table 2), to assess reproducibility of the data for these compounds as described in literature. While warfarin transport was slightly lower in our experiments than reported in literature, the same trends were observed (i.e., high transport for warfarin versus low transport for atenolol), so for comparisons of the apparent permeability coefficients (Papp) between the three models, the data from literature were used (Ayehunie et al. 2018).
Model compound absorption (A–B direction) in the EpiIntestinal model, hiPSC-derived IEC layers and Caco-2 layers. A Cumulative amount of high-permeable compounds in the basolateral compartment during the course of the transport study upon apical exposure to 10 µM of the model compounds at t = 0. Samples were taken from the basolateral compartment at settled time points and model compounds were quantified using LC/MS–MS. B Cumulative amount of low permeable compounds in the basolateral compartment during the course of the transport study upon apical exposure to 10 µM of the model compounds at t = 0. Samples were taken from the basolateral compartment at settled time points and model compounds were quantified using LC/MS–MS. In the EpiIntestinal model, only transport studies for warfarin and atenolol were conducted. Data are mean values ± SEM from triplicate wells
Of the high-permeable drugs, Papp values of A–B transport of warfarin were not significantly different between the three models, but those of A–B transport of propranolol were significantly higher in the hiPSC-derived IEC layer model than in the EpiIntestinal model. Also, the Papp values of A–B transport of quinidine were higher in the hiPSC-derived IEC layer model than in both the EpiIntestinal model and Caco-2 cell layers (Fig. 6A; Table 2). The Papp values of all low permeable drugs were significantly higher in the hiPSC-derived IEC layer model as compared to Caco-2 cell layers and the EpiIntestinal model (Fig. 6B; Table 2). There was a general trend of higher Papp values in the hiPSC-derived IEC model, but the A–B transport of low- and high-permeable compounds can be clearly discriminated. The A–B transport of enalapril and acebutolol in hiPSC-derived IEC layers has been assessed previously (Kabeya et al. 2020) and showed comparable Papp values as compared to our hiPSC-derived IEC layers, indicating that these higher Papp values are a characteristic of the model (Table 2). The transport of the model compounds was also assessed in basolateral to apical (B–A) direction (Supplementary Fig. 2), again showing significantly higher Papp values for almost all model compounds in the hiPSC-derived IEC layers as compared to Caco-2 cell layers. The exception was the lower Papp value for warfarin in hiPSC-derived IEC layers, for which the B–A transport was higher across the Caco-2 cell layers compared to the hiPSC-derived IEC layers (Supplementary Table 8). To further characterize the transport properties of the hiPSC-derived IEC layer model, the efflux ratio (Papp B–A/Papp A–B) of the model compounds was calculated and compared with the efflux ratio derived from the Caco-2 cell model (Table 3). According to the FDA guidance (US Food and Drug Administration 2020), any investigational drug with an efflux ratio greater than 2 is indicative of a potential substrate of active efflux transporters. The mean efflux ratio for all model compounds, except acebutolol, were below 2, suggesting no involvement of active efflux transporters which is in accordance to information from the literature for these drugs. For acebutolol, the efflux ratio was 7.37 in the Caco-2 cell layers and 3.67 in the hiPSC-derived IEC layers, suggesting active efflux transport of acebutolol in both models. Indeed, acebutolol has been described to be a substrate of the efflux transporter P-glycoprotein (Terao et al. 1996). Taken together, the hiPSC-derived IEC layer model can clearly discriminate between low- and high-permeable compounds, and shows the presence of active transport.
Transport of PFASs
The main aim of the current study was to get insight into the transport of some well-known PFASs across the human intestinal barrier in vitro. To that end, the PFASs, PFOA, PFOS, PFNA, PFHxS and HFPO-DA were selected for bidirectional studies (Fig. 7A).
PFASs absorption (A–B direction) in the EpiIntestinal model, hiPSC-derived IEC layers and Caco-2 layers. A Chemical structures of the PFASs tested in the present study. PFOA perfluorooctanoic acid, PFOS perfluorooctane sulfonate, PFNA perfluorononanoic acid, PFHxS perfluorohexane sulfonate, HFPO-DA hexafluoropropylene oxide-dimer acid. B Cumulative amount of PFOA, PFOS, PFNA, PFHxS and HFPO-DA in the basolateral compartment during the course of the transport study upon apical exposure to a mixture containing 1 µM of each PFAS. Samples were taken from the basolateral compartment at settled time points and quantified using LC/MS–MS. Data are mean values ± SEM from triplicate wells
PFAS transport was evaluated in the hiPSC-derived IEC layers, Caco-2 cell layers and in the EpiIntestinal model. Mass balance calculations showed a general recovery of PFASs between 75 and 100% in all three models (Supplementary Fig. 3), indicating some adsorption to the plasticware or degradation of the PFASs. Specifically for PFOS, it was apparent that there was a higher recovery in the cell lysate fractions. The Papp values of PFAS transport in the hiPSC-derived IEC model in the A–B direction were in the following order: PFHxS > PFOA > HFPO-DA > PFNA > PFOS (Fig. 7B). The order of PFAS transport was highly similar between all models, but differences in the Papp values were noted. In general, the Papp values were highest in the EpiIntestinal model (ranging from 1.16 to 12.48 × 10–6 cm/s) and lowest in the hiPSC-derived IEC model (ranging from 0.63 to 3.95 × 10–6 cm/s) (Fig. 7B; Table 4). Transport in B–A direction was also studied (Supplementary Fig. 4; Supplementary Table 9) and generally reflected the results of transport in the A–B direction. The permeability of PFOS was relatively low compared to the transport of the other PFASs with only small differences between the three intestinal models, with the permeability being highest in the EpiIntestinal model in the A–B and B–A directions (Fig. 7B; Supplementary Fig. 4). Interestingly, there was limited to no transport of HFPO-DA in the EpiIntestinal and Caco-2 cell models, whereas transport could be readily detected in the hiPSC-IEC model (Fig. 7B; Supplementary Fig. 4). The efflux ratios were comparable between the three models and were all < 2, suggesting no active transport of PFASs (Table 5). Overall, PFHxS transport was highest in all models and HFPO-DA and PFOS transport was rather limited depending on the intestinal model.
Discussion
In the present study, we aimed to get insight into the transport of PFASs across the intestinal barrier using hiPSC-derived IEC layers in comparison with the human primary IEC-based EpiIntestinal model, which has been described to closely resemble human small intestinal tissue (Ayehunie et al. 2018), and the human colonic adenocarcinoma cell line Caco-2, which is commonly used for permeability studies. Characterization of the hiPSC-derived IEC layers showed that they had a polarized epithelium, consisting of enterocytes, intestinal stem cells, enteroendocrine cells, goblet cells and Paneth cells and thus mimic the cellular make-up of intestinal tissue in vivo. RNA sequencing was performed to benchmark the hiPSCs-derived IEC and Caco-2 layer models against the human EpiIntestinal model. All models showed a significant amount of differentially regulated genes compared to the EpiIntestinal model, but the Caco-2 model had the least differentially expressed genes, which was surprising considering that this model is a cancerous cell line and consists only of epithelial cells. Interestingly, comparison of 492 intestine-specific genes showed that hiPSC-derived IEC layers have the highest expression of intestine-specific genes of all models. The hiPSCs-derived IEC layers and HIOs, and the Caco-2 cell layers showed a higher expression of genes related to immune function and signaling pathways suggesting that these models might be more sensitive in their responses to immune stimuli or toxicants. Furthermore, they also showed a lower expression of genes related to DNA replication and DNA repair pathways. Since the main focus of this study lies on transport of PFASs, the expression of intestinal transporters was evaluated in more detail. The hiPSC-derived IEC layers expressed substantial levels of relevant intestinal transporters at gene level. In fact, of the 37 evaluated genes 20 genes were significantly higher expressed in the hiPSC-derived IEC layers than in the EpiIntestinal model, whereas 8 genes had a significantly lower expression. In Caco-2 cell layers, there were 18 significantly higher and 9 lower expressed genes compared with the EpiIntestinal model. Comparison of the Caco-2 cell model with the hiPSC-derived IEC layers showed a generally lower differential expression of the transporter genes in the Caco-2 model. This suggests that the IEC model is likely well suited for transport studies, including studies on active transport. To validate this, the expression of transporters in the hiPSC-derived IEC layers should be confirmed, for instance using proteomics, immunohistochemistry or by measuring the activity of the transporters, which was out of the scope of this study.
In the present study, hiPSCs were differentiated into IEC layers for bidirectional transport studies. The transport data of the model compounds in the iPSC-derived IEC layers were consistent with the Biopharmaceuticals Classification System, showing higher transport for warfarin, propranolol and quinidine and lower transport for atenolol, enalapril and acebutolol, indicating that the hiPSC-derived IEC layers can clearly discriminate between high and low permeable compounds. When comparing the three different models, the order of transport of the low- and high-permeable compounds was largely comparable, but differences in the Papp values were seen. In general, the apparent permeability was higher in the hiPSC-derived IEC layers than in the other two models. Apart from the hydrophobicity and size of a compound, the intestinal apparent permeability of a compound is mainly determined by the barrier integrity, the cellular composition and the presence of uptake and efflux transporters (Markus et al. 2021; Vanuytsel et al. 2021; Franco et al. 2021) implicating that several factors or a combination of factors could be the cause of the observed differences. Caco-2 cell layers are known to generally have a high barrier integrity as indicated by high TEER values ranging from 300 to 2400 Ω cm2, which is dependent on clonal type and culture parameters (Markus et al. 2021). In contrast, the TEER values of the EpiIntestinal model have been reported to lie between 90 and 300 Ω cm2 (Markus et al. 2021). In the present study, only Caco-2 cell layers with TEER values > 400 Ω cm2 were used and the average TEER values of the hiPSC-derived IEC models was 330 Ω cm2 during the transport studies (data not shown). Another contributor to the apparent permeability is the cellular composition. While the Caco-2 model consists only of epithelial cells, the other two models consist of multiple cell types, but the exact ratio of the different cell types in the models is unknown and may differ between the models. For example, Caco-2 cell layers do not possess goblet cells, while hiPSC-derived IEC layers do, and the EpiIntestinal model has been described to contain mucus secreting granules which may point toward the presence of goblet cells as well. The presence of goblet cells, and as a result a mucus layer, also influences the apparent permeability of compounds. It has been reported that hydrophobic compounds have a reduced ability to migrate through mucus in comparison with hydrophilic compounds (Larhed et al. 1997). Additionally, the EpiIntestinal model contains a layer of fibroblasts that the other two models lack, which may also influence the apparent permeability. A last contributor to the apparent permeability is the presence of uptake and efflux transporters. The gene expression data on the expression of transporter genes (Fig. 5) showed that there is a distinct possibility that the presence of uptake and efflux transporters varies between the three models. This could influence the apparent permeability values for some of the compounds that have been described to involve active transport mechanisms (e.g., quinidine and acebutolol) (Terao et al. 1996; Ishida et al. 2006). Clearly, all three models have their own specific differences influencing the transport of chemicals which makes it hard to directly compare the intestinal models studied in the present study. The selection of the model compounds was based on the availability of transport data in literature. As can be expected, most transport data is available for the Caco-2 cell model. Comparing reported Papp values from literature using the Caco-2 cell model for the compounds used in this study (Supplementary Table 10) clearly illustrates that with the Caco-2 cell model large differences in Papp values have been reported. These variations in Papp values could be related to robustness of the model or to variation in study set-ups which would call for harmonization of study designs to reduce interlaboratory variation between Papp values. For the hiPSC-derived IEC layers, it is interesting to note that in the study by Kabeya et al. (2020), that first described the differentiation method to obtain hiPSC-derived IEC layers which was applied in this study, the transport data on enalapril and acebutolol were comparable to the data from this study (see Table 2). These initial findings are promising, but an extensive interlaboratory comparison on both models using harmonized protocols would be needed to conclude whether this method is more robust than the Caco-2 model. Nevertheless, the strength of these types of in vitro models might not lie in obtaining a quantitative Papp value, but in evaluation of relative transport properties of compounds, like was done in the experiment with PFASs in the present study.
Little is known about transport kinetics of PFASs and underlying mechanisms in the intestine. In the present study, the order of PFAS transport was highly similar between the three models, the Papp values in the hiPSC-derived IEC layers in A–B direction were in the following order PFHxS > PFOA > HFPO-DA > PFNA > PFOS. Currently, there are no quantitative PFASs absorption data from humans available, but based on animal studies it is estimated that some PFASs are rapidly and well absorbed (Gannon et al. 2011; Lupton et al. 2012). Several studies have examined active uptake and clearance mechanisms of PFASs in liver and kidney and have identified transporters that appear to be involved in active transport of PFASs. It is still unknown whether transporters are involved in the transport of PFAS in the intestine. Of the identified transporters so far, many are not present in the intestine, except for OATP2B1 and ASBT. These two transporters were shown to transport PFOS, PFHxS, PFOA and PFNA (Zhao et al. 2015, 2017; Kimura et al. 2017, 2020) and both transporters are expressed at mRNA levels in the tested models in this study. This may indeed suggest that these transporters played a role in the observed transport, but the efflux ratios suggest that there was no active transport of PFAS in the three models.
Currently, there are no data available about human exposure or the transport kinetics of HFPO-DA. In the present study, HFPO-DA transport was highest in the hiPSC-derived IEC model, while there was limited transport in the EpiIntestinal model and no transport in the Caco-2 model. Mass balance calculations showed that there were no indications that HFPO-DA was extensively metabolized, degraded or adsorbed to the plasticware in the Caco-2 samples in comparison with the other two models (Supplementary Fig. 3). A previous study (Kotlarz et al. 2020) measured HFPO-DA in serum of residents living near fluorochemical manufacturing facilities. HFPO-DA was present in water samples near the manufacturing facilities as well as other PFASs, such as PFHxS, PFOA, PFOS and PFNA. The latter were detected in serum of most participants at higher levels than the U.S. national levels for the 2015–2016 National Health and Nutrition Examination Survey, indicating exposure to PFASs which potentially included HFPO-DA, but HFPO-DA was not detected in serum. While this may indicate that HFPO-DA is not or limitedly absorbed, it might also be that serum HFPO-DA levels were below the limit of quantification or that HFPO-DA is quickly eliminated from the human body resulting in lower serum concentrations in humans compared to PFASs that are slowly eliminated. Clearly, more research is required to get better insights into the pharmacokinetics of HFPO-DA in the human body, for example by using isotope labeled HFPO-DA to allow easier tracing of HFPO-DA.
In summary, in the current study transport properties of the hiPSC-derived IEC model, the Caco-2 model and the EpiIntestinal model were evaluated. Our data showed similar functional properties as the conventional Caco-2 model and the EpiIntestinal model, with some differences in absolute measured values. In addition, our data indicated that the hiPSC-derived IEC model highly resembles human intestinal physiology, due to the presence of multiple intestinal cell types, and is therefore a promising novel in vitro model to study transport of chemicals across the intestinal barrier. Furthermore, the present study provides insight into the intestinal transport kinetics of PFASs, which was highest for PFHxS in all models and lowest for PFOS and HFPO-DA.
Data availability
All datasets used and/or analyzed to support the findings of this study are available in this article or in the Supplementary Information. All the raw data supporting the results of this study are available from the corresponding author upon request.
Abbreviations
- HFPO-DA:
-
Hexafluoropropylene oxide-dimer acid
- hiPSCs:
-
Human induced pluripotent stem cells
- IECs:
-
Intestinal epithelial cells
- PFASs:
-
Per- and polyfluoroalkyl substances
- PFHxS:
-
Perfluorohexane sulfonate
- PFNA:
-
Perfluorononanoic acid
- PFOA:
-
Perfluorooctanoic acid
- PFOS:
-
Perfluorooctane sulfonate
References
ATSDR (2021) Toxicological profile for perfluoroalkyls. Version May 2021 https://www.atsdr.cdc.gov/toxprofiles/tp200.pdf. Accessed 19 Jan 2023
Ayehunie S, Landry T, Stevens Z et al (2018) Human primary cell-based organotypic microtissues for modeling small intestinal drug absorption. Pharm Res. https://doi.org/10.1007/s11095-018-2362-0
Brusseau ML, Anderson RH, Guo B (2020) PFAS concentrations in soils: background levels versus contaminated sites. Sci Total Environ 740:140017. https://doi.org/10.1016/j.scitotenv.2020.140017
Cui Y, Claus S, Schnell D et al (2020) In-depth characterization of epiintestinal microtissue as a model for intestinal drug absorption and metabolism in human. Pharmaceutics. https://doi.org/10.3390/pharmaceutics12050405
Death C, Bell C, Champness D et al (2021) Per- and polyfluoroalkyl substances (PFAS) in livestock and game species: a review. Sci Total Environ 774:144795. https://doi.org/10.1016/j.scitotenv.2020.144795
EFSA Contam Panel (2018) Scientific opinion. Risk to human health related to the presence of perfluorooctane sulfonic acid and perfluorooctanoic acid in food. EFSA J 16:5194. https://doi.org/10.2903/j.efsa.2018.5194
EFSA Contam Panel (2020) Scientific opinion. Risk to human health related to the presence of per fluoroalkyl substances in food. EFSA J 18:6223. https://doi.org/10.2903/j.efsa.2020.6223
EPA (2021) Human health toxicity values for hexafluoropropylene oxide (HFPO) dimer acid and its ammonium salt (CASRN 13252-13-6 and CASRN 62037-80-3) Also Known as “ GenX Chemicals.” October 2021 EPA Document Number: 822R-21-010
EU (2019) Regulation (EU) 2019/1021 of the European Parliament and of the Council of 20 June 2019 on persistent organic pollutants. Official Journal of the European Union: L 169/45
EU (2020) Commission Delegated Regulation (EU) 2020/784 of 8 April 2020 amending Annex I to Regulation (EU) 2019/1021 of the European Parliament and of the Council as regards the listing of perfluorooctanoic acid (PFOA), its salts and PFOA-related compounds. Official Journal of the European Union: L 188 I/1
Fenton SE, Ducatman A, Boobis A et al (2021) Per- and polyfluoroalkyl substance toxicity and human health review: current state of knowledge and strategies for informing future research. Environ Toxicol Chem 40:606–630. https://doi.org/10.1002/etc.4890
Franco YL, Da Silva L, Cristofoletti R (2021) Navigating through cell-based in vitro models available for prediction of intestinal permeability and metabolism: are we ready for 3D? AAPS J 24:2. https://doi.org/10.1208/s12248-021-00665-y
Gannon SA, Johnson T, Nabb DL et al (2011) Absorption, distribution, metabolism, and excretion of [1-14C]-perfluorohexanoate ([14C]-PFHx) in rats and mice. Toxicology 283:55–62. https://doi.org/10.1016/j.tox.2011.02.004
Gluge J, Scheringer M, Cousins IT et al (2020) An overview of the uses of per- and polyfluoralkyl substances (PFAS). Environ Sci Process Impacts 22:2345–2373. https://doi.org/10.1039/d0em00291g
Grouls M, Janssen AWF, Duivenvoorde LPM et al (2022) Differential gene expression in iPSC - derived human intestinal epithelial cell layers following exposure to two concentrations of butyrate, propionate and acetate. Sci Rep. https://doi.org/10.1038/s41598-022-17296-8
Haug LS, Huber S, Becher G, Thomsen C (2011) Characterisation of human exposure pathways to perfluorinated compounds—comparing exposure estimates with biomarkers of exposure. Environ Int 37:687–693. https://doi.org/10.1016/j.envint.2011.01.011
Ishida K, Takaai M, Hashimoto Y (2006) Pharmacokinetic analysis of transcellular transport of quinidine across monolayers of human intestinal epithelial Caco-2 cells
Janssen AWF, Duivenvoorde LPM, Rijkers D et al (2021) Cytochrome P450 expression, induction and activity in human induced pluripotent stem cell-derived intestinal organoids and comparison with primary human intestinal epithelial cells and Caco-2 cells. Arch Toxicol 95:907–922. https://doi.org/10.1007/s00204-020-02953-6
Jian JM, Chen D, Han FJ et al (2018) A short review on human exposure to and tissue distribution of per- and polyfluoroalkyl substances (PFASs). Sci Total Environ 636:1058–1069. https://doi.org/10.1016/j.scitotenv.2018.04.380
Kabeya T, Qiu S, Hibino M et al (2018) Cyclic AMP signaling promotes the differentiation of human induced pluripotent stem cells into intestinal epithelial cells. Drug Metab Dispos 46:1411–1419
Kabeya T, Mima S, Imakura Y et al (2020) Pharmacokinetic functions of human induced pluripotent stem cell-derived small intestinal epithelial cells. Drug Metab Pharmacokinet 35:374–382. https://doi.org/10.1016/j.dmpk.2020.04.334
Kimura O, Fujii Y, Haraguchi K et al (2017) Uptake of perfluorooctanoic acid by Caco-2 cells: involvement of organic anion transporting polypeptides. Toxicol Lett 277:18–23. https://doi.org/10.1016/j.toxlet.2017.05.012
Kimura O, Fujii Y, Haraguchi K et al (2020) Effects of perfluoroalkyl carboxylic acids on the uptake of sulfobromophthalein via organic anion transporting polypeptides in human intestinal Caco-2 cells. Biochem Biophys Rep 24:100807. https://doi.org/10.1016/j.bbrep.2020.100807
Kotlarz N, McCord J, Collier D et al (2020) Measurement of novel, drinking water-associated pfas in blood from adults and children in Wilmington, North Carolina. Environ Health Perspect 128:1–12. https://doi.org/10.1289/EHP6837
Küblbeck J, Hakkarainen JJ, Petsalo A et al (2016) Genetically modified Caco-2 cells with improved cytochrome P450 metabolic capacity. J Pharm Sci 105:941–949. https://doi.org/10.1016/S0022-3549(15)00187-2
Larhed AW, Artursson P, Gråsjo J et al (1997) Diffusion of drugs in native and purified gastrointestinal mucus. J Pharm Sci 86:660–665. https://doi.org/10.1021/js960503w
Li Y, Fletcher T, Mucs D et al (2018) Half-lives of PFOS, PFHxS and PFOA after end of exposure to contaminated drinking water. Occup Environ Med 75:46–51. https://doi.org/10.1136/oemed-2017-104651
Lupton SJ, Huwe JK, Smith DJ et al (2012) Absorption and excretion of 14C-perfluorooctanoic acid (PFOA) in angus cattle (Bos taurus). J Agric Food Chem 60:1128–1134. https://doi.org/10.1021/jf2042505
Markus J, Landry T, Stevens Z et al (2021) Human small intestinal organotypic culture model for drug permeation, inflammation, and toxicity assays. In Vitro Cell Dev Biol Anim 57:160–173
OECD (2018) Toward a new comprehensive global database of per- and polyfluoroalkyl substances (PFASs): Summary report on updating the OECD 2007 list of per- and polyfluoroalkyl substances (PFASs). Series on Risk Management No. 39. ENV/JM/MONO(2018)7
Olsen GW, Burris JM, Ehresman DJ et al (2007) Half-life of serum elimination of perfluorooctanesulfonate, perfluorohexanesulfonate, and perfluorooctanoate in retired fluorochemical production workers. Environ Health Perspect 115:1298–1305. https://doi.org/10.1289/ehp.10009
Onozato D, Yamashita M, Nakanishi A et al (2018) Generation of intestinal organoids suitable for pharmacokinetic studies from human induced pluripotent stem cells. Drug Metab Dispos. https://doi.org/10.1124/dmd.118.080374
Ozawa T, Takayama K, Okamoto R et al (2015) Generation of enterocyte-like cells from human induced pluripotent stem cells for drug absorption and metabolism studies in human small intestine. Sci Rep 5:1–11. https://doi.org/10.1038/srep16479
Panieri E, Baralic K, Djukic-Cosic D et al (2022) PFAS molecules: a major concern for the human health and the environment. Toxics 10:1–55. https://doi.org/10.3390/toxics10020044
Pizzurro DM, Seeley M, Kerper LE, Beck BD (2019) Interspecies differences in perfluoroalkyl substances (PFAS) toxicokinetics and application to health-based criteria. Regul Toxicol Pharmacol 106:239–250. https://doi.org/10.1016/j.yrtph.2019.05.008
Poothong S, Papadopoulou E, Padilla-Sánchez JA et al (2020) Multiple pathways of human exposure to poly- and perfluoroalkyl substances (PFASs): from external exposure to human blood. Environ Int 134:105244. https://doi.org/10.1016/j.envint.2019.105244
Prueksaritanont T, Gorham L, Hochman J et al (1996) Comparative studies of drug-metabolizing enzymes in dog, monkey, and human small intestines, and in Caco-2 cells. Drug 24:634–642
Rogers-Broadway KR, Karteris E (2015) Amplification efficiency and thermal stability of qPCR instrumentation: current landscape and future perspectives. Exp Ther Med 10:1261–1264. https://doi.org/10.3892/etm.2015.2712
Sedlak MD, Benskin JP, Wong A et al (2017) Per- and polyfluoroalkyl substances (PFASs) in San Francisco Bay wildlife: temporal trends, exposure pathways, and notable presence of precursor compounds. Chemosphere 185:1217–1226. https://doi.org/10.1016/j.chemosphere.2017.04.096
Spence JR, Mayhew CN, Rankin SA et al (2011) Directed differentiation of human pluripotent stem cells into intestinal tissue in vitro. Nature 470:105–109. https://doi.org/10.1038/nature09691.Directed
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 102:15545–15550
Sun H, Chow ECY, Liu S et al (2008) The Caco-2 cell monolayer: usefulness and limitations. Expert Opin Drug Metab Toxicol 4:395–411. https://doi.org/10.1517/17425255.4.4.395
Takayama K, Negoro R, Yamashita T et al (2019) Generation of human iPSC-derived intestinal epithelial cell monolayers by CDX2 transduction. Cmgh 8:513–526. https://doi.org/10.1016/j.jcmgh.2019.06.004
Tamminen K, Balboa D, Toivonen S et al (2015) Intestinal commitment and maturation of human pluripotent stem cells is independent of exogenous FGF4 and R-. PLoS ONE. https://doi.org/10.1371/journal.pone.0134551
Terao T, Hisanaga E, Sai Y et al (1996) Active secretion of drugs from the small intestinal epithelium in rats by P-glycoprotein functioning as an absorption barrier. J Pharm Pharmacol 48:1083–1089. https://doi.org/10.1111/j.2042-7158.1996.tb05904.x
Uhlén M, Fagerberg L, Hallström BM et al (2015) Tissue-based map of the human proteome. Science (1979). https://doi.org/10.1126/science.1260419
UNEP (2020) Stockholm Convention on persistent organic pollutants (POPS)—text and annexes revised in 2019. UNEP-POPS-COP-CONVTEXT-2021.English
UNEP (2022) Stockholm Convention on persistent organic pollutants. Amendment to Annex A: Listing of perfluorohexane sulfonic acid (PFHxS), its salts and PFHxS-related compounds. Decision SC-10/13: C.N.401.2022.TREATIES-XXVII.15
US Food and Drug Administration (2020) In vitro drug interaction studies-cytochrome P450 enzyme-and transporter-mediated drug interactions guidance for industry. Center for Drug Evaluation and Research, Beltsville
van der Hee B, Loonen LMP, Taverne N et al (2018) Optimized procedures for generating an enhanced, near physiological 2D culture system from porcine intestinal organoids. Stem Cell Res 28:165–171. https://doi.org/10.1016/j.scr.2018.02.013
van der Hee B, Madsen O, Vervoort J et al (2020) Congruence of transcription programs in adult stem cell-derived jejunum organoids and original tissue during long-term culture. Front Cell Dev Biol. https://doi.org/10.3389/fcell.2020.00375
Vanuytsel T, Tack J, Farre R (2021) The role of intestinal permeability in gastrointestinal disorders and current methods of evaluation. Front Nutr 8:717925. https://doi.org/10.3389/fnut.2021.717925
Vestergren R, Cousins IT (2009) Tracking the pathways of human exposure to perfluorocarboxylates. Environ Sci Technol 43:5565–5575. https://doi.org/10.1021/es900228k
Xu Y, Fletcher T, Pineda D et al (2020) Serum half-lives for short-and long-chain perfluoroalkyl acids after ceasing exposure from drinking water contaminated by firefighting foam. Environ Health Perspect 128:1–11. https://doi.org/10.1289/EHP6785
Zhao W, Zitzow JD, Ehresman DJ et al (2015) Na+/taurocholate cotransporting polypeptide and apical sodium-dependent bile acid transporter are involved in the disposition of perfluoroalkyl sulfonates in humans and rats. Toxicol Sci 146:363–373. https://doi.org/10.1093/toxsci/kfv102
Zhao W, Zitzow JD, Weaver Y et al (2017) Organic anion transporting polypeptides contribute to the disposition of perfluoroalkyl acids in humans and rats. Toxicol Sci 156:84–95. https://doi.org/10.1093/toxsci/kfw236
Acknowledgements
This research was supported by the Dutch Ministry of Agriculture, Nature and Food Quality (Grant: KB37-001-003 and KB-37-002-036) and by Topsectoren TKI Agri & Food (Grant: LWV19125).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Janssen, A.W.F., Duivenvoorde, L.P.M., Beekmann, K. et al. Transport of perfluoroalkyl substances across human induced pluripotent stem cell-derived intestinal epithelial cells in comparison with primary human intestinal epithelial cells and Caco-2 cells. Arch Toxicol (2024). https://doi.org/10.1007/s00204-024-03851-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00204-024-03851-x