Abstract
The dynamics of the backbone and side-chains of protein are routinely studied by interpreting experimentally determined 15N spin relaxation rates. R1(15N), the longitudinal relaxation rate, reports on fast motions and encodes, together with the transverse relaxation R2, structural information about the shape of the molecule and the orientation of the amide bond vectors in the internal diffusion frame. Determining error-free 15N longitudinal relaxation rates remains a challenge for small, disordered, and medium-sized proteins. Here, we show that mono-exponential fitting is sufficient, with no statistical preference for bi-exponential fitting up to 800 MHz. A detailed comparison of the TROSY and HSQC techniques at medium and high fields showed no statistically significant differences. The least error-prone DD/CSA interference removal technique is the selective inversion of amide signals while avoiding water resonance. The exchange of amide with solvent deuterons appears to affect the rate R1 of solvent-exposed amides in all fields tested and in each DD/CSA interference removal technique in a statistically significant manner. In summary, the most accurate R1(15N) rates in proteins are achieved by selective amide inversion, without the addition of D2O. Importantly, at high magnetic fields stronger than 800 MHz, when non-mono-exponential decay is involved, it is advisable to consider elimination of the shortest delays (typically up to 0.32 s) or bi-exponential fitting.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In complex molecular systems such as proteins, structure, function and dynamics are closely linked. It is now widely accepted that intramolecular movements in proteins are one of the key determinants of their biological activity (Teilum et al. 2009; Micheletti 2013; Dong et al. 2017; Pacini et al. 2021; Yuan et al. 2021). Nuclear magnetic relaxation analysis provides insight into the molecular motions of proteins. The distinctive ability to detect and visualize alternative conformations at atomic level positions the dynamics studies an emerging approach in the structure-based drug discovery (Emwas et al. 2020; Kharchenko et al. 2022a, b; Hansen et al. 2023). Since the first application of magnetic relaxation measurements of 15N nuclei applied to a protein, staphylococcal nuclease (Kay et al. 1989), this method has become essential in determining molecular motions in proteins (Kempf and Loria 2003; Palmer III 2004; Jarymowycz and Stone 2006; Reddy and Rayney 2010; Nowakowski et al. 2011, 2013; Lisi and Loria 2016; Charlier et al. 2016; Jaremko et al. 2011, 2015, 2016, 2018; Stetz et al. 2019).
Three relaxation parameters - longitudinal (R1) and transverse (R2) relaxation rates together with 15N{1H} nuclear Overhauser effect (NOE) - have been most commonly used in studies of backbone mobility in proteins (Jaremko et al. 2015; Hoch et al. 2023). To interpret experimental relaxation data in terms of molecular mobility, the following complex tasks need to be performed: trustworthy experimental techniques for measuring relaxation parameters and reliable data reduction methods must be used.
As a continuation of our research focused on reliable measurements of 15N{1H} NOEs (Kharchenko et al. 2020) the aim of this study is to investigate the most important experimental methods and faithful data analysis procedures to determine the longitudinal relaxation rate R1 for 15N backbone nuclei in proteins.
Experimental
Samples
Two different samples of uniformly labelled U-[15N] human ubiquitin (purchased as a lyophilized powder from Cambridge Isotope Laboratories, Inc) were used in the study; one sample containing 100% H2O and a sealed capillary filled with 99.9% D2O inserted into an NMR tube and the other containing a mixture of H2O/D2O 90%/10% (v/v). These are indicated in the text of manuscript by the letters H or D. Both samples contain 0.8 mM concentration of protein in 10 mM sodium phosphate buffer at pH 6.65 (pH meter read-out), 0.01% (m/v) NaN3 and DSS-d6 of 0.1% (m/v).
NMR measurements
Measurements of R1 were carried out at three magnetic field strengths, 16.4, 18.8, and 22.3 T. The corresponding 1H resonance frequencies, 700, 800, and 950 MHz, were inserted into the names of the appropriate experiments. The experimental temperature was 298 K for each NMR measurement calibrated with an ethylene glycol reference sample (Raiford et al. 1979). The maximum detected temperature deviation before and after the experiment was ± 0.3 K.
The 15N R1 pulse sequences used here are optimized for both perdeuterated and protonated samples and based on the HSQC or TROSY method encoded with I or T, respectively. The following Bruker-coded pulse programs were used: for the I type experiments: hsqct1etf3gptcwg3d and for T type experiments: trt1etf3gpsitc3d for 1H 180° hard pulses and trt1etf3gpsitc3d.3 for IBURP-2 shaped inversion pulses (Lakomek et al. 2012). All pulse programs contain the temperature compensation (tc) block. Interference between 1H and 15 N DD and 15N CSA mechanisms was suppressed by applying a sequence of 1H 180° pulses during the variable evolution period of the longitudinal 15N relaxation, τ. Three types of pulses were used: non-selective broadband 180°-shaped inversion pulses (Smith et al. 2001) equivalent to rectangular hard inversion pulses (denoted HP), band selective amide proton IBURP-2 pulses of 0.48 W centered at 8.5 ppm, 5.5 ppm wide (labelled AM) (Geen and Freeman 1991), and cosine-modulated IBURP-2 pulses selectively inverting amide and aliphatic protons (power 1.55 W, centered at 8.5 and 0.9 ppm, both 5.5 ppm wide; labelled AMAL). The IBURP-2 proton inversion pulses were generated using the WaveWaker (vwm), an automated pulse shaping tool in Topspin ver. 4.0.7 (Bruker). The HP inversion pulses for T type experiments were spaced 5 ms apart, while for AM and AMAL, the pulses were spaced 40 ms apart. For I type experiments, all HP, AM, and AMAL pulses were spaced at 40 ms intervals during the evolution period.
Example of abbreviations used in the manuscript: 800-ID-AM describes an experiment performed on an 800 MHz spectrometer and a ubiquitin sample containing 10% of D2O. The pulse sequence was based on HSQC (I) while the interference between the DD and CSA mechanisms was suppressed with an IBURP-2 pulse train centered at 8.5 ppm and therefore in the amide region (AM).
In the current study, to optimize the experimental conditions of R1(15N) measurements in proteins for backbone amides, 42 independent experiments were collected. All experiments, unless stated otherwise, used a recycle delay (RD) of 3 s. A list of all experiments is given in Table S3 (supplementary material). Two additional 700-IH-AM R1 measurements to determine the temperature gradient were performed at 288 K and 308 K.
Data processing
The chemical shifts in the 1H NMR spectra were reported relative to DSS-d6, while the chemical shifts of the 15N signals were referenced indirectly (Wishart et al. 1995). The spectral widths were set to 12 ppm and 22 ppm for 1H and 15N, respectively. The complex data points collected for 1H and 15N dimensions were 2048 and 256, respectively. Eight scans per FID were accumulated in each experiment. A double zero filling and a 90° shifted squared sine filter were applied prior to Fourier transform. Data were processed with nmrPipe (Delaglio et al. 1995) and analyzed with SPARKY (Goddard and Kneller).
Resonance intensities were used to calculate relaxation rates from a non-linear least-squares analyses performed using Fortran routines written in-house, based on Newton–Raphson or Levenberg-Marquardt algorithms (Press et al. 1986).
Results and discussion
Elimination of systematic errors from the procedure for determining R1
Relaxation times are determined by sampling a relaxing magnetization M after set of evolution times, τ (Kowalewski and Mäler 2006). For the pulse sequences commonly used to study proteins, an exponential decrease in signal intensity is observed. No special attention is paid to possible deviations from the exponential decay. However, a number of different effects interfere with measurements of the longitudinal relaxation rate R1 of amide backbone 15N nuclei in proteins: spin saturation of water proton affecting HN magnetization through exchange or NOE mechanisms (Grzesiek and Bax 1993; Chen and Tjandra 2011), incompletely suppressed DD/CSA interference (Boyd et al. 1990; Kay et al. 1992), or imperfect pulses (Lakomek et al. 2012; Ishima 2014; Gairì et al. 2015). Another source of perturbation in FID detection may be the non-ideal behavior of NMR transmitter and probe electronics exhibiting frequency-dependent interference (Uribe and Martin 2024). The mentioned above fast fading artifacts are difficult or impossible to eliminate or prevent. Consequently, the longitudinal 15N magnetization decay is no longer mono-exponential, and the initial experimental data points obtained for the shortest evolution times, τ, are the most disturbed. Consequently, the apparent R1 values are artificially increased and their standard deviations are enlarged. The anomalous effects become stronger at high magnetic fields. This phenomenon has usually been overlooked for two reasons. Deviations from mono-exponential decays can be hidden when spectra show a poor signal-to-noise ratio and/or R1 are derived from automatic procedures without careful inspection of the input data and intermediate results. Most of the artifacts can be quantitatively eliminated using a bi-exponential model rather than a mono-exponential decay model. However, the bi-exponential fitting procedure is prone to numerical instability due to strong correlations between the fitted parameters. Ishima (2014) recommended removing such artifacts by discarding the initial data points in mono-exponential fitting. She recommends using a τ values longer than 0.2 s for experiments performed at 21.1 T. However, the cutoff τ value can be expected to depend not only on the magnetic field strength but also on the setup of the individual NMR spectrometer and the properties of the specific sample.
To define more stringent conditions for R1 data processing, we measured R1 at 16.4, 18.8, and 22.3 T using a list of ten evolution times, τ (cf. Table S3). In addition to bi-exponential fitting, the R1 data were analyzed with mono-exponential decays, sequentially discarding the initial values of τ.
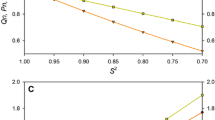
A visualization of the data obtained at 22.3 T for the 100% H2O sample is shown in Fig. 1. The R1 values determined for evolution times τ ≥ 0.32 s with a mono-exponential fit are equivalent to a bi-exponential fit. It leads to the conclusion that, in this particular case, the data for τ ≥ 0.32 s represent mono-exponential decay. On the other hand, too many rejected initial data points reduce the accuracy of the fit, as can be seen by comparing the data for 0.32, 0.48, and 0.64 s. Their average relative errors successively increase: 0.11%, 0.17%, and 0.26%. The site-specific differences between R1 values obtained from the bi-exponential and mono-exponential fits, calculated for different values of the initial τ value, display uniform distribution. Their mean value obtained for the shortest τ = 0.32 s is close to zero (Fig. 2). The numerical R1 values and their standard deviations shown in Fig. 2 are given in Table S1.
Mean R1 values fitted to mono-exponential decay for increasing values of the shortest evolution time τ at 22.3 T (950-IH-AM: see the code description in the section NMR measurements). Error bars represent the average standard deviations of individual R1 values. The horizontal solid line represents the mean R1 value obtained by fitting data to bi-exponential decays and dashed lines represent the average standard deviations of individual R1 values
Site-specific differences between mono-exponential and bi-exponential R1 values are calculated for initial τ value: 0.08 s and 0.32 s in the upper and lower parts of the Figure (code 950-IH-AM). Dashed lines correspond to the means of differences. Their numerical values for the upper and lower parts of the Figure are equal to 0.0144 s− 1 and 0.0003 s− 1, respectively
Similar results were obtained for the remaining measurements at 22.3 T (Figures S1 - S7). The bi-exponentially derived R1 values are on average 1% smaller than the R1s calculated mono-exponentially at 22.3 T for the 100% H2O sample. Consequently, the local mobility of the protein backbone can be erroneously estimated if evaluated from a model-free approach (Lipari and Szabo 1982) with inaccurate R1 values as input data (Chen and Tjandra 2011; Lakomek et al. 2012).
In contrast to the data obtained at 22.3 T, measurements obtained with the same method (HSQC, IBURP-2) for the 100% H2O sample at weaker magnetic fields (16.4 and 18.8 T) do not show a regular dependence of R1 value on the initial value of the evolution time τ (Figures S8 and S9) suggesting frequency dependent hardware problem. Such data can be safely processed using a mono-exponential decay model without discarding the shortest τ values. Three cases of the largest values of the Fisher-Snedecor statistics, F, in measurements taken at 16.4 and 18.8 T give the same R1 values for mono- and bi-exponential fits within the experimental error (Table 1). Therefore, the mono-exponential analysis of R1 is suitable for spectrometers with a magnetic field 18.8 T or weaker, at least for samples dissolved in H2O and the HSQC/IBURP-2 method.
Based on our research, we recommend an initial check of the mono-exponential fitting model with subsequent rejection of the shortest evolution time data points. If the rejection of one or two initial data points does not regularly change the results, the full data set and the mono-exponential decay model can be used with caution. Otherwise, mainly for measurements in high magnetic fields, two options are possible: to use a bi-exponential fit of the R1 data, if the calculations are stable and the derived parameter errors are acceptable, or to discard the initial data points and then analyze a mono-exponential decay until no further changes in R1 values are observed.
Reproducibility of duplicated experiments
Knowing the reproducibility of the individual experiments is crucial for investigating very small effects, such as, for example, possible differences between HSQC and TROSY detection or differences between interference suppression methods. For this reason, several types of experiments were duplicated at intervals of several months. The statistical analysis of these measurements is given in Table 2.
The mean differences between duplicate experiments, <diff>, do not exceed 0.01 s− 1, and in many cases are even an order of magnitude smaller. The rms values of the differences are also less than 0.01 s− 1 with the exception of one case of 950-TH-HP experiments. These values limit the meaningless differences between the two experiments.
However, it should be born in mind that the changing the temperature of a sample usually results in a significant, often unrecognized source of experimental error. To assess temperature-induced errors, the temperature gradient of R1 was determined. Its value was equal to 0.028±0.003 s− 1/K at 16.4 T (details of the determination of the temperature gradient are given in the Supplementary Material). Given the maximum detected temperature deviation of 0.3 K in our work, temperature can cause R1 to vary by up to 0.008 s− 1. All < diff > values given in Table 2 do not exceed this value.
Comparison of HSQC and TROSY techniques applied to R1 measurements
Two direct comparisons of HSQC and TROSY techniques can be found in the literature. Zhu at all. (2000) compared relaxation measurements of holo-calmoduline (17 kDa) sample at 17.6 T (750 MHz) removing interference effects during relaxation evolution times using 1H 180° HN-band selective pulses at 5–10 ms interval. The authors found that excellent agreement was obtained between TROSY-based experiments and the corresponding HSQC-based experiments. Lakomek et al. (2012) measured the small perdeuterated GB3 protein (6.2 kDa) at 14.1 T (600 MHz) using HN-band selective IBURP pulses every 40 ms to suppress interference effects during relaxation evolution times. These authors also found that no systematic differences were observed between R1 relaxation rates measured for GB3 using TROSY-detection and those obtained using HSQC-detection.
We conducted systematic comparison experiments based on TROSY and HSQC on two samples (100% H2O and 90% H2O/10% D2O) using three different interference suppression schemes at high magnetic fields of 22.3 T (950 MHz) and 18.8 T (800 MHz).
All twelve pairs of experiments given in Table 3 show very good statistical agreement between the TROSY and HSQC-based methods. The result shows that the two techniques produce virtually identical results, and the differences between them are comparable to those in the duplicated experiments.
Site specific differences for the 800/TH-AM/IH-AM pair are shown in Fig. 3. Another example is shown in Fig. S10. Well-defined ranges of anomalies cannot be recognized.
Site-specific differences between TROSY-based and HSQC-based experiments at 18.8 T (800 MHz); codes 800-TH-AM and 800-IH-AM. The band HN selective pulses were used for interference suppression; solvent − 100% H2O. Solid line represents mean value of differences and dashed lines correspond to the mean ± 3σ. All differences are within this range
DD/CSA interference removal in R1 measurements
Interference effects between dipolar and chemical-shift anisotropy relaxation interactions in the measurement of relaxation times can cause significant errors in a simple mono-exponential analysis of the raw data (Goldman 1984; Werbelow 1996). Initially, interference was neglected in the analysis of 15N protein relaxation data (Kay et al. 1989; Clore et al. 1990). Shortly thereafter, Boyd et al. (1990) showed that broadband decoupling of 1H nuclei applied during the evolution delay, τ, removed interference effects. Kay et al. (1989) introduced a different approach, using 1H 180° pulses every 5–10 ms. For the staphylococcal nuclease protein (18 kDa), they observed an increase in R1 relaxation rate up to 9%. Moreover, the errors increase rapidly with decreasing molecular weight of the molecules studied. 1H 180° pulses were also used for the suppression of interference effects in the determination of transverse relaxation rates (Palmer III et al. 1992).
Another source of error in R1 measurements is due to the varying number of non-selective 180° 1H pulses used to eliminate relaxation interference effects during evolution times. Progressive suppression of water magnetization due to an increasing number of 180° 1H pulses for longer relaxation delays, τ, manifests itself in an increase in apparent R1. Elimination of this experimental defect has been achieved by replacing non-selective 180° 1H pulses with flip-back pulses (Grzesiek and Bax 1993; Chen and Tjandra 2011,) or HN-band selective pulses (Geen and Freeman 1991; Chill et all. 2006; Lakomek et al. 2012).
Gairí et al. (2015), studying small GB3 protein in two magnetic fields (14.1 and 18.8 T), observed that HN-band selective pulses (IBURP-2) resulted in smaller R1 values than the cosine-modulated IBURP-2 method, which selectively inverts two spectral bands, amide and aliphatic protons. The authors concluded that the differences in R1 values obtained with the different pulse trains were small but systematic and significant (rms diff = 0.075 s− 1) and greater than the reproducibility of the individual experiments (rms diff = 0.030 s− 1). They concluded that the source of this discrepancy could be either ineffective interference suppression or scheme-dependent saturation effects differently affecting HN magnetization at the start of each scan. Differential saturation effects should be sensitive to the recycle delay, whereas not fully effective interference suppression should not be affected by the delay. Rates of 15N R1 were measured using IBURP-2 and cosine-modulated IBURP-2 at recycle delay values of 1.7 and 3.5 s. The mean relaxation rates R1 measured by the cosine-modulated IBURP-2 method did not depend on the length of the recycle delay and were very similar to those obtained using IBURP-2 pulses with a longer recycle delay, whereas this method with a shorter recycle delay resulted in smaller values of R1. Therefore, the authors concluded that the cosine-modulated IBURP-2 pulses were superior to the IBURP-2 approach. However, in the experimental part the authors stated that cosine-modulated IBURP-2 pulses were 1.26 times longer and required four times more power (6 dB) than IBURP-2 pulses. It can be expected that stronger microwave heating without temperature compensation can increase the relaxation rate R1 based on the cosine-modulation of IBURP-2 compared to R1 derived from IBURP-2.
Our results include measurements of two ubiquitin samples (100% H2O and 90% H2O/10% D2O) at three magnetic field strengths, 16.4, 18.8, and 22.3 T. The interference suppression by the HN-band selective pulses (AM) almost never shows lower mean relaxation rates < R1 > compared to < R1 > measured with cosine-modulated IBURP-2 pulses (AMAL) (Fig. 4 and Figs. S11 and S12). The mean rms difference calculated for the 10 pairs AM/AMAL measurements is 0.012 s-1 (Table 4), while the reproducibility of the 10 experiments gives a comparable mean rms difference of 0.009 s-1. Differences between the two suppression methods are comparable to the reproducibility of the experiment and several times smaller than those observed by Gairí et al. (2015). The largest differences compared to Gairí’s data are: rms difference (Gairí) = 0.075 s-1, rms difference (22.3 T) = 0.010 s-1, rms difference (18.8 T) = 0.015 s-1, and rms difference (16.4 T) = 0.014 s-1. These can be attributed to temperature fluctuations and stronger microwave heating in the cosine-modulated IBURP-2 method.
Mean R1 values measured with HSQC or TROSY based sequences at B0 = 22.3 T. DD/CSA interference was suppressed by IBURP-2 (AM), cosine modulated IBURP-2 (AMAL), or nonselective 180° (HP) pulses. Error bars represent average standard deviations of individual R1 values. Boxes mark corresponding AM/AMAL pairs. Dashed lines represent averages of all data points in appropriate parts of the figure. Difference of R1 values between AM and AMAL approaches observed for the mixed H2O/D2O solvent, <ΔR1 > = 0.02 s− 1, rms diff = 0.01 s− 1, might be attributed to 0.5 K temperature divergence
In addition, we determined R1 relaxation data using the IBURP-2 method for 100% H2O sample at 22.3 T with 3 recycle delays: 2, 4, and 6 s. The averaged R1 values are virtually identical (Fig. 5). Therefore, the difference between IBURP-2 and the cosine-modulated IBURP-2 cannot be explained by differential saturation effects. In conclusion, the IBURP-2 method seems to be superior to its cosine-modulated counterpart.
One more observation reported by Gairí et al. (2015) requires comment. They found that the use of non-selective 180° 1H pulses, causing strong water saturation, resulted in an overestimation of R1. Furthermore, errors from water saturation effects varied along the protein sequence, being larger in the most solvent-exposed regions of the GB3 protein.
A close inspection of Fig. 4, S11, and S12 reveals, that suppressing interference with non-selective 180° 1H pulses has no effect on the averaged R1. Incidentally, this is a good example of how the the analysis of R1 averaged differences often gives a deceptively simple but false picture. Analysis of individual, residue-specific differences reveals a non uniform pattern. Site-specific differences between measurements using cosine-modulated IBURP-2 or non-selective 180° pulses for 100% H2O sample in three magnetic fields were calculated twice. In Fig. 6 all R1 values were obtained in a way that rejects fast fading artifacts, i.e. the data with the shortest evolution times τ = 0.32 s were used in the fitting procedures. The differences show no significant variation along the protein sequence for the use of non-selective 180° 1H pulses. On the other hand, in Fig. 7 all R1 values were obtained without discarding these artifacts. The highlighted rapidly exchanging HN residues are different from the other residues.
Site specific differences between measurements applying the cosine-modulated IBURP-2 and nonselective 180° pulses for 100% H2O sample at three magnetic fields. The shortest evolution time τ = 0.48 s. Negligible rms diff values are equal to 0.006, 0.012, and 0.010 s− 1 for 22.3, 18.8, and 16.4 T, respectively. Solid lines represent mean values of differences and dashed lines represent the average standard deviations of individual R1 values
Site specific differences between measurements applying the cosine-modulated IBURP-2 and nonselective 180° pulses for 100% H2O sample at three magnetic fields. The shortest evolution time τ = 0.08 s. Rms diff values are equal to 0.008, 0.012, and 0.010 s− 1 for 22.3, 18.8, and 16.4 T, respectively and do not differ from those given in Fig. 6. Amino acid residues with fast HN exchange are shown in red squares. Solid lines represent mean values of differences and dashed lines represent the average standard deviations of individual R1 values
Such an effect can be observed in certain situations, and it is discussed in the following section. It should be noted, however, that the data shown in Figs. 6 and 7 were obtained for a sample of 100% H2O. The differences between the samples of H2O and H2O/D2O are analyzed in detail in the following section.
Exchangeable deuterons perturb the apparent longitudinal relaxation rates 15N for solvent-exposed amides
Magnetic field stability is one of the key factors in NMR spectroscopy. For this purpose, a deuterium lock system is generally used. To make it work, a mixed H2O/D2O solvent (usually 90%/10% or 95%/5%) is used instead of pure H2O for NMR measurements of proteins. As the protons of HN exchange with both protons and deuterons of the solvent, a reduction in the signal intensity of HN is observed due to partial H→D substitution. This effect reduces the sensitivity but does not affect the R1 values. On the contrary, other mechanisms mentioned in the section “Elimination of systematic errors…” modulate the longitudinal relaxation. In magnetic relaxation measurements, HN exchange leads to erroneous values of the apparent relaxation parameters, provided that these exchange rates are not significantly slower than the relaxation rates.
There are nine amino acid residues in ubiquitin molecule that show increased rates of HN exchange: L8-T12, A46, and L73-G75 (Cordier and Grzesiek 2002). Moreover, these residues show reduced temperature gradients of the longitudinal relaxation rate R1 (Fig. 8). Such residues can be identified by a strong deviation from the mono-exponential fit of the experimental decay of R1. There are two methods to identify such residues.
Experimental exchange HN values, k(HN) taken from Cordier and Grzesiek (2002); lower part. Temperature gradients of R1 relaxation rates (upper part) determined from the ubiquitin spectra measured at 288 and 308 K and 16.4 T; solvent 90% H2O/10% D2O, method HSQC, interference DD/CSA suppressed with nonselective 180° 1H pulses
Firstly, the mono-exponential determination of R1 values with and without the shortest evolution times, τ, which are most strongly disrupted, was analyzed. This perturbation of R1 values clearly appears in samples containing some D2O. It disappears in samples without D2O, as can be seen in Fig. 9 and S13. However, it can be, partially preserved in 100% H2O when non-selective 180° 1H pulses are used to suppress interference (Figures S14 and S6).
Both plots display residue specific differences between R1 values determined with data comprising evolution times τ ≥ 0.08 s and data after rejecting the four shortest τ values (τ ≥ 0.48 s). Upper part - solvent 90% H2O/10% D2O, lower part - solvent 100% H2O. Interference suppression - HN selective band (AM). Similar comparisons for other field strengths and other techniques are presented in Supplementary Materials
Secondly, the bi-exponential fits of the experimental data, which show the largest values of the F statistics with respect to the mono-exponential fits and the largest amplitudes of the faster decay components, indicate a fast HN exchange (Tables 5 and 6).
An example of a bi-exponential fit to experimental data for fast exchangeable HN residue A46 is shown in Fig. 10. The deviation of the mono-exponential R1 value from the slow component in bi-exponential fit is equal to 6.7% and 0.3% for H2O/D2O and H2O solutions, respectively. For less demanding applications, the mono-exponential fit for H2O solution data is sufficiently accurate, and its variation from the bi-exponential fit is comparable to the accuracy of the measurement. In many cases, this allows a simplification of experimental determination of R1. Such solvent-induced effect is independent of magnetic field strength, detection method (HSQC, TROSY), or interference suppression method (selective or nonselective HN pulses), (Supplementary Material). As before, a key element in such a distinction is the presence of D2O.
R1 data for A46 residue measured in different solvent (with 10% D2O - upper panel and without D2O - lower panel) at 22.3 T using HSQC method and HN band selective irradiation. Parameters determined in bi-exponential fit for upper panel: F = 278, fast component (Af=181, R1f = 10.8 s− 1), slow component (As=1033, R1s = 1.399 ± 0.009 s− 1), mono-exponential fit (A = 1098, R1 = 1.493 ± 0.030 s− 1). Parameters determined for lower panel: F = 40, fast component (Af=34, R1f = 13.8 s− 1) slow component (As=1110, R1s = 1.446 ± 0.003 s− 1), mono-exponential fit (A = 1114, R1 = 1.451 ± 0.002 s− 1)
In summary, R1 measurements made in the 100% H2O using HN-band selective pulses for interference suppression appear to be the least susceptible to experimental artifacts.
Conclusions
In the present work measurements of the longitudinal relaxation rate R1 for backbone nuclei 15N in proteins have focused on identifying sources of error occurring at the experimental stage, as well as suppressing data reduction. Starting with sample preparation, D2O should be avoided as much as possible, as the presence of deuterons disturbs the rates of R1 for rapidly exchanging HN backbone protons. Rigorous temperate control is necessary because a change in sample temperature usually results in a significant, often unrecognized source of experimental error. With regard to the measurement pulse sequence, it has been shown that DD/CSA interference suppression must be performed using HN-band selective pulses. Our result confirms that the TROSY and HSQC-based methods give virtually identical results. If experimental data do not decay mono-exponentially, two options are possible: using bi-exponential fitting of the R1 data, if the calculations are stable and the derived parameter errors are acceptable, or discarding the initial data points and then analyzing the mono-exponential decay until no further changes in the R1 values are observed.
Data availability
No datasets were generated or analysed during the current study.
References
Boyd J, Hommel U, Campbell ID (1990) Influence of cross-correlation between dipolar and anisotropic chemical shift relaxation mechanisms upon longitudinal relaxation rates of 15N in macromolecules. Chem Phys Lett 175:477–482
Charlier C, Cousin SF, Ferrage F (2016) Protein dynamics from nuclear magnetic relaxation. Chem Soc Rev 45:2410–2422
Chen K, Tjandra N (2011) Water Proton spin saturation affects measured protein backbone 15N spin relaxation rates. J Magn Reson 213:151–157
Chill JH, Louis JM, Baber JL, Bax A (2006) Measurement of 15N relaxation in the detergent solubilized tetrameric KcsA potassium channel. J Biomol NMR 36:123–136
Clore GM, Driscoll PC, Wingfield PT, Gronenborn AM (1990) Analysis of the Backbone dynamics of Interleukin-β using two-dimensional Inverse detected Heteronuclear 15N-1H NMR spectroscopy. Biochemistry 29:7387–7401
Cordier F, Grzesiek S (2002) Temperature-dependence of Protein Hydrogen Bond Properties as studied by high-resolution NMR. J Mol Biol 715:739–752
Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J Biomol NMR 6:277–293
Dong Z, Zhou H, Tao P (2017) Combining protein sequence, structure, and dynamics: a novel approach for functional evolution analysis of PAS domain superfamily. Prot Sci 27:421–430
Emwas AH, Szczepski K, Poulson BG, Chandra K, McKay RT, Dhahri M, Alahmari F, Jaremko L, Lachowicz JI, Jaremko M (2020) NMR as a gold standard method in Drug Design and Discovery, vol 25. Molecules, p 4597
Gairì M, Dyachenko A, González MT, Feliz M, Pons M, Giralt E (2015) An optimized method for 15N R1 relaxation rate measurements in non-deuterated proteins. J Biomol NMR 62:209–220
Geen H, Freeman R (1991) Band-Selective Radiofrequency pulses. J Magn Reson 93:93–141
Goddard TD, Kneller DG, SPARKY 3 : University of California, San Francisco, http://www.cgl.ucsf.edu/home/sparky
Goldman M (1984) Interference effects in the relaxation of a pair of unlike Spin-½ nuclei. J Magn Reson 60:437–452
Grzesiek S, Bax A (1993) The importance of not saturating H2O in protein NMR–application to sensitivity enhancement and NOE measurements. J Am Chem Soc 115:12593–12593
Hansen AL, Xiang X, Yuan C, Bruschweiler-Li L, Brüschweiler R (2023) Excited-state observation of active K-Ras reveals differential structural dynamics of wild-type versus oncogenic G12D and G12C mutants. Nat Struct Mol Biol 30:1446–1455
Hoch JC, Baskaran K, Burr H, Chin J, Eghbalnia HR, Fujiwara T, Gryk MR, Iwata T, Kojima C, Kurisu G, Maziuk D, Miyanoiri Y, Wedell JR, Wilburn C, Yao H, Yokochi M (2023) Biological Magnetic Resonance Data Bank. Nucleic Acids Res 51:D368–D376
Ishima R (2014) A probe to monitor performance of 15N longitudinal relaxation experiments for proteins in solution. J Biomol NMR 58:113–122
Jaremko Ł, Jaremko M, Elfaki I, Mueller JW, Ejchart A, Bayer P, Zhukov I (2011) Structure and dynamics of the first archaeal parvulin reveal a new functionally important loop in parvulin-type prolyl isomerases. J Biol Chem 286:6554–6565
Jaremko Ł, Jaremko M, Nowakowski M, Ejchart A (2015) The Quest for simplicity: remarks on the Free-Approach models. J Phys Chem B 119:11978–11987
Jaremko M, Jaremko Ł, Villinger S, Schmidt CD, Griesinger C, Becker S, Zweckstetter M (2016) High-resolution NMR determination of the dynamic structure of membrane proteins. Angew Chem Int Ed Engl 22:55:10518–10521
Jaremko Ł, Jaremko M, Ejchart A, Nowakowski M (2018) Fast evaluation of protein dynamics from deficient 15 N relaxation data. J Biomol NMR 70:219–228
Jarymowycz VA, Stone MJ (2006) Fast time scale dynamics of protein backbones: NMR relaxation methods, applications, and functional consequences. Chem Rev 106:1624–1671
Kay LE, Torchia DA, Bax A (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to Staphylococcal nuclease. Biochemistry 28:8972–8979
Kay LE, Nicholson LK, Delaglio F, Bax A, Torchia DA (1992) Pulse sequences for removal of the effects of cross correlation between dipolar and chemical-shift anisotropy relaxation mechanisms on the measurement of heteronuclear T1 and T2 values in proteins. J Magn Reson 97:359–375
Kempf JG, Loria JP (2003) Protein dynamics from solution NMR. Cell Biochem Biophys 37:187–211
Kharchenko V, Nowakowski M, Jaremko M, Ejchart A, Jaremko Ł (2020) Dynamic 15N{1H} NOE measurements: a tool for studying protein dynamics. J Biomol NMR 74:707–716
Kharchenko V, Linhares BM, Borregard M, Czaban I, Grembecka J, Jaremko M, Cierpicki T, Jaremko Ł (2022a) Increased slow dynamics defines ligandability of BTB domains. Nat Commun 13(1):6989. https://doi.org/10.1038/s41467-022-34599-6
Kharchenko V, Ejchart A, Jaremko L (2022b) in NMR Spectroscopy for Probing Functional Dynamics at Biological Interfaces, ed. A. Bhunia, H. S. Atreya, and N. Sinha, The Royal Society of Chemistry, 2022, ch. 3, pp. 56–81
Kowalewski J, Mäler L (2006) Nuclear relaxation in liquids: theory, experiments, and applications. CRC, Boca Raton, FL, pp 177–185
Lakomek N-A, Ying J, Bax A (2012) Measurement of 15N relaxation rates in perdeuterated proteins by TROSY-based methods. J Biomol NMR 53:209–221
Lipari G, Szabo A (1982) Model-Free Approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of Validity. J Am Chem Soc 104:4546–4559
Lisi GP, Loria JP (2016) Using NMR spectroscopy to elucidate the role of molecular motions in enzyme function. Prog NMR Spectrosc 92–93:1–17
Micheletti C (2013) Comparing proteins by their internal dynamics: exploring structure–function relationships beyond static structural alignments. Phys Life Rev 10:1–26
Nowakowski M, Jaremko Ł, Jaremko M, Zhukov I, Belczyk A, Bierzyński A, Ejchart A (2011) Solution NMR structure and dynamics of human apo-S100A1 protein. J Struct Biol 174:391–399
Nowakowski M, Ruszczyńska-Bartnik K, Budzińska M, Jaremko L, Jaremko M, Zdanowski K, Bierzyński A, Ejchart A (2013) Impact of calcium binding and thionylation of S100A1 protein on its nuclear magnetic resonance-derived structure and backbone dynamics. Biochemistry 52:1149–1159
Pacini L, Dorantes-Gilardi R, Vuillon L, Lesieur C (2021) Mapping function from Dynamics: Future challenges for Network-based models of protein structures. Front Mol Biosci 8:744646
Palmer AG III (2004) NMR characterization of the dynamics of biomacromolecules. Chem Rev 104:3623–3640
Palmer AG III, Skelton NJ, Chazin WJ, Wright PE, Rance M (1992) Suppression of the effects of cross-correlation between dipolar and anisotropic chemical shift relaxation mechanisms in the measurement of spin-spin relaxation rates. Mol Phys 75:699–711
Press WH, Flannery BP, Teukolsky SA, Vetterling WT (1986) Numerical recipes: the art of Scientific Computing. Cambridge University Press, New York. Chap. 9
Raiford DS, Fisk CL, Becker ED (1979) Calibration of methanol and ethylene glycol nuclear magnetic resonance thermometers. Anal Chem 51:2050–2051
Reddy T, Rayney JK (2010) Interpretation of biomolecular NMR spin relaxation parameters. Biochem Cell Biol 88:131–142
Smith MA, Hu H, Shaka AJ (2001) Improved Broadband Inversion performance for NMR in liquids. J Magn Reson 151:269–283
Stetz MA, Caro JA, Kotaru S, Yao X, Marques BS, Valentine KG, Wand AJ (2019) Characterization of internal protein dynamics and conformational entropy by NMR relaxation. Method Enzymol 615:237–284
Teilum K, Olsen JG, Kragelund BB (2009) Functional aspects of protein flexibility. Cell Mol Life Sci 66:2231–2247
Uribe JL, Martin RW (2024) A practical introduction to radio frequency electronics for NMR probe builders. JMRO 19:100153
Werbelow LG (1996) Relaxation Processes: Cross Correlation and Interference Terms. Encyclopedia of Nuclear Magnetic Resonance 6:4072–4078 eds. DM Grant, RK Harris, Wiley, Chichester
Wishart DS, Bigam CG, Yao CG, Abildgaad F, Dyson HJ, Oldfield E, Markley JL, Sykes BD (1995) 1H, 13 C and 15 N chemical shift referencing in biomolecular NMR. J Biomol NMR 6:135–140
Yuan G, Flores NM, Hausmann S, Lofgren SM, Kharchenko V, Angulo-Ibanez M, Sengupta D, Lu X, Czaban I, Azhibek D, Vicent S, Fischle W, Jaremko M, Fang B, Wistuba II, Chua KF, Roth JA, Minna JD, Shao NY, Jaremko Ł, Mazur PK, Gozani O (2021) Elevated NSD3 histone methylation activity drives squamous cell lung cancer. Nature 590:504–508
Zhu G, Xia Y, Nicholson LK, Sze KH (2000) Protein Dynamics measurements by TROSY-Based NMR experiments. J Magn Reson 143:423–426
Author information
Authors and Affiliations
Contributions
All authors contributed to the experiment, data analysis and manuscript writing. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Kharchenko, V., Al-Harthi, S., Ejchart, A. et al. Pitfalls in measurements of R1 relaxation rates of protein backbone 15N nuclei. J Biomol NMR (2024). https://doi.org/10.1007/s10858-024-00449-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10858-024-00449-4