Abstract
Genetic improvement of local rabbit breeds using modern approaches such as marker-assisted selection requires accurate and precise information about marker‒trait associations in animals with different genetic backgrounds. Therefore, this study was designed to estimate the association between two mutations located in the Neuropeptide Y (NPY, g.1778G > C) and Phosphoglycerate Mutase 2 (PGAM2, c.195 C > T) genes in New Zealand White (NZW), Baladi (BR), and V-line rabbits. The first mutation was genotyped using high-resolution melting, and the second mutation was genotyped using the PCR-RFLP method. The results revealed significant associations between the NPY mutation and body weight at 10 (V-line) and 12 weeks of age (NZW, BR, and V-line), body weight gain (BWG) from 10 to 12 weeks of age (BR), BWG from 6 to 12 weeks of age (NZW, BR, and V-line), average daily gain (NZW, BR, and V-line, and BR), growth rate (GR) from 8 to10 weeks (V-line), 10 to 12 weeks (BR), and GR from 6 to 12 weeks of age (BR, and V-line). The PGAM2 mutation was associated with body weight at 10 (V-line) and 12 (NZW, and V-line) weeks of age, with significant positive additive effects at 12 weeks of age in all breeds, and was associated with BWG from 8 to 10 and 10 to 12 in BR, and BWG from 6 to 12 weeks of age (NZW, and BR), and average daily gain (NZW, and BR), and was associated with GR form 8 to 10 weeks (BR), from10 to 12 weeks (BR, and V-line) and from 6 to 12 weeks (BR). The results highlighted the importance of the two mutations in growth development, and the possibility of considering them as candidate genes for late growth in rabbits.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Breeding practices and domestication have impacted growth performance (El-Sabrout and Soliman 2018). Alves et al. (2015) examined the effects of domestication on the genetic diversity of rabbits and discovered evidence for two significant genetic diversity reductions, one of which was linked to breed creation. Due to feeding costs and consumer preferences for lean meat, selection based on growth characteristics has been extremely important to the rabbit business (El-Sabrout and Soliman 2018). Body weight gain is one of the most economically important traits in rabbits (Cartuche et al. 2014), and it has been a selection criterion in different selection programs (Mínguez et al. 2016). However, traditional rabbit breeding is challenged by many factors, including the poor data and information about relationships between relatives, the limited number of animals, and the sensitivity of rabbits to high-temperature weather (Khalil 2020; Sabry et al. 2021).
Marker-assisted selection (MAS) is a powerful tool in animal breeding programs to improve the efficiency and accuracy of selection (ABDEL-KAFY et al. 2016). Additionally, in MAS, genetic markers such as microsatellites and single-nucleotide polymorphisms (SNPs) are considered as bioinformatics indicators to provide detailed information regarding polymorphism in allelic genotypes at a certain locus (Bovo et al. 2021). This information enables in-depth analyses and evaluations of genetic variants as well as the identification of genes impacting economically significant features in farm animals (Carneiro et al. 2011). Single-nucleotide polymorphisms of numerous candidate genes have been used to identify genes related to body weight and meat quality in rabbits (Fontanesi et al. 2006; Wu et al. 2015; Helal et al. 2022). According to Seidel (2010), using SNP markers in genomic selection is a powerful new tool, because SNPs can be detected using a variety of simple techniques, such as PCR-RFLP, and therefore SNPs can be used for mass screening of many samples in a short time (Fontanesi et al. 2006). Moreover, SNPs are the most recent contribution to the study of DNA sequence variation, and SNPs represent the most innovative molecular marker in genotyping studies.
The complex neuroendocrine system that regulates energy balance includes both central and peripheral signals, including neuropeptides (Hillebrand et al. 2002). Feeding-related neuropeptides are a part of this intricate control system, and orexigenic neuropeptides such as Neuropeptide Y (NPY) are crucial for controlling energy balance and feed intake (Bazhan and Zelena 2013). The 36 amino acid neuropeptide NPY was initially identified in the pig brain in 1982 and is a plentiful and well-characterized neuropeptide (Mercer et al. 2011). The stimulation of food intake and correlation with weight gain in mammals are the strongest effects of this powerful orexigenic factor (Hillebrand et al. 2002; Ding et al. 2005). In animals, intracerebroventricular (ICV) NPY injection boosted appetite in a dose-dependent manner (Clark et al. 1997). As feed intake increases, NPY eventually decreases after initially increasing just before the start of eating (Kalra et al. 1991). In animal obesity models, tubby mice and rats without brown adipose tissue showed overexpression of the NPY gene (Guan et al. 1998). Additionally, it has been discovered that the bovine NPY gene is positioned in the QTL region for carcass characteristics, marbling, and lean meat (Casas et al. 2004; Mizoshita et al. 2004). In rabbits, SNPs in the NPY gene were significantly associated with eviscerated slaughter percentage and semi-eviscerated slaughter percentage in rabbits (Liu et al. 2014).
Phosphoglycerate mutase (PGAM) is an enzyme in the glycolytic pathway that converts 3-phosphoglycerate into 2-phosphoglycerate to release energy during glycolysis in muscle tissues (Fontanesi et al. 2006). The PGAM dimer in mammalian tissues is composed of the brain form (B form, also known as PGAM1), which is extensively expressed, and the muscle form (M form, also known as PGAM2), which is exclusively present in adult skeletal and cardiac muscle. According to Qiu et al. (2008), PGAM2 is connected to growth, feed conversion, and slaughter quality and is highly expressed in skeletal muscle during all periods of pig development. Additionally, PGAM2 plays crucial functions in the glycolysis process that regulates postnatal growth and associated meat quality indices (Sevane et al. 2013).
To our knowledge, there is little information on the relationship between growth traits in domestic rabbit breeds and genetic variations in Neuropeptide Y (NPY) and Phosphoglycerate Mutase 2 (PGAM2) genes. Therefore, the goal of this work was to determine the association between growth features in the New Zealand White (NZW), Valencian line V (V-line), and Baladi rabbit (BR) breeds and single nucleotide polymorphisms in the NPY and PGAM2 genes.
Materials and methods
Animals and management
Three rabbit breeds were used for the current study, including two commercial lines and one local breed. The commercial lines were New Zealand White (NZW) rabbits, which is a commercial rabbit breed that originated and developed in the USA and was raised in Egypt at the commercial level for many decades. The other commercial line was the Valencian line V (V-line), which was developed in Spain, and has been selected for litter size at weaning (Ragab and Baselga 2011), and then imported and bred in Egypt in the past few decades. Concerning the local breed, improved Baladi rabbit Breeds (BR), which were developed by crossbreeding for several generations between native Egyptian rabbits and Flemish Giant (Khalil 1999). The number of rabbits was 92, 60, and 76 for NZW (40 males and 52 females), BR (26 males and 34 females), and V-line rabbits (33 and 43 individuals), respectively.
The kits of the different genetic groups were weaned at the same age (35 ± 1 d). Rabbits were collected from different farms in Giza, Fayoum, and Zagazig governorates in Egypt, and all rabbits were genetically unrelated. After receiving rabbits, they were kept for one week for an adaptation period. The rabbits were housed in the same room and received the same management and environmental conditions. The housing system was a semi-open housing system including cages (60 × 50 × 40 cm) supplied with a nipple system and feeding hoppers. A commercial fattening diet (18% protein, 2750 ME, and 12% crude fiber) was provided for all rabbits, and water was provided ad libitum. At the end of the experimental procedures animals were released.
Individual body weights (BW, g) were recorded every two weeks until marketing age (From 6 to 12 weeks of age), and body weight gains for different periods (BWG, g) were calculated as the difference between body weight at the beginning and end of the period. Relative growth rates (GR, %) were calculated as BWG for a certain period divided by the average BW at the start and end of the period.
Samples and genotyping
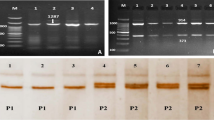
Blood samples were collected from all rabbits at the end of the fattening period. Samples were collected in tubes containing ethylene diamine tetra-acetic acid (EDTA) and stored at -20 °C until use. DNA was extracted using a whole genomic extraction kit (Wiz-bio-solution Inc., South Korea). Two SNPs were genotyped. The first SNP (g.1778G > C), located in the intron 1 of the NPY gene, was genotyped using high-resolution melting (HRM). Primers (Table 1) were designed to amplify a fragment of 98 bp containing the SNP, which was also described previously (Liu et al. 2014). The HRM analysis was performed in a total volume of 10 µl using HOT FIREPol EvaGreen HRM Mix (Solis BioDyne, Estonia). The reaction mixture included 5 µl of EvaGreen HRM mix, 1 µl of DNA template, 0.5 µl of each primer, and 3 µl of ddH2O. The HRM reaction profile included initial denaturation at 95 °C for 12 min to activate HOT FIREPOL DNA polymerase; followed by 40 cycles of 95 °C for 20 s, 48 °C for 20 s, and 72 °C for 20 s; and an HRM step from 65 to 95 °C rising at 0.4 °C.
The second mutation was the c.195 C > T SNP of the PGAM2 gene, which was genotyped using restriction fragment length polymorphism-polymerase chain reaction (RFLP-PCR) analysis as described previously (Wu et al. 2015). The primers were used to amplify 855 bp of the 5´ UTR to intron 2 of the PGAM2 gene (Table 1). PCR was performed in a total reaction volume of 20 µl, containing 1 µl of each of the two primers, 2 µl of DNA template, 10 µl of PCR master mix, and 6 µl of PCR grade water. The reaction was carried out with an initial denaturation cycle at 95 °C for 5 min, 34 cycles of 95 °C for 40 s, 62 °C for 40 s, and 72 °C for 40 s, and an additional 10 min at 72 °C as an extension cycle. The PCR products (10 µl) were then incubated with 10 U of Csp6I restriction enzyme (Vivantis, Malaysia) at 37 °C, followed by electrophoresis on a 2.5% agarose gel.
Statistical analysis
Genetic parameters including heterozygosity (He), effective number of alleles (Ne) and polymorphic information content (PIC) were calculated according to Botstein et al. (Botstein et al. 1980). The association analysis was performed using the general linear model (GLM) of SAS (SAS 1999), using the following model:
where Yijk is the trait recorded of animal lth, µ is the overall mean of trait, Gi is the genotype effect (three levels; p2, 2pq, and q2 ), Sj is the sex effect (two levels; male and female), Bk is the breed effect (three levels; NZW, BR, and V-line), Rl is the source of rabbits (three levels; farm A, farm B, and farm C), and εijkl is the random residual of the model.
Results
SNP identification
The c.195 C > T SNP was detected via PCR-Csp6I digestion of 855 kb of the PGAM2 gene. Table 2 showed the allelic and genotypic frequencies, as well as population genetic parameters, including He, PIC, and Ne. The three genotypes were found in the NZW, V-line, and BR breeds, with frequencies of 0.50, 0.42, and 0.61 for the heterozygous genotype (CT) being predominant in each breed. The c.195 C > T SNP locus was classified by PIC as having moderate genetic diversity, which suggested that this SNP has a sizable amount of selection potential. Additionally, for this SNP, no rabbit breeds showed any deviation from HWE.
The three genotypes of the NPY gene were detected by HRM g.1778G > C SNP analysis (Table 2). In NZW and V-line rabbits, the heterozygous genotype (CG) predominated with frequencies of 0.41 and 0.47, respectively. However, with a frequency of 0.47, the homozygous genotype AA predominated in BR rabbits. The g.1778G > C SNP locus was classified by PIC as having considerable genetic diversity, which suggested that this SNP had a sizable selection potential. Similarly, for this SNP, no rabbit breeds showed deviation from HWE.
Association of NPY and PGAM2 genes with growth traits in rabbits
The SNP g.1778G > C of the NPY gene has impacts on growth features in all breeds, as evidenced by the data in Table 3. In NZW rabbits, individuals with the G allele had higher body weight at 12 weeks of age (BW12), body weight gain from 10 to 12 weeks of age (BWG1012), body weight gain from 6 to 12 weeks of age (BWG612), and average daily gain (ADG) than those with the CC genotype. The additive effect of the SNP was significant for BW12, BWG810, and BWG612; however, the dominance effect was significant for BW12, and BWG612. For the BR rabbits, the GG and CG genotypes had higher BW12, BWG1012, BWG612, ADG, GR1012, and GR612 than the CC genotype. The additive and dominance effects of the SNP were significant for BW12, BW1012, BW612 and GR612. Meanwhile, the GG and GC rabbits exhibited higher BW10, BW12, BWG612, ADG, GR810 and GR612 than the CC genotype for the V-line breed. However, there was no discernible difference in the different traits between GG and CG individuals. The additive effect was significant for BW10, BW12, BW1012 and BWG612, and the dominance effect was significant for BW10, and BW12.
The data presented in Table 4 showed that the three rabbit breeds’ growth traits are influenced by the SNP c.195 C > T in the PGAM2 gene. The homozygous TT and heterozygous CT genotypes had higher BW12, BWG612, and ADG than the CC genotype in the NZW rabbits, with a significant additive and dominance effects for BW12, and BWG612 and a significant negative additive effect for BW6. The heterozygous genotype CT in the V-line rabbits, however, showed higher values for BW10 and BW12 compared with the CC genotype, with significant additive (BW10, BW12, and BWG612) and dominance effects (BW12 and BWG612) effects. In BR individuals, CC genotype rabbits had lower BWG1012, BW612, and ADG values than the CT rabbits, and had lower BWG810 than the TT genotype, with highly significant additive (BW12 and BWG810) and dominance (BW12 and BWG612) effects.
Discussion
Genetic improvement of rabbits using qualitative genetics approaches was successful and resulted in the development of new breeds and lines (Khalil and Bolet 2010). Nevertheless, there is a need for new approaches to speed up the selection process with more accuracy and to overcome the limitations of traditional selection. Marker-assisted selection (MAS) offers an accurate, reliable, and low-cost approach that can be used for the genetic improvement of local breeds that exist in limited numbers. When the marker effect is known for a given trait, animals can be evaluated without a phenotype based on the existence of markers (Meuwissen and Goddard 1996; Helal and El-Gendy 2023). This strategy can increase the rate of genetic gain (De Roos et al. 2011; Helal et al. 2024), and cut the cost of breeding schemes by approximately 92% (Schaeffer 2006). Moreover, it is very useful in the improvement of traits that are difficult to measure in live animals such as carcass traits, and traits that are expressed in a single sex, such as reproductive traits. Additionally, in the case of expensive phenotypic recording for numerous economic traits, the use of genotype information as a selection aid can be a reasonable alternative (Yang et al. 2013).
One of the main requirements for MAS practice is the estimation of the effect of different markers on the traits of interest. The estimations of SNP effects can theoretically be applied for a few generations with just a minimal loss in prediction accuracy; hence, it is not required to test the crossbred phenotypes every generation (Szkiba et al. 2014). In this regard, there are some recent efforts to estimate the effect of different SNPs of different candidate genes that affect the important traits in rabbits (Fontanesi 2016; Safaa et al. 2023).
The goal of this research was to identify DNA markers whose variability may be related to rapid growth features using the candidate gene technique in portions of two growth-related genes (NPY and PGAM2) that are located on the same chromosome. PCR-Csp6I digestion of 855 kb was used for genotyping the PGAM2 gene and HRM was used for genotyping the g.1778G > C SNP. The three genotypes for the two investigated genes were detected. The different genotypes were distributed differently among the three rabbit populations. All genotypic frequencies followed the Hardy-Weinberg distribution, indicating the goodness of animal sampling.
The results revealed significant associations between NPY and PGAM2 genotypes and growth traits at different levels in the three breeds. The polymorphisms found in the three breeds of high growth-rate rabbits may modify the activity or functionality of the NPY and PGAM2 genes in rabbits. To the best of our knowledge, there is very little information about investigating polymorphisms in the NPY and PGAM2 genes and their association with growth traits in rabbits, as reported previously (Khalil 2020; Helal et al. 2022). The two studied SNPs showed high PIC and heterozygosity values, indicating high levels of genetic variation, and therefore, a high genetic progress through selection is expected. Additionally, many traits associated with the SNPs of the two genes showed significant additive and dominance variance that could be consumed by breeding programs. The observed oscillations in the additive and dominance effect may be attributed to two main reasons, the first reason is the pleiotropic effect of the two genes, where the relative contribution may change with age, leading to oscillations in the observed genetic effects (Charmantier et al. 2006). The second reason is that the same allele may have an opposite effects on the same trait across ages (Maklakov et al. 2015; Everman and Morgan 2018).
Liu et al. (2014) sequenced the NPY gene in Ira, Champagne, and Tianfu black rabbits. According to the findings, intron 1 of the rabbit NPY gene contains four SNPs, namely, g.1491G > A, g.1525G > T, g.1530 C > T, and g.1778G > C. The g.1778G > C SNP was genotyped further, but it was not significantly associated with body weight at different ages. However, the eviscerated slaughter % and semi-eviscerated slaughter percentage were substantially connected with that SNP, and those with the CC genotype performed better than those with the CG genotype. The results of the current study, on the contrary, indicated that allele G is preferred in growth traits over allele C, and GG genotype rabbits had better performance than the CC genotype. These differences in the results of our study and previous work may be attributed to the genetic background differences between the studied animals (Ateya et al. 2021). Furthermore, the results of the current study also indicated that this SNP seems to affect late growth traits rather than post-weaning growth traits, and therefore, it is more likely to affect carcass traits.
The link between the NPY gene and growth performance has been addressed in different farm animals. In beef cattle, three SNPs in intron 2 of the NPY gene were strongly correlated with body weight, ADG, feed conversion ratio (FCR), and marbling (Sherman et al. 2008). In Chinese indigenous cattle, polymorphisms of the NPY gene also significantly affected body length and chest girth (Zhang et al. 2011). We hypothesized that NPY may also be a candidate gene and relevant given the substantial conservation of the NPY protein structure and DNA sequence in rabbits and other livestock.
Ateya et al. (2021) characterized SNPs of the PGAM2 gene in three breeds of rabbits (V-line, Baladi Black, and Baladi Red rabbits) concerning the association of PGAM2 gene polymorphisms in rabbits. The nucleotide sequence alignment revealed no differences between the sequences of the tested breeds, indicating that the PGAM2 gene is highly conserved in rabbits. Genetic polymorphisms were examined on various PGAM2 gene fragments, which may explain the differences between our study and earlier research. Additionally, Wu et al. (Wu et al. 2015) sequenced part of the PGAM2 gene in Tianfu black, Ira, and Champagne rabbit breeds, and detected three SNPs. c.195 C > T was genotyped further to investigate the association between growth traits, and the heterozygous (CT) genotype of c.195 C > T was associated with body weight at 84 days of age and with average daily weight gain. These results were in agreement with the results obtained in the current study, where the SNP c.195 C > T significantly affected body weight at 10 and 12 weeks of age, and did not affect the post-weaning weights. Additionally, the results indicated the advantage of the mutant allele T, in favor of the heterozygous genotype in BR and V-line rabbits. In contrast, contrary, the same mutation of the PGAM2 gene was genotyped in V-line, Alexandria rabbits (El-Sabrout and Aggag 2017), and in meat line P91 rabbits (Nahácky et al. 2018), but the association with growth traits was insignificant. In ducks, PGAM2 was considered a candidate gene for the rigorous selection of growth performance (Ghanem et al. 2023).
In conclusion, the results in the present study revealed the evaluation of the genetic variations in the NPY and PGAM2 genes among NZW, BR, and V-line rabbit breeds. The two SNPs were significantly correlated with growth traits in the different breeds, with special emphasis on late growth traits. The NPY and PGAM2 genes may therefore serve as candidates and proxies for indicators of rabbit growth features.
Data availability
Data available upon request from the authors.
References
Abdel-Kafy E, Larbi G, Ahmed Ms, Badawi SS, Hassan Y, N.S (2016) Genetic diversity and phenotype characterization of native rabbit in Middle-Egypt. J New Sci IABC (16), 1312–1320
Alves JM, Carneiro M, Afonso S, Lopes S, Garreau H, Boucher S, Allain D, Queney G, Esteves PJ, Bolet G, Ferrand N (2015) Levels and patterns of genetic diversity and population structure in domestic rabbits. PLoS ONE 10:e0144687
Ateya AI, Hendam BM, Radwan HA, Abo Elfadl EA, Al-Sharif MM (2021) Using Linear Discriminant Analysis to characterize Novel single nucleotide polymorphisms and expression Profile Changes in genes of three breeds of rabbit (Oryctolagus cuniculus). Comp Med 71:222–234
Bazhan N, Zelena D (2013) Food-intake regulation during stress by the hypothalamo-pituitary-adrenal axis. Brain Res Bull 95:46–53
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Bovo S, Schiavo G, Utzeri VJ, Ribani A, Schiavitto M, Buttazzoni L, Negrini R, Fontanesi L (2021) A genome-wide association study for the number of teats in European rabbits (Oryctolagus cuniculus) identifies several candidate genes affecting this trait. Anim Genet 52:237–243
Carneiro M, Afonso S, Geraldes A, Garreau H, Bolet G, Boucher S, Tircazes A, Queney G, Nachman MW, Ferrand N (2011) The genetic structure of domestic rabbits. Mol Biol Evol 28:1801–1816
Cartuche L, Pascual M, Gómez EA, Blasco A (2014) Economic weights in rabbit meat production. World Rabbit Sci 22:165–177
Casas E, Keele JW, Shackelford SD, Koohmaraie M, Stone RT (2004) Identification of quantitative trait loci for growth and carcass composition in cattle. Anim Genet 35:2–6
Charmantier A, Perrins C, McCleery RH, Sheldon BC (2006) Quantitative genetics of age at reproduction in wild swans: support for antagonistic pleiotropy models of senescence. Proc Natl Acad Sci 103:6587–6592
Clark JT, Kalra PS, Kalra SP (1997) Neuropeptide Y stimulates feeding but inhibits sexual behavior in rats. Obes Res 5:275–283
De Roos APW, Schrooten C, Veerkamp RF, van Arendonk JAM (2011) Effects of genomic selection on genetic improvement, inbreeding, and merit of young versus proven bulls. J Dairy Sci 94:1559–1567
Ding B, Kull B, Liu Z, Mottagui-Tabar S, Thonberg H, Gu HF, Brookes AJ, Grundemar L, Karlsson C, Hamsten A, Arner P, Östenson CG, Efendic S, Monné M, Von Heijne G, Eriksson P, Wahlestedt C (2005) Human neuropeptide Y signal peptide gain-of-function polymorphism is associated with increased body mass index: possible mode of function. Regul Pept 127:45–53
El-Sabrout K, Aggag SA (2017) Associations between single nucleotide polymorphisms in multiple candidate genes and body weight in rabbits. Veterinary World 10:136–139
El-Sabrout K, Soliman F (2018) Association of single-nucleotide polymorphism of melanocortin gene with feed intake in rabbit (Oryctolagus cuniculus). J Anim Physiol Anim Nutr 102:564–567
Everman ER, Morgan TJ (2018) Antagonistic pleiotropy and mutation accumulation contribute to age-related decline in stress response. Evolution 72:303–317
Fontanesi L (2016) The rabbit in the genomics era: applications and prespectives in rabbit biology and breeding. In: Proc. 11th World Rabbit Congress
Fontanesi L, Tazzoli M, Scotti E, Russo V (2006) Analysis of Candidate Genes for Meat Production Traits in Domestic Rabbit Breeds. In: Proceedings of the 9th World Rabbit Congress, Verona, Italy, 10–13 June 2008
Ghanem H, Ibrahim S, Sharif M, Abdo M, Ateya A (2023) Efficiency of candidate Gene Approach, Gene expression and economic evaluation for rigorous selection of growth performance in ducks. Alexandria J Veterinary Sci 76:68
Guan XM, Yu H, Van Der Ploeg LHT (1998) Evidence of altered hypothalamic pro-opiomelanocortin/neuropeptide Y mRNA expression in tubby mice. Mol Brain Res 59:273–279
Helal MM, El-Gendy EA (2023) Marker-assisted selection for improving body weight in local chickens in Egypt. J Agric Sci 161:135–147
Helal M, Hany N, Maged M, Abdelaziz M, Osama N, Younan YW, Ismail Y, Abdelrahman R, Ragab M (2022) Candidate genes for marker-assisted selection for growth, carcass and meat quality traits in rabbits. Animal Biotechnol 33:1691–1710
Helal M, Sameh J, Gharib S, Merghany RM, Bozhilova-Sakova M, Ragab M (2024) Candidate genes associated with reproductive traits in rabbits. Trop Anim Health Prod 56:94
Hillebrand JJG, De Wied D, Adan RAH (2002) Neuropeptides, food intake and body weight regulation: a hypothalamic focus. Peptides 23:2283–2306
Kalra SP, Dube MG, Sahu A, Phelps CP, Kalra PS (1991) Neuropeptide Y secretion increases in the paraventricular nucleus in association with increased appetite for food. Proc Natl Acad Sci USA 88:10931–10935
Khalil MH (1999) Rabbit genetic resources of Egypt. Anim Genetic Resour Inform 26:95–111
Khalil MH (2020) Molecular applications of candidate genes in genetic improvement programs in livestock. Egypt J Anim Prod 57:1–23
Khalil MH, Bolet G (2010) Sustainable rabbit breeding and genetic improvement programs achieved in developing countries. In: In Proceedings of the World Congress on Genetics Applied to Livestock Production. Leipzig, Germany
Liu WC, Chen SY, Jia XB, Wang J, Lai SJ (2014) Effects of variants in proopiomelanocortin and neuropeptide Y genes on growth, carcass, and meat quality traits in rabbits. Asian-Australasian J Anim Sci 27:609–615
Maklakov AA, Rowe L, Friberg U (2015) Why organisms age: evolution of senescence under positive pleiotropy? BioEssays 37:802–807
Mercer RE, Chee MJS, Colmers WF (2011) The role of NPY in hypothalamic mediated food intake. Front Neuroendocr 32:398–415
Meuwissen THE, Goddard ME (1996) The use of marker haplotypes in animal breeding schemes. Genet Selection Evol 28:161–176
Mínguez C, Sanchez JP, El Nagar AG, Ragab M, Baselga M (2016) Growth traits of four maternal lines of rabbits founded on different criteria: comparisons at foundation and at last periods after selection. J Anim Breed Genet 133:303–315
Mizoshita K, Watanabe T, Hayashi H, Kubota C, Yamakuchi H, Todoroki J, Sugimoto Y (2004) Quantitative trait loci analysis for growth and carcass traits in a half-sib family of purebred Japanese Black (Wagyu) cattle. J Anim Sci 82:3415–3420
Nahácky J, Židek R, Gábor M, Miluchová M, Bučko O, Kováčik A (2018) MC4R and PGAM2 genes polymorphism association with production traits in rabbit (Oryctolagus cuniculus). J Microbiol Biotechnol Food Sci 7:493–495
Qiu H, Zhao S, Xu X, Yerle M, Liu B (2008) Assignment and expression patterns of porcine muscle-specific isoform of phosphoglycerate mutase gene. J Genet Genomics 35:257–260
Ragab M, Baselga M (2011) A comparison of reproductive traits of four maternal lines of rabbits selected for litter size at weaning and founded on different criteria. Livest Sci 136:201–206
Sabry MIE, Zaki MM, Elgohary FA, Helal MM (2021) Sustainable rabbit production under the global warming conditions in Southern Mediterranean Region. World’s Veterinary J 11:543–548
Safaa HM, Ragab M, Ahmed M, El-Gammal B, Helal M (2023) Influence of polymorphisms in candidate genes on carcass and meat quality traits in rabbits. PLoS ONE, 18, e0294051
SAS (1999) SAS/STAT user’s guide: statistics. SAS Institute Inc, Cary
Schaeffer LR (2006) Strategy for applying genome-wide selection in dairy cattle. J Anim Breed Genet 123:218–223
Seidel GE (2010) Brief introduction to whole-genome selection in cattle using single nucleotide polymorphisms. Reprod Fertility Dev 22:138–144
Sevane N, Armstrong E, Cortés O, Wiener P, Wong RP, Dunner S (2013) Association of bovine meat quality traits with genes included in the PPARG and PPARGC1A networks. Meat Sci 94:328–335
Sherman EL, Nkrumah JD, Murdoch BM, Li C, Wang Z, Fu A, Moore SS (2008) Polymorphisms and haplotypes in the bovine neuropeptide Y, growth hormone receptor, ghrelin, insulin-like growth factor 2, and uncoupling proteins 2 and 3 genes and their associations with measures of growth, performance, feed efficiency, and carcass meri. J Anim Sci 86:1–16
Szkiba D, Kapun M, von Haeseler A, Gallach M (2014) SNP2GO: functional analysis of genome-wide association studies. Genetics 197:285–289
Wu ZL, Chen SY, Jia XB, Lai SJ (2015) Association of a synonymous mutation of the PGAM2 gene and growth traits in rabbits. Czech J Anim Sci 60:139–144
Yang W, Kang X, Yang Q, Lin Y, Fang M (2013) Review on the development of genotyping methods for assessing farm animal diversity. J Anim Sci Biotechnol 4:1–6
Zhang L, Zhang AL, Zhang LZ, Zhang Q, Zhang CL, Lan XY, Chen H (2011) Association analysis between variants in bovine NPY gene and growth traits in Nanyang cattle (Bos Tarus). Gen Comp Endocrinol 170:189–192
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB).
Author information
Authors and Affiliations
Contributions
All authors have made considerable contributions to this work. Conceptualization: Mostafa Helal, Mohamed Ragab; Methodology: Mostafa Helal, Marwa Ahmed, Mohamed Ragab, Ahmed Ateya, Shimaa Sakr; Formal analysis and investigation: Mostafa Helal, Marwa Ahmed, Mohamed Ragab; Original draft preparation: Mostafa Helal, Marwa Ahmed, Ahmed Ateya; Writing – review and editing: Mostafa Helal, Marwa Ahmed, Mohamed Ragab, Ahmed Ateya, Shimaa Sakr.
Corresponding author
Ethics declarations
Ethical approval
All the experimental procedures were approved by the institutional animal care and use committee at Cairo University (CU-IACUC), approval number: CU-IACUC/ I /F/46/22.
Competing interests
The authors report there are no competing interests to declare.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Helal, M., Ahmed, M., Ragab, M. et al. Association of single nucleotide polymorphisms in Neuropeptide Y (NPY) and Phosphoglycerate Mutase 2 (PGAM2) genes with growth traits in rabbits. Trop Anim Health Prod 56, 239 (2024). https://doi.org/10.1007/s11250-024-04085-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11250-024-04085-w