Abstract
In a parasitological survey of fishes from Moreton Bay (southeastern Queensland, Australia), 169 teleost fishes, representing 54 species from 28 families, were examined for larval cestodes. Of these 54 species, 36 were found to be infected by metacestodes. Metacestodes were characterised by morphological and molecular data (the D1-D3 region of the 28S rDNA gene); these data were analysed in parallel to inform larval type allocation. Metacestodes collected represented eight morphological types, seven previously reported (Types I, II, IV, V, VI, VII, and X) and one novel type (Type XVI). Phylogenetic analyses were conducted to genetically match larval types to adult cestodes. Six of the eight larval types found were matched to adult forms: Type I metacestodes matched species of Phoreiobothrium Linton, 1889 (Onchobothriidae); Type II metacestodes matched species of Acanthobothrium van Beneden, 1849 (Onchobothriidae); Type IV metacestodes matched species of Scyphophyllidium Woodland, 1927 and Alexandercestus Ruhnke & Workman, 2013 (Phyllobothriidae); Type VI metacestodes matched species of Anthobothrium van Beneden, 1850 (Tetraphyllidea incertae sedis); Type X metacestodes matched species of Ambitalveolus Caira & Jensen, 2022 (Tetraphyllidea incertae sedis); and Type XVI metacestodes matched species of Platybothrium Linton, 1890 (Onchobothriidae). Based on phylogenetic topology, Type V metacestodes are inferred to match Pedibothrium Linton, 1909 (Balanobothriidae) and Type VII metacestodes are inferred to match Spongiobothrium Linton, 1889 (Rhinebothriidae). These findings support and extend the unified morphological type system proposed previously, but suggest that morphological types will ultimately be informative to identify metacestodes to a group of related genera rather than any distinct genus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since the original descriptions of Scolex pleuronectis Müller, 1788 and Scolex polymorphus Rudolphi, 1819, there has been disorder in the identification of “tetraphyllidean” metacestodes. The lack of development in scolex morphology at this stage, and the usually complete absence of strobila development, has rendered metacestodes notoriously challenging to identify. While identification has occasionally been attempted using morphological techniques (e.g., Cake, 1976; Carvajal et al., 1982; Chambers et al., 2000; Hamilton & Byram, 1974), more often than not, identification to species or genus was not attempted at all and specimens have simply been reported as metacestode morphotypes. Chambers et al. (2000) provided the only comprehensive Australian report of metacestodes infecting teleosts, describing 11 metacestode types, from teleosts off Heron Island on the Great Barrier Reef. These authors assigned putative generic identities to the metacestode types using morphological features and, in some cases, on the basis of some in vitro development (Chambers et al., 2000).
Over the last two decades, however, there have been substantial advances in our understanding of metacestode identities, with molecular data enabling the definitive association of metacestode types to their adult forms (e.g., Agustí et al., 2005; Aznar et al., 2007; Brickle et al., 2001; Gordeev & Sokolov, 2016; Holland & Wilson, 2009; Jensen & Bullard, 2010; Randhawa, 2011; Tedesco et al., 2020). In the most comprehensive review of marine metacestodes to date, Jensen & Bullard (2010) drew on both morphological and molecular data to present a classification of “tetraphyllidean” and rhinebothriidean metacestodes. These authors proposed a classification scheme of 15 larval types (Types I–XV), eight of which were characterised by partial 28S rDNA sequence data. Seven of these eight types were genetically matched to known genera: Larval Type I was linked to Phoreiobothrium Linton, 1889 and Triloculatum Caira & Jensen, 2009; Type II to Acanthobothrium van Beneden, 1849; Type III to Duplicibothrium Williams & Campbell, 1978; Type IV to Paraorygmatobothrium Ruhnke, 1994 (now Scyphophyllidium Woodland, 1927); Type VI to Anthobothrium van Beneden, 1850; Type VII to Rhinebothrium Linton, 1890 and Spongiobothrium Linton, 1889; and Type VIII to Rhodobothrium Linton, 1889. Despite the major advances represented by the work of Jensen & Bullard (2010), there is clearly more that remains to be clarified in the identification and biology of these metacestodes.
In this study we integrate morphological and molecular data to expand on the work of Jensen & Bullard (2010). This study focuses on piscivorous elasmobranchs of the orders Carcharhiniformes and Orectolobiformes as definitive hosts and the infraclass Teleostei as intermediate hosts, all within a single locality, Moreton Bay, in southeastern Queensland, Australia. The cestode fauna of Moreton Bay, although incompletely described, is rich and provides an ideal location for a study of this type as several genera absent from the analysis of Jensen & Bullard (2010) have been reported, and genetically characterised, from the region (Cutmore et al., 2010; Cutmore et al., 2017; Cutmore et al., 2018; Cutmore et al., 2011).
Methods
Sample collection
Teleost fishes belonging to 29 families (Table 1) were collected from eastern Moreton Bay (27°26'S, 153°24'E) and western Moreton Bay (27°22'S, 153°13'E) by baited lines, seine nets, cast nets or sourced from the commercial fishery. Systematics of host fishes follows that of FishBase (Froese & Pauly, 2023). Intestines were removed, opened longitudinally and examined under a dissecting microscope. Metacestodes found were washed and subsequently killed in near-boiling vertebrate saline (0.85% NaCl solution) and fixed 10% formalin for morphological examination and in 100% ethanol for molecular analysis. Larval cestodes were analysed as paragenophore pairs (hologenophores sensu Pleijel et al., 2008), where two morphologically identical specimens were processed for parallel morphological (one specimen) and molecular (one specimen) analyses.
Carcharhiniform and orectolobiform sharks (Table 2) were collected from eastern Moreton Bay (27°26'S, 153°24'E), western Moreton Bay (27°22'S, 153°13'E) and the Brisbane River (27°31'S, 152°59'E), Queensland, using gill nets, seine nets and baited lines or sourced from the commercial fishery. All shark hosts were identified to species using Last & Stevens (2009). Specimens of Chiloscyllium punctatum Müller & Henle are reported in this study as Chiloscyllium cf. punctatum, as the status of this shark species in Australian waters remains ambiguous (see Cutmore et al., 2010; Naylor et al., 2012). Sharks were euthanised by neural pithing and spiral intestines were removed, opened longitudinally and examined under a dissecting microscope. Cestodes were removed, washed and subsequently killed in near-boiling saline solution (0.85% NaCl solution). Worms were fixed in 10% formalin for morphological examination and in 100% ethanol for molecular analysis. Some individual worms were fixed for both morphological and molecular analysis (hologenophores sensu Pleijel et al., 2008). For these specimens the anterior two-fifths and posterior two-fifths of the worm were fixed in 10% formalin and the middle fifth in 100% ethanol.
Morphological analysis
Specimens for morphological analysis were washed in fresh water, stained in Mayer's haematoxylin, destained in a solution of 1.0% hydrochloric acid, and neutralised in 1.0% ammonium hydroxide solution. Specimens were then dehydrated through a graded ethanol series, cleared in methyl salicylate, and mounted in Canada balsam. Measurements were made using an Olympus SC50 digital camera mounted on an Olympus BX-53 compound microscope using cellSens Standard imaging software. Measurements are in micrometres unless otherwise stated and are given as the range followed by the mean in parentheses. Drawings were made using an Olympus BX-53 compound microscope and drawing tube. Voucher specimens are lodged in the Queensland Museum (QM), Brisbane, Australia.
Molecular and phylogenetic analysis
Specimens for molecular analyses were processed according to the protocols used by Cutmore et al. (2017), with total genomic DNA extracted using phenol/chloroform extraction techniques (Sambrook & Russell, 2001) and the partial D1-D3 region of the large (28S) ribosomal subunit RNA coding region amplified and sequenced using the primers LSU5 (5'-TAG GTC GAC CCG CTG AAY TTA AGC-3'; Littlewood, 1994), 300F (5'-CAA GTA CCG TGA GGG AAA GTT-3'; Littlewood et al., 2000), ECD2 (5'-CTT GGT CCG TGT TTC AAG ACG GG-3'; Littlewood et al., 1997), and 1200R (5'-GCA TAG TTC ACC ATC TTT CGG-3'; Lockyer et al., 2003). Geneious® version 10.2.3 (Kearse et al., 2012) was used to assemble and edit contiguous sequences.
The partial 28S rDNA data generated during this study were aligned with relevant sequence data available on GenBank (Tables 3 and 4) using MUSCLE version 3.7 (Edgar, 2004) run on the CIPRES portal (Miller et al., 2010), with ClustalW sequence weighting and UPGMB clustering for iterations 1 and 2. The resultant alignments were refined by eye using Maddison & Maddison (2024); the ends of the alignments were trimmed, and indels constituting more than three base positions and present in greater than 5% of the sequences in the dataset were removed. Maximum likelihood analyses were conducted to explore relationships among cestode taxa, using RAxML version 8.2.12 (Stamatakis, 2014), run on the CIPRES portal. Nodal support in the maximum likelihood analyses was estimated by performing 1,000 bootstrap pseudoreplicates. Functional outgroup taxa for each analysis were chosen based on the most recent phylogenetic topologies for the relevant group.
Results
Metacestodes
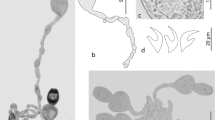
A total of 169 individual teleosts, representing 54 species from 29 families, were examined for metacestodes. All larval cestodes were collected from the intestine and pyloric caeca. Of the 169 individual teleosts examined, 101 were infected (Table 1). The metacestodes collected represent eight morphological types (Fig. 1), seven corresponding to those of Jensen & Bullard (2010) (Types I, II, IV, V, VI, VII, and X) and one new type, which we describe as Type XVI. Each of the metacestode types found in this study are reported below, with images and measurements provided for the new material. Morphological vouchers and genetic data for metacestode specimens are lodged in the Queensland Museum (G233050–186) and in GenBank (PQ146203–226), respectively.
Adult cestodes
Sixteen species of carcharhiniform and orectolobiform sharks, representing five families and eight genera, were examined for the presence of adult cestodes. Of the 103 individuals examined, 79 were infected with adult cestodes (Table 2), belonging to 13 genera: Alexandercestus Ruhnke & Workman, 2013, Acanthobothrium, Ambitalveolus Caira & Jensen, 2022, Anthobothrium, Caulopatera Cutmore, Bennett & Cribb, 2010, Hemipristicola Cutmore, Theiss, Bennett & Cribb, 2011, Megalonchos Baer & Euzet, 1962, Phoreiobothrium, Platybothrium, Scyphophyllidium, Spiniloculus Southwell, 1925, Thysanocephalum and Yorkeria Southwell, 1927. Morphological and molecular identification of adult worms belonging to Alexandercestus, Phoreiobothrium, Scyphophyllidium, and Thysanocephalum were reported in Cutmore et al. (2017), Caulopatera in Cutmore et al. (2010) and Caira & Jensen (2022), Hemipristicola in Cutmore et al. (2011), and Acanthobothrium, Megalonchos, Spiniloculus, and Yorkeria in Cutmore et al. (2018). Specimens of the remaining genera (Ambitalveolus, Anthobothrium, Phoreiobothrium, and Platybothrium) were not identified to species, and are reported only to genus; morphological vouchers and genetic data for these four genera are lodged in the Queensland Museum (G232778–855, G232871–936, and G233024) and in GenBank (PQ146187–202), respectively.
Phylogenetic results
The partial 28S rDNA sequences generated for adult and larval cestodes were initially analysed with all available cestode taxa on GenBank (phylogenetic tree not shown). Metacestode specimens matched adult specimens in several unrelated clades across the Eucestoda phylogenetic tree. Incorporation of the massive amount of data across the entire Eucestoda resulted in substantial alignment-induced data loss. Thus, to increase species-level resolution, the six clades with matching metacestode data were each aligned and analysed separately.
Types I, II and XVI metacestodes were identified as onchoproteocephalideans. Based on the topology of Caira et al. (2020), the Onchoproteocephalidea analyses incorporated published sequence data of Types I and II metacestodes from teleost fishes of the Gulf of Mexico, Type II metacestode sequences from an octopus from the Mediterranean, adult worms of relevant onchobothriid genera, Matticestus Caira, Jensen & Fyler, 2018, and a select few proteocephalid genera. Sequences of Type I metacestodes from Moreton Bay form four clades, three of which form well-supported clades with species of Phoreiobothrium (Fig. 2). The fourth clade of Type I metacestodes resolve as basal to all the included taxa excluding species of Platybothrium; however, nodal support for this position is poor, and notably species of Phoreiobothrium do not form monophyletic clade in the analysis. Sequences of Type II metacestodes from Moreton Bay form four clades, all of which resolve in a large well-supported clade of adult Acanthobothrium and Type II metacestodes from the Gulf of Mexico and the Mediterranean. Sequences of Type XVI metacestodes from Moreton Bay form two clades which both resolve in a well-supported clade with sequences of adult Platybothrium. Although Jensen & Bullard (2010) matched Type I and Type II metacestodes to adult genera in the Gulf of Mexico, they did not encounter Type XVI, which is here clearly identified as relating to Platybothrium.
Phylogenetic tree from the Maximum likelihood analysis of the Onchoproteocephalidea dataset, incorporating Types I, II and XVI metacestodes. Strongly supported nodes (>80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site. Abbreviations: GoM, Gulf of Mexico; MB, Moreton Bay.
Type IV metacestodes were identified as phyllobothriids. Based on the topology of Caira et al. (2020), the Phyllobothriidae analysis incorporated sequence data for species of Alexandercestus, Hemipristicola, Scyphophyllidium, and Thysanocephalum from across the Pacific, Atlantic and Indian Oceans, and Type IV larval data from teleost fishes from the Gulf of Mexico. Sequences of Type IV metacestodes from Moreton Bay form three clades, all of which matched adult sequences, two species of Scyphophyllidium and one species of Alexandercestus (Fig. 3). Jensen & Bullard (2010) matched Type IV metacestodes to adult Scyphophyllidium (then Paraorygmatobothrium) in the Gulf of Mexico.
Phylogenetic tree from the Maximum likelihood analysis of the Phyllobothriidae dataset, incorporating Type IV metacestodes. Strongly supported nodes (>80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site. Abbreviations: GoM, Gulf of Mexico; MB, Moreton Bay.
Type V metacestodes were identified as balanobothriids. Based on the topology of Caira et al. (2017), the Balanobothriidae analysis incorporated sequence data of Type V metacestodes from teleost fishes from the Gulf of Mexico and adult Balanobothrium Hornell, 1911, Pachybothrium, Pedibothrium, Spiniloculus and Yorkeria. The sequences of Type V metacestodes from Moreton Bay represent three genotypes, each of which form a clade with sequences of adult Pedibothrium (Fig. 4); however, none of these larval sequences were an identical match to those of adult cestodes. Although Jensen & Bullard (2010) did not find an exact match for larval Type V, they inferred that it relates to species of Pachybothrium and/or Pedibothrium. It is clear from the current study that larval Type V relates to Pedibothrium and, given the close phylogenetic relationships of Pedibothrium and Pachybothrium, it seems plausible that this type might also represent Pachybothrium. Notably, Pedibothrium did not resolve as monophyletic, with a single sequence of an undescribed species of Balanobothrium rendering the genus paraphyletic.
Phylogenetic tree from the Maximum likelihood analysis of the Balanobothriidae dataset, incorporating Type V metacestodes. Strongly supported nodes (>80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site. Abbreviations: GoC, Gulf of California; GoM, Gulf of Mexico; MB, Moreton Bay.
Type VI metacestodes were identified as Anthobothrium species. The analysis of these data incorporated sequences of Type VI metacestodes from teleost fishes United States waters, from a squid from Japanese waters and an octopus from the Mediterranean, and from adult Anthobothrium worms from carcharhinid sharks from Australia and the United States. Sequences of Type VI metacestodes from Moreton Bay form five clades, four of which match sequences of adult Anthobothrium specimens (Fig. 5). These findings support those of Jensen & Bullard (2010), who matched Type VI metacestodes to adult Anthobothrium in the Gulf of Mexico. Notably, based on results from in vitro cultivation, Chambers et al. (2000) also predicted that this larval type would represent species of Anthobothrium.
Phylogenetic tree from the Maximum likelihood analysis of the Anthobothrium dataset, incorporating Type VI metacestodes. Strongly supported nodes (>80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site. Abbreviations: GoM, Gulf of Mexico; MB, Moreton Bay.
Type VII metacestodes were identified as rhinebothiids. Based on the topology of Herzog et al. (2023), the Rhinebothriidae analysis incorporated sequences of Type VII metacestodes from fishes and bivalves of the Gulf of Mexico and of adult worms of the genera Rhabdotobothrium Euzet, 1953, Rhinebothrium, Rhinebothroides Mayes, Brooks & Thorson, 1981, Rhodobothrium, Scalithrium Ball, Neifar & Euzet, 2003 and Spongiobothrium. Sequences of Type VII metacestodes collected during this study form a clade with those of adult Spongiobothrium and Type VII metacestodes from the Gulf of Mexico (Fig. 6). Jensen & Bullard (2010) identified this larval type as relating to Rhinebothrium or Spongiobothrium.
Phylogenetic tree from the Maximum likelihood analysis of the Rhinebothriidae dataset, incorporating Type VII metacestodes. Strongly supported nodes (>80) are indicated by a filled circle. The scale-bar indicates expected number of substitutions per site. Abbreviations: GoC, Gulf of California; GoM, Gulf of Mexico; MB, Moreton Bay.
Type X metacestodes were identified as belonging to ‘tetraphyllidean Clade 3’ (sensu Caira et al., 2017). Based on the topology of Caira & Jensen (2022), the Clade 3 analysis incorporated sequences for adults of Ambitalveolus, Carpobothrium and Caulopatera from orectolobiform sharks of the tropical Indo-west Pacific. The sequences of Type X metacestodes represent a single genotype and match an undescribed species of Ambitalveolus collected from Orectolobus ornatus (De Vis), also from Moreton Bay (Fig. 7). This undescribed species differs from the only other Ambitalveolus for which sequence data are available, Ambitalveolus penghuensis Caira & Jensen, 2022 (also described form a species of Orectolobus Bonaparte), at 22 base positions. Jensen & Bullard (2010) speculated that Larval Type X related to Carpobothrium. While it is clear that the new specimens of this larval type relate to species of Ambitalveolus, it is plausible that all three genera in this clade will share the larval morphotype given the close phylogenetic relationship and similarities in bothridial structure.
Metacestode types
Larval Type I sensu Jensen & Bullard (2010) (Fig. 1a)
Generic identity: Phoreiobothrium Linton, 1889 and Triloculatum Caira & Jensen, 2009 (Onchoproteocephalidea: Onchobothriidae).
New hosts: 21 species of Teleostei, see Table 1.
Material deposited: 20 voucher specimens (QM G 233050–69).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, 13 sequences representing four genotypes (four sequences submitted to GenBank, PQ146203–06).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body elongate, undivided, tapering posteriorly, length highly variable, 327–1,960 (905) long, 109–370 (217) wide. Apical sucker present, often funnel shaped, 47–141 (83) long, 58–130 (93) wide. Bothridia oval, 46–171 (115) long, 33–126 (81) wide, divided into two loculi; posterior loculus generally larger than anterior loculus.
Remarks: Notably, there was variation regarding body size and the shape of the apical sucker between specimens of this type; however, these differences did not align with genetic differences.
Larval Type II sensu Jensen & Bullard (2010) (Fig. 1b, c)
Generic identity: Acanthobothrium van Beneden, 1849 (Onchoproteocephalidea: Onchobothriidae).
New hosts: 11 species of Teleostei, see Table 1.
Material deposited: 48 voucher specimens (QM G233070–88, QM G233158–86).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, 17 sequences representing five genotypes (five sequences submitted to GenBank, PQ146207–11).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body elongate, undivided or divided, tapering posteriorly, length highly variable, 681–5,040 (1,527) long, 142–327 (219) wide. Apical sucker present, 42–100 (65) long, 51–119 (78) wide. Bothridia elongate, 101–254 (156) long, 58–139 (89) wide, divided into anterior pad and posterior loculus; posterior loculus usually subdivided by horizontal septa forming two to three loculi; posteriormost septa often inconspicuous.
Remarks: There was clear morphological variability within this type, with specimens of differing in the division of the bothridia (weak, inconspicuous septa vs strong, conspicuous septa), body length and the body divisions (undivided vs divided). Notably, all Type II metacestodes with some body division represent a single genotype, which is an identical molecular match to Acanthobothrium margieae Fyler, 2011.
Larval Type IV sensu Jensen & Bullard (2010) (Fig. 1d)
Generic identity: Alexandercestus Ruhnke & Workman, 2013 and Scyphophyllidium Woodland, 1927 (Phyllobothriidea: Phyllobothriidae).
Hosts: 16 species of Teleostei, see Table 1.
Material deposited: 20 voucher specimens (QM G 233089–108).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, seven sequences representing three genotypes (three sequences submitted to GenBank, PQ146212–14).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body tiny, elongate, undivided, tapering posteriorly, 192–351 (274) long, 78–137 (103) wide. Apical sucker present, 28–53 (38) long, 35–77 (51) wide. Bothridia circular to oval, undivided, 33–61 (47) long, 28–57 (40) wide.
Larval Type V sensu Jensen & Bullard (2010) (Fig. 1f)
Generic identity: Pedibothrium Linton, 1909 (Tetraphyllidea: Balanobothriidae).
New hosts: six species of Teleostei, see Table 1.
Material deposited: seven voucher specimens (QM G 233109–15).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, three sequences representing three genotypes (three sequences submitted to GenBank, PQ146215–17).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body tiny, elongate, undivided, tapering posteriorly, 276–334 (307) long, 120–174 (138) wide. Apical sucker absent. Bothridia oval, 74–114 (87) long, 46–77 (58) wide, divided into anterior and posterior loculi; division between loculi sometimes inconspicuous.
Larval Type VI sensu Jensen & Bullard (2010) (Fig. 1g)
Generic identity: Anthobothrium van Beneden, 1850 (Tetraphyllidea incertae sedis).
New hosts: 33 species of Teleostei, see Table 1.
Material deposited: 20 voucher specimens (QM G 233129–48).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, 33 sequences representing five genotypes (five sequences submitted to GenBank, PQ146218–22).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body elongate, undivided, tapering posteriorly, 518–1,009 (767) long, 147–292 (216) wide. Apical sucker present, 32–63 (51) long, 43–86 (64) wide. Bothridia roughly circular, undivided, 58–125 (94) long, 53–117 (86) wide.
Larval Type VII sensu Jensen & Bullard (2010) (Fig. 1h)
Generic identity: Likely Spongiobothrium Linton, 1889 (Rhinebothriidea: Rhinebothriidae).
New hosts: Labridae: Thalassoma jansenii (Bleeker), Jansen's wrasse. Paralichthyidae: Pseudorhombus arsius (Hamilton), Largetooth flounder.
Material deposited: 3 voucher specimens (QM G 233149–51).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, three sequences representing one genotype (one sequence submitted to GenBank, PQ146223).
Measurements: Morphology consistent with diagnosis provided by Jensen & Bullard (2010). Body elongate, undivided, tapering posteriorly, 861–987 (929) long, 241–275 (257) wide. Apical sucker present, 102–108 (105) long, 106–125 (117) wide. Bothridia elongate, facially loculated, 174–235 (197) long, 106–134 (118) wide; loculation consisting of two columns of rectangular loculi.
Larval Type X sensu Jensen & Bullard (2010) (Fig. 1e)
Generic identity: Ambitalveolus Caira & Jensen, 2022 (Tetraphyllidea incertae sedis).
New hosts: Labridae: Pseudolabrus guentheri Bleeker, Günther's wrasse; Thalassoma lunare (Linnaeus), Moon wrasse.
Material deposited: Six voucher specimens (QM G 233152–57).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, four sequences representing one genotype (one sequence submitted to GenBank, PQ146224).
Measurements: Morphology consistent with characterisation provided by Chambers et al. (2000) and Jensen & Bullard (2010). Body elongate, undivided, tapering slightly posteriorly, 1,819–3,536 (2,478) long, 484–578 (512) wide. Apical sucker present, 100 long (measurable for only one specimen), 95–140 (110) wide. Bothridia large, undivided, pouch-shaped, with muscular bands on anterior and posterior margins of aperture, 201–238 (217) long, 216–304 (258) wide.
Larval Type XVI (Fig. 1i)
Generic identity: Platybothrium Linton, 1890 (Onchoproteocephalidea: Onchobothriidae).
New hosts: 12 species of Teleostei, see Table 1.
Material deposited: 13 voucher specimens (QM G 233116–28).
Molecular sequence data: D1-D3 region of the 28S rDNA gene, four sequences representing two genotypes (two sequences submitted to GenBank, PQ146225–26).
Measurements: Body tiny, elongate, undivided, tapering posteriorly, 172–453 (314) long, 110–168 (128) wide. Apical sucker present, small, sometimes poorly defined. Bothridia oval, 66–114 (83) long, 49–75 (60) wide, divided into anterior and posterior loculi with division between loculi sometimes inconspicuous; anterior loculus smaller than posterior loculus.
Remarks: Type XVI metacestodes are highly morphologically similar to Type V metacestodes (Pedibothrium) but differ from this type in possessing a small (albeit sometimes inconspicuous) apical sucker.
Discussion
The results of the present study both support and extend the unified morphological type system proposed by Jensen & Bullard (2010), in that almost all metacestodes collected from Moreton Bay were identifiable to one of the 15 types proposed in their study. However, the results of this study clearly demonstrate that morphological types will ultimately be best used to identify metacestodes to a group of related genera rather than any distinct genus. Type IV metacestodes were found to represent species of at least two genera (Alexandercestus and Scyphophyllidium) and the reliable topology of this clade permits the cautious prediction that this type likely also represents Hemipristicola (which resolves between Alexandercestus and Scyphophyllidium). While all Type I metacestodes matched species of Phoreiobothrium, based on the topology of the onchoproteocephalidean analysis it seems likely that, despite the poor nodal support, Type I metacestodes will also represent species of Triloculatum. Based on the current topology of the balanobothriid analyses, and topology of this group previously published (Caira et al., 2014), it seems likely that Type V metacestodes represent both Pedibothrium and Balanobothrium and may represent Pachybothrium. This similarity of morphology between closely related genera is not unexpected, however, given the simplified form of metacestodes. More rigorous examination of samples within these types (enabled by the incorporation of hologenophore specimens) may reveal distinctions between genera, but we suspect this level of distinction, and thus an expansion of the number of types, would not be advantageous for most researchers attempting identification of these cestode larvae.
The larval type of several tetraphyllidean genera prominent in Moreton Bay remains unknown, with no metacestodes matches found for Caulopatera, Hemipristicola, Megalonchos, Spiniloculus, Thysanocephalum or Yorkeria. Based on the findings that larvae of a single type can represent species of multiple genera, more extensive genetic sequencing of the current types may reveal matches for some of the unmatched adult genera from Moreton Bay. However, given the highly morphologically distinct adult forms of Spiniloculus, Thysanocephalum and Yorkeria, there are potentially multiple types still to be discovered in the region. The restriction of intermediate host collection to teleost fishes may explain the failure to find metacestodes of Caulopatera, Spiniloculus and Yorkeria, as species of these genera are almost exclusively reported from species of Chiloscyllium (Hemiscylliidae) (Caira et al., 2007a; Cutmore et al., 2010; Cutmore et al., 2018). The diet of Chiloscyllium species comprises primarily benthic invertebrates (Gauthier et al., 2019; Last & Stevens, 2009; Lowry & Motta, 2007) and it is likely that the intermediate stages for these three genera will be found there. Failure to identify the larval form of Megalonchos may be due to the distribution of the definitive host. Megalonchos species infect hemigaleid sharks of the genera Chaenogaleus Gill and Hemipristis Agassiz (Caira et al., 2007b), with both Megalonchos species sequenced from Moreton Bay collected from Hemipristis elongata (Klunzinger) (Cutmore et al., 2018). Hemipristis elongata is rarely found in Moreton Bay (Taylor & Bennett, 2013), and is more commonly encountered in tropical regions of Australia (Last & Stevens, 2009). The absence of a metacestode match for Thysanocephalum thysanocephalum (Linton, 1889) Braun, 1900 is more surprising, however, as Galeocerdo cuvier (Péron & Lesueur) is a partial piscivore and is considered common in the region (Johnson, 2010).
During this study we examined just 54 of 700+ teleost species known from Moreton Bay (Johnson, 2010). The number of individual fishes examined for most species were far too low to draw any decisive inferences on prevalence and richness of larvae. Despite the low dissection numbers, there were some notable patterns. Some fish species [e.g., Microcanthus strigatus (Cuvier), Ostorhinchus limenus Randall & Hoese, Saurida undosquamis (Richardson)] had both a high richness and high prevalence of metacestodes; all three of these species had 100% infection prevalence and were infected by five of the eight larval types. Some species [e.g., Lethrinus genivittatus Valenciennes and Monodactylus argenteus (Linnaeus)] had a high prevalence but low richness. And some species [e.g., Atherinomorus vaigiensis (Quoy & Gaimard) and Gerres subfasciatus Cuvier] were not infected at all, despite respectable number of examinations (12 and 13, respectively). While we do not think that all 700+ teleost species in Moreton Bay will harbour metacestodes, based on the findings above, it is clear that there are likely hundreds of individual host/parasite combinations yet to be identified.
Similar to prevalence and richness, the number of individual fishes examined for most species were far too low to allow definitive inferences on the host specificity of the larval types. However, of the eight larval types, seven were found in more than one order of fishes. Larval Types I, II, IV, and VI exhibit strikingly low host-specificity; Type VI metacestodes, which relate to species of Anthobothrium, were recovered from 21 families and 33 species of teleost fishes. There were five genotypes of Type VI larvae, of which four matched data for adult worms. Although the number of definitively identified intermediate hosts for each of these genotypes was relatively low (1–11 teleost species), even within these genotypes the host-specificity was clearly low. The four matched genotypes were recovered from three, five, six and 10 families of teleosts. In contrast, the Type X larvae appear to be highly host-specific, found only in two species of labrids (two of five Pseudolabrus guentheri and two of four T. lunare examined). An additional 15 teleost species were examined from the same location at which these infections were found, all of which were infected with several metacestode types but not by Type X.
In the only other broad survey of metacestodes infecting Australian fishes, Chambers et al. (2000) found a similar metacestode fauna, also identifying Types I, II, V, VI, VII, and X from a wide range of teleosts off Heron Island on the southern Great Barrier Reef. In a more taxonomically restricted study, Muñoz & Cribb (2005) and Muñoz et al. (2007) identified Types I, IV, V, VI, and VII from 14 species of labrids from Lizard Island, on the northern Great Barrier Reef. Several differences were observed between the present survey and those from the Great Barrier Reef. Chambers et al. (2000) did not collect any larval Type IV from teleosts off Heron Island; this absence is surprising as it is likely that several Scyphophyllidium species infect the many carcharhiniform and orectolobiform elasmobranchs found at Heron Island. Type IV larvae are by far the smallest metacestodes encountered in this study and it is possible that low infection levels were overlooked. However, Muñoz & Cribb (2005) identified what appears to be Type IV larvae from Lizard Island. One morphological type identified by Chambers et al. (2000) was absent from the present study; no specimens of larval Type IX (Chambers et al. Type 9) were collected from Moreton Bay. Muñoz & Cribb (2005) also identified several metacestode types not collected during this survey, identifying over 30 tetraphyllidean larval morphotypes from 14 species of labrids. There was no voucher material deposited by Muñoz et al. (2007) but based on the known fauna of elasmobranch hosts at Lizard Island, it seems unlikely that 30 genetically different larval types would be present at this location.
Notably, two larval types found during this study, Type X and Type XVI, were not collected by Jensen & Bullard (2010). Type X larvae were matched to Ambitalveolus and given the close phylogenetic relationship and similarities in bothridial structure, it seems plausible that this larval morphotype will also represent Carpobothrium and Caulopatera. All three of these genera have been reported from only orectolobiform sharks of the genera Brachaelurus (Brachaeluridae), Chiloscyllium (Hemiscylliidae), and Orectolobus (Orectolobidae). The absence of Type X in the Gulf of Mexico study by Jensen & Bullard (2010) is thus expected as species of Brachaelurus, Chiloscyllium, and Orectolobus are restricted to the Indo-west Pacific (Last & Stevens, 2009). The absence of Type XVI, identified as Platybothrium, in the collections of Jensen & Bullard (2010) is more perplexing, as there have been numerous Platybothrium species reported form sharks of the Atlantic (Healy, 2003), including from shark species examined as part of the Gulf of Mexico study. In contrast, Jensen & Bullard (2010) found two larval types, Type III and Type VIII, that were not collected from Moreton Bay. Jensen & Bullard (2010) identified Type III as Duplicibothrium and Type VIII as Rhodobothrium. The definitive hosts of these two genera, myliobatiform rays of the genera Dasyatis, Myliobatis and Rhinoptera, are abundant in Moreton Bay, and we suspect species of these genera will be found in the region. We predict that the absence in the current study is due to the restriction of our examinations to teleosts; Jensen & Bullard (2010) found both Type III and Type VIII in bivalve and gastropod hosts.
The low host-specificity of most of the larval types reported here is consistent with effective transmission to sharks that themselves have low dietary specificity. Carcharhiniform sharks identified as hosts of adult cestodes reported in this study have been widely shown to have exceptionally low dietary specificity (e.g., Salini et al., 1992; Simpfendorfer et al., 2001; Tillett et al., 2014; White et al., 2004). Dietary ranges of some orectolobiforms are similarly broad, with Huveneers et al. (2007) identifying five orders of teleosts, three orders of elasmobranchs and one order of cephalopod in the diet of the Orectolobus maculatus and six orders of teleosts and two orders of cephalopods in the diet of the O. ornatus. Evidently, there is typically no evolutionary pressure or advantage in cestode specialisation for a more restricted range of intermediate hosts than the dietary range of the definitive host. Overall, it is noteworthy that the generally high prevalence and low host-specificity of tetraphyllidean metacestodes in teleost fishes implies that transmission success of metacestodes is quantitatively very low.
Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.
References
Adán-Torres B., Oceguera-Figueroa A., Martínez-Flores G., & García-Prieto L. (2022). Phylogenetic position of Acanthobothrium cleofanus (Cestoda: Onchoproteocephalidea) using molecular evidence. Parasitology International, 86, 102473. https://doi.org/10.1016/j.parint.2021.102473
Agustí C., Aznar F. J., Olson P. D., Littlewood D. T. J., Kostadinova A., & Raga J. A. (2005). Morphological and molecular characterization of tetraphyllidean merocercoids (Platyhelminthes: Cestoda) of striped dolphins (Stenella coeruleoalba) from the Western Mediterranean. Parasitology, 130, 461–474. https://doi.org/10.1017/S0031182004006754
Aznar F. J., Agustí C., Littlewood D. T. J., Raga J. A., & Olson P. D. (2007). Insight into the role of cetaceans in the life cycle of the tetraphyllideans (Platyhelminthes: Cestoda). International Journal for Parasitology, 37, 243–255. https://doi.org/10.1016/j.ijpara.2006.10.010
Brickle P., Olson P. D., Littlewood D. T. J., Bishop A., & Arkhipkin A. I. (2001). Parasites of Loligo gahi from waters off the Falkland Islands, with a phylogenetically based identification of their cestode larvae. Canadian Journal of Zoology, 79, 2289–2296. https://doi.org/10.1139/z01-189
Bueno V. M., & Caira J. N. (2017). Redescription and molecular assessment of relationships among three species of Echeneibothrium (Rhinebothriidea: Echeneibothriidae) parasitizing the Yellownose skate, Dipturus chilensis, in Chile. Journal of Parasitology, 103, 268–284. https://doi.org/10.1645/16-177
Caira J. N., & Jensen K. (2022). Diversity and phylogenetic relationships of 'tetraphyllidean' Clade 3 (Cestoda) based on new material from orectolobiform sharks in Australia and Taiwan. Folia Parasitologica, 69, 010. https://doi.org/10.14411/fp.2022.010
Caira J. N., Jensen K., & Fyler C. A. (2018). A new genus of tapeworm (Cestoda: Onchoproteocephalidea) from sawfish (Elasmobranchii: Pristidae). Journal of Parasitology, 104, 133–144. https://doi.org/10.1645/17-165
Caira J. N., Jensen K., Hayes C., & Ruhnke T. R. (2020). Insights from new cestodes of the crocodile shark, Pseudocarcharias kamoharai (Lamniformes: Pseudocarchariidae), prompt expansion of Scyphyophyllidum and formal synonymization of seven phyllobothriidean genera – at last! Journal of Helminthology, 94, e132. https://doi.org/10.1017/S0022149X20000036
Caira J. N., Jensen K., & Rajan C. (2007a). Seven new Yorkeria species (Cestoda: Tetraphyllidea) from Borneo and Australia and their implications for identification of Chiloscyllium (Elasmobranchii: Orectolobiformes) species. Journal of Parasitology, 93, 357–376. https://doi.org/10.1645/GE-801R.1
Caira J. N., Jensen K., & Ruhnke T. R. (2017). “Tetraphyllidea” van Beneden, 1849 relics. In Caira JN and Jensen K (eds), Planetary Biodiversity Inventory (2008–2017): Tapeworms from Vertebrate Bowels of the Earth. Lawrence: University of Kansas, Natural History Museum, 371–400.
Caira J. N., Jensen K., Waeschenbach A., Olson P. D., & Littlewood D. T. J. (2014). Orders out of chaos – molecular phylogenetics reveals the complexity of shark and stingray tapeworm relationships. International Journal for Parasitology, 44, 55–73. https://doi.org/10.1016/j.ijpara.2013.10.004
Caira J. N., Reyda F. B., & Mega J. D. (2007b). A revision of Megalonchos Baer & Euzet, 1962 (Tetraphyllidea: Onchobothriidae), with the description of two new species and transfer of two species to Biloculuncus Nasin, Caira & Euzet, 1997. Systematic Parasitology, 67, 211–223. https://doi.org/10.1007/s11230-006-9085-z
Cake E. W. (1976). A key to larval cestodes of shallow-water, benthic mollusks of the northern Gulf of Mexico. Proceedings of the Helminthological Society of Washington, 43, 160–171.
Carvajal J., Barros C., & Santander G. (1982). In vitro culture of Rhodobothrium mesodesmatum (Cestoda: Tetraphyllidea), parasite of a Chilean clam. Proceedings of the Helminthological Society of Washington, 49, 226–230.
Chambers C. C., Cribb T. H., & Jones M. K. (2000). Tetraphyllidean metacestodes of teleosts of the Great Barrier Reef, and the use of in vitro cultivation to identify them. Folia Parasitologica, 47, 285–292. https://doi.org/10.14411/fp.2000.050
Cutmore S. C., Bennett M. B., & Cribb T. H. (2010). A new tetraphyllidean genus and species, Caulopatera pagei n. g., n. sp. (Tetraphyllidea: Phyllobothriidae), from the grey carpetshark Chiloscyllium punctatum Müller & Henle (Orectolobiformes: Hemiscylliidae). Systematic Parasitology, 77, 13–21. https://doi.org/10.1007/s11230-010-9252-0
Cutmore S. C., Bennett M. B., Miller T. L., & Cribb T. H. (2017). Patterns of specificity and diversity in species of Paraorygmatobothrium Ruhnke, 1994 (Cestoda: Phyllobothriidae) in Moreton Bay, Queensland, Australia, with the description of four new species. Systematic Parasitology, 94, 941–970. https://doi.org/10.1007/s11230-017-9759-8
Cutmore S. C., Cribb T. H., Bennett M. B., & Beveridge I. (2018). Tetraphyllidean and onchoproteocephalidean cestodes of elasmobranchs from Moreton Bay, Australia: description of two new species and new records for seven described species. Systematic Parasitology, 95, 807–827. https://doi.org/10.1007/s11230-018-9817-x
Cutmore S. C., Theiss S. M., Bennett M. B., & Cribb T. H. (2011). A new phyllobothriid genus and species from the snaggletooth shark, Hemipristis elongata (Carcharhiniformes: Hemigaleidae), from Moreton Bay, Australia. Folia Parasitologica, 58, 187–196. https://doi.org/10.14411/fp.2011.019
Edgar R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Research, 32, 1792–1797. https://doi.org/10.1093/nar/gkh340
Froese R., & Pauly D. (2023). FishBase. World Wide Web electronic publication. www.fishbase.org, version (11/2023).
Fyler C. A., & Caira J. N. (2010). Phylogenetic status of four new species of Acanthobothrium (Cestoda: Tetraphyllidea) parasitic on the wedgefish Rhynchobatus laevis (Elasmobranchii: Rhynchobatidae): implications for interpreting host associations. Invertebrate Systematics, 24, 419–433. https://doi.org/10.1071/IS10034
Fyler C. A., Caira J. N., & Jensen K. (2009). Five new species of Acanthobothrium (Cestoda: Tetraphyllidea) from an unusual species of Himantura (Rajiformes: Dasyatidae) from northern Australia. Folia Parasitologica, 56, 107–128. https://doi.org/10.14411/fp.2009.016
Gallagher K. A., & Caira J. N. (2020). A new species of Acanthobothrium (Cestoda: Onchoproteocephalidea) from the Smalleye pygmy shark, Squaliolus aliae (Chondrichthyes: Squaliformes: Dalatiidae), from Taiwan. Journal of Parasitology, 106, 818–827. https://doi.org/10.1645/20-15
Gauthier A. R. G., Whitehead D. L., Tibbetts I. R., & Bennett M. B. (2019). Comparative morphology of the electrosensory system of the epaulette shark Hemiscyllium ocellatum and brown-banded bamboo shark Chiloscyllium punctatum. Journal of Fish Biology, 94, 207–348. https://doi.org/10.1111/jfb.13893
Golzarianpour K., Malek M., Golestaninasab M., Sarafrazi A., & Kochmann J. (2020). Two new enigmatic species of Rhinebothrium (Cestoda: Rhinebothriidae) from the Persian Gulf: notes on generic traits and host specificity. Systematics and Biodiversity, 19, 273–295. https://doi.org/10.1080/14772000.2020.1832606
Gordeev I. I., & Sokolov S. G. (2016). Parasites of the Antarctic toothfish (Dissostichus mawsoni Norman, 1937) (Perciformes, Nototheniidae) in the Pacific sector of the Antarctic. Polar Research, 35, 29364. https://doi.org/10.3402/polar.v35.29364
Hamilton K. A., & Byram J. E. (1974). Tapeworm development: the effects of urea on a larval tetraphyllidean. Journal of Parasitology, 60, 20–28. https://doi.org/10.2307/3278672
Healy, C. J. (2003). A revision of Platybothrium Linton, 1890 (Tetraphyllidea: Onchobothriidae), with a phylogenetic analysis and comments on host-parasite associations. Systematic Parasitlogy, 56, 85–139. https://doi.org/10.1023/A:1026135528505
Healy C. J., Caira J. N., Jensen K., Webster B. L., & Littlewood D. T. J. (2009). Proposal for a new tapeworm order, Rhinebothriidea. International Journal for Parasitology, 39, 497–511. https://doi.org/10.1016/j.ijpara.2008.09.002
Herzog K. S., Caira J. N., Ker P. K., & Jensen K. (2023). Novelty and phylogenetic affinities of a new family of tapeworms (Cestoda: Rhinebothriidea) from endangered sawfish and guitarfish. International Journal for Parasitology, 53, 347–362. https://doi.org/10.1016/j.ijpara.2023.02.007
Holland N. D., & Wilson N. G. (2009). Molecular identification of larvae of a tetraphyllidean tapeworm (Platyhelminthes: Eucestoda) in a razor clam as an alternative intermediate host in the life cycle of Acanthobothrium brevissime. Journal of Parasitology, 95, 1215–1217. https://doi.org/10.1645/GE-1946.1
Huveneers C., Otway N. M., Gibbs S. E., & Harcourt R. G. (2007). Quantitative diet assessment of wobbegong sharks (genus Orectolobus) in New South Wales, Australia. ICES Journal of Marine Science, 64, 1272–1281. https://doi.org/10.1093/icesjms/fsm111
Jensen K., & Bullard S. A. (2010). Characterization of a diversity of tetraphyllidean and rhinebothriidean cestode larval types, with comments on host associations and life-cycles. International Journal for Parasitology, 40, 889–910. https://doi.org/10.1016/j.ijpara.2009.11.015
Johnson J. W. (2010). Fishes of the Moreton Bay Marine Park and adjacent continental shelf waters, Queensland, Australia. Memoirs of the Queensland Museum, 54, 299–353.
Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S., Buxton S., Cooper A., Markowitz S., Duran C., Thierer T., Ashton B., Mentjies P., & Drummond A. (2012). Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics, 28, 1647–1649. https://doi.org/10.1093/bioinformatics/bts199
Last P. R., & Stevens J. D. (2009). Sharks and Rays of Australia, 2nd edn. Collingwood: CSIRO Publishing.
Littlewood D. T. J. (1994). Molecular phylogenetics of cupped oysters based on partial 28S rRNA gene sequences. Molecular Phylogenetics and Evolution, 3, 221–229. https://doi.org/10.1006/mpev.1994.1024
Littlewood D. T. J., Curini-Galletti M., & Herniou E. A. (2000). The interrelationships of Proseriata (Platyhelminthes: Seriata) tested with molecules and morphology. Molecular Phylogenetics and Evolution, 16, 449–466. https://doi.org/10.1006/mpev.2000.0802
Littlewood D. T. J., Rohde K., & Clough K. A. (1997). Parasite speciation within or between host species?—Phylogenetic evidence from site-specific polystome monogeneans. International Journal for Parasitology, 27, 1289–1297. https://doi.org/10.1016/S0020-7519(97)00086-6
Lockyer A. E., Olson P. D., & Littlewood D. T. J. (2003). Utility of complete large and small subunit rRNA genes in resolving the phylogeny of the Neodermata (Platyhelminthes): implications and a review of the cercomer theory. Biological Journal of the Linnean Society, 78, 155–171. https://doi.org/10.1046/j.1095-8312.2003.00141.x
Lowry D., & Motta P. J. (2007). Ontogeny of feeding behavior and cranial morphology in the whitespotted bambooshark Chiloscyllium plagiosum. Marine Biology, 151, 2013–2023. https://doi.org/10.1007/s00227-007-0642-z
Maddison W. P., & Maddison D. R. (2024). Mesquite: a modular system for evolutionary analysis. Version 3.31 http://mesquiteproject.org.
Miller M. A., Pfeiffer E., & Schwartz T. (2010). Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In: Proceedings of the Gateway Computing Environments Workshop (GCE), 14 Nov. 2010, New Orleans, LA, pp. 1–8. https://doi.org/10.1109/GCE.2010.5676129
Muñoz G., & Cribb T. H. (2005). Parasite communities and diet of Coris batuensis (Pisces: Labridae) from Lizard Island, Great Barrier Reef. Memoirs of the Queensland Museum, 52, 191–198.
Muñoz G., Grutter A. S., & Cribb T. H. (2007). Structure of the parasite communities of a coral reef fish assemblage (Labridae): testing ecological and phylogenetic host factors. Journal of Parasitology, 93, 17–30. https://doi.org/10.1645/GE-969R.1
Naylor G. J. P., Caira J. N., Jensen K., Rosana K. A. M., White W. T., & Last P. R. (2012). A DNA sequence-based approach to the identification of shark and ray species and its implications for global elasmobranch diversity and parasitology. Bulletin of the American Museum of Natural History, 367, 1–262. https://doi.org/10.1206/754.1
Nitta M., Matsumoto Y., & Urabe M. (2023). Larval cestodes of Anthobothrium sp. (Cestoda: “Tetraphyllidea”) parasitic on gills of Sepioteuthis lessoniana (Cephalopoda: Loliginidae) from Wakayama Prefecture, Japan. TAXA, Proceedings of the Japanese Society of Systematic Zoology, 55, 9–14. https://doi.org/10.19004/taxa.55
Olson P. D., Littlewood D. T. J., Bray R. A., & Mariaux J. (2001). Interrelationships and evolution of the tapeworms (Platyhelminthes: Cestoda). Molecular Phylogenetics and Evolution, 19, 443–467. https://doi.org/10.1006/mpev.2001.0930
Pleijel F., Jondelius U., Norlinder E., Nygren A., Oxelman B., Schander C., Sundberg P., & Thollesson M. (2008). Phylogenies without roots? A plea for the use of vouchers in molecular phylogenetic studies. Molecular Phylogenetics and Evolution, 48, 369–371. https://doi.org/10.1016/j.ympev.2008.03.024
Randhawa H. S. (2011). Insights using a molecular approach into the life cycle of a tapeworm infecting great white sharks. Journal of Parasitology, 97, 275–280. https://doi.org/10.1645/GE-2530.1
Ruhnke T. R., & Workman R. E. (2013). Two new species and a new phyllobothriid cestode genus from sharks of the genus Negaprion Whitley (Carcharhiniformes). Systematic Parasitology, 85, 37–48. https://doi.org/10.1007/s11230-013-9411-1
Salini J. P., Blaber S. J. M., & Brewer D. T. (1992). Diets of sharks from estuaries and adjacent waters of the north-eastern Gulf of Carpentaria, Australia. Australian Journal of Marine and Freshwater Research, 43, 87–96. https://doi.org/10.1071/MF9920087
Sambrook J., & Russell D. W. (2001). Molecular Cloning: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York.
Simpfendorfer C. A., Goodreid A., & McAuley R. B. (2001). Diet of three commercially important shark species from Western Australian waters. Marine and Freshwater Research, 52, 975–985. https://doi.org/10.1071/MF01017
Stamatakis A. (2014). RAxML Version 8: A tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313. https://doi.org/10.1093/bioinformatics/btu033
Stephan D., & Caira J. N. (2022). Three new species of Duplicibothrium (Cestoda: ‘Tetraphyllidea’) from cownose rays in Senegal with a phylogenetic analysis of the genus. Journal of Helminthology, 96, e8. https://doi.org/10.1017/S0022149X21000766
Taylor S. M., & Bennett M. B. (2013). Size, sex and seasonal patterns in the assemblage of Carcharhiniformes in a sub-tropical bay. Journal of Fish Biology, 82, 228–241. https://doi.org/10.1111/jfb.12003
Tedesco P., Caffara M., Gustinelli A., Fiorito G., & Fioravanti M. L. (2020). Metacestodes of elasmobranch tapeworms in Octopus vulgaris (Mollusca, Cephalopoda) from Central Mediterranean—SEM and molecular data. Animals, 10, 2038. https://doi.org/10.3390/ani10112038
Tillett B. J., Meekan M. G., & Field I. C. (2014). Dietary overlap and partitioning among three sympatric carcharhinid sharks Endangered Species Research, 25, 283–293. https://doi.org/10.3354/esr00615
Waeschenbach A., Webster B. L., Bray R. A., & Littlewood D. T. J. (2007). Added resolution among ordinal level relationships of tapeworms (Platyhelminthes: Cestoda) with complete small and large subunit nuclear ribosomal RNA genes. Molecular Phylogenetics and Evolution, 45, 311–325. https://doi.org/10.1016/j.ympev.2007.03.019
White W. T., Platell M. E., & Potter I. C. (2004). Comparisons between the diets of four abundant species of elasmobranchs in a subtropical embayment: implications for resource partitioning. Marine Biology, 144, 439–448. https://doi.org/10.1007/s00227-003-1218-1
Acknowledgements
We thank John Page, Dave Thompson, Steve Taylor, and the past members of the Marine Parasitology Laboratory at the University of Queensland for their continued support and assistance in the field. We also thank staff at the Moreton Bay Research Station for their support. This project represents a contribution to Taxonomy Australia (2020), a national initiative organised under the auspices of the Australian Academy of Science that brings together the taxonomic community to develop approaches that will significantly increase the rate at which new species are discovered, resolved and named, with a view to completely documenting the Australian biota within a generation.
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions. This work was supported by funding from the Australian Government’s Australian Biological Resources Study National Taxonomy Research Grants Program (4-H04JDSM).
Author information
Authors and Affiliations
Contributions
All authors collected and prepared the material. S.C. wrote the manuscript and prepared the figures. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable institutional, national and international guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is registered in the Official Register of Zoological Nomenclature (ZooBank) as urn:lsid:zoobank.org:pub:9827FA42-3E1E-448C-8642-C8C39D59EFB1.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cutmore, S.C., Bennett, M.B. & Cribb, T.H. Morphological and molecular identification of metacestodes infecting teleost fishes of Moreton Bay, Australia. Syst Parasitol 101, 57 (2024). https://doi.org/10.1007/s11230-024-10183-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11230-024-10183-y