Abstract
Background
Cancer immunotherapy approaches that elicit immune cell responses, including T and NK cells, have revolutionized the field of oncology. However, immunosuppressive mechanisms restrain immune cell activation within solid tumors so additional strategies to augment activity are required.
Methods
We identified the co-stimulatory receptor NKG2D as a target based on its expression on a large proportion of CD8+ tumor infiltrating lymphocytes (TILs) from breast cancer patient samples. Human and murine surrogate NKG2D co-stimulatory receptor-bispecifics (CRB) that bind NKG2D on NK and CD8+ T cells as well as HER2 on breast cancer cells (HER2-CRB) were developed as a proof of concept for targeting this signaling axis in vitro and in vivo.
Results
HER2-CRB enhanced NK cell activation and cytokine production when co-cultured with HER2 expressing breast cancer cell lines. HER2-CRB when combined with a T cell-dependent-bispecific (TDB) antibody that synthetically activates T cells by crosslinking CD3 to HER2 (HER2-TDB), enhanced T cell cytotoxicity, cytokine production and in vivo antitumor activity. A mouse surrogate HER2-CRB (mHER2-CRB) improved in vivo efficacy of HER2-TDB and augmented NK as well as T cell activation, cytokine production and effector CD8+ T cell differentiation.
Conclusion
We demonstrate that targeting NKG2D with bispecific antibodies (BsAbs) is an effective approach to augment NK and CD8+ T cell antitumor immune responses. Given the large number of ongoing clinical trials leveraging NK and T cells for cancer immunotherapy, NKG2D-bispecifics have broad combinatorial potential.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
More than 2 million women worldwide in 2020 were diagnosed with breast cancer according to Global Cancer Statistics 2020 [1]. HER2, a transmembrane glycoprotein overexpressed in 20–30% of breast cancers, represents a crucial therapeutic target. Trastuzumab, the first FDA approved drug targeting HER2, was developed more than twenty years ago and radically improved the prognosis of this disease. Pertuzumab was approved as an adjuvant therapy in 2017 and prevents HER2 heterodimer formation with HER1, HER3 and HER4 [2]. Both drugs exhibit antibody-dependent cellular cytotoxicity (ADCC) activating the innate immune system to kill tumor cells [3, 4]. Margetuximab, a HER2-targeted antibody with increased Fc domain affinity for CD16A, was developed for enhanced ADCC activity [5]. Despite the synergistic effect of trastuzumab with adjuvants and chemotherapy treatments, a large number of patients relapse indicating the need for additional therapies [6].
In the last decade, immunotherapy has become a successful strategy for treating various cancer indications and include inhibitors of cytotoxic T-lymphocyte associated protein 4 (CTLA-4), programmed cell death protein 1 (PD-1) and its corresponding ligand, programmed death-ligand 1 (PD-L1) [7]. Such therapies are limited by the requirement of tumor neo-epitopes and the presence of tumor-specific T cells that recognize tumor cells via T cell receptor (TCR) and peptide-major histocompatibility complex (MHC) interactions [8]. In contrast, TDBs, that simultaneously bind tumor-associated antigens on tumor cells and CD3 on T cells, are capable of MHC-independent redirected lysis of cancer cells engaging both tumor-specific and non-specific T cells. Similar approaches for NK cells, such as AFM13 which targets CD30 for T cell lymphoma, are also in clinical development [9]. BsAbs and chimeric antigen receptor T (CART) cells provide impressive benefit for hematological cancer patients including blinatumomab, a CD19-bispecific for the treatment of B cell acute lymphoblastic leukemia [10]. However, numerous challenges remain in the context of solid tumors including insufficient tumor T cell infiltration, immunosuppression and low/heterogeneous tumor antigen expression [11]. Therefore, identifying strategies to increase cytotoxic activity and function of immune cells is necessary for such therapeutics.
NKG2D is a C-type lectin-like receptor constitutively expressed on a large portion of human natural killer (NK), CD8+ T, invariant NKT and γδ T cells [12, 13]. NKG2D ligands are expressed under cellular stress conditions such as viral infection or cellular transformation. While NKG2D agonism can lead to NK cell activation or co-stimulation depending on species, NKG2D acts as a co-stimulatory receptor for T cells [14,15,16]. NKG2D plays a critical role in tumor immune surveillance and eradication as shown by NKG2D ligand overexpression [17, 18], antibody-mediated neutralization of NKG2D [19] and NKG2D-deficient mice [20]. NKG2D co-expression with CD39 and CX3CR1 is a cell surface signature for tumor-specific CD8+ T cells in mice and patients [21]. Together with NKG2D’s expression on NK and CD8+ T cells makes it an attractive target for augmenting antitumor effector immune responses.
In this study, we identified NKG2D as a co-stimulatory receptor target by its expression on a large proportion of CD8+ breast cancer TILs and its association with improved survival of atezolizumab-treated, anti-PD-L1, triple negative breast cancer (TNBC) patients. HER2-CRB, a BsAb bearing a HER2-targeting arm and an NKG2D agonist arm, was developed. We found that HER2-CRB was able to form immunological synapses (IS), induce cytokine production and increase cytotoxicity by primary NK cells and TDB-stimulated CD8+ T cells. HER2-CRBs were efficacious in both humanized and immunocompetent mouse models in combination with TDBs and enhanced effector function of NK and CD8+ T cells. Altogether, HER2-CRBs engage NK and CD8+ T cells leading to enhanced antitumor activity and has potential for broad combinability with immunotherapeutics.
Materials and methods
Flow cytometric analysis
Human DTCs from Discovery Life Sciences were thawed as indicated by the manufacturer. Cells were stained with fixable viability dye eFluor780 (eBioscience, 65-0865-14) to exclude dead cells for 30 min at 4 °C. To avoid non-specific binding to Fc receptors, cells were incubated for 10 min at 4 °C with human FcR blocking reagent (Miltenyi Biotec, 130-059-901). Cells were then stained as indicated using fluorescently-labeled antibodies (Table S1). For intracellular staining, the FoxP3 buffer kit (eBioscience, 00-5123-43) was used according to manufacture’s protocol. HER2-CRB binding was detected with an anti-human Fc-A488 antibody (Biolegend, 409,322). NKG2D ligand expression was detected using human NKG2D-Fc and detected with anti-human Fc-A488 antibody (Biolegend, 409,322). MICA/B was detected using an anti-MICA/B antibody. All antibody reagents for flow cytometry can be found in Table S1.
Primary NK cells and BT474 target cells were preloaded with 2 µM Cell Trace Violet (CTV) and 10 µM 5-(and-6)-(((4-chloromethyl)benzoyl)amino)tetramethylrhodamine (CMTMR), respectively. NK cells and BT474 target cells were conjugated by short centrifugation at a 1:1 ratio at 2 × 106 cells/ml in the presence of HER2-CRB at 1 or 10 µg/ml. After 10 min co-culture, cells were fixed with 3% paraformaldehyde. Conjugate formation was evaluated by flow cytometry using the BD LSR Fortessa X20.
Fo5 tumors were harvested and processed as previously described [22]. Cells were resuspended in FACS buffer and counted using ViCell WX (Beckman Coulter). For degranulation and intracellular cytokine analysis, 3 × 106 tumor cells were incubated for 5 h at 37 °C in complete media supplemented with CD107a/b-Alexa647 (BioLegend 121,610 and 108,512), Golgi Plug (BD Pharmingen 555,029), GolgiStop, (BD Pharmingen 55,474) and PMA-ionomycin cocktail (BioLegend 423,304). Cells were surface and intracellularly stained as described above and antibodies are listed in Table S1. Samples were analyzed using Symphony Flow Cytometers (BD Biosciences) and data analyzed using FlowJo Software (Tree Star).
Patient outcome analysis
The effect of KLRK1expression on patient survival was assessed retrospectively by stratifying patients into two groups based on median expression. Hazard ratios, confidence intervals and significance were calculated by a univariate Cox proportional hazards model on the binarized expression in R. IMPassion130 (NCT02425891) trial was previously described [23].
Antibodies and reagents
Anti-human HER2 clones 4D5 and 2C4 as well as anti-human NKG2D clones 26F3, 16F8 and 20E6 and anti-mouse NKG2D clone 19G8 were generated at Genentech. The National Institute of Standards and Technology (NIST) clone that targets a respiratory syncytial virus fusion protein was generated at Genentech [24]. BsAbs were produced as described previously [25]. TDBs and CRBs were diluted in vehicle (20 mM histidine acetate, 240 mM sucrose, 0.02% Tween 20, pH 5.5 buffer). Recombinant MICA extracellular domain (ECD) was produced at Genentech as described previously [26].
Monovalent binding kinetic measurement
We used a BIAcore T200 instrument for both HER2-TDB and mHER2-CRB. BIAcore research grade CM5 chips were activated with 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) and N-hydroxysuccinimide (NHS) reagents according to the supplier’s instructions, followed by coupling of NKG2D proteins. Human (R&D Systems 1299-NK) and mouse (R&D Systems 10,960-NK) NKG2D-Fc fusion antigens were coupled at a density of approximately 240 response units (RU). Threefold serial dilutions of HER2-TDB or mHER2-CRB were injected in HBS-EP buffer with a flow rate of 100µL/min at a temperature of 37 °C. Sensorgrams and maximum binding response units (RMax) were evaluated to determine level of binding to each antigen. Association rates (ka) and dissociation rates (kd) for antibodies that exhibited a binding response were calculated using a 1:1 Langmuir binding model (BIAcore T200 Evaluation Software version 2.0). For these, the equilibrium dissociation constant (KD) was calculated as the ratio kd/ka.
Cell culture
HCC2218 cells (ATCC, CRL-2343) were maintained with RPMI 1640 media supplemented with 10% FCS and 2 mM L-Glutamine. BT474 cells (ATCC) containing a stable nucLight™ (Essen Instruments, Inc.) red fluorescent protein (RFP) were maintained in RPMI 1640 media supplemented with 10% FCS, 2 mM L-Glutamine. Jurkat-NFAT luciferase cells (BP Bioscience, 60,621) were engineered to overexpress mouse and human NKG2D. Cells were maintained and selected in RPMI 1640 + 10% FCS + 1000 µg/ml Geneticin + 0.4 µg/ml puromycin. NK92-6F5 cell line was modified from the original NK92 cell line (ATCC, CRL-2407). Fcgr3a (encoding CD16A) was introduced by transfection using vector originated from Glycart/Roche with puromycin selection. Cells were maintained in RPMI 1640 + 10% FBS + L-Glu + 1:1000 ß-Mercaptoethanol + 20U/ml Il2 (Millipore Sigma, 11,011,456,001).
Peripheral blood mononuclear cells (PBMCs) were separated from healthy volunteer blood through the Genentech employee donation program. Blood was collected with written informed consent from donors and performed under an Institutional Review Board-approved protocol. PBMCs were harvested using 50 ml Leucosep Tubes (Greiner, 22-7290) and lymphocyte separation medium (MP Biomedicals, 0850494). CD8+ cells were isolated from PBMCs using human CD8+ Isolation Kit from Miltenyi Biotec (130-094-156). NK cells were isolated from PBMCs using NK cell isolation kit from Miltenyi Biotec (130-092-657). RPMI 1640 + 10% FBS + L-glutamine + 2-b-Mercaptoethanol + Na/Py + MEM: Non-essential amino acids were added to media for all CD8 + and NK cell assays. Purified cells with purity of > 90% and viability > 95% were used for assays. NK cells were used fresh while CD8 + T cells were used fresh or after cryopreservation. For NK cell conjugation and immune synapse assays, PBMCs were isolated from healthy donor samples obtained from the French blood bank (EFS). Primary NK cells were isolated as described above and were frozen directly post-isolation or after 14-17d of expansion in the NK MACS medium (Miltenyi Biotec, 130–114-429) supplemented by Human AB serum (5%, Sigma, H3667), 50 ng/ml IL-2 (Miltenyi Biotec, 130-097-746) and 10 ng/ml IL-15 (Miltenyi Biotec, 130-097-746). Primary NK cells were used after an overnight resting phase in NK MACS medium supplemented by Human AB serum. For some experiments using preactivated or expanded (activated) NK cells, 50 ng/ml IL-2 and 10 ng/ml IL-15 were added during the resting phase.
Immunological synapse visualization
Primary NK cells preloaded with CTV (2 µM) and BT474 target cells were conjugated by short centrifugation at a 1:1 ratio (1 NK for 1 target cell) at 2 × 106 cells/ml in the presence of HER2-CRB at 1 µg/ml. Cells were seeded on poly-L-lysine pre-coated slides and incubated at 37 °C. After 10 min of co-culture, cells were fixed with 3% paraformaldehyde, permeabilized with 0.1% saponin in PBS with 3% BSA and stained in the permeabilization buffer with phalloidin-Alexa546 (1/40, Fisher,#A22283) and anti-phospho-tyrosine (mouse IgG2b, 1/20, Santa Cruz, #sc-7020) followed by incubation with goat-anti-mouse IgG2b-secondary antibody conjugated to Alexa488 (1/200, Fisher, #A21141). Slides were mounted in the EverBrite Hardset Mounting Medium (Biotium, #23,003). 30 conjugates per condition were acquired using the microscope confocal spinning disk Axioobserver Z1 CSUW1. Definition of the IS region and quantification of pTyr and phalloidin signal intensities in the IS area were performed with Metamorph software.
Luciferase assays
BT474 cells (10,000cells /well) were seeded one day prior to co-culture with Jurkat-NFAT luciferase mouse or human NKG2D overexpressing cells at an E:T ratio 1:1 in the presence of BsAbs at a final volume of 100 µl. After 6 h incubation at 37 °C at 5% CO2, 100µL of ONE-Step luciferase assay system (BPS Bioscience, 60690) was added and samples were gently agitated for 20 min at room temperature. A luminometer was used to measure luminescence.
In vitro cytotoxicity assays
HCC2218 cells were co-cultured with NK92-6F5 cells in the presence of BsAbs. Tumor cells were enumerated by HER2 + gated cell counts after acquisition of a defined volume of single-cell suspension by flow cytometry. Primary NK cells were stimulated with IL-15 at 10 ng/ml (Peprotech, 200-15) overnight at 37 °C at 5% CO2 prior to use. BT474-nucLight RFP cells were seeded one day before the experiment in a clear bottom black well microplate (Corning, 3603) and CD8+ T or aforementioned stimulated NK cells were added in the presence of molecules as indicated. For MICA-ECD competition experiments, BT474-nucLight RFP cells were co-cultured with purified normal donor CD8+ T cells at an E:T ratio of 1:1 (10,000:10,000 cells). No TDB or 10 ng/ml of HER2.2C4-TDB with 0, 0.6 or 6.0 nM HER2-CRBs were added to co-cultures. Plates were placed in Incucyte S3 instrument (Essen Bioscience) at 37 °C at 5% CO2 and were scanned every 4 h for 7d.
Western blot
Primary NK cells were prestimulated overnight with 10 ng/ml IL-15. Dynabeads™ M-450 tosylactivated beads (Thermo Fisher, 14,013) were incubated overnight with human HER2-Fc (R&D, 1129-ER) at 100 µg/ml at room temperature with gentle rotation. 3 × 106 NK cells were co-cultured at a 1:1 ratio with HER2-coated beads and incubated at 37 °C for 5 or 15 min. The reaction was stopped by adding RIPA supplemented with protease and phosphatase inhibitors (ThermoFisher, 78,440). Cells were centrifugated for 15 min at 14000 rpm at 4 °C, supernatants were stored at − 80 °C. The Jess Western blot system from Protein Simple was used according to manufacturer’s instructions. 2 µg of total protein was added and anti-phospho-ERK antibody (Cell Signaling, 4370S, 1:25 dilution), anti-ERK (Cell Signaling, 46,955, 1:50 dilution) and anti-Hsp60 (R&D, AF1800, 1:50) were used as primary antibodies. The anti-rabbit-HRP kit was used for detection (Protein Simple, DM-001).
RNA-seq and analysis
After 24 h in the stated conditions, human CD8+ T cells were directly sorted into lysis buffer provided by the RNeasy micro kit (Qiagen, 74,004). The RNA was extracted according to manufacturer’s instructions. For Founder #5 mouse ex vivo studies, 4d following the stated in vivo treatment CD45+ CD8+ CD3+ CD4- viability dye cells were directly sorted into lysis buffer and RNA was extracted as described above. The libraries were sequenced on Illumina HiSeq4000 (Illumina). An average of 30 million single-end RNA-seq reads (50 base pairs (bp)) per sample were obtained. Sequencing reads were aligned to the human genome (GRCh38) or the mouse genome (NCBI Build 38) using GSNAP version ‘2013-10-10’, allowing for a maximum of two mismatches per 50 bp sequence (parameters: ‘-M 2 -n 10 -B 2 -i 1 -N 1 -w 200,0000-E1–pairmax-rna = 200,000–clip-overlap’). Lastly, the HTSeqGenie R package was used to quantify gene expression levels by calculating the number of reads mapped to the exons of each RefSeq gene. Transcript annotation was based on the RefSeq database NCBI Annotation Releases 104 for mouse. Read counts were scaled by library size, quane normalized and precision weights calculated using the 613 “voom” R package. Subsequently, differential expression analysis on the normalized count data was performed using the “limma” R package by contrasting treated samples with control samples using Partek Flow software. Low expressed genes were filtered out if the mean of the group was ≤ 1.0. Gene expression was considered significantly different across groups with an FDR-adjusted p-value ≤ 0.05. In addition, gene expression was obtained in the form of normalized Reads Per Kilobase gene model per Million total reads (nRPKM).
We tested for the enrichment of particular gene categories to identify relevant biological processes associated with differential expression by using functions from the “clusterProfiler” R package and the MSigDB gene set collection. In the case of differential expression, treated samples were compared to control samples, genes were thresholded by p-value ≤ 0.05 and ranked based on the signed LogFC for directionality. Subsequently gene set enrichment was performed using the clusterProfiler::GSEA function using Partek Flow software.
Mice and mouse models
Animal protocols were reviewed and approved by Institutional Animal Care and Use Committee (IACUC) at Genentech prior to experimentation. Female NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ (NSG) mice at 6–8 weeks of age were inoculated subcutaneously with five million JimT-1 cells in HBSS/Matrigel in a volume of 0.1 ml per mouse. Human PBMCs obtained from a human donor leukopack were cultured overnight in non-activating conditions. The next day, mice were not inoculated with 1 × 107 human PBMCs intra-peritoneally. When tumor volumes ranged from 100 to 200 mm3, mice were treated intravenously by tail vein with 0.5 mg/kg HER2-TDB (anti-HER2.4D5xanti-CD3e.40G5c) diluted in vehicle with or without 2 mg/kg HER2-CRB. The vehicle buffer was used as a negative control.
The MMTV-Her2-transgenic Founder #5 tumor model was developed from MMTV-HER2 transgenic mice [27]. Founder #5 tumors were expanded in vivo in female FVB WT mice at 6–8 weeks of age. Tumors were removed and cut into tumor fragments that were surgically implanted into the mammary fat pad of tumor naive female FVB WT mice at 6–8 weeks of age. When mean tumor volume was 350–570 mm3, 6–16 mice, based on efficacy or pharmacodynamic studies, were treated with vehicle or 0.5 mg/kg of mHER2-TDB (anti-HER2.4D5xanti-CD3e.2C11) in combination with vehicle or 2 mg/kg of mHER2-CRB. For all mouse studies, mice were removed from study when tumors reached a volume of ≥ 1500 mm3 or exhibited deterioration of heath.
Cytokine and chemokine analysis
Mouse blood was collected from the retroorbital sinus and centrifuged at 10,000 rpm for 10 min. Sera were transferred into a clean 96 V-bottom plate and stored in − 80 °C freezer until cytokine analysis was performed using Premix Panel I 32-Plex (Millipore) as previously described [22].
Results
NKG2D is expressed on intratumoral CD8+ T cells and is prognostic in TNBC
To identify a co-stimulatory receptor to enhance NK and CD8+ T cell activity in breast cancer, we analyzed expression of the co-stimulatory receptors 4-1BB, GITR, ICOS, NKG2D and OX40 on TILs from human breast cancer dissociated tumor cells (DTCs). A greater proportion of CD8+ T cells expressed NKG2D compared to 4-1BB, GITR, ICOS or OX40. (Fig. 1A). NKG2D was expressed primarily on CD8+ T cells with little expression on CD4+ T cell subpopulations (Fig. 1B–C). In contrast, other co-stimulatory receptors exhibited expression on effector CD4+FoxP3- and regulatory CD4+FoxP3+ (Treg) T cells. Expression of 4-1BB, GITR, ICOS and OX40 on Tregs made these targets less favorable as signaling through these receptors can elicit negative antitumoral effects [28]. NK cells were too few within DTC samples to accurately assess co-stimulatory receptor expression.
NKG2D is expressed on CD8 + T cells from human breast cancer samples and is associated with better prognosis in atezolizumab-treated TNBC patients. Percentage of co-stimulatory receptor positive A CD8 + , B FoxP3- CD4 + and C FoxP3 + CD4 + T cells from human-dissociated breast cancer tumor cells. D Kaplan–Meier analysis of Overall Survival (OS, left) and Progression-free Survival (PFS, right) among patients with TNBC treated with placebo + nanoparticle albumin-bound (Nab)-paclitaxel (top) or with Atezolizumab + Nab-Paclitaxel (bottom) stratified by high and low KLRK1 (encoding NKG2D) expression normalized to a CD8 signature shown above. (E) Frequency of NKG2D + CD4 + and CD8 + T cells (top) and corresponding NKG2D mean fluorescence intensities (MFI, bottom) from matched PBMCs and human-dissociated HER2 + breast cancer DTCs. Mann–Whitney test used for statistics; *, p ≤ 0.05; **p, ≤ 0.01
TNBC patients from the IMPassion130 trial were separated based on high and low expression of KLRK1 (encoding NKG2D) with no normalization or normalization to CD8+ T and NK cell signatures to rule out the contribution of these cell types. Patients stratified by high KLRK1 expression exhibited significantly improved outcome for non-normalized (Fig. S1A) and data normalized to NK (Fig. S1B) or CD8+ T cell (Fig. 1D) signatures for patients treated with atezolizumab, anti-PD-L1, in combination with nab-paclitaxel. Importantly, KLRK1 expression was not prognostic in the chemotherapy only treated population regardless of normalization demonstrating an association between KLRK1 expression and atezolizumab treatment. Considering TNBC lacks defined tumor antigens and TILs share similarities between TNBC and HER2+ breast cancer [29], we analyzed NKG2D expression on TILs from HER2+ tumors and matched PBMCs. We confirmed NKG2D was expressed on CD8+ , but not CD4+ TILs, in HER2+ tumors and matched PBMC samples (Fig. 1E).
HER2-CRB induces NK-tumor cell immune synapse formation and is signaling competent
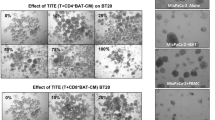
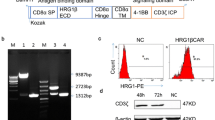
A HER2-targeted (anti-HER2 clone 4D5) anti-NKG2D (clone 26F3)-bispecific (HER2-CRB) was produced using knob-into-hole methodology (Fig. 2A). Fc receptor binding was attenuated via introduction of an N297G mutation. HER2-CRB bound to hNKG2D with an equilibrium dissociation constant of 1.752E-7 M (Fig. S2A). We developed a non-tumor-targeted control, NIST-CRB (anti-NISTxanti-NKG2D; NIST, National Institute of Standards and Technology [24]) and an NKG2D non-binding control, HER2/NIST (Fig. 2A). BT474 cells that express high levels of HER2 (Fig. S2B) and low levels of NKG2D ligands (Fig. S2C) were used for the following assays. HER2-CRB bound to BT474 cells, but not NIST-CRB as expected (Fig. S2D). Both HER2- and NIST-CRB bound to hNKG2D overexpressing Jurkat-NFAT luciferase cells (Jurkat-hNKG2D+ NFAT-Luc, Fig. S2E) that express high levels of NKG2D (Fig. S2F). HER2-CRB formed conjugates between BT474 and primary NK cells that were unstimulated, IL-2 and IL-15 preactivated to increase NKG2D expression (Fig. S2G) or expanded (activated) with the greatest proportion of conjugate formation observed for activated NK cells (Fig. 2B). Confocal microscopy confirmed that HER2-CRB significantly improved immune synapse (IS) formation and phosphotyrosine staining on NK cells within the IS when compared to NIST-CRB (Fig. 2C–D). Using human HER2-coated beads, HER2-CRB increased pERK in primary NK cells, a downstream kinase of the NKG2D signaling pathway (Fig. 2E). HER2-CRB induced transient NKG2D internalization, in contrast to NIST-CRB (Fig. S2H), consistent with ERK signaling [30]. Therefore, HER2-CRB bridges tumor cells to NK cells and exhibits signaling competency.
HER2-CRB induces NK cell immunological synapse formation and signaling. A Diagram of HER2-CRB, NIST-CRB and HER2/NIST molecules. B Flow cytometry-based conjugate quantification following 10 min incubation of primary NK cells conjugated with BT474 cells in the absence or presence of 1 or 10 µg/mL HER2-CRB. Results obtained from two independent healthy donors. Representative dot plot (left) and quantified data (right). C Representative images of cell trace violet (CTV)-labeled primary NK cells conjugated with CMTMR-labeled BT474 cells in the presence of NIST- or HER2-CRB. Quantified area depicting immunological synapse used to quantify phosphotyrosine (pTyr) and phalloidin signals. D Quantification of actin and pTyr intensities by confocal imaging of 30 individual interactions after 10 min incubation of conjugation of primary NK cells with BT474 cells in the presence of NIST- or HER2-CRB. Similar results were obtained from another independent healthy donor. E Western blot of phosphorylated ERK, total ERK and loading control Hsp60 after 5 and 15 min incubation of NK cells, HER2-coated beads and either HER2-CRB or NIST-CRB. Mann–Whitney test used for statistics; ****, p ≤ 0.0001
HER2-CRB promotes NK cell activation
To validate the functional activity of HER2-CRB, first we utilized NK92.6DF5 cells which express NKG2D (Fig. S3A) and HCC2218 cells with high HER2 positivity (Fig. S3B) and low NKG2D ligand expression (Fig. S3C). Although NK92.6DF5 cells exhibited high spontaneous killing compared to primary NK cells (Fig. S3D), cytotoxicity was increased with HER2-CRB (Fig. 3A). To study HER2-CRB activity in a more relevant cell type, we used IL-15-stimulated primary NK cells as this cytokine induces higher levels of NKG2D and cytotoxic potential [31]. Similar to NK92.6DF5 cells, HER2-CRB reduced target cell numbers when compared to negative controls (Fig. 3B). Cytokines and chemokines were elevated with HER2-CRB treatment compared to negative controls (Fig. 3C). HER2-CRB increased CD69+ and Ki67+ NK cell frequencies and higher expression levels of these proteins (Fig. S3E–F). To assess how proinflammatory conditions influence HER2-CRB activity, we performed a cell killing assay using 24 h conditioned media from co-cultures of PBMCs containing T cells with BT474 cells in the presence of HER2-TDB (anti-HER2 clone 4D5xanti-CD3) or NIST-TDB (anti-NISTxanti-CD3, negative control). NK cells were co-cultured with BT474 cells in the presence of HER2-CRB, negative controls or trastuzumab as a positive control with or without conditioned media. Conditioned media from co-culture with HER2-TDB induced greater NK cell cytotoxic activity with HER2-CRB compared to NIST-CRB or conditioned media with NIST-TDB (Fig. 3D–E). Altogether, these data support that HER2-CRB enhanced NK cell effector function that is augmented by proinflammatory conditions.
HER2-CRB elicits NK cell cytotoxicity. A Percentage of killing normalized to NIST-CRB for NK92.6DF5 cells co-cultured with HER2 + HCC2218 cells for three days in the presence of titrated NIST-l or HER2-CRB. B Fold-change of BT474 cell killing normalized to time 0 h. BT474 cells were co-cultured with IL-15 prestimulated primary NK cells with an E:T ratio of 1:1 in the absence or presence of HER2-CRB. Data derived from three independent donors. C NK cell secretion of interferon-gamma (IFN-γ), tumor necrosis factor-alpha (TNF-α) as well as chemokines CXC chemokine motif ligand 10 (CXCL10), macrophage inflammatory protein-1α (MIP-1α) and MIP-1β from experiment depicted in (B) on day 4. Three independent donors illustrated. D Fold-change of BT474 cell killing normalized by time 0 h. BT474 cells were co-cultured with primary NK cells at an E:T ratio of 1:1 in the presence of HER2-CRB, negative controls or trastuzumab with conditioned media obtained from 24 h co-culture of PBMCs and BT474 cells in the presence of HER2-4D5-TDB or NIST-TDB. Three independent donors are represented for donor conditioned media mix cultured with NK cells from 3 separate donors (9 total samples). E Area under the curve quantification from (D). Mann–Whitney test was used for statistical significance; *p, ≤ 0.05
HER2-CRB augments HER2-TDB-mediated CD8+ T cell activation
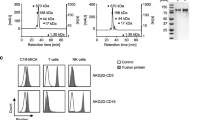
The variable domain sequence of two anti-HER2 clones, 4D5 and 2C4, have been used to generate TDBs [32]. We used both HER2-TDBs in combination with HER2-CRB to assess activity on T cells. Jurkat-HuNKG2D + NFAT-Luc cells were co-cultured with BT474 cells in the presence of sub-optimal concentrations of HER2.2C4-TDB, HER2-TDB or NIST-TDB in combination with HER2-CRB or negative control molecules. HER2-CRB induced greater luciferase activity when combined with HER2.2C4-TDB and HER2-TDB compared to NIST-CRB (Fig. 4A). For primary CD8+ T cell assays, donors were screened for NKG2D expression and exhibited similar proportions of NKG2D+ CD8+ T cells as patient DTCs and PBMCs (Fig. S4)A. NKG2D frequencies and MFI did not vary dramatically across CD8+ T cell subtypes (Fig. S4B-C). Treatment of CD8+ T and BT474 cell co-cultures with HER2-TDB resulted in a narrow concentration window for killing activity (Fig. S4D). In contrast, HER2.2C4-TDB provided a discernable dose titration (Fig. S4E) and a concentration of 10 ng/mL was selected to assess the effects of HER2-CRB on T cell activity. Both HER2- and HER2.2C4-TDBs increased percentages of NKG2D+ CD8+ T cells following stimulation (Fig. S4F). HER2-CRB enhanced HER2.2C4-TDB-mediated CD8+ T cell cytotoxicity against BT474 cells in a dose dependent manner (Fig. 4B) while HER2-CRB combined with the negative control NIST-TDB exhibited no activity (Fig. S4G). 600 nM of recombinant MICA extracellular domain (ECD) did not interfere with activity of HER2-CRB with anti-NKG2D clone 26F3. This was in contrast to CRBs made with alternate anti-NKG2D clones where MICA-ECD blocked activity which was more pronounced at lower CRB concentrations (Fig. S4H). HER2-CRB led to increased secretion of proinflammatory mediators (Fig. 4C). The CRB negative controls did not alter cytokine levels compared to HER2.2C4-TDB alone (Fig. S4I). To understand how NKG2D signaling influences CD8+ T cells, we performed bulk RNA-seq analysis 24 h after co-culture with BT474 cells. HER2.2C4-TDB by itself elicited robust transcriptomic changes and gene set enrichment analysis (GSEA) was consistent with TCR engagement (Fig. S4J–K). Addition of HER2-CRB enhanced expression of transcripts associated with T cell activation (Fig. 4D). HER2-CRB enhanced multiple biological processes including oxidative phosphorylation, cytokine pathway signaling, translation, transcription and proliferation (Fig. 4E) and a gene signature of acutely activated CD8+ T cells (Fig. 4F) highlighting NKG2D’s multifaceted biological effects upon human CD8+ T cells.
HER2-CRB enhances HER2-TDB-mediated T cell activation and cytotoxic activity. A Fold-change of luciferase activity normalized to untreated Jurkat-NKG2D + NFAT-Luc cells. Jurkat cells were co-cultured with BT474 cells for 6 h with either HER2.2C4-TDB or HER2-TDB or NIST-TDB in combination with either HER2-CRB or controls. B BT474 target cell killing normalized to time 0 h. BT474 cells were co-cultured for over five days with primary CD8 + T cells at an E:T ratio of 1:1 in the presence of 10 ng/ml of HER2-2C4-TDB alone or in combination with titrated HER2-CRB. Representative of four independent donors. C Cytokine quantification of CD8 + T cell and BT474 cell co-culture at an E:T of 1:1 in the presence of 10 ng/ml of HER2.2C4-TDB alone or in combination with HER2-CRB after 96 h. D Differential gene expression analysis from bulk RNA-seq on CD8 + T cells co-cultured for 24 h with BT474 cells in the presence of 10 ng/mL of HER2.2C4-TDB alone or in combination with HER2-CRB represented as a volcano plot. Blue dots represent Log(fold-change) of ≤ -0.7 and red dots depict Log(fold-change) ≥ 0.7, p ≤ 0.05. Black points are below stated thresholds. E GSEA data from Hallmark (top) and Reactome (bottom) gene sets from results in (D). F GSEA from immunologic gene sets (C7) from data in (D)
HER2-CRB improves in vivo activity of HER2-TDB
To assess in vivo activity of HER2-CRB, immunodeficient mice were inoculated with a HER2 + trastuzumab and pertuzumab resistant cell line, JIMT-1 [33]. Tumor bearing mice were or were not inoculated with human PBMCs and treated with HER2-CRB and/or HER2-TDB. Although HER2-CRB and HER2-TDB are composed of the same anti-HER2 clone and compete for the same HER2 epitope, HER2-TDB is efficacious in vivo on high HER2 expressing tumors when co-dosed with trastuzumab (bivalent clone 4D5) [34]. Combination of HER2-CRB with HER2-TDB significantly reduced tumor growth compared to HER2-TDB alone (Figs. 5A and S5A). The negative control cohorts treated with vehicle, HER2-CRB alone or conditions without PBMCs had no effect on tumor growth.
HER2-CRB improves HER2-TDB in vivo activity. A JimT-1 tumor volume growth over-time in NSG mice (left) with or without adoptive transfer of human PBMCs treated with HER2-TDB alone, HER2-CRB alone or in combination (combo). Individual mouse tumor volumes are represented in right panel. Slash represents a mouse death due to health conditions. (B, left) Change in tumor volumes of the Fo5 tumor model. Mice were treated with mHER2-TDB, mHER2-CRB or in combination (combo). Vehicle treatment was used as a negative control. (Right) Individual tumor volumes at day 4 and day 7 from. Mann–Whitney test used for statistical analysis; *p, ≤ 0.05; **, p ≤ 0.01
To study antitumor activity of HER2-CRB in an immunocompetent system, we developed a mouse-reactive HER2-CRB (anti-HER2 clone 4D5xanti-mouse NKG2D, mHER2-CRB). mHER2-CRB had a monovalent equilibrium dissociation constant of 7.183E-9 M for murine NKG2D (Fig. S5B). mHER2-CRB bound to the surface of Jurkat-NFAT luciferase cells overexpressing murine NKG2D (Fig. S5C–D). mHER2-CRB in combination with a murine surrogate HER2-TDB (anti-HER2 clone 4D5xanti-mCD3ε clone 2C11, mHER2-TDB) enhanced luciferase activity in the aforementioned cell line compared to mHER2-TDB alone (Fig. S5E). To assess mHER2-CRB activity on mouse CD8+ T cells in vitro, we confirmed human HER2 overexpressing MC38 cells were sensitive to mHER2-TDB and selected a sub-optimal dose of 72 ng/mL (Fig. S5F). Using this assay, we confirmed that mHER2-CRB enhanced mouse CD8+ T cell cytotoxicity (Fig. S5G). To evaluate the biological consequences of mHER2-CRB in an immunocompetent in vivo model, we used the Founder #5 (Fo5) tumor model developed from MMTV-HER2 transgenic mice that exhibits inherent resistance to trastuzumab [27]. NKG2D expression was confirmed on CD8 + T cells following mHER2-TDB treatment in Fo5 tumors (Fig. S5H). Mice were treated with either vehicle, mHER2-TDB alone, mHER2-CRB alone or both molecules combined. Combination treatment significantly decreased tumor volume compared to single-agent cohorts (Figs. 5B and S5I). Therefore, HER2-CRBs enhance HER2-TDB in vivo efficacy in both humanized and murine immunocompetent models.
NKG2D-CRB augments NK and CD8+ T cell effector function in combination with HER2-TDB
Next we interrogated the pharmacodynamic consequences of mHER2-CRB and mHER2-TDB combination in the Fo5 model. Lymphocyte numbers were similar between HER2-TDB alone and combination cohorts (Fig. S6A–B). Significant increases in frequencies of CD8 + T and/or NK cells expressing Ki67, Granzyme B, CD107a/b, IFN-γ and TNF-α were detected in combination treatment compared to mHER2-TDB treatment alone (Fig. 6A–C). Proportions of CD8+ T cells expressing markers of dysfunction were similar between mHER2-TDB-treated and combination cohorts (Fig. S6C). Considering cytokine release syndrome (CRS) is a complication with TDBs in clinic, it is important to note that systemic proinflammatory cytokines/chemokines and body weights were similar between the mHER2-TDB and combination groups (Fig. S6D–E). Differential gene expression analysis on sorted CD8+ T cells from the combination treatment exhibited elevated expression of cell activation and cytotoxicity genes including Ccr5, Ccl5, Ifng, Prf1 and Gzmb (Fig. 6D). GSEA analysis showed enrichment of pathways associated with proinflammatory responses, cytokine signaling (Fig. 6E) and effector T cells (Fig. 6F) compared to mHER2-TDB alone. Altogether, engaging NKG2D with a HER2-targeted-BsAb led to improved antitumor activity and enhanced CD8+ and NK cell effector function when combined with HER2-TDB in vivo.
HER2-CRB promotes NK cell and CD8 + T cell effector function in vivo. Fo5 tumor-bearing mice were treated with NIST- or mHER2-TDB with NIST- or mHER2-CRB. Tumors were harvested 4 days following treatment for analysis. A Frequency of Ki67 + CD8 + T and NK cells. B Percent Granzyme B + and CD107a/b + CD8 + T and NK cells. C Percentages of IFN-γ + and TNF-α + CD8 + T and NK cells. D Differential gene expression analysis of bulk RNA-seq from sorted CD8 + TILs. Blue dots represent Log(fold-change) of ≤ − 0.7 and red dots depict Log(fold-change) ≥ 0.7, p ≤ 0.05. Black points are below stated thresholds. (E) GSEA data from Hallmark (top) and Reactome (bottom) gene sets from results in (D). F GSEA from immunologic gene sets (C7) from data in (D). Mann–Whitney test used for statistical analysis; *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001
Discussion
Given the advent of immunotherapeutic strategies that leverage NK and T cells, it is critical to develop additional agonists to boost therapeutic responses to overcome hurdles associated with the solid tumor microenvironment. Targeting co-stimulatory receptors is a clear option considering their potent activity and expression by immune cell subpopulations that elicit antitumor responses. CD28 as well as tumor necrosis factor superfamily (TNFRSF) members including 4-1BB, GITR and OX40 are a focus of both academia and industry. Broad expression of such targets on immune cells, both immunosuppressive and proinflammatory, and expression on non-hematopoietic cells could potentially hamper their application by acting as a therapeutic sink or causing unforseen biological consequences. For example, agonist antibodies against CD28 and 4-1BB have exhibited serious adverse events in human providing caution for agonizing such pathways [35, 36]. Mechanistically it has been proposed that the adverse effects of the CD28 agonist were driven by superagonist activity, CD4+ T effector memory cells and bivalency of the antibody [37, 38]. For 4-1BB, adverse effects have been attributed to FcγR biding of the antibody and liver myeloid cells [39,40,41]. Antigen-targeted bispecifics for both receptors have improved preclinical safety due to tumor-specific targeting and monovalency of effector arm or FcgR null mutations [42, 43]. CD28 and 4-1BB agonists engage both CD4+ and CD8+ T cells possibly making them more prone to robust cytokine production and may elicit CD4+ Treg immunosuppressive activity as both of these co-stimulatory receptors are expressed by this cell type [28, 44]. NKG2D’s expression profile is restricted to NK and CD8+ T cells, while it is also expressed on γδ-T and NKT cells that also contribute to antitumor immunity [13, 45]. Further, NKG2D, as opposed to TNFRSF members evaluated, was expressed on a large proportion of CD8+ TILs from breast cancer patients and had little to no expression on CD4+ T cell subsets. Therefore, we selected NKG2D for the development of a tumor-targeted-bispecific agonist to elicit CD8+ T and NK cell responses. This is a strategy that has been explored before; however, our study provides novel biological insight, and our approach is differentiated.
Few studies have directly evaluated the effects of NKG2D-bispecifics on CD8 + T cell responses. A study by Stamova et al. [46] demonstrated that an anti-CD33xULBP2 NKG2D ligand-based-bispecific enhanced in vitro CD8+ T cell cytotoxicity mediated by an anti-CD33xanti-CD3 TDB. An anti-HER2-RAE-1β fusion antibody with an effector competent Fc was shown to be efficacious in a human HER2 overexpressing syngeneic model and dependent on CD8+ T cells [47]. However, it is unclear if this dependency was mediated by NKG2D signaling on CD8+ T cells or induction of an adaptive immune response via Fc-directed NK cell-mediated killing and subsequent antigen presentation resulting in de novo tumor-specific responses. The latter was supported by tumor growth inhibition of HER2-negative tumors co-implanted with HER2 + tumors. Other studies have provided indirect evidence for CD8+ T cells following NKG2D-bispecific treatment using immunodeficient mouse xenograft models inoculated with human PBMCs in which few NK cells engraft [48,49,50]. In total, these studies provided little functional and mechanistic understanding. Our study provides greater insight into the functional role of NKG2D signaling on CD8+ T cells in the context of synthetic immunity. CD8+ T cell numbers were not increased with mHER2-CRB treatment in combination with mHER2-TDB; however, later timepoints may be required to observe such effects. NKG2D engagement enhanced CD8+ T cell effector function as demonstrated by increased GZMB, degranulation and cytokine production after ex vivo stimulation. Moreover, the combination resulted in improved tumor growth control without altering mouse body weight and circulating proinflammatory cytokines associated with CRS that are seen with TDB therapies highlighting potential low toxicity of HER2-CRB. Bulk RNA-Seq analysis of human CD8+ T cells following in vitro stimulation demonstrated enhanced metabolism, transcription, translation and activation. Mouse CD8+ TILs further supported enhanced inflammatory processes, but provided little evidence for other alterations observed in human CD8+ T cells. These differences may be due to timing of RNA-seq analysis, species disparities or in vitro versus in vivo conditions. Nonetheless, synthetic NKG2D signaling enhanced effector CD8+ T cell functionality that ultimately bolsters antitumor immune responses.
Our results confirmed that an antibody-based NKG2D-bispecific elicits NK cell responses in vitro [51] and shows for the first time engagement of both NK and CD8+ T cells by this class of molecule. Moreover, it provides in vivo mechanistic understanding of NK cells targeted with an NKG2D-bispecific demonstrating greater effector molecule expression and ex vivo cytokine production. However, improved NK cell effector responses may be mediated indirectly by greater CD8+ T cell activation and cytokine production. Conditioned media from PBMCs stimulated with TDB improved NK cell cytotoxic function in the presence of HER2-CRB supporting cytokines may contribute to enhanced NK cell responses in vivo. Regardless, NKG2D-CRB induced NK cell-target cell IS formation with enhanced pTYR at effector-target cell contacts. In addition, increased pERK signaling in NK cells co-cultured with hHER2-coated beads was observed with HER2-CRB demonstrating direct NK cell signaling with this molecule. Therefore, it is plausible that both mHER2-CRB and mHER2-TDB-induced cytokines contributed to augmented NK cell effector function in vivo.
Most studies have interrogated NKG2D ligand tumor antigen-targeted fusion proteins in vitro and in vivo with primary or immortalized NK cells [48,49,50, 52,53,54,55,56]. Tumor cells release soluble NKG2D ligands that can inhibit immunosurveillance and potentially compete with ligand-based-bispecifics. [57]. Antibody-based approaches can be tailored to not compete with human NKG2D ligand binding or have greater affinity for NKG2D thereby displacing weak affinity (0.16–25 µM) soluble ligands [58]. Our monovalent anti-hNKG2D arm has a stronger affinity (0.175 µM) than most NKG2D ligands. HER2-CRB blocked MICA-ECD binding, bound to NKG2D when MICA-ECD was prebound (data not shown) and its activity was not inhibited when MICA-ECD was added to tumor cell plus CD8+ T cell co-cultures in combination with HER2.2C4-TDB, in contrast to other HER2-CRBs with alternate anti-NKG2D arms. Some NKG2D ligands are highly polymorphic which could lead to development of anti-drug antibodies against ligand-based-bispecifics in polymorphism mismatched patients [59]. Therefore, NKG2D antibody approaches have greater suitability for development of therapeutic tumor-targeted-bispecifics for both biological and exposure-related concerns.
While we demonstrated proof of concept with HER2, a highly expressed tumor antigen in a subset of breast cancer patients, such an approach is applicable across indications given the availability of tumor-specific antigens. We have observed NKG2D-positivity on CD8+ T cells within patient samples from colorectal, ovarian and prostate cancers (data not shown) highlighting application for additional cancer types. Considering HER2-CRB enhances NK and CD8+ T cell functionality, this approach can be combined with checkpoint inhibitors, CART/-NK cells, ADCC-competent targeted antibodies and additional immunotherapeutic approaches. Overall, NKG2D-bispecifics represent a class of molecules with great applicability and combinability with immunotherapeutics that elicit NK and CD8+ T cell antitumor activity.
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Abbreviations
- ADCC:
-
Antibody-dependent cellular cytotoxicity
- BsAb:
-
Bispecific antibody
- CART:
-
Chimeric antigen receptor T
- CRB:
-
Co-stimulatory receptor-bispecific
- DTC:
-
Dissociated tumor cells
- FACS:
-
Flow cytometry
- hNKG2D:
-
Human NKG2D
- IACUC:
-
Institutional Animal Care and Use Committee
- IFN-γ:
-
Interferon gamma
- IS:
-
Immunological synapse
- Jurkat-HuNKG2D + NFAT-Luc:
-
Jurkat-NFAT luciferase cells that overexpress hNKG2D
- Jurkat-mNKG2D + NFAT-Luc:
-
Jurkat-NFAT luciferase cells overexpressing mNKG2D
- mNKG2D:
-
Mouse NKG2D
- NIST:
-
National Institute of Standards and Technology
- NK:
-
Natural killer
- TAA:
-
Tumor-associated antigen
- TDB:
-
T cell-dependent-bispecific
- TNBC:
-
Triple negative breast cancer
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249. https://doi.org/10.3322/caac.21660
Eiger D, Ponde NF, de Azambuja E (2019) Pertuzumab in HER2-positive early breast cancer: current use and perspectives. Future Oncol 15:1823–1843. https://doi.org/10.2217/fon-2018-0896
Lewis GD, Figari I, Fendly B, Wong WL, Carter P, Gorman C, Shepard HM (1993) Differential responses of human tumor cell lines to anti-p185HER2 monoclonal antibodies. Cancer Immunol Immunother 37:255–263. https://doi.org/10.1007/BF01518520
Scheuer W, Friess T, Burtscher H, Bossenmaier B, Endl J, Hasmann M (2009) Strongly enhanced antitumor activity of trastuzumab and pertuzumab combination treatment on HER2-positive human xenograft tumor models. Cancer Res 69:9330–9336. https://doi.org/10.1158/0008-5472.CAN-08-4597
Bang YJ, Giaccone G, Im SA et al (2017) First-in-human phase 1 study of margetuximab (MGAH22), an Fc-modified chimeric monoclonal antibody, in patients with HER2-positive advanced solid tumors. Ann Oncol 28:855–861. https://doi.org/10.1093/annonc/mdx002
O’Shaughnessy J, Gradishar W, O’Regan R, Gadi V (2023) Risk of recurrence in patients with HER2+ early-stage breast cancer: literature analysis of patient and disease characteristics. Clin Breast Cancer 23:350–362. https://doi.org/10.1016/j.clbc.2023.03.007
Ribas A, Wolchok JD (2018) Cancer immunotherapy using checkpoint blockade. Science 359:1350–1355. https://doi.org/10.1126/science.aar4060
Sharma P, Siddiqui BA, Anandhan S, Yadav SS, Subudhi SK, Gao J, Goswami S, Allison JP (2021) The next decade of immune checkpoint therapy. Cancer Discov 11:838–857. https://doi.org/10.1158/2159-8290.CD-20-1680
Sasse S, Brockelmann PJ, Momotow J et al (2022) AFM13 in patients with relapsed or refractory classical Hodgkin lymphoma: final results of an open-label, randomized, multicenter phase II trial. Leuk Lymphoma 63:1871–1878. https://doi.org/10.1080/10428194.2022.2095623
Jen EY, Xu Q, Schetter A et al (2019) FDA approval: blinatumomab for patients with B-cell precursor acute lymphoblastic leukemia in morphologic remission with minimal residual disease. Clin Cancer Res 25:473–477. https://doi.org/10.1158/1078-0432.CCR-18-2337
Middelburg J, Kemper K, Engelberts P, Labrijn AF, Schuurman J, van Hall T (2021) Overcoming challenges for CD3-bispecific antibody therapy in solid tumors. Cancers. https://doi.org/10.3390/cancers13020287
Bauer S, Groh V, Wu J, Steinle A, Phillips JH, Lanier LL, Spies T (1999) Activation of NK cells and T cells by NKG2D, a receptor for stress-inducible MICA. Science 285:727–729. https://doi.org/10.1126/science.285.5428.727
Kuylenstierna C, Bjorkstrom NK, Andersson SK et al (2011) NKG2D performs two functions in invariant NKT cells: direct TCR-independent activation of NK-like cytolysis and co-stimulation of activation by CD1d. Eur J Immunol 41:1913–1923. https://doi.org/10.1002/eji.200940278
Lanier LL (2015) NKG2D receptor and its ligands in host defense. Cancer Immunol Res 3:575–582. https://doi.org/10.1158/2326-6066.CIR-15-0098
Groh V, Rhinehart R, Randolph-Habecker J, Topp MS, Riddell SR, Spies T (2001) Costimulation of CD8alphabeta T cells by NKG2D via engagement by MIC induced on virus-infected cells. Nat Immunol 2:255–260. https://doi.org/10.1038/85321
Jamieson AM, Diefenbach A, McMahon CW, Xiong N, Carlyle JR, Raulet DH (2002) The role of the NKG2D immunoreceptor in immune cell activation and natural killing. Immunity 17:19–29. https://doi.org/10.1016/s1074-7613(02)00333-3
Diefenbach A, Jensen ER, Jamieson AM, Raulet DH (2001) Rae1 and H60 ligands of the NKG2D receptor stimulate tumour immunity. Nature 413:165–171. https://doi.org/10.1038/35093109
Cerwenka A, Baron JL, Lanier LL (2001) Ectopic expression of retinoic acid early inducible-1 gene (RAE-1) permits natural killer cell-mediated rejection of a MHC class I-bearing tumor in vivo. Proc Natl Acad Sci USA 98:11521–11526. https://doi.org/10.1073/pnas.201238598
Smyth MJ, Swann J, Cretney E, Zerafa N, Yokoyama WM, Hayakawa Y (2005) NKG2D function protects the host from tumor initiation. J Exp Med 202:583–588. https://doi.org/10.1084/jem.20050994
Guerra N, Tan YX, Joncker NT et al (2008) NKG2D-deficient mice are defective in tumor surveillance in models of spontaneous malignancy. Immunity 28:571–580. https://doi.org/10.1016/j.immuni.2008.02.016
Pauken KE, Shahid O, Lagattuta KA et al (2021) Single-cell analyses identify circulating anti-tumor CD8 T cells and markers for their enrichment. J Exp Med. https://doi.org/10.1084/jem.20200920
Li J, Ybarra R, Mak J et al (2018) IFNgamma-induced chemokines are required for CXCR3-mediated T-cell recruitment and antitumor efficacy of anti-HER2/CD3 bispecific antibody. Clin Cancer Res 24:6447–6458. https://doi.org/10.1158/1078-0432.CCR-18-1139
Schmid P, Adams S, Rugo HS et al (2018) Atezolizumab and nab-paclitaxel in advanced triple-negative breast cancer. New Engl J Med 379:2108–2121. https://doi.org/10.1056/NEJMoa1809615
Trina Formolo ML, Michaella L, Lisa K, Scott L, Karen P, Lisa M, Kurt B, Michael B, Darryl D, John S (2015) Determination of the NISTmAb Primary Structure. In: John E. Schiel DLD, Oleg V. Borisov (eds) State-of-the-art and emerging technologies for therapeutic monoclonal antibody characterization Volume 2. Biopharmaceutical Characterization: The NISTmAb Case Study. ACS Publications. pp. 1–62
Spiess C, Merchant M, Huang A et al (2013) Bispecific antibodies with natural architecture produced by co-culture of bacteria expressing two distinct half-antibodies. Nat Biotechnol 31:753–758. https://doi.org/10.1038/nbt.2621
Lombana TN, Matsumoto ML, Berkley AM et al (2019) High-resolution glycosylation site-engineering method identifies MICA epitope critical for shedding inhibition activity of anti-MICA antibodies. MAbs 11:75–93. https://doi.org/10.1080/19420862.2018.1532767
Lewis Phillips GD, Li G, Dugger DL et al (2008) Targeting HER2-positive breast cancer with trastuzumab-DM1, an antibody-cytotoxic drug conjugate. Cancer Res 68:9280–9290. https://doi.org/10.1158/0008-5472.CAN-08-1776
Toker A, Ohashi PS (2019) Expression of costimulatory and inhibitory receptors in FoxP3(+) regulatory T cells within the tumor microenvironment: Implications for combination immunotherapy approaches. Adv Cancer Res 144:193–261. https://doi.org/10.1016/bs.acr.2019.05.001
Pal B, Chen Y, Vaillant F et al (2021) A single-cell RNA expression atlas of normal, preneoplastic and tumorigenic states in the human breast. EMBO J 40:e107333. https://doi.org/10.15252/embj.2020107333
Quatrini L, Molfetta R, Zitti B et al (2015) Ubiquitin-dependent endocytosis of NKG2D-DAP10 receptor complexes activates signaling and functions in human NK cells. Sci Signal 8:ra108. https://doi.org/10.1126/scisignal.aab2724
Zhang C, Zhang J, Niu J, Zhang J, Tian Z (2008) Interleukin-15 improves cytotoxicity of natural killer cells via up-regulating NKG2D and cytotoxic effector molecule expression as well as STAT1 and ERK1/2 phosphorylation. Cytokine 42:128–136. https://doi.org/10.1016/j.cyto.2008.01.003
Junttila TT, Li J, Johnston J et al (2014) Antitumor efficacy of a bispecific antibody that targets HER2 and activates T cells. Cancer Res 74:5561–5571. https://doi.org/10.1158/0008-5472.CAN-13-3622-T
Tanner M, Kapanen AI, Junttila T, Raheem O, Grenman S, Elo J, Elenius K, Isola J (2004) Characterization of a novel cell line established from a patient with Herceptin-resistant breast cancer. Mol Cancer Ther 3:1585–1592
Junttila TT, Lutzker S (2020) Treatment of cancer with her2xcd3 bispecific antibodies in combination with anti-her2 mab. GENENTECH INC (US), HOFFMANN LA ROCHE (CH)
Segal NH, Logan TF, Hodi FS et al (2017) Results from an integrated safety analysis of urelumab, an agonist Anti-CD137 monoclonal antibody. Clin Cancer Res 23:1929–1936. https://doi.org/10.1158/1078-0432.CCR-16-1272
Suntharalingam G, Perry MR, Ward S, Brett SJ, Castello-Cortes A, Brunner MD, Panoskaltsis N (2006) Cytokine storm in a phase 1 trial of the anti-CD28 monoclonal antibody TGN1412. N Engl J Med 355:1018–1028. https://doi.org/10.1056/NEJMoa063842
Wu L, Seung E, Xu L et al (2020) Trispecific antibodies enhance the therapeutic efficacy of tumor-directed T cells through T cell receptor co-stimulation. Nat Cancer 1:86–98. https://doi.org/10.1038/s43018-019-0004-z
Eastwood D, Findlay L, Poole S, Bird C, Wadhwa M, Moore M, Burns C, Thorpe R, Stebbings R (2010) Monoclonal antibody TGN1412 trial failure explained by species differences in CD28 expression on CD4+ effector memory T-cells. Br J Pharmacol 161:512–526. https://doi.org/10.1111/j.1476-5381.2010.00922.x
Ho SK, Xu Z, Thakur A et al (2020) Epitope and Fc-mediated cross-linking, but not high affinity, are critical for antitumor activity of CD137 agonist antibody with reduced liver toxicity. Mol Cancer Ther 19:1040–1051. https://doi.org/10.1158/1535-7163.MCT-19-0608
Bartkowiak T, Jaiswal AR, Ager CR et al (2018) Activation of 4–1BB on liver myeloid cells triggers hepatitis via an interleukin-27-dependent pathway. Clin Cancer Res 24:1138–1151. https://doi.org/10.1158/1078-0432.CCR-17-1847
Zhang J, Jiao Y, Hou S, Tian T, Yuan Q, Hao H, Wu Z, Bao X (2017) S100A4 contributes to colitis development by increasing the adherence of Citrobacter rodentium in intestinal epithelial cells. Sci Rep 7:12099. https://doi.org/10.1038/s41598-017-12256-z
Claus C, Ferrara C, Xu W et al (2019) Tumor-targeted 4–1BB agonists for combination with T cell bispecific antibodies as off-the-shelf therapy. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aav5989
Skokos D, Waite JC, Haber L et al (2020) A class of costimulatory CD28-bispecific antibodies that enhance the antitumor activity of CD3-bispecific antibodies. Sci Transl Med. 12:eaaw7888. https://doi.org/10.1126/scitranslmed.aaw7888
Wing JB, Tay C, Sakaguchi S (2019) Control of regulatory T cells by Co-signal molecules. Adv Exp Med Biol 1189:179–210. https://doi.org/10.1007/978-981-32-9717-3_7
Park JH, Lee HK (2021) Function of gammadelta T cells in tumor immunology and their application to cancer therapy. Exp Mol Med 53:318–327. https://doi.org/10.1038/s12276-021-00576-0
Stamova S, Cartellieri M, Feldmann A et al (2011) Simultaneous engagement of the activatory receptors NKG2D and CD3 for retargeting of effector cells to CD33-positive malignant cells. Leukemia 25:1053–1056. https://doi.org/10.1038/leu.2011.42
Cho HM, Rosenblatt JD, Tolba K, Shin SJ, Shin DS, Calfa C, Zhang Y, Shin SU (2010) Delivery of NKG2D ligand using an anti-HER2 antibody-NKG2D ligand fusion protein results in an enhanced innate and adaptive antitumor response. Cancer Res 70:10121–10130. https://doi.org/10.1158/0008-5472.CAN-10-1047
von Strandmann EP, Hansen HP, Reiners KS et al (2006) A novel bispecific protein (ULBP2-BB4) targeting the NKG2D receptor on natural killer (NK) cells and CD138 activates NK cells and has potent antitumor activity against human multiple myeloma in vitro and in vivo. Blood 107:1955–1962. https://doi.org/10.1182/blood-2005-05-2177
Vyas M, Schneider AC, Shatnyeva O et al (2016) Mono- and dual-targeting triplebodies activate natural killer cells and have anti-tumor activity in vitro and in vivo against chronic lymphocytic leukemia. Oncoimmunology 5:e1211220. https://doi.org/10.1080/2162402X.2016.1211220
Wang Y, Li H, Xu W, Pan M, Qiao C, Cai J, Xu J, Wang M, Zhang J (2020) BCMA-targeting bispecific antibody that simultaneously stimulates NKG2D-enhanced efficacy against multiple myeloma. J Immunother 43:175–188. https://doi.org/10.1097/CJI.0000000000000320
Raynaud A, Desrumeaux K, Vidard L, Termine E, Baty D, Chames P, Vigne E, Kerfelec B (2020) Anti-NKG2D single domain-based antibodies for the modulation of anti-tumor immune response. Oncoimmunology 10:1854529. https://doi.org/10.1080/2162402X.2020.1854529
Wang T, Sun F, Wang Y et al (2018) NKG2D Immunoligand rG7S-MICA enhances NK Cell-mediated immunosurveillance in colorectal carcinoma. J Immunother 41:109–117. https://doi.org/10.1097/CJI.0000000000000215
Rothe A, Jachimowicz RD, Borchmann S et al (2014) The bispecific immunoligand ULBP2-aCEA redirects natural killer cells to tumor cells and reveals potent anti-tumor activity against colon carcinoma. Int J Cancer 134:2829–2840. https://doi.org/10.1002/ijc.28609
Han Y, Sun F, Zhang X et al (2019) CD24 targeting bi-specific antibody that simultaneously stimulates NKG2D enhances the efficacy of cancer immunotherapy. J Cancer Res Clin Oncol 145:1179–1190. https://doi.org/10.1007/s00432-019-02865-8
Kellner C, Hallack D, Glorius P et al (2012) Fusion proteins between ligands for NKG2D and CD20-directed single-chain variable fragments sensitize lymphoma cells for natural killer cell-mediated lysis and enhance antibody-dependent cellular cytotoxicity. Leukemia 26:830–834. https://doi.org/10.1038/leu.2011.288
Zou Y, Luo W, Guo J, Luo Q, Deng M, Lu Z, Fang Y, Zhang CC (2018) NK cell-mediated anti-leukemia cytotoxicity is enhanced using a NKG2D ligand MICA and anti-CD20 scfv chimeric protein. Eur J Immunol 48:1750–1763. https://doi.org/10.1002/eji.201847550
Xing S, Ferrari de Andrade L (2020) NKG2D and MICA/B shedding: a ‘tag game’ between NK cells and malignant cells. Clin Transl Immunology 9:e1230. https://doi.org/10.1002/cti2.1230
Zuo J, Willcox CR, Mohammed F et al (2017) A disease-linked ULBP6 polymorphism inhibits NKG2D-mediated target cell killing by enhancing the stability of NKG2D ligand binding. Sci Signal. https://doi.org/10.1126/scisignal.aai8904
Zuo JM, Mohammed F, Moss P (2018) The Biological Influence and Clinical Relevance of Polymorphism Within the NKG2D Ligands. Front Immunol. 9:ARTN1820
Acknowledgements
We thank A. Tan, C.K. Poon, T. Ho, G. Tweet and J. Borneo for flow cytometry and FACS support. A.P. Martinez for cytokine and chemokine analysis. Scot Liu for sharing the NK92-6DF5 cell line. Kelsey Homyk for organizing and providing BsAbs. Members of the Ye laboratory for discussion and feedback on the project. Graphical abstract created with BioRender.com. This work has not been presented or published as an abstract at a conference.
Funding
Authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Project and molecule conceptualization were done by KBW, CS, AH, and GL. Molecule design, production and affinity determination were done by MD, DE, JW, DS, and CS. Reagent production was done by SJ. In vitro experiment conceptualization, design and execution were done by AF, JC, KRK, JM, CCK, and EC. Design and execution of NK cell conjugate and IS experiments were done by MP and MG. Design and execution of bulk RNA-seq experiments were done by YL and ZM. In vivo model conceptualization, design and execution were done by MG, EC, RC, EC, GG, KT, and SY. Animal model and ex vivo assay support were done by JM and SY. Bioinformatic analysis was done by TN, RP, VC, and KBW. Manuscript preparation was done by AH and KBW. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
Authors employed by Genentech declare that they have ownership interests in and/or patents with Genentech.
Ethical approval
All studies were conducted in accordance with the American Association of Laboratory Animal Care guidelines. Experimental studies were conducted with approval of the IACUC at Genentech.
Consent for publication
This manuscript does not contain patient personal data.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Herault, A., Mak, J., de la Cruz-Chuh, J. et al. NKG2D-bispecific enhances NK and CD8+ T cell antitumor immunity. Cancer Immunol Immunother 73, 209 (2024). https://doi.org/10.1007/s00262-024-03795-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00262-024-03795-2