Abstract
Genetic gain in potato breeding is limited by the heterozygous tetraploid genome of cultivated potato. Recent efforts to breed potato at the diploid level promise to improve genetic gain and allow more straightforward genetics and introgression breeding. Diploid F1 hybrid potato breeding relies on the ability to create diploid inbred lines via repeated self-fertilization. However, self-fertilization of diploid potato is hampered by a gametophytic self-incompatibility system encoded by the S-locus that prevents fertilization by self-pollen. Nonetheless, self-compatible diploid potato genotypes exist and have been used to create inbred lines. The S-locus inhibitor (Sli) gene is a dominant gene that provides strong self-compatibility in diploid potato and was previously mapped to Chromosome 12. While the Sli gene has already been identified and characterized, the most tedious challenge was to develop the optimal phenotyping methods and genetic populations preceding the cloning of this gene. To this end, we developed an effective phenotyping protocol to identify suitable parents and create diploid populations segregating for Sli. We show that an accurate phenotyping method is crucial to discriminate between confounding fertility factors and self-compatibility. In addition, we found that the Sli locus shows extreme segregation distortion on Chromosome 12. Finally, we used these insights to develop three F1 populations that segregate for Sli, which we later used for the identification of the Sli gene.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Potato (Solanum tuberosum) is the third most important food crop after rice and wheat. Although the yield in these cereals has increased annually with 1–2%, genetic gain in potato has been limited or absent over the last century. This is mainly due to characteristics of the current potato breeding system. A typical breeding programme starts with an initial cross between two tetraploid parents. The F1 progeny populations are vegetatively multiplied as clones and phenotypically assessed over multiple years, resulting in a limited number of well-performing genotypes. However, due to the high allelic diversity, including deleterious alleles that are hidden in the multiple genomes in tetraploid potato, the F1 populations show tremendous segregation of traits and large populations are required to identify clones that combine the desired traits of the parents, and show a yield performance that is at least similar to if not better than the parental genotypes (Lindhout et al. 2011).
Recently, an alternative breeding strategy has been developed based on the self-fertilization of self-compatible (SC) diploid genotypes to create homozygous inbred lines, which can then be crossed to produce homogenous F1 hybrids (Lindhout et al. 2011; Jansky et al. 2016). Near-homozygous diploid inbred lines are the result of multiple rounds of self-fertilization, each of which offers the opportunity to expose homozygous deleterious alleles, while meiotic recombination may break undesired negative linkages and lead to desired combinations of linked traits enabling the selection of favourable genotypes. This has sparked interest in the mechanisms of sexual reproduction in potato (Plantenga et al. 2019; Seibert et al. 2020; Bethke and Jansky 2021).
Most diploid potato lines are naturally self-incompatible (SI) due to the Gametophytic Self-Incompatibility (GSI) system, encoded by the S-locus. The potato S-locus encodes S-RNases that are expressed in stylar tissues, which are toxic to pollen tubes unless the pollen expresses S-locus F-Box (SLF) proteins that can recognize and detoxify the S-RNases. Each S-allele encodes one S-RNase and multiple SLFs that together can recognize all S-RNases except the one encoded on the same S-allele (Kubo et al. 2010).
While most diploid potatoes are self-incompatible, most tetraploid potatoes are self-compatible due to the heteroallelic pollen effect (McClure et al. 2011). Tetraploid genotypes that have at least two different S-alleles produce diploid pollen, of which at least 25% express two different S-alleles and are thus able to detoxify all S-RNases, resulting in self-compatibility. When diploids are generated from such tetraploids via anther culture or prickle pollination, they lose the heteroallelic pollen effect and become self-incompatible because diploids produce haploid pollen that express only one S-allele. However, rare examples of self-compatible diploid potatoes have been described by several authors. Olsder and Hermsen identified two self-compatible dihaploids, G254 and B16, derived from tetraploid cultivars Gineke and Black 4495, respectively (Olsder and Hermsen 1976). They concluded that these genotypes harbour a genetic system that overcomes self-incompatibility. Dihaploid G254 has since then been extensively used in diploid breeding programmes at Wageningen University & Research, and one of the many descendants of G254, RH89-039–16, has been used as reference genotype around the world for genetic studies and genome sequencing (Xu et al. 2011; Peterson et al. 2016; Zhang et al. 2019; Clot et al. 2020; Zhou et al. 2020). Similarly, Peloquin and Hougas obtained a self-compatible dihaploid, US-W4, from the tetraploid clone Minn. 20–20-34 (Peloquin and Hougas 1960). This diploid genotype, US-W4, has also been widely used in breeding and research programmes (De Jong and Rowe 1971; Jansky 2011; Braun et al. 2017; Marand et al. 2017; Kaiser et al. 2021; Bamberg et al. 2021; Song and Endelman 2023).
Later, Hosaka and Hanneman identified a self-compatible accession of S. chacoense (chc 525–3) and mapped the “S-locus inhibitor (Sli) gene” on Chromosome 12 in an F1 population derived from a complex interspecific pedigree (Hosaka and Hanneman 1998a, b). They mapped the Sli locus at the telomeric end of the long arm of Chromosome 12, at 10-cm distance from markers CT156E5120 and 89–1320. Interestingly, they observed segregation distortion in this region and suggested that pollen carrying Sli may have a gametophytic advantage during the fertilization process. However, based on the observation of phenotypic segregation for self-berry set in the S8 generation, Hosaka and Hanneman suggested that Sli acts sporophytically, meaning that the genotype of the sporophyte rather than the haploid genotype of the gametophyte determines the outcome of the pollination, and that homozygosity for Sli itself is lethal or that it is closely linked to a lethal allele.
The location of Sli was later confirmed by Peterson et al. (2016) who mapped self-compatibility in an F1 population derived from the cross DM × RH. Remarkably, they observed highly significant associations between self-compatibility and 88 SNPs on the long arm of Chromosome 12, but this association was absent in the S3 generation, suggesting that this locus was fixed in the S37. Further confirmation of the location of Sli was provided by Clot et al., who used Comparative Subsequence Sets Analysis (CoSSA) to narrow down the location of Sli to a 333 kB interval on the distal end of the long arm of Chromosome 12. Furthermore, Clot et al. showed that the Sli specific haplotype is present in a wide variety of tetraploid potato cultivars, a feature that they attribute to the presence of the clone Rough Purple Chili in pedigrees of many American and European varieties (Clot et al. 2020).
Using the Sli gene, Hosaka et al. generated nearly homozygous potato inbred lines (Phumichai et al. 2005; Hosaka and Sanetomo 2020). Similarly, Jansky et al. generated the M6 line that has since then been used as a reference genotype and parent for diploid mapping populations (Jansky et al. 2014; Endelman and Jansky 2016; Leisner et al. 2018; Marand et al. 2019). Recently, Solynta and Wageningen University & Research released the genome sequence of the inbred line Solyntus and Solynta released this genotype to the academic community where it is now being used in labs around the world for potato genetic research (van Lieshout et al. 2020; Freire et al. 2021; Hosaka et al. 2022).
Most results so far point into the direction of the Sli locus at the distal end of Chromosome 12. However, the exact position and the identity of the Sli gene still remain obscure. Here we report on the quest for Sli in a detailed genetic study. As a first step, we define the most accurate protocol to phenotype the self-compatibility that is associated with the Sli allele. Next, we select the most suitable parents for genetic studies. Lastly, we show that Sli inherits gametophytically, and we develop populations segregating for Sli. In this way we show the gametophytic inheritance of the trait which opened the route to the molecular cloning and characterization of the responsible gene (Sli) from the distal end of Chromosome 12.
Materials and Methods
Plant Materials
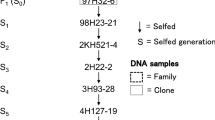
In this study, we used a total of 19 populations of diploid potato material from S. tuberosum background derived from five parental clones from the Solynta breeding germplasm (Lindhout et al. 2011; Meijer et al. 2018). All parental genotypes and populations are listed in Supplementary Tables 1 and 2, and the pedigree information is shown in Fig. 1.
Greenhouse Conditions
Potato seeds were sown in seed trays in the greenhouse, and 5-week-old seedlings were transplanted into pots. All plants were grown in greenhouses that were heated when the temperature dropped below 14 °C and cooled by opening the windows when the temperature rose above 19 °C. Artificial lighting was used to supplement the natural light when the light intensity outside dropped below 85 W/m2. Plants were grown in a special potato substrate mix from Lentse Potgrond (Lentse Potgrond B.V, Katwijk, the Netherlands). The used substrate mix is composed of a peat-mixture for balanced water uptake, basic slow-release fertilizer and lime to ensure the required pH level. The substrate mix was fertilized using a 20:20:20 nitrogen:phosphorus:potassium solution with an electrical conductivity (EC) of 1.5 which was supplied to the plants using a drip system. The amount of the solution given via the drip irrigation was adjusted to the growing stage of the plants, with small plants receiving about 60 ml per day and large plants receiving up to 720 ml per day. Depending on the greenhouse climate conditions, additional manual watering was applied when required.
Evaluation of Self-Compatibility
Male fertility was assessed by collecting pollen from multiple flowers per plant in a micro-centrifuge tube and scoring the amount of released pollen on a scale from 0 to 3 (where a score of 0 means no pollen was released, and a score of 3 means abundant pollen was released). Female fertility was assessed by bulking pollen from at least three unrelated plants and using this bulk pollen to pollinate the plants under investigation. Successful fertilization requires that pollen tubes grow through the stylar tissue until the ovules. Therefore, we analysed pollen tube growth in pollinated pistils using fluorescence microscopy.
Styles from self- and cross-pollinated flowers were detached from the ovaries 48 h after pollination, and pollen tube growth was imaged using a fluorescence microscope. Plants that set more than one self-berry containing at least 35 seeds per berry were considered self-compatible. This lower limit is based on rare observations of seeds in berries in self-incompatible controls, which were set without manual pollination. Plants were declared self-incompatible when:
-
1.
No self-berries were set after ten manual self-pollinations.
-
2.
Berries were set after crossing with bulked pollen.
-
3.
Pollen tube growth arrested before the ovaries had been reached in self-pollinated styles as observed by using fluorescence microscopy (Eggers et al. 2021).
DNA Extraction, SNP Selection and KASP Analysis
Leaf discs from population S1(P1) were sampled into 96 well plates. DNA was extracted using sbeadex™ (LGC Genomics GmbH, Berlin, Germany) by VHLGenetics (Wageningen, the Netherlands) according to the manufacturer’s protocol. To design KASP markers, P1 was sequenced with Illumina 150 nt PE with approximately 20 × coverage, reads were mapped to DM4.03 and variant calling was performed by ServiceXS (Leiden, the Netherlands). For selected SNPs on Chromosomes 1 and 12, Kompetitive allele-specific PCR (KASP™) was performed by VHLGenetics (Wageningen, the Netherlands) according to the protocol provided by the manufacturer (LGC Genomics GmbH, Berlin, Germany). The quality of the resulting KASP marker data was assessed using SNPviewer (available at lgcgroup.com/products/genotyping-software/snpviewer); markers that did not segregate or showed unexpected segregation were discarded from the analysis. An overview of the markers and their genotypes can be found in Supplementary Table 3.
Linkage Analysis and QTL Mapping
Each genotypic call was assigned to either haplotype 1 or 2 of parent P1 using the recombination rates between the markers. All the marker data was converted to ahb coding (where a means homozygous haplotype 1, h means heterozygous and b means homozygous haplotype 2), and genetic maps of Chromosomes 1, 2 and 12 of P1 were generated using Joinmap4.1 (Van Ooijen 2006) using the F2 population type.
Results
Development of an Accurate Phenotyping Protocol
In the early breeding programme of Solynta, we observed segregation for self-berry set in the first inbred populations. At that time, we assumed that this was due to segregation of the Sli locus; however, initial analyses revealed only a weak QTL on Chromosome 2 (data not shown). Later we realized that these populations may instead have segregated for fertility rather than compatibility.
Therefore, we improved the phenotyping protocol by accurately assessing the female and male fertility. Plants with little or no pollen were regarded as male sterile whereas plants that set neither self nor cross berries were regarded as female sterile. Sterile plants were excluded from the analysis of self-compatibility. Self-compatibility was defined as the ability of fertile plants to produce berries and seeds after self-pollination. Self-incompatibility was defined as the inability of fertile plants to set berries and seeds after self-pollination. Furthermore, to exclude pseudo self-compatibility from the analysis, we removed plants that set self-berries with fewer than 35 seeds per berry from analysis by classifying them as Not Determined (N.D.). While this approach allowed us to efficiently assess self-compatibility, not all plants of each population produced enough flowers to do both self and cross pollinations. For such poorly flowering plants, we visualized pollen tube growth with fluorescence microscopy of self-pollinated pistils to assign the compatibility phenotype. In our germplasm, we found that all SI plants showed pollen tube growth arrest before reaching the ovules.
Selection of Optimal Parents for Mapping the S-locus Inhibitor Gene
Initially, the materials used for the localization of the Sli locus were from the diploid breeding program of Solynta. These materials were useful to select genotypes carrying the Sli locus and were used to generate dedicated segregating populations for our quest for the Sli locus. To map the Sli gene, we needed to identify and select genotypes with high vigour, good fertility and clear compatibility phenotypes. These were used as parents for our mapping populations.
Selection of Self-Incompatible Parents: D2, D14 and D16 Are Fertile and Self-Incompatible
The identification of good SI parents was hampered by the fact that gradually the majority of the plants in the research and breeding programmes of Solynta were self-compatible. This prompted us to use the founders of the Solynta breeding germplasm as the best sources for SI genotypes (Lindhout et al. 2011). The founders D2, D14 and D16, showed particularly good vigour and male fertility. Clone D2 is a vigorous, highly fertile and strictly self-incompatible plant. Clone D14 is also vigorous, highly male fertile and strictly self-incompatible but has poor female fertility. Lastly, Clone D16 is vigorous and highly fertile, however, showed occasional spontaneous self-berries with some seeds; we therefore designated this clone as pseudo self-compatible.
We crossed D16 as a male to D2 and analysed the resulting F1 population to establish whether the pseudo self-compatibility of D16 is caused by a single dominant allele. We analysed the resulting population (F1(D2 × D16), n = 150) with the accurate phenotyping protocol. While this population consisted of 150 plants, only 51 were both male and female fertile and segregated into five SC, 21 SI and 25 plants as Not Determined (ND) as these did not unambiguously comply with the criteria to assess SC. In conclusion, the pseudo self-compatibility of D16 is different from the dominant self-compatibility allele of the Sli gene identified by Hosaka and Hanneman (Hosaka et al. 1998), it does not contain the dominant Sli allele and it can be used as an SI parent for mapping the Sli gene.
Selection of Self-Compatible Parents: P1 and P2 Are Fertile and Self-Compatible
Next, we searched for optimal SC parents. We used the experience with the F1(D2 × D16) population and aimed to generate highly fertile populations, in which the majority of individuals could be assigned unambiguous phenotypes for SC / SI. Genotypes P1 and P2 set many berries without manual pollination. Upon vibration with an electronic toothbrush, flowers of P1 and P2 released abundant pollen and set berries containing many seeds (Fig. 2). These characters made them good candidates as SC parents. Nevertheless, we preferred to use mapping parents that were highly fertile, vigorous and more homozygous than P1. Therefore, we continued inbreeding the P1 derived progenies selecting for high self-fertility. We selected the three most self-fertile genotypes of population S1(P1), cultivated three S2 populations, S2-1(P1), S2-2(P1) and S2-3(P1) and assessed self-compatibility according to our phenotyping protocol. Interestingly, self-fertility was generally higher in the S2 populations than in the S1 population as larger percentages of plants were able to set self-berries. Furthermore, only two genotypes from population S2-1(P1) and one plant from population S2-2(P1) were classified as SI, indicating that self-compatibility was already fixed in these populations (Table 1). Finally, we selected eight of the most self-fertile genotypes from S2-1(P1) and proceeded to grow eight small S3 populations (n = 32 per population). Among these populations, all flowering male and female fertile plants were unambiguously SC, indicating that by selecting the most self-fertile plants in each generation, we could obtain highly fertile Sli homozygous plants.
Selection of Optimal Populations for Mapping the S-Locus Inhibitor Gene
Self-Fertilization of P1 Results in Extreme Segregation Distortion on Chromosome 12
In the publications identifying and mapping the Sli gene, Hosaka and Hanneman concluded that the Sli gene inherits sporophytically, meaning that the diploid genotype of the pollen donor plant determines the outcome of the pollination and implying that in a Sli/sli heterozygote all pollen will be able to self-fertilize and that Sli would segregate as a normal dominant trait (Hosaka and Hanneman 1998b, a). However, we hypothesized that the Sli gene acts gametophytically and that only pollen that contains the dominant Sli allele is able to self-fertilize. Consequently, all plants derived from the self-fertilization of a Sli/sli heterozygote would have at least one copy of the dominant allele and would be genotypically SC. To study the mode of inheritance of Sli, we generated a population from self-fertilized seeds from genotype P1 (Population S1(P1), n = 222, Table 1). We genotyped 186 plants with 10 KASP markers on Chromosome 1, because of the presence of the S-locus, and with 15 KASP markers on Chromosome 12 because of the mapping of the Sli gene by Hosaka and Hanneman (Hosaka and Hanneman 1998a) (Supplementary Table 3). We observed a complete absence of one homozygous genotype class on the bottom of Chromosome 12 in the same location where the Sli gene had previously been mapped by Hosaka and Hanneman and Peterson et al. (Hosaka and Hanneman 1998a; Peterson et al. 2016), suggesting that Sli inherits gametophytically and that it will not segregate in populations derived from self-fertilization (Table 2).
F1 Populations Derived from Reciprocal Crosses of P1 and D16 Segregate for Self-Compatibility
Based on our observations during the inbreeding of P1, we hypothesized that Sli acts gametophytically and cannot be mapped in inbred populations. Thus, we set out to generate segregating F1 populations by crossing P1 and P2 to SI genotypes. We hypothesized that the high fertility and vigour of the two genotypes would be inherited in the F1 populations, allowing us to score self-compatibility in a large part of the resulting progeny. We crossed P1 with D16 and grew 32 seedlings from population F1(P1 × D16) and the reciprocal population F1(D16 × P1). Both populations clearly segregated for self-compatibility, but the phenotypical segregation among those plants of which a compatibility phenotype could be assigned was significantly distorted in both populations (χ2 = 3.841 for F1(P1 × D16)*, χ2 = 10.6667 for F1(D16 × P1)***) (Table 3).
We visualized the pollen tube growth in the styles of all SI plants of populations F1(P1 × D16) and F1(D16 × P1), all of which showed a typical incompatible interaction, where no pollen tubes were able to grow into the ovaries (Fig. 3). By eliminating confounding sterility issues and by visualization of self-pollen tube growth of SI plants, we concluded that the two F1 populations derived from P1 and D16 segregate for the Sli gene, and that we could proceed with analysis of larger populations.
Pollen tube growth in self-pollinated pistils. White arrows point to the site where the style is attached to the ovary. Most pollen tubes of self-compatible genotypes grow through this point, whereas most pollen tubes from self-incompatible genotypes do not. a. Parental genotype P1 exhibits a typical self-compatible interaction, where the majority of pollen tubes are able to grow completely through the style towards the ovaries. b. Genotype F1(D16 × P1)-27 exhibits self-compatible interaction similar to its parent P1. c. Genotype F1(P1 × D16)-21 exhibits a typical incompatible interaction, the growth of the majority of pollen tubes is arrested in the style and only a few pollen tubes reach the ovaries
F1 Populations Derived from P2, D14 and F1(P1 × D16) Segregate for Self-Compatibility
To generate an F1 population from P2, we crossed it as female to SI genotype D14. All 32 individuals of population F1(P2 × D14) were able to set self-berries with seeds, indicating that P2 is homozygous Sli/Sli (Table 3). We backcrossed the most fertile individual, F1(P2 × D14)-8 (afterwards referred to as P3) to D14 and assessed (self-)fertility in the resulting BC1 population BC1(P3 × D14) (n = 220). Unfortunately, the BC1 showed severely reduced fertility compared to the F1. Merely 173 plants flowered, and from those, the self-compatibility status of only 35 individuals could be determined based on berry set (Table 3). We decided not to proceed with further analysis of population BC1(P3 × D14), but instead chose to generate two new populations by crossing P3 to two of the most fertile SI individuals from population F1(P1 × D16), F1(P1 × D16)-27 and F1(P1 × D16)-21, referred to as P4 and P5, respectively (Fig. 1). The resulting F1 populations F1(P3 × P4) and F1(P3 × P5) were generally more fertile and segregated for self-compatibility. The genetic analysis of more individuals from population F1(P1 × D16), and of populations F1(P3 × P4) and F1(P3 × P5) allowed us to narrow down the location of the Sli gene on Chromosome 12, confirming the original map position of the Sli locus but at a smaller interval of only 169 kB (Fig. 1 and Supplementary data 1 of Eggers et al. (Eggers et al. 2021)). This has paved the way to further research on narrowing down the mapping and eventually cloning the Sli gene (Eggers et al. 2021).
Discussion
Phenotyping Protocol to Distinguish between Fertility and Compatibility
Here, we show the accurate mapping of the Sli gene in diploid potato, whereby accurate phenotyping methods and segregating populations are crucial. In order to map the Sli gene, it was critical to discriminate between general fertility effects and self-compatibility. While studying self-incompatibility in inbreeding progenies of the DM × RH population, Peterson et al. observed the same phenomenon and made a distinction between self-compatibility and self-fertility (Peterson et al. 2016). We measured male fertility by vibrating flowers with an electronic toothbrush and quantifying the released pollen. Generally, obtaining a visible amount of pollen indicates good male fertility, as in our material we did not observe plants that produced visible amounts of dead pollen due to for example tetrad sterility (Sanetomo and Nashiki 2021; Sanetomo et al. 2022). Determination of female fertility is somewhat more laborious. If a plant sets self-berries and seeds after manual pollination with collected pollen, this reliably indicates female fertility. However, for plants that are SI, cross pollinations using pollen bulked from at least three unrelated genotypes must be used to increase the chance that the bulk contains pollen with different S-alleles. One important consequence of this is that it is easier to score a plant as SC than it is to score one as SI, leading to an underestimation of the number of SI plants in a segregating population. This underestimation of SI plants can be alleviated by visualization of self-pollen tube growth using fluorescence microcopy. With this visualization, male fertility and self-compatibility can be evaluated at the same time, but it requires the removal of the styles from the flowers 48 h after self-pollination, which may affect berry set due to mechanical stress on the pistil.
Well-Defined Parents Are a Prerequisite for Genetic Mapping
For all mapping studies in bi-parental populations, selection of suitable contrasting parents is critical. In the mapping of the Sli gene, parent selection is complicated by its gametophytic inheritance and the required fertility in the segregating populations. By definition, all individuals of a selfed progeny of a genotype carrying at least one copy of the Sli allele will inherit this Sli allele. Thus, selfed progenies are not useful for classical genetic mapping of the Sli locus, although it is possible to use the segregation distortion itself as a signal (Table 2), as was done by Ma et al. (Ma et al. 2021).
Nonetheless, our aim was to identify suitable contrasting parents to generate populations segregating for the Sli locus. Fertility is a crucial factor in SI parent selection, and populations whereby the majority of individual plants were highly fertile were rarely identified. Still, we identified good female fertility in population F1(P2 × D14), as all 32 individuals, including individual P3, readily set self-berries.
We observed poor fertility in BC1(P3 × D14), possibly due to inbreeding depression or specific recessive sterility factors in the genome of D14, and as such, D14 is not a perfect SI parent. In contrast, D16 is a sub-optimal parent not because of sterility but because its pseudo self-compatibility, although ultimately this did not interfere with the genetic analysis as Sli provides much stronger self-compatibility in terms of self-seed and berry set.
For the selection of SC parents, high fertility was also crucial. We selected two parents, P1 and P2 that showed remarkable self-berry set even without manual pollination. However, because Sli inherits gametophytically, it is essential that the SC parent is heterozygous for Sli. The only way to determine whether our SC parents were heterozygous was to make the crosses with the SI parents and observe the compatibility phenotypes in the resulting populations. Indeed, for P1, we found that its F1 populations (F1(P1 × D16) and F1(D16 × P1)) segregate for self-compatibility. In contrast, for P2, we found that all individuals of population F1(P2 × D14) were able to set self-berries, requiring that we cross one of the progenies to other SI genotypes.
Dedicated Mapping Populations
The genotypic analysis of population S1(P1) revealed the extreme segregation distortion on Chromosome 12, supporting the hypothesis that Sli inherits gametophytically. However, several studies have reported the presence of lethal alleles on Chromosome 12 (Endelman et al. 2019; Zhang et al. 2019; Hosaka and Sanetomo 2020; Kaiser et al. 2021), providing an alternative explanation for the observed segregation distortion. In the founder that we used to introduce Sli into our breeding program (DS (Lindhout et al. 2011)), linkage between Sli and a lethal allele was already broken (Meijer et al. 2018). Furthermore, in another publication, we show that, in our material, segregation distortion on Chromosome 12 is indeed caused by Sli and that homozygosity for Sli in itself is not lethal (Eggers et al. 2021).
The Quest for Sli: Controlling for Confounding Genetic Effects and Accounting for its Gametophytic Inheritance
Sli based self-compatibility is a simple monogenic trait. However, its functional expression may be obscured by several other factors, mainly related to fertility: the plants must set flowers and be male and female fertile and should preferably be reasonably vigorous. These other factors are largely determined by the genetic background of the plants, although environmental effects may have influence plant growth and fertility as well. It is the art of the geneticist to identify the effect of the desired gene among dozens of epistatic effects of other genes often each of which may only have small or moderate effects on the phenotype under investigation.
One of the main challenges of marker assisted introgression breeding is to determine the phenotypic effects of seemingly monogenic traits in multiple genetic backgrounds. Genes for which their functional expression have been clearly established in specific genetic backgrounds may not produce the same phenotypic effects when introgressed into different genetic backgrounds (Koornneef et al. 1994; Ray et al. 1995; Krüger et al. 2002; Sun et al. 2004; Hurni et al. 2014). Our quest for Sli was ultimately successful because we used an improved phenotyping protocol to screen parental genotypes and experimental populations for true segregation of self-compatibility instead of self-fertility by avoiding confounding fertility effects. Furthermore, the realization that Sli inherits gametophytically refocused our research efforts on developing segregating F1 populations. We developed three good mapping populations derived from two SC and three SI parents. With these populations we mapped the gene to a 169 kB interval; we then used a recombinant screening approach to narrow down the interval even further and finally identified the gene by using CRISPR-Cas9 and transgenic complementation (Eggers et al. 2021). In conclusion, this study is a clear example of how the effect of a single gene can be revealed among confounding epistatic effects of other genes by the development of an accurate phenotyping protocol, the analysis of segregation distortion and the development of dedicated segregating populations. This approach has been critical in the subsequent cloning of the Sli gene (Eggers et al. 2021) .
Data Availability
All relevant data are in the main figures and tables and in the supplementaries.
References
Bamberg J, Kielar A, del Rio A, Douches D (2021) Making hybrids with the wild potato Solanum jamesii. Am J Potato Res 98:187–193. https://doi.org/10.1007/S12230-021-09828-1/FIGURES/2
Bethke PC, Jansky SH (2021) Genetic and environmental factors contributing to reproductive success and failure in potato. Am J Potato Res 98:24–41. https://doi.org/10.1007/S12230-020-09810-3/TABLES/7
Braun SR, Endelman JB, Haynes KG, Jansky SH (2017) Quantitative trait loci for resistance to common scab and cold‐induced sweetening in diploid potato. Plant Genome 10. https://doi.org/10.3835/plantgenome2016.10.0110
Clot CR, Polzer C, Prodhomme C et al (2020) The origin and widespread occurrence of Sli-based self-compatibility in potato. Theor Appl Genet. https://doi.org/10.1007/s00122-020-03627-8
De Jong H, Rowe PR (1971) Inbreeding in cultivated diploid potatoes. Potato Res 14:74–83. https://doi.org/10.1007/BF02355931
Eggers EJ, van der Burgt A, van Heusden SAW et al (2021) Neofunctionalisation of the Sli gene leads to self-compatibility and facilitates precision breeding in potato. Nat Commun 12:1–9. https://doi.org/10.1038/s41467-021-24267-6
Endelman JB, Jansky SH (2016) Genetic mapping with an inbred line - derived F2 population in potato. Theor Appl Genet 129:935–943. https://doi.org/10.1007/s00122-016-2673-7
Endelman J, Jansky SH, Butler N, Christensen G (2019) Genetic evidence of a recessive lethal allele on potato chromosome 12-annual report of the Potato Association of America. Am J Potato Res 96:331
Freire R, Weisweiler M, Guerreiro R et al (2021) Chromosome-scale reference genome assembly of a diploid potato clone derived from an elite variety. G3 Genes|Genomes|Genetics 11. https://doi.org/10.1093/G3JOURNAL/JKAB330
Hosaka K, Hanneman RE Jr (1998a) Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 2. Localization of an S locus inhibitor (Sli) gene on the potato genome using DNA markers. Euphytica 103:265–271. https://doi.org/10.1023/a:1018353613431
Hosaka K, Hanneman RE Jr (1998b) Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 1. Detection of an S locus inhibitor (Sli) gene. Euphytica 99:191–197. https://doi.org/10.1023/a:1018353613431
Hosaka K, Sanetomo R (2020) Creation of a highly homozygous diploid potato using the S locus inhibitor (Sli) gene. Euphytica 216:1–16. https://doi.org/10.1007/s10681-020-02699-3
Hosaka K, Hanneman REE, Drive L, Swaminathan SI (1998) Genetics of self-compatibility in a self-incompatible wild diploid potato species Solanum chacoense. 1. Detection of an S locus inhibitor (Sli) gene. Euphytica 99:191–197. https://doi.org/10.1023/a:1018353613431
Hosaka AJ, Sanetomo R, Hosaka K (2022) A de novo genome assembly of Solanum verrucosum Schlechtendal, a Mexican diploid species geographically isolated from other diploid A-genome species of potato relatives. G3 Genes|Genomes|Genetics 12. https://doi.org/10.1093/G3JOURNAL/JKAC166
Hurni S, Brunner S, Stirnweis D et al (2014) The powdery mildew resistance gene Pm8 derived from rye is suppressed by its wheat ortholog Pm3. Plant J 79:904–913. https://doi.org/10.1111/TPJ.12593
Jansky S (2011) Parental effects on the performance of cultivated × wild species hybrids in potato. Euphytica 178:273–281. https://doi.org/10.1007/S10681-010-0323-8/TABLES/4
Jansky SH, Chung YS, Kittipadukal P (2014) M6: a diploid potato inbred line for use in breeding and genetics research. J Plant Regist 8:195–199. https://doi.org/10.3198/jpr2013.05.0024crg
Jansky SH, Charkowski AO, Douches DS et al (2016) Reinventing potato as a diploid inbred line-based crop. Crop Sci 56:1412–1422. https://doi.org/10.2135/cropsci2015.12.0740
Kaiser NR, Jansky S, Coombs JJ et al (2021) Assessing the contribution of Sli to self-compatibility in North American diploid potato germplasm using KASP™ markers. Am J Potato Res 98:104–113. https://doi.org/10.1007/S12230-021-09821-8/METRICS
Koornneef M, Vries HB, Hanhart C et al (1994) The phenotype of some late-flowering mutants is enhanced by a locus on chromosome 5 that is not effective in the Landsberg erecta wild-type. Plant J 6:911–919. https://doi.org/10.1046/J.1365-313X.1994.6060911.X
Krüger J, Thomas CM, Golstein C et al (2002) A tomato cysteine protease required for Cf-2-dependent disease resistance and suppression of autonecrosis. Science (1979) 296:744–747. https://doi.org/10.1126/SCIENCE.1069288/SUPPL_FILE/1069288S2_THUMB.GIF
Kubo K, Entani T, Takara A et al (2010) Collaborative non-self recognition system in S-RNase–based self-incompatibility. Science (1979) 330:796–799
Leisner CP, Hamilton JP, Crisovan E et al (2018) Genome sequence of M6, a diploid inbred clone of the high-glycoalkaloid-producing tuber-bearing potato species Solanum chacoense, reveals residual heterozygosity. Plant J 1967:562–570. https://doi.org/10.1111/tpj.13857
Lindhout P, Meijer D, Schotte T et al (2011) Towards F 1 hybrid seed potato breeding. Potato Res 54:301–312. https://doi.org/10.1007/s11540-011-9196-z
Ma L, Zhang C, Zhang B et al (2021) A nonS-locus F-box gene breaks self-incompatibility in diploid potatoes. Nat Commun 12:1–8. https://doi.org/10.1038/s41467-021-24266-7
Marand AP, Jansky SH, Zhao H et al (2017) Meiotic crossovers are associated with open chromatin and enriched with Stowaway transposons in potato. Genome Biol 18:1–16. https://doi.org/10.1186/S13059-017-1326-8/FIGURES/6
Marand AP, Jansky SH, Gage JL et al (2019) Residual heterozygosity and epistatic interactions underlie the complex genetic architecture of yield in diploid potato. Genetics 212:317–332. https://doi.org/10.1534/genetics.119.302036
McClure B, Cruz-Garcia F, Romero C et al (2011) Compatibility and incompatibility in S-RNase-based systems. Ann Bot 108:647–658. https://doi.org/10.1093/aob/mcr179
Meijer D, Viquez-Zamora M, van Eck HJ et al (2018) QTL mapping in diploid potato by using selfed progenies of the cross S. tuberosum × S. chacoense. Euphytica 214:121. https://doi.org/10.1007/s10681-018-2191-6
Olsder J, Hermsen JGT (1976) Genetics of Self-Compatibility in dihaploids of Solanum tuberosum L. 1. Breeding behaviour of two self-compatible dihaploids. Euphytica 25:597–607
Peloquin SJ, Hougas RW (1960) Genetic variation among haploids of the common potato. Am Potato J 37:289–297. https://doi.org/10.1007/BF02855072
Peterson BA, Holt SH, Laimbeer FPE et al (2016) Self-fertility in a cultivated diploid potato population examined with the Infinium 8303 potato single-nucleotide polymorphism array. Plant Genome. https://doi.org/10.3835/plantgenome2016.01.0003
Phumichai C, Mori M, Kobayashi A et al (2005) Toward the development of highly homozygous diploid potato lines using the self-compatibility controlling Sli gene. Genome 48:977–984. https://doi.org/10.1139/g05-066
Plantenga FDM, Bergonzi S, Abelenda JA et al (2019) The tuberization signal StSP6A represses flower bud development in potato. J Exp Bot 70:937–948. https://doi.org/10.1093/JXB/ERY420
Ray JD, Hinson K, Mankono JE, Malo MF (1995) Genetic control of a long-juvenile trait in soybean. Crop Sci 35:1001–1006. https://doi.org/10.2135/CROPSCI1995.0011183X003500040012X
Sanetomo R, Nashiki A (2021) Identification of the tetrad-sterility-causing Solanum stoloniferum Schltdl. & Bouché cytoplasm in interspecific hybrids with S. tuberosum L. Genet Resour Crop Evol 68:3383–3397. https://doi.org/10.1007/s10722-021-01197-2
Sanetomo R, Akai K, Nashiki A (2022) Discovery of a novel mitochondrial DNA molecule associated with tetrad pollen sterility in potato. BMC Plant Biol 22:1–16. https://doi.org/10.1186/s12870-022-03669-8
Seibert T, Abel C, Wahl V (2020) Flowering time and the identification of floral marker genes in Solanum tuberosum ssp. andigena. J Exp Bot 71:986–996. https://doi.org/10.1093/JXB/ERZ484
Song L, Endelman JB (2023) Using haplotype and QTL analysis to fix favorable alleles in diploid potato breeding. Plant Genome 16:e20339. https://doi.org/10.1002/TPG2.20339
Sun X, Cao Y, Yang Z et al (2004) Xa26, a gene conferring resistance to Xanthomonas oryzae pv. oryzae in rice, encodes an LRR receptor kinase-like protein. Plant J 37:517–527. https://doi.org/10.1046/J.1365-313X.2003.01976.X
van Lieshout N, van der Burgt A, de Vries ME et al (2020) Solyntus, the new highly contiguous reference genome for potato (Solanum tuberosum). G3: Genes Genomes, Genetics 10:3489–3495. https://doi.org/10.1534/g3.120.401550
Van Ooijen JW (2006) JoinMap®4, Software for the calculation of genetic linkage maps in experimental populations. Kyazma BV, Wageningen 33. https://doi.org/10.1111/j.1365-313X.1993.00739.x
Xu X, Pan S, Cheng S et al (2011) Genome sequence and analysis of the tuber crop potato. Nature 475:189–195. https://doi.org/10.1038/nature10158
Zhang C, Wang P, Tang D et al (2019) The genetic basis of inbreeding depression in potato. Nat Genet 51:374–378. https://doi.org/10.1038/s41588-018-0319-1
Zhou Q, Tang D, Huang W et al (2020) Haplotype-resolved genome analyses of a heterozygous diploid potato. Nat Genet 52:1018–1023. https://doi.org/10.1038/s41588-020-0699-x
Acknowledgements
We acknowledge Eyup Oztutuncu, Marc Schreurs, Eveline Heynen, Kayleigh van Til, Lisette van Noord, Siti Rosidah and Megiel van Sloten for their help with phenotyping of the populations. We further acknowledge Jarno Sinnige and the Solynta Ressen greenhouse team for their help with sowing, transplanting, plant maintenance and seed extraction of all populations in this research.
Funding
This project received partial financial support from the Dutch Research Council (NWO, Grant ID: NWA.17.023).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing Interests
Authors EJE, YS, SAWvH, MEdV and PL were affiliated with the company Solynta during this research.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Co-author Sjaak A. W. van Heusden has sadly passed away before publication of this article.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eggers, EJ., Su, Y., van Heusden, S.A.W. et al. The Quest for the Sli Locus. Potato Res. (2024). https://doi.org/10.1007/s11540-024-09792-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11540-024-09792-3