Abstract
Liver cancer stem cells (LCSCs) are responsible for recurrence, metastasis, and drug resistance in liver cancer. However, the genes responsible for inducing LCSCs have not been fully identified. Based on our previous study, we found that tescalcin (TESC), a calcium-binding EF hand protein that plays a crucial role in chromatin remodeling, transcriptional regulation, and epigenetic modifications, was up-regulated in LCSCs of spheroid cultures. By searching the Cancer Genome Atlas, International Cancer Genome Consortium, Human Protein Atlas, and Kaplan–Meier Plotter databases, we found that TESC expression was significantly elevated in liver cancer compared with that in normal liver tissue and was predictive of a decreased overall survival rate. Multivariate Cox analysis revealed TESC to be an independent prognostic factor for survival. High TESC expression was positively associated with cancer stem cell pathways, cancer stem cell surface markers, stemness transcription factors, epithelial–mesenchymal transition (EMT) factors, immune checkpoint proteins, and various cancer-related biological processes in liver cancer. Furthermore, TESC was implicated as promoting cancer stem cell properties through its influence on EMT. We demonstrated that TESC is a novel stemness-related gene that can serve as an independent prognostic factor for liver cancer.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatocellular carcinoma (HCC) is one of the most prevalent types of liver cancer, ranking third in terms of associated mortality among all malignant tumors [1]. Treatment modalities such as surgical resection, transplantation, radiation therapy, chemotherapy, transcatheter arterial chemoembolization, tyrosine kinase inhibitor use, and immunotherapy are available for HCC [2]. However, even when complex treatment strategies aimed at improving overall survival (OS) are implemented, HCC management remains challenging, with many cases exhibiting treatment resistance and recurrence [3]. Liver cancer stem cells (LCSCs), a small subset of oncogenic cells with self-renewal ability and drug resistance, play a pivotal role in the recurrence and metastasis of HCC. However, the mechanisms regulating LCSCs have not been fully elucidated.
The EF hand calcium-binding protein tescalcin (TESC) plays a crucial role in the regulation of processes such as chromatin remodeling, transcriptional regulation, and epigenetic modification through interactions with the cytosolic domain of the Na+/H+ exchanger isoform type-1 (NHE1) [4]. TESC has been implicated as promoting malignant progression in various cancers, including HCC [5], papillary thyroid microcarcinoma [6], colorectal cancer [7], gastric cancer [8], and cholangiocarcinoma [9]. However, few studies have explored the association of TESC with cancer stemness properties. TESC was demonstrated to promote epithelial–mesenchymal transition (EMT) and thereby contribute to cancer progression in esophageal squamous cell cancer [10]. In non–small cell lung cancer (NSCLC), TESC overexpression was demonstrated to mediate STAT3 function and consequently enhance EMT and CSC characteristics, ultimately leading to increased cellular resistance to gamma radiation. These findings indicate that targeting TESC may be a strategy for enhancing the efficacy of radiation therapy in NSCLC [11]. In another study, we enriched LCSCs by culturing Huh7 cells in ultra-low-attachment surface plates to form spheroid culture cells and performed RNA-seq. The results revealed that TESC expression was upregulated in LCSCs from spheroid cultures [12]. However, the precise function of TESC in LCSCs remains uncertain.
This study evaluated the association between TESC expression in HCC tissues and patient prognosis by using publicly available databases. Subsequently, the relationship between TESC mRNA levels and clinicopathological characteristics as well as tumor-infiltrating immune cells was assessed in patients with HCC. The results revealed a positive association between TESC expression and cancer stem cell pathways as well as various biological processes. Furthermore, cellular experiments were conducted to elucidate the role of TESC in promoting tumor proliferation and metastasis, potentially facilitating the progression of EMT and enhancing the properties of LCSCs. The study findings highlight the substantial involvement of TESC in LCSCs and provide new perspectives on treatment of this malignancy type.
Materials and methods
Data source and software
The data of patients with HCC were obtained from multiple databases, namely, the Cancer Genome Atlas Liver HCC cohort (TCGA-LIHC; https://tcga-data.nci.nih.gov/tcga/), International Cancer Genome Consortium (ICGC) dataset (https://dcc.icgc.org/releases/current/Projects), and Clinical Proteomic Tumor Analysis Consortium (CPTAC) in the UALCAN database (http://ualcan.path.uab.edu/cgi-bin/ualcan-res.pl). The TCGA-LIHC cohort was categorized into high and low expression groups on the basis of median values for CD133, CD90, and EpCAM expression. Differential expression analysis of mRNAs was conducted between the two groups by using the limma package in R software (version 4.0.3), with the threshold set as adjusted P < 0.05 and log2 (fold change) > 1.5 or log2 (fold change) < − 1.5. Gene set enrichment analysis (GSEA) was performed using GSEA software (version 4.3.2;http://software.broadinstitute.org/gsea). The Hallmark gene set (h.all.v2023.2.Hs.symbols.gmt) from the Molecular Signatures Database was used as the reference gene set. The number of permutations was set at 1000. RNA-seq data of our previous study about spheroid cultures were deposited in the GEO (www.ncbi.nlm.nih.gov/geo/, accessed on 1 July 2022) with the GEO series number GSE199940.
Kaplan–Meier plot of survival analysis
Correlations between mRNA levels of the TESC gene and the survival of patients with liver cancer were assessed using the Kaplan–Meier plotter database (https://kmplot.com/, accessed on December 1, 2023) [13].
Correlations between TESC expression and immune cells and immune-checkpoint–relevant transcripts
The TIMER2.0 database (http://timer.cistrome.org/) [14] was used to analyze the expression of TESC and its correlation with the abundance of six types of infiltrating immune cells in patients with liver cancer, namely B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells. Additionally, the association between the expression level of TESC and tumor purity was analyzed.
Gene correlation analysis was employed to identify significant associations between TESC expression and immune-checkpoint–relevant transcripts, namely SIGLEC15, TIGIT, CD274, HAVCR2, PDCD1, CTLA4, LAG3, and PDCD1LG2. The potential response to immune checkpoint blockade (ICB) was predicted using the TIDE algorithm. TIDE uses a set of gene expression markers to evaluate two mechanisms of tumor immune evasion: dysfunction of tumor-infiltrating cytotoxic T lymphocytes (CTLs) and rejection of CTLs by immunosuppressive factors. A high TIDE score indicates poor efficacy of ICB, which leads to a shorter survival period following ICB therapy [15].
Cell line culture
All cell lines used in this study were purchased from the cell bank of Shanghai Institute for Biological Science, Chinese Academy of Sciences (Shanghai, China). The cells were authenticated through short tandem repeat profiling and subjected to regular checks for mycoplasma contamination. HEK-293 T cells and human HCC Huh7 and HepG2 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; BI, Israel). The media for all cell lines were supplemented with 10% inactivated fetal bovine serum (04–001-1ACS; BI) and 1% penicillin–streptomycin (Cat. 15,140,122; Thermo Fisher Scientific, Inc.). Incubation for all cells was performed in a 37 °C incubator under 5% carbon dioxide.
Plasmid construction products
The pLKO.1 shNRA plasmids and overexpression plasmid were synthesized by Qingke Biotechnology (Qingke, Beijing, China). The shRNA-TESC#1 sequence was 5′-CCTGACCATCATGTCCTACTT-3′, The shRNA-TESC#2 sequence was 5′- CGCATCACTCTGGAAGAATAT-3′.
Real-time quantitative polymerase chain reaction
Total RNA was extracted from the cells by using a total RNA isolation kit (RC101-01, Vazyme) in accordance with the manufacturer’s instructions. Complementary DNA (cDNA) was synthesized using the RevertAid Master Mix reagent (M1631, Thermo Fisher Scientific, Inc.). Real-time quantitative polymerase chain reaction (PCR) was performed on the CFX96 Real-Time PCR System (Bio-rad, USA) by using TB Green Premix Ex Taq II (Cat. RR820A, TaKaRa). The PCR primer sequences were as follows: TESC forward primer: CCTACCATTCGGAACCTGCG and TESC reverse primer: AGCTCCTCGACCACATTTCG.
Western blot analysis
A Western blot analysis was conducted to visualize antibody binding by using luminol reagent (sc-2048, Santa Cruz) on a Tanon-5200 chemiluminescent imaging system (Tanon, China). The following antibodies were used in this study: TESC (Cat. 11,125–1-AP, 1:1000, Proteintech), Snail (Cat.3879S, 1:1000, Cell Signaling Technology), Vimentin (Cat.10366–1-AP, 1:5000, Proteintech), N-cadherin (Cat. 13,116, 1:1000, Cell Signaling Technology), SOX2 (Cat. GTX101507, 1:5000, GeneTex), CD133 (Cat. ab19898, 1:1000, Abcam), β-actin (Cat.HRP-60008, 1: 5000, Proteintech), and goat antirabbit (Cat. 7074S, 1:3000, Cell Signaling Technology, Inc.).
Immunohistochemistry
Human liver cancer with matched cancer adjacent liver tissue array was obtained from TissueArray.Com (Cat. LV1505a, USA). Immunohistochemical staining was performed on liver cancer and adjacent liver tissue using TESC antibody (Cat. 11,125–1-AP, 1:200, Proteintech), SOX2 antibody (Cat. GTX101507, 1:200, GeneTex) and CD133 antibody (Cat. ab19898, 1:200, Abcam). Protein expression in the immunohistochemical staining was quantified according to the Histoscore (H-score) method. H-score was evaluated by a semi-quantitative assessment of both the intensity of staining and the percentage of positive cells. Cases with H-score higher than average were considered as high expression and those less than average as low expression.
Sphere formation assay
For spherical culture, a serum-free medium was prepared using DMEM/F12 supplemented with 2% B27 (Gibco, USA), 20 ng/mL epidermal growth factor (Peprotech, USA), and 20 ng/mL basic fibro-blast growth factor (Peprotech, USA).
Statistical analysis
The Wilcoxon test was employed to analyze the TESC mRNA levels in tumors compared with those in normal tissues. The Spearman correlation coefficient was used to evaluate the correlation of gene expression or pathway score. To compare survival curves, we used the log-rank test to calculate the HR and log-rank P value in the Kaplan–Meier Plotter. Both univariate and multivariate Cox regression analyses were performed using the “survival” package in R software (version 4.0.3). Only variables with a P value of less than 0.05 in the univariate analysis were further analyzed by multivariable Cox regression. Fisher’s exact test was used to compare dichotomous variables. Statistical analyses were performed using GraphPad Prism software (version 10.0.0). Experimental data are presented as means ± standard deviations, and significant differences were analyzed using a two-tailed independent Student’s t test. A P value of < 0.05 was considered significant.
Results
TESC is highly expressed and associated with poor prognosis in HCC
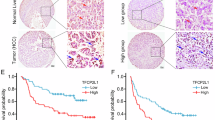
To identify a novel gene regulating cancer stemness in HCC, RNA-sequencing expression profiles and clinical data from 374 HCCs and 50 normal liver tissues obtained from the TCGA-LIHC dataset (https://portal.gdc.cancer.gov /projects/TCGA-LIHC) were analyzed. CSC biomarkers of HCC, such as CD133, CD90, EpCAM, CD44, and SRY-box transcription factor 2 (SOX2), play a crucial role in maintaining stem cell stability [16]. In the current study, on the basis of median CD133, CD90, and EpCAM expression values, the TCGA-LIHC cohort was categorized into high expression and low expression groups. The limma package in R software (version 4.0.3) was used to conduct differential expression analysis, and 13 common, differentially upregulated genes were identified by intersecting the 3 groups of differentially upregulated genes (Fig. 1A). In our previous study, through analyzing RNA sequencing data (GEO database, GSE199940), we found the TESC and QSOX1 genes were upregulated in LCSCs from spheroid cultures [12]. Therefore, in the present study, we investigated the prognostic value of TESC and QSOX1 by using the Kaplan–Meier Plotter. The results revealed that TESC was a detrimental prognostic factor in HCC (OS: HR = 1.57, 95% CI = 1.11 − 2.23, log-rank P = 0.011) (Fig. 1B). However, no significant correlation between QSOX1 expression and prognosis. Furthermore, univariate and multivariate Cox regression analyses were conducted. We found that pathologic stage III (HR = 2.734, 95% CI = 1.792 − 4.172, P < 0.001), pathologic stage IV (HR = 5.597, 95% CI = 1.726 − 18.148, P = 0.004) and high TESC expression (HR = 1.701, 95% CI = 1.197− 2.416, P = 0.003) were risk factors for HCC patients in the univariate Cox regression analysis. Consistently, pathologic stage III (HR = 2.748, 95% CI = 1.800 − 4.196, P < 0.001), pathologic stage IV (HR = 5.131, 95% CI = 1.580 − 16.669, P = 0.007) and high TESC expression (HR = 1.683, 95% CI = 1.155− 2.451, P = 0.007) were independent prognostic factors according to the multivariate Cox regression. These indicated that TESC was an independent prognostic factor for OS in liver cancer (Table 1).
Association between TESC gene expression and clinical prognosis. A High and low expression groups, established by dividing the TCGA-LIHC on the basis of median expression of CD133, CD90, and EpCAM. Thirteen intersecting differentially expressed genes were identified, with high expression of TESC and QSOX1 being noted in LCSCs from spheroid cultures. B TESC’s high expression in the TCGA-LIHC was significantly correlated with shorter overall survival (OS). C TESC expression in various cancers compared with that in adjacent normal tissues (if available), assessed using TIMER2.0. D RNA expression of the TESC gene in liver cancer and normal liver tissues from the ICGC database. E Protein expression of the TESC gene in liver cancer and normal liver tissues from the CPTAC database. F Immunohistochemistry (IHC) image of TESC in liver cancer and adjacent liver tissues of tissue microarray (Cat. LV1505a, TissueArray.Com). Scale bar, 60 μm. *P < 0.05; **P < 0.01; ***P < 0.001
We completed a comprehensive assessment of TESC expression across various cancers by using TIMER2.0 to analyze RNA-sequencing data from TCGA. The different expression patterns of TESC in tumors compared with those in adjacent normal tissues are depicted in (Fig. 1C). Notably, TESC expression was significantly upregulated in HCCs compared with in normal tissues (P < 0.001). A similar trend was observed in a Japanese liver cancer cohort from the ICGC, where TESC expression was significantly increased in tumors compared with in normal tissues (P = 0.012) (Fig. 1D). Furthermore, we analyzed the protein expression of TESC by using data from the CPTAC in the UALCAN database and revealed that TESC was highly expressed in HCC (P < 0.001) (Fig. 1E). To determine the protein expression of TESC in clinical specimens, immunohistochemistry (IHC) staining was performed on liver cancer and adjacent liver tissue of tissue microarray. These findings indicate a clear elevation in TESC protein levels in HCC tissue compared with in adjacent normal liver tissue (Fig. 1F).
We explored the potential role of TESC expression in cancer and the underlying mechanisms of this role by using the Kaplan–Meier Plotter to integrate clinical and pathological data to explore the associations between TESC expression and various clinical characteristics in patients with liver HCC. In terms of OS, TESC exhibited a negative effect on specific subgroups of patients with HCC, including male patients (n = 246, HR = 1.98, 95% CI = 1.25 − 3.12, P = 0.0029), those consuming alcohol (n = 115, HR = 1.99, 95% CI = 1.03 − 3.82, P = 0.0365), and those without hepatitis virus infection (n = 167, HR = 1.90, 95% CI = 1.20 − 2.99, P = 0.0053; Supplementary Fig. S1). These findings indicate that high TESC expression levels are associated with poor prognosis in HCC.
TESC overexpression is associated with cancer stem cell-related pathways in HCC
On the basis of mean TESC mRNA levels, we divided the TCGA-LIHC cohort into two groups and investigated the associations with clinicopathological characteristics. As presented in Table 2, TESC expression exhibited an inverse association with age (P = 0.008) and a positive association with histologic grade (P < 0.001) and adjacent hepatic tissue inflammation (P = 0.004) in patients with HCC. A total of 274 differentially upregulated genes and 167 downregulated genes (relative to those in the group with low TESC expression levels) were identified in the group with high TESC expression levels [P < 0.05, log2 (fold change) > 2] (Fig. 2A). The results of GSEA of the data in the TCGA-LIHC revealed that pathways traditionally associated with CSCs, that is, the Wnt pathway (P = 0.002, NES = 1.86) and Notch pathway (P = 0.006, NES = 1.74), were enriched in the group with high TESC expression levels (Fig. 2B). To further elucidate the underlying function of TESC, differentially upregulated genes were analyzed using Gene Ontology and the Kyoto Encyclopedia of Genes and Genomes (Fig. 2C, D). The results confirmed the enrichment of some crucial CSC-related pathways, such as the Wnt pathway, TNF signaling pathway, and PI3K-Akt signaling pathway. Additionally, we employed the GSVA package in R software to analyze the correlations between TESC expression and pathway scores [17, 18]. The results indicated a positive correlation between TESC and EMT markers (P < 0.001, R = 0.42), the PI3K-Akt-mTOR pathway (P < 0.001, R = 0.21), TGF-beta signaling (P < 0.001, R = 0.35), angiogenesis (P < 0.001, R = 0.42), apoptosis (P < 0.001, R = 0.28), and inflammatory response (P < 0.001, R = 0.31) (Fig. 2E). These findings indicate that TESC may promote cancer stemness in HCC.
Association of high TESC expression with cancer stem cell–related pathways. A Volcano plot depicting differential expression genes (DEGs) between groups with high and low TESC expression levels in the TCGA-LIHC. Red dots represent upregulated genes, and blue dots represent downregulated genes. B GSEA results revealing enrichment of the Wnt pathway (P = 0.002, NES = 1.86) and Notch pathway (P = 0.006, NES = 1.74) in the group with high TESC expression levels in the TCGA-LIHC. C Bubble plot of the Kyoto Encyclopedia of Genes and Genomes enrichment analysis of upregulated DEGs. The ordinate represents the top 20 upregulated pathways, the bubble colors indicate the size of the P value, and the bubble sizes indicate the number of enriched differential genes. D Bubble plot of the Gene Ontology term enrichment analysis of upregulated DEGs. The ordinate represents the top 20 terms, the bubble colors indicate the size of the P value, and the bubble sizes indicate the number of enriched differential genes. E Correlation analysis between the TESC gene and EMT, the PI3K-Akt-mTOR pathway, TGF-beta signaling, angiogenesis, apoptosis, and inflammatory response. *P < 0.05; **P < 0.01; ***P < 0.001
TESC gene expression is positively correlated with immune infiltration and m6A methylation
To investigate the immunological implications of TESC in tumor immunotherapy, we employed the TIDE algorithm to predict potential responses to ICB. Our findings revealed that patients with high TESC expression levels had higher TIDE scores, indicating a prediction of poor efficacy of ICB therapy and short survival rates (Supplementary Fig. S2A). N6-methyladenosine (m6A) RNA methylation–related genes were previously implicated as being involved in the initiation, progression, metastasis, treatment resistance, immune evasion, and self-renewal of cancer stem cells [19]. In the current study, the results of gene correlation analysis indicated a positive correlation between TESC expression and most m6A modifiers (writers, erasers, and readers), indicating that TESC might interfere with m6A processes and thereby affect tumor immunity and stemness (Supplementary Fig. S2B). Furthermore, we explored the association between TESC expression and immune checkpoints, such as SIGLEC15, TIGIT, CD274, HAVCR2, PDCD1, CTLA4, LAG3, and PDCD1LG2 [20]. A high TESC expression was demonstrated to be positively correlated with CTLA4 (P < 0.001), HAVCR2 (P < 0.001), PDCD1 (P < 0.001), and TIGIT (P < 0.01) (Supplementary Fig. S2C). Additionally, we investigated the association between TESC expression and 6 types of infiltrating immune cells (i.e., B cells, CD4+ T cells, CD8+ T cells, neutrophils, macrophages, and dendritic cells) by using TIMER2.0. The results revealed significantly positive correlations between TESC expression levels and infiltration levels of B cells (P < 0.001, R = 0.0.39), CD4+ T cells (P < 0.001, R = 0.37), neutrophils (P < 0.001, R = 0.19), macrophages (P < 0.001, R = 0.17), and dendritic cells (P < 0.001, R = 0.38) (Supplementary Fig. S2D). Further exploration is required to understand the impact of TESC expression on immune cell infiltration and m6A RNA modification.
Downregulation of TESC reduces cancer stemness characteristics in liver cancer cell lines
Considering the association between TESC expression and cancer stem cell–related pathways (Fig. 2E) and the significant positive correlation of TESC expression with biomarkers of CSCs (CD24, CD44, CD90, CD133, EpCAM, SOX2, and OCT4) and EMT (VIM, TWIST1, and Snail) in HCC (Fig. 3A) [21, 22], we investigated whether alterations in TESC expression influence the stemness characteristics of HCC cells. First, TESC was knocked down by using short hairpin RNA (shRNA) in HepG2 and Huh7 liver cancer cell lines (Fig. 3B). This knockdown led to a reduction in the protein levels of CSC biomarkers CD133 and EpCAM as well as stemness-related transcription factor SOX2 (Fig. 3C). Besides, IHC staining was performed in tissue microarray to determine the relationship between TESC expression and biomarkers of CSCs (Fig. 3D). The result revealed that TESC expression was positively associated with stem cell marker CD133 (P = 0.0002, Fig. 3E) and SOX2 (P = 0.0482, Fig. 3F). Moreover, CD133 was also positively associated with SOX2 expression (P < 0.0001, Fig. 3G). Furthermore, the results from sphere formation assays revealed that TESC downregulation led to a reduction in the sphere-forming efficiency of HepG2 and Huh7 cells (Fig. 3H). Taken together, these findings suggest involvement of TESC in promoting or maintaining the stemness of LCSCs.
Reduced stemness characteristics in liver cancer cell lines due to downregulation of TESC. A Correlation between TESC expression and CSC biomarkers. B Knockdown of the TESC gene in HepG2 and Huh7 cell lines. C Western blot revealing the protein expression of CSC biomarkers CD133, EpCAM, and SOX2 after TESC knockdown. D IHC staining of liver cancer microarray (Cat. LV1505a, TissueArray.Com) for the correlation between TESC, CD133 and SOX2. Scale bar, 60 μm. E–G Fisher’s exact test showed a positive correlation between TESC and CD133 (E), TESC and SOX2 (F), and CD133 and SOX2 (G). H Sphere formation assay in stable TESC knockdown cell lines and TESC-overexpressing cell lines (left panel). The statistical bar graphs present the sphere numbers (right panel). *P < 0.05; **P < 0.01; ***P < 0.001
Downregulation of TESC reduces the expression of EMT inducers and cell migration in liver cancer cell lines
EMT is believed to generate CSCs and be a key contributor to cancer cell metastasis [22]. Additionally, TESC expression was discovered to be positively correlated with the biomarkers of EMT and CSCs. Therefore, we hypothesized that TESC would be involved in CSC formation or maintenance through the promotion of EMT processes. According to our results, in HepG2 and Huh7 cells, knockdown of TESC resulted in reduced protein levels of the EMT inducers N-cadherin, vimentin, and Snail (Fig. 4A). We assessed the impact of TESC on tumor migration and colony formation abilities, and transwell migration assay revealed that TESC knockdown in HepG2 and Huh7 cells impaired their migration ability (Fig. 4B). Moreover, downregulation of TESC led to a decrease in the number of colonies formed in soft agar (Fig. 4C). These findings indicate that TESC may enhance the EMT process to promote cancer stemness and malignancy in HCC.
Downregulation of expression of key EMT genes and impairment of migration ability of cancer cells due to TESC knockdown. A Western blot revealing the association between TESC knockdown and protein expression of EMT key genes. B Transwell test demonstrating that TESC knockdown reduces the migration ability of cancer cells in HepG2 and Huh7 cell lines. C Colony formation experimental results indicating that colony formation ability was reduced after TESC gene knockdown. *P < 0.05; **P < 0.01; ***P < 0.001
Discussion
HCC is a cancer with high mortality, recurrence, and metastasis rates, mainly because of its resistance to radiotherapy and chemotherapy [3]. Clinical observations indicated that CSCs play a pivotal role in influencing these characteristics of HCC [16]. Although numerous therapeutic drugs targeting CSCs have been developed, their clinical efficacy remains limited [23]. Therefore, identification of potential additional mechanisms that can be used to control CSCs could help with the development of novel HCC treatment strategies. This study identified TESC as a novel gene associated with cancer stemness that is highly expressed in LCSCs from spheroid cultures [12]. Few reports have explored the association between TESC and CSCs. One study demonstrated that TESC can enhance cancer stemness by mediating the IGF1R–STAT3 signaling axis in NSCLC [11]. However, the precise regulatory mechanism by which TESC induces and sustains LCSCs remain unclear. TESC was also reported to promote malignant progression in HCC, including proliferation and apoptosis, making it an independent prognostic factor for short OS in patients with HCC [5]. However, these studies have not investigated the mechanisms by which TESC regulates the biological behaviors of HCC, including tumor invasion, metastasis, and tumor stemness. The present study demonstrated that TESC enhances migratory and stemness properties in HCC (Fig. 4B, C). Additionally, classical CSCs pathways, such as the Wnt and Notch pathways, were enriched in the group exhibiting high TESC expression levels (Fig. 2B), and TESC expression was significantly positively correlated with CSC biomarkers in patients with HCC (Fig. 3A). These findings indicate that TESC may be a novel cancer stemness–related gene, potentially serving as a promising prognostic predictor and therapeutic target for HCC.
We noted a significant increase in TESC expression across various cancers compared with that in normal tissues (Fig. 1C), indicating that TESC may serve as a crucial biomarker or oncogene in cancers. TESC has been reported to promote tumor progression in multiple cancers, including papillary thyroid microcarcinoma [6], colorectal cancer [7], gastric cancer [8], and cholangiocarcinoma [9]. However, the specific mechanisms through which TESC promotes tumor development remain unclear. Therefore, further research is warranted to comprehensively investigate the detailed mechanisms of TESC across different cancers. Another notable aspect of this study is the correlation between TESC and cancer immune infiltration. Analyses of public databases revealed that high TESC expression levels are associated with high TIDE scores and positively correlated with some immune checkpoints and infiltrating immune cells, which are considered predictors of poor efficacy in ICB therapy and shorter survival times (Fig. 2A-D).
The EMT process, which is crucial in cancer cell metastasis, was reported to be associated with cancer stemness [22]. Various signaling pathways, such as TGF-β, Wnt, Notch, and PI3K-AKT, participate in the EMT regulatory network [24, 25]. Considering the positive correlations that have been observed between TESC and genes involved in both EMT and cancer stemness, we propose that TESC may sustain malignant properties through modulation of the EMT process. EMT progression is regulated by the expression of EMT-translational factors (e.g., Snail, vimentin, and TWIST). Our study demonstrated that knockdown of TESC led to reduced protein levels of N-cadherin, vimentin, and Snail (Fig. 4A). These findings indicate that TESC may enhance cancer stemness properties by promoting EMT in liver cancer.
Although our in vitro experiments demonstrated that TESC downregulation inhibits CSCs and impedes their migratory properties in liver cancer cell lines, this study has some limitations that should be considered when the results are interpreted. First, the detailed molecular mechanism through which TESC regulates LCSCs requires further exploration through additional in vivo and in vitro experiments. Second, although an associated was observed between TESC expression and immune cell infiltration into tumors as well as patient survival, we were unable to establish a direct link between TESC and patient survival through immune infiltration. Future studies specifically focusing on TESC expression and immune infiltration in cancers could provide more definitive evidence. Third, the potential of TESC as an effective therapeutic target for patients with HCC warrants further investigation.
Conclusion
The present study highlights TESC as a novel gene associated with liver cancer stemness with a high expression in tumor tissues and that can serve as an independent prognostic factor for HCC. In addition, TESC may promote cancer stemness by enhancing the EMT process. Furthermore, elevated TESC expression is positively associated with cancer immune infiltration and therefore has potential to predict poor efficacy of ICB therapy in HCC. The use of TESC as a biomarker or therapeutic target in patients with liver cancer warrants further evaluation.
Data availability
All available data are reported in the manuscript and supplementary file.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: A Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Nault JC, Cheng AL, Sangro B, Llovet JM. Milestones in the pathogenesis and management of primary liver cancer. J Hepatol. 2020;72(2):209–14. https://doi.org/10.1016/j.jhep.2019.11.006.
Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477-91.e1. https://doi.org/10.1053/j.gastro.2018.08.065.
Takamatsu G, Katagiri C, Tomoyuki T, Shimizu-Okabe C, Nakamura W, Nakamura-Higa M, et al. Tescalcin is a potential target of class I histone deacetylase inhibitors in neurons. Biochem Biophys Res Commun. 2017;482(4):1327–33. https://doi.org/10.1016/j.bbrc.2016.12.036.
Zhou ZG, Chen JB, Zhang RX, Ye L, Wang JC, Pan YX, et al. Tescalcin is an unfavorable prognosis factor that regulats cell proliferation and survival in hepatocellular carcinoma patients. Cancer commun (London, England). 2020;40(8):355–69. https://doi.org/10.1002/cac2.12069.
Zou X, Zhou Q, Nie Y, Gou J, Yang J, Zhu J, et al. Tescalcin promotes highly invasive papillary thyroid microcarcinoma by regulating FOS/ERK signaling pathway. BMC Cancer. 2022;22(1):595. https://doi.org/10.1186/s12885-022-09643-9.
Kang J, Kang YH, Oh BM, Uhm TG, Park SY, Kim TW, et al. Tescalcin expression contributes to invasive and metastatic activity in colorectal cancer. Tumour Biol. 2016;37(10):13843–53. https://doi.org/10.1007/s13277-016-5262-0.
Kim TW, Han SR, Kim JT, Yoo SM, Lee MS, Lee SH, et al. Differential expression of tescalcin by modification of promoter methylation controls cell survival in gastric cancer cells. Oncol Rep. 2019;41(6):3464–74. https://doi.org/10.3892/or.2019.7099.
Hsieh CH, Chu CY, Lin SE, Yang YSH, Chang HS, Yen Y. TESC promotes TGF-α/EGFR-FOXM1-mediated tumor progression in cholangiocarcinoma. Cancers (Basel). 2020;12(5):1105. https://doi.org/10.3390/cancers12051105.
Dong Y, Fan B, Li M, Zhang J, Xie S, Di S, et al. TESC acts as a prognostic factor and promotes epithelial-mesenchymal transition progression in esophageal squamous carcinoma. Pathol Res Pract. 2024;253:154964. https://doi.org/10.1016/j.prp.2023.154964.
Lee JH, Choi SI, Kim RK, Cho EW, Kim IG. Tescalcin/c-Src/IGF1Rβ-mediated STAT3 activation enhances cancer stemness and radioresistant properties through ALDH1. Sci Rep. 2018;8(1):10711. https://doi.org/10.1038/s41598-018-29142-x.
Ye P, Chi X, Yan X, Wu F, Liang Z, Yang WH. Alanine-glyoxylate aminotransferase sustains cancer stemness properties through the Upregulation of SOX2 and OCT4 in hepatocellular carcinoma cells. Biomolecules. 2022;12(5):668. https://doi.org/10.3390/biom12050668.
Menyhárt O, Nagy Á, Győrffy B. Determining consistent prognostic biomarkers of overall survival and vascular invasion in hepatocellular carcinoma. Royal Soc open sci. 2018;5(12):181006. https://doi.org/10.1098/rsos.181006.
Li T, Jingxin F, Zeng Z, Cohen D, Li J, Chen Q, Bo Li X, Liu S. TIMER2.0 for analysis of tumor-infiltrating immune cells. Nucleic Acids Res. 2020;48(W1):W509–14. https://doi.org/10.1093/nar/gkaa407.
Jiang P, Gu S, Pan D, Fu J, Sahu A, Hu X, et al. Signatures of T cell dysfunction and exclusion predict cancer immunotherapy response. Nat Med. 2018;24(10):1550–8. https://doi.org/10.1038/s41591-018-0136-1.
Makena MR, Ranjan A, Thirumala V, Reddy AP. Cancer stem cells: Road to therapeutic resistance and strategies to overcome resistance. Biochim Et Biophys Acta-Mol Basis Dis. 2020;1866(4):165339. https://doi.org/10.1016/j.bbadis.2018.11.015.
Wei J, Huang K, Chen Z, Hu M, Bai Y, Lin S, et al. Characterization of glycolysis-associated molecules in the tumor microenvironment revealed by pan-cancer tissues and lung cancer single cell data. Cancers (Basel). 2020;12(7):1788. https://doi.org/10.3390/cancers12071788.
Hänzelmann S, Castelo R, Guinney J. GSVA: gene set variation analysis for microarray and RNA-seq data. BMC Bioinform. 2013;14:7. https://doi.org/10.1186/1471-2105-14-7.
Deng X, Qing Y, Horne D, Huang H, Chen J. The roles and implications of RNA m(6)A modification in cancer. Nat Rev Clin Oncol. 2023;20(8):507–26. https://doi.org/10.1038/s41571-023-00774-x.
Ravi R, Noonan KA, Pham V, Bedi R, Zhavoronkov A, Ozerov IV, et al. Bifunctional immune checkpoint-targeted antibody-ligand traps that simultaneously disable TGFβ enhance the efficacy of cancer immunotherapy. Nat Commun. 2018;9(1):741. https://doi.org/10.1038/s41467-017-02696-6.
Babaei G, Aziz SG, Jaghi NZZ. EMT, cancer stem cells and autophagy; The three main axes of metastasis. Biomed Pharmacother. 2021;133:110909. https://doi.org/10.1016/j.biopha.2020.110909.
Verstappe J, Berx G. A role for partial epithelial-to-mesenchymal transition in enabling stemness in homeostasis and cancer. Semin Cancer Biol. 2023;90:15–28. https://doi.org/10.1016/j.semcancer.2023.02.001.
Saygin C, Matei D, Majeti R, Reizes O, Lathia JD. Targeting cancer stemness in the clinic: From hype to hope. Cell Stem Cell. 2019;24(1):25–40. https://doi.org/10.1016/j.stem.2018.11.017.
Puthdee N, Sriswasdi S, Pisitkun T, Ratanasirintrawoot S, Israsena N, Tangkijvanich P. The LIN28B/TGF-β/TGFBI feedback loop promotes cell migration and tumour initiation potential in cholangiocarcinoma. Cancer Gene Ther. 2022;29(5):445–55. https://doi.org/10.1038/s41417-021-00387-5.
Huang Y, Hong W, Wei X. The molecular mechanisms and therapeutic strategies of EMT in tumor progression and metastasis. J Hematol Oncol. 2022;15(1):129. https://doi.org/10.1186/s13045-022-01347-8.
Acknowledgements
We thank Novogene (Beijing, China) for the technical support of RNA sequencing.
Funding
The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this work was supported by the Science and Technology Project of Guangzhou Health Commission (20241A011117), the Science and Technology Project of Panyu District, Guangzhou (2023-Z04-021), the Internal Scientific Research Fund of Guangzhou Panyu Central Hospital (PY-2023–025), the National Natural Science Foundation of China (82172789), Project of Educational Commission of Guangdong Province of China (2021KCXTD023), the YingTsai Young Scholar Award (CMU108-YTY-04), and the National Science and Technology Council, Taiwan (NSTC 112–2320-B-039–024 and 113–2320-B-039–017).
Author information
Authors and Affiliations
Contributions
P.Y., L.S. and J.H. designed and carried out the experiments, and interpreted data. P.Y., and J.H. extracted the information from the databases. P.Y., X.F., X.C. Y.M. and Y.C. analyzed the data. P.Y., J.-H.C., K.-W.H., X.Y. and W.-H.Y. provided scientific ideas. P.Y., X.Y. and W.-H.Y. prepared the manuscript. K.-W.H., X.Y. and W.-H.Y. supervised the entire project. All authors reviewed the manuscript.
Corresponding authors
Ethics declarations
Conflict of interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ye, P., Luo, S., Huang, J. et al. TESC associated with poor prognosis enhances cancer stemness and migratory properties in liver cancer. Clin Exp Med 24, 206 (2024). https://doi.org/10.1007/s10238-024-01469-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10238-024-01469-y