Abstract
Patients with EGFR-mutated non-small cell lung cancer (NSCLC) benefit from treatment with tyrosine kinase inhibitors (TKI) targeting EGFR. Despite improvements in patient care, especially with the 3rd generation TKI osimertinib, disease relapse is observed in all patients. Among the various processes involved in TKI resistance, epithelial-to-mesenchymal transition (EMT) is far from being fully characterized. We hypothesized that the cellular prion protein PrPC could be involved in EMT and EGFR-TKI resistance in NSCLC. Using 5 independent lung adenocarcinoma datasets, including our own cohort, we document that the expression of the PRNP gene encoding PrPC is associated with EMT. By manipulating the levels of PrPC in different EGFR-mutated NSCLC cell lines, we firmly establish that the expression of PrPC is mandatory for cells to maintain or acquire a mesenchymal phenotype. Mechanistically, we show that PrPC operates through an ILK-RBPJ cascade, which also controls the expression of EGFR. Our data further demonstrate that PrPC levels are elevated in EGFR-mutated versus wild-type tumours or upon EGFR activation in vitro. In addition, we provide evidence that PRNP levels increase with TKI resistance and that reducing PRNP expression sensitizes cells to osimertinib. Finally, we found that plasma PrPC levels are increased in EGFR-mutated NSCLC patients from 2 independent cohorts and that their longitudinal evolution mirrors that of disease. Altogether, these findings define PrPC as a candidate driver of EMT-dependent resistance to EGFR-TKI in NSCLC. They further suggest that monitoring plasma PrPC levels may represent a valuable non-invasive strategy for patient follow-up and warrant considering PrPC-targeted therapies for EGFR-mutated NSCLC patients with TKI failure.

Similar content being viewed by others
Introduction
Lung cancer remains the deadliest cancer worldwide with 1.8 million deaths in 2020, representing 18% of the overall cancer-related deaths [1]. Because the majority of cases are detected at late stages, either non-resectable or metastatic diseases at diagnosis, the 5-year survival rate for non-small cell lung cancer (NSCLC), the most common subtype, remains beyond 25% and drops below 5% for stage 4 disease (https://seer.cancer.gov/statfacts/html/lungb.html). Advances in the molecular characterization of NSCLC tumours have led to the identification of targetable genomic alterations, among which EGFR mutations are the most frequently encountered, occurring in 15–25% of cases in Caucasians, and up to 40 to 55% cases in East Asians [2]. Despite the advent of tyrosine kinase inhibitors (TKI) targeting mutant EGFR, including the third generation TKI osimertinib, all patients bearing EGFR mutations eventually develop resistance, typically after 9–12 months of treatment [3, 4]. Resistance mechanisms include secondary genomic alterations, activation of bypass signalling pathways, epigenetic changes or histological transformation [3, 4]. Among the latter notably features epithelial-to-mesenchymal transition (EMT), initially identified on re-biopsy in relapsed EGFR NSCLC [5, 6], validated in vitro [7], and more recently found as a hallmark of drug tolerant states in single-cell experiments [8]. Accordingly, several preclinical studies have shown that cells undergoing EMT acquire resistance to EGFR-TKIs (reviewed in [9, 10], while others have demonstrated that counteracting EMT may allow to overcome resistance to anti-EGFR therapies in NSCLC [7, 11,12,13]. However, the signals and effectors that orchestrate EMT in NSCLC remain imperfectly understood [14].
Over the last years, the cellular prion protein PrPC has emerged as an important contributor to EMT in various types of cancers [15]. For instance, we documented that the expression of the PrPC-encoding gene PRNP is specifically enriched in the mesenchymal subtype of colon cancer and that it controls the expression of the master EMT transcription factor ZEB1 [16]. Furthermore, several studies have reported on the pro-migratory and pro-invasive properties of PrPC in cancer cells [15] for review. In lung cancer, Lin et al. have shown that PrPC is more abundantly expressed in invasive versus non-invasive cell lines and enhances lamellipodium formation [17]. However, our understanding of the interplay between PrPC and EMT in lung cancer and more globally the contribution of PrPC to hallmarks of lung cancer cells is far from complete.
Here, our goal was to investigate the role played by PrPC in lung adenocarcinoma (LUAD). We performed a comprehensive analysis aimed at identifying transcriptomic and proteomic signatures linked to PRNP expression and EMT in LUAD. We employed cell-based assays to assess the contribution of PRNP to the regulation of the EMT process and study the interplay between PrPC and EGFR. Finally, we combined in silico analyses, in vitro assays and plasma measurements of circulating PrPC to evaluate the contribution of PrPC to TKI resistance in EGFR-mutated LUAD. Altogether, this work delineates the role of the PrPC as a driver of mesenchymal transition and associated resistance to EGFR-TKI. It suggests that PRNP or its related activated pathways could be potential biomarkers or therapeutic targets to overcome resistance in LUAD.
Results
The cellular prion protein correlates with EMT in NSCLC
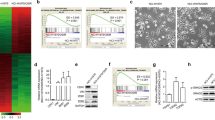
As a first step to assess whether the expression of PRNP may be associated with EMT, we performed Gene set Enrichment Analysis (GSEA) in different cell and patient datasets of lung adenocarcinoma (LUAD). These include the LUAD cell lines (n = 45) from the Cancer Cell Line Encyclopedia (CCLE) [18], LUAD from the Cancer Genome Atlas (TCGA) (n = 511), LUAD from the Onco-HEGP cohort (n = 107) [19], the tumours from the proteogenomic study by Chen et al. (n = 90) [20], and the LUAD tumours of the proteogenomic study by Lehtiö et al. (n = 90) [21]. Remarkably, we found that PRNP mRNA expression was consistently associated with EMT in all five studies (Fig. 1A, B and Supplementary Fig. S1A). In accordance, the levels of PRNP transcripts were highly correlated with those of EMT transcription factors (TF), most notably SNAI2, ZEB1 and ZEB2, across the various datasets (Fig. 1C and Supplementary Fig. S1B). Likewise, we found very significant correlations between PrPC and SLUG, encoded by the SNAI2 gene, ZEB1 or ZEB2 at protein levels using studies by Chen et al. [20] and Lehtiö et al. [21] (Supplementary Fig. S1C). Furthermore, in all patient cohorts, PRNP expression was significantly anti-correlated with the epithelial (EMT_epi) score and positively correlated with the mesenchymal (EMT_mes) score, and thereby with the patient-derived pan-cancer EMT (EMT_score) score designed by Mak et al. [22] (Fig. 1D and Supplementary Fig. S1D) as well as with a mesenchymal score (mes_score) derived from the study by de Reyniès et al. [23] (Fig. 1D and Supplementary Fig. S1E). Finally, in the Onco-HEGP cohort, PRNP mRNA levels were higher in mixed or mesenchymal tumours, as compared with epithelial tumours (Supplementary Fig. S1F). They were also higher in the group of tumours with a low miR-200 signature as compared to those with a high miR-200 signature (Supplementary Fig. S1G), the former being associated with a poor outcome [24]. Altogether, these in silico data provide compelling evidence for a link between PRNP gene expression and EMT in lung adenocarcinoma.
GSEA analysis showing enrichment of the EMT signature in the genes most correlated to PRNP in cell lines of the CCLE LUAD (A) and in patients from the Onco-HEGP LUAD cohort (B). C Heatmap summarizing the correlation indexes between the expression of PRNP and that of EMT TF in multiple datasets. D Heatmap summarizing the correlation indexes between the expression of PRNP and that of the pan-cancer epithelial (EMT-epi), mesenchymal (Epi-mes) and EMT scores from [22] and the mesenchymal score derived from [23] in multiple datasets.
Loss of PrPC hinders EMT in NSCLC
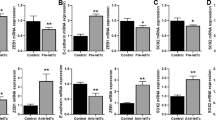
We next employed cell-based assays to probe a functional relationship between PrPC and EMT. We deliberately selected EGFR-mutant LUAD cell lines with the view to study the relationship between PrPC expression, EMT and resistance to EGFR-TKIs. H1975, HCC827 and H1650 cell lines display EGFR genetic alterations detailed in Supplementary Fig. S2A. In the H1975 cell line that expresses high amounts of PrPC (Supplementary Fig. S2B,C) and possesses mesenchymal features in its basal state, siRNA-mediated silencing of PrPC caused a marked reduction in the expression of the EMT TF SNAI2, TWIST1, ZEB1 and ZEB2 (Fig. 2A), which we confirmed at the protein level for ZEB1 (Fig. 2B) and SLUG, encoded by the SNAI2 gene (Fig. 2C). Transcriptomic analyses followed by GSEA further indicated that PrPC silencing in H1975 cells mitigates their EMT signature (Fig. 2D). Corroborating these overall findings, PrPC-silenced H1975 cells were found to exhibit a more flattened morphology as compared to control conditions (Fig. 2E). In addition, cell counting with the CASY TT instrument indicated that the knockdown of PrPC in H1975 cells was associated swith a decrease in cell number (Fig. 2F), while cell volume was increased (Fig. 2G), in full agreement with the switch in cell shape observed in Fig. 2E. We then performed functional assays through real-time monitoring of cell migration and invasion using the xCELLigence technology. SiPRNP-pre-treated H1975 exhibited drastically reduced migratory and invasive capacities, as shown in Fig. 2H and I, respectively.
A qRT-PCR analysis of the expression of the EMT TF SNAI1, SNAI2, TWIST, ZEB1, ZEB2 in PRNP-silenced versus control H1975 cells. Western blot analysis of the expression of ZEB1 (B) and SLUG (C) in PRNP-silenced versus control H1975 cells. D GSEA analysis highlighting the EMT signature as one of the most affected pathway in H1975 cells in response to PRNP silencing. E Immunofluorescence images showing PrPC and F-actin staining in PRNP-silenced versus control H1975 cells. Analysis with the CASY cell counter revealing a decrease in proliferation (F) and an increase in cell volume (G) in PRNP-silenced versus control H1975 cells. Cell index measurements of PRNP-silenced versus control H1975 cells in xCELLigence migration (H) or invasion (I) assay. qRT-PCR analysis of the expression of the EMT TF SNAI1, SNAI2, ZEB1, ZEB2 (J) and EMT genes VIM, CDH1 and CDH2 (K) in HCC827 cells exposed to siRNA against PRNP and recombinant TGFβ1. L Western blot analysis of the expression of SNAIL, ZEB1, ZEB2, Vimentin, E-cadherin and N-cadherin in HCC827 cells exposed to siRNA against PRNP and recombinant TGFβ1. Results are expressed as means ± s.e.m of n = 2 independent triplicates (A–C, J) or n = 1 triplicate (F–I, L) of cell preparations. *p < 0.05, **p < 0.01 ***p < 0.001 vs. control (siScramble), #p < 0.05, ###p < 0.001 vs. TGFβ1 treated, siScramble cells, §p < 0.05 vs. TGFβ1 untreated, PRNP-silenced cells. Protein levels in western blots were normalized to α-tubulin (α-tub) with quantifications summarized in Supplementary Fig. S2F.
Having shown that PrPC expression in the mesenchymal H1975 cell line is required for the maintenance of mesenchymal features, we next investigated whether PrPC is necessary for cells to acquire mesenchymal hallmarks in response to growth factors. To this aim, we first turned to the HCC827 cell line that is a commonly used model of growth factor-induced EMT – more precisely in response to TGFβ. Using the dataset from Sun et al. (GSE49644) [25], we first observed that PRNP and EMT TF were highly induced in HCC827 cells having acquired a mesenchymal state upon long-term exposure to TGFβ (Supplementary Fig. S3). In our experiment, following shorter-term exposure to TGFβ (10 ng/ml, 24 h), HCC827 cells were found to upregulate among EMT markers, SNAI1, ZEB1 and ZEB2, VIM and CDH2 (Fig. 2J, K). Of note, when cells were depleted of PrPC, TGFβ completely failed to induce the expression of EMT TF (Fig. 2J), and mitigated the upregulation of VIM and CDH2 (Fig. 2K) PrPC-silenced cells also exhibited higher levels of E-cadherin mRNA (CDH1) and protein (Fig. 2K, L). Western blot analyses further confirmed that the TGFβ-induced increases in SNAIL, ZEB1, ZEB2, Vimentin and N-cadherin protein levels were abrogated upon PrPC silencing (Fig. 2L and Supplementary Fig. S2F). Immunofluorescence staining with antibodies against Vimentin, E-cadherin and N-cadherin in TGFβ-treated HCC827 cells that were PrPC-silenced versus control cells corroborated these findings (Supplementary Fig. S2G).
We may note that transient TGFβ treatment had no impact on mRNA or protein levels of PrPC (Supplementary Fig. S2H, I), suggesting that the increase in PRNP mRNA observed in the Sun study is a late TGFβ-dependent event. This observation also suggests that an increase in PrPC expression is not mandatory for HCC827 to switch on EMT features in response to TGFβ but that the basal expression of PrPC, although much lower as compared to the very abundant level found in H1975 cells (Supplementary Fig. S2B, C), endows cells with a permissive TGFβ-responsive state. In addition, RNAseq followed by GSEA performed on control and PrPC-depleted HCC827, either untreated or exposed to TGFβ, revealed an activation of the TGFβ pathway after exposure to TGFβ in both conditions (Supplementary Fig. S2J). We may thus consider that PrPC silencing does not directly alters the TGFβ signalling pathway itself but rather affects a cooperative pathway. This hypothesis is further supported by the TGFβ-mediated increase in TGFBI transcripts in PrPC-depleted HCC827 exposed to TGFβ, albeit to lower levels than in control cells (Supplementary Fig. S2K). This observation contrasts with the failure of TGFβ treatment to induce EMT TF in PrPC-silenced HCC827 cells (Fig. 2J). These findings collectively indicate that PrPC expression is mandatory for lung cancer cells to maintain (H1975) or undergo (HCC827) EMT, in line with in silico observations displayed in Fig. 1.
PrPC controls the NOTCH pathway in NSCLC via ILK
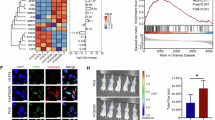
The above results demonstrate that the expression of PrPC in NSCLC cells is required to switch on the expression of EMT TF in response to TGFβ, and suggest that PrPC sustains the cooperative crosstalk of the TGFβ pathway with another EMT-promoting signalling cascade. Based on several observations, we suspected the action of PrPC to involve the NOTCH pathway. First, we have previously documented a control of PrPC on NOTCH signalling, both in cellular models and in mouse embryos [26], and Wang et al. have reported a similar link in pancreatic cancer cells [27]. Second, the involvement of the NOTCH pathway in the acquisition and/or maintenance of EMT features in NSCLC is well established [10]. NOTCH and TGFβ signalling cascades are known to cooperate to coordinate EMT [28] and, accordingly, we found that silencing of the ligand JAG1 mitigated the EMT-promoting effect of TGFβ in HCC827 cells (Supplementary Fig. S4). Third, our RNAseq experiments followed by GSEA indicated that the NOTCH pathway is downregulated upon PRNP silencing in H1975 cells (Fig. 3A). Fourth, in mixed or mesenchymal tumours from the Onco-HEGP cohort, we found significantly higher expression of JAG1, the receptor NOTCH2, as well as the effector RBPJ [29], as compared with epithelial tumours (Supplementary Fig. S5A–C). Moreover, expression levels between these three genes and PRNP were correlated (Supplementary Fig. S5D–F). Similar findings were obtained with the Chen and Lehtiö datasets (Supplementary Fig. S5G–R), with correlations exceeding 0.50 between PrPC and NOTCH2 and RBPJ protein levels in the Chen study (Supplementary Fig. S5K, L).
A GSEA analysis showing that the NOTCH signalling pathway is affected in response to PRNP silencing in H1975 cells. B qRT-PCR analysis of the expression of JAG1, JAG2, NOTCH1, NOTCH2 and RBPJ in PRNP-silenced versus control H1975 cells. Western blot analysis of the expression of JAGGED1 (C) and RBPJ (D) in PRNP-silenced versus control H1975 cells. qRT-PCR analysis of the expression of JAG1, JAG2, NOTCH1, NOTCH2 and RBPJ (E) and Western blot analysis of the expression of JAGGED1 (F) and RBPJ (G) in HCC827 cells exposed to siRNA against PRNP and recombinant TGFβ1. H Venn diagram highlighting ILK as a common protein most correlated with PrPC levels in the Chen and Lehtiö proteogenomic studies and a PrPC partner in melanoma. Scatter plots illustrating the correlation between ILK and PrPC levels in the Lehtiö proteogenomic study I. Western blot analysis of the expression of ILK in HCC827 cells exposed to siRNA against PRNP and recombinant TGFβ1 (J). qRT-PCR analysis of the expression of SNAI1, ZEB1 and ZEB2 EMT TF (K) and JAG1, JAG2, NOTCH1, NOTCH2 and RBPJ (L) in QLT0267-treated versus control H1975 cells. qRT-PCR (M) and Western blot (N) analysis of the expression of RBPJ in QLT0267-treated versus control MDST8 cells. O Schematic diagram illustrating the proposed regulation of EMT TF downstream from PrPC. Results from qRT-PCR and western blots are expressed as means ± s.e.m of n = 2 independent triplicates of cell preparations. *p < 0.05, **p < 0.01, ***p < 0.001 vs. control (siScramble or vehicle), #p < 0.05, ###p < 0.001 vs. TGFβ1 treated, siScramble cells, §p < 0.05, §§p < 0.01 vs. TGFβ1 untreated, PRNP-silenced cells. Protein levels in western blots were normalized to α-tubulin (α-tub).
As a validation of these observations, we showed that PrPC-silenced H1975 cells expressed significantly lower NOTCH ligands JAG1 and JAG2, receptors NOTCH1 and NOTCH2, and effector RBPJ mRNA levels as compared to control H1975 cells (Fig. 3B). This was confirmed at protein level for JAG1 (Fig. 3C) and RBPJ (Fig. 3D). This PrPC-dependent control over NOTCH pathway effectors in mesenchymal cancer cells seems to be a generic feature. Indeed, the above observations were recapitulated in PC3 prostate cancer cells after PrPC depletion, which caused both a reversal of EMT and a reduction in the expression of Notch pathway effectors (Supplementary Fig. S6A-I). Furthermore, in HCC827 cells, basal mRNA and protein levels of JAG1 were reduced in PrPC-silenced cells (Fig. 3E, F). As already shown by others [30], TGFβ induced robust increases in JAG1 mRNA and protein levels (Fig. 3E, F), and most importantly, these upregulations were much milder in PrPC-depleted cells (Fig. 3E, F). In the study by Sethi et al. [30], the induction of JAG1 by TGFβ was shown to be relayed by the canonical SMAD pathway. If the binding of the TGFβ-dependent SMAD effector to the JAG1 promoter was completely dependent on PrPC expression, then we would have expected a complete absence of response of JAG1 to TGFβ in PrPC-depleted, which was not the case. We may instead hypothesize that PrPC induces a second signalling cascade that cooperates with TGFβ to upregulate JAG1 expression. Among the transcriptional regulators known to bind the JAG1 promoter features RPBJ itself [31]. As for H1975 lung (Fig. 3D) and PC3 prostate (Supplementary Fig. S6I) cancer cells, RBPJ mRNA and protein levels were strongly reduced upon PrPC silencing in HCC827 cells, both at basal levels and after TGFβ treatment (Fig. 3E, G). Thus, the PrPC-dependent control on RBPJ expression may possibly account for the subsequent control on JAG1. This observation now raises the question as to the mechanisms through which PrPC controls RBPJ. Focusing on proximal effectors of PrPC, we hypothesized a potential contribution of the Integrin Linked Kinase ILK since we recently reported that ILK relays the action of PrPC in mesenchymal colon cancer cells [32]. In addition, ILK has recently been found in the interactome of PrPC in two melanoma cell lines [33]. Directly supporting our hypothesis, we found that PrPC and ILK protein levels were highly correlated in two lung cancer proteogenomic datasets (Fig. 3H, I and Supplementary Fig. S5S). We further observed that ILK protein levels were reduced in PrPC-silenced HCC827 cells and failed to increase in response to TGFβ (Fig. 3J). Then, by exposing H1975 cells to QLT0267, a specific ILK inhibitor [34], we assessed whether ILK may regulate the expression of EMT TF and NOTCH pathway effectors. QLT0267-treated H1975 cells exhibited significantly reduced levels of the EMT TF SNAI1, ZEB1 and ZEB2 (Fig. 3K). In addition, QLT0267 exposure promoted a robust decrease in the expression of most actors of the Notch pathway, most prominently JAG1 (Fig. 3L). Likewise, in MDST8 colon cancer cells where ILK acts as a key downstream effector of PrPC [32], we confirmed that QLT0267 treatment was associated with reduced expression of RBPJ, at both the mRNA and protein levels (Fig. 3M, N). Of note, RBPJ was reported to directly bind the promoter of ZEB1 [35] and silencing RBPJ was shown to reduce both SNAIL and ZEB1 expression [36]. Altogether, our results thus argue that PrPC mobilizes an ILK-RBPJ axis that further cooperates with the TGFβ pathway to control the expression of EMT TF as well as that of JAG1 (Fig. 3O).
Reciprocal functional interactions occur between PrPC and EGFR signalling in NSCLC
Further exploring our GSEA analyses, we observed that PRNP gene expression was consistently associated with an enrichment of a signature associated with EGFR activation (EGFR_UP.V1_UP) across the cell line panel and all cohorts (Fig. 4A). This may mirror an EGFR-dependent induction of PrPC expression and/or a PrPC-dependent activation of EGFR signalling. To shed light on this issue, we queried the LUAD cell lines of the CCLE and found that PRNP transcripts were increased in EGFR-mutated cell lines as compared with EGFR-wild-type (WT) cells (Fig. 4B). This was also true in the patients of the Onco-HEGP LUAD cohort (Fig. 4B). Furthermore, mining the proteogenomic studies by Chen et al. and Lehtiö et al. [20, 21] revealed a very significant increase in the protein levels of PrPC in patients harbouring EGFR mutations vs EGFR-WT cases (Fig. 4C). Likewise, Prnp levels are increased in lungs of mice bearing tumours harbouring the L858R, the T790M or both mutations, as compared with normal lung tissue (GSE17373 dataset, [37]) (Fig. 4D). NOTCH2 and ILK expression were also found to be significantly increased in EGFR mutated vs WT patients in 2 out of 3 datasets (Supplementary Fig. S7).
A GSEA analyses showing enrichment of the activated EGFR pathway (EGFR_up.V1_up) signature in the genes most correlated to PRNP in cell lines of the CCLE LUAD and in patients from the TCGA LUAD, Onco-HEGP LUAD, Chen LUAD or Lehtiö LUAD studies. B Boxplots showing the distribution of PRNP mRNA levels according to the EGFR mutational status in the CCLE and Onco-HEGP LUAD studies. C Boxplots showing the distribution of the PrPC protein levels according to the EGFR mutational status in the Chen and Lehtiö LUAD studies. D Boxplot showing the distribution of mouse Prnp mRNA in lung tissue of several genetically modified mouse models of LUAD bearing the EGFR L858R or T790M mutation or combining the two mutations.
Further supporting the notion that PrPC may be induced upon EGFR activation, we observed an upregulation of PrPC at the mRNA and protein levels upon exposure of H1650 cells to EGF (25 ng/ml, 24 h) (Fig. 5A, B), which was shown to also induce EMT [38]. Next, we probed whether PrPC may reciprocally control the expression of EGFR. In the 4 cell lines studied, namely, H1650, HCC827, H1975 and PC3, we found that PrPC-silencing caused a reduction in EGFR mRNA (Fig. 5C and Supplementary Fig. S6J). PrPC-silenced H1975 and PC3 cells also exhibited reduced EGFR protein levels (Fig. 5D and Supplementary Fig. S6K) and PrPC-silenced HCC827 cells were unable to upregulate EGFR mRNA or protein levels in response to TGFβ (Fig. 5E, F), as observed with EMT TF. Finally, there was a drastic reduction in EGFR mRNA levels in H1975 cells exposed to the ILK inhibitor QLT0267 (Fig. 5G). Hence, we surmise that the PrPC-ILK-RBPJ axis controls EGFR expression (Fig. 5H), which aligns well with the NOTCH-dependent regulation of EGFR that we [26] and others [39] previously documented.
qRT-PCR (A) and Western blot (B) analysis of the expression of PRNP / PrPC in EGF-treated versus control H1650 cells. C qRT-PCR analysis of the expression of EGFR mRNA in H1650, HCC827 and H1975 cells after PRNP silencing as compared to control cells. D Western blot analysis of the expression of EGFR protein levels in H1975 cells after PRNP silencing as compared to control cells. qRT-PCR (E) and western blot (F) analysis of the expression of EGFR in HCC827 cells exposed to siRNA against PRNP and recombinant TGFβ1. G qRT-PCR analysis of the expression of EGFR mRNA in QLT0267-treated versus control H1975 cells. H Schematic diagram illustrating the proposed regulation of EGFR downstream from the PrPC-ILK-RBPJ axis. I Confocal microscopy images showing the staining of PrPC and EGFR in H1975 cells. White arrows indicate co-localization regions. Scale bar = 20 µm. J Heatmap showing the most differentially expressed genes in H1650 cells exposed to siRNA against PRNP and recombinant EGF. qRT-PCR analysis of the expression of SNAI2 (K), ILK (L), JAG1 and RBPJ (M) mRNA in H1650 cells exposed to siRNA against PRNP and recombinant EGF. N Western blot analysis of the expression of RBPJ in H1650 cells exposed to siRNA against PRNP and recombinant EGF. Results from qRT-PCR and Western blots are expressed as means ± s.e.m of n = 2 independent triplicates of cell preparations. *p < 0.05, **p < 0.01, ***p < 0.001 vs. control (siScramble or vehicle), # p < 0.05, ###p < 0.001 vs. TGFβ1 or EGF treated, siScramble cells, §p < 0.05 vs. EGF untreated, PRNP-silenced cells. Protein levels in Western blots were normalized to α-tubulin (α-tub).
Having highlighted a positive regulatory loop between PrPC and EGFR, we went on to examine whether PrPC may modulate the activity of EGFR. We first gained evidence that PrPC co-localized with EGFR in H1975 cells (Fig. 5I), in agreement with the report by Atkinson et al. in HT29 colon cancer cells [40]. We next investigated the impact of PrPC siRNA-mediated silencing on H1650 cells treated with EGF. Through RNAseq analysis, we found that the response of H1650 cells to EGF (25 ng/ml, 24 h) was drastically affected in PRNP-silenced cells, as illustrated in the heatmap displayed in Fig. 5J. Notably, while H1650 engaged an EMT programme when exposed to EGF as already reported [38], we found that PRNP silencing affected EGF-induced mesenchymal transition as shown by lower basal levels and a decrease in SNAI2 induction, as compared to control cells (Fig. 5K). PrPC-silenced H1650 cells also failed to upregulate the expression of ILK and JAG1 in response to EGF, and RBPJ transcript and protein levels were decreased in PrPC-silenced versus control H1650 cells at basal levels (Fig. 5L-N). Altogether, these data suggest that the induction of PRNP expression (i) is enhanced by EGFR activation, either constitutive (mutant EGFR) or in response to EGF, (ii) potentiates the activity of EGFR itself and (iii) contributes to the onset and/or maintenance of EMT.
PrPC is associated with resistance to EGFR-TKI in NSCLC
While PrPC has been broadly implicated in resistance to anti-cancer therapies in various cancer types (reviewed in [15]), it has not been assessed for EGFR-targeted therapies in lung cancer. This question is all the more relevant since our findings substantiate that PrPC fosters EMT, an important mechanism of resistance to anti-EGFR therapies [41]. Mining the study by Nilsson et al. [13], we first found that PRNP transcript levels were robustly increased in three erlotinib-resistant HCC827 clones, as compared to parental cells (Fig. 6A). These observations could be extended to the minimal residual cell population obtained after combined treatment of HCC827 cells with osimertinib and trametinib (Fig. 6B) [42]. Likewise, PRNP mRNA levels were found to be elevated in HCC827 cells exposed to osimertinib in two different experimental paradigms, acute exposure or generation of drug tolerant persister cells (Fig. 6C) [43]. Of note, ILK mRNA levels were also elevated in all TKI-resistant conditions as compared to controls (Supplementary Fig. S8). Next, we tested the impact of PrPC silencing on sensitivity to osimertinib in H1975. Beyond a reduced cell number observed under basal conditions, in line with Fig. 2E, cell counting demonstrated that PrPC depletion led to a significantly more potent effect of osimertinib in H1975 cells (Fig. 6D). Finally, we found in the single-cell patient dataset by Maynard et al. that PRNP levels were enriched in cancer cells from EGFR-mutant NSCLC patients with progressive disease after EGFR-TKI treatment (Fig. 6E) [44].
Boxplots showing the distribution of PRNP mRNA in (A) erlotinib-resistant HCC827 clones versus parental cells (B) the minimal residual cell population obtained after combined treatment of HCC827 cells with osimertinib and trametinib (C) HCC827 cells acutely exposed to osimertinib (left) or HCC827 persister cells after long-term exposure osimertinib (right). D Quantification of cell numbers in H1975 cells pre-treated or not with PRNP siRNA and exposed to different doses of osimertinib for 72 h, according to the schematic workflow (top). E Differential expression of PRNP is shown in violin plots for single cancer cells collected from a patient treated with erlotinib (left) or a patient treated with osimertinib (right). TN treatment naive, RD residual disease, PD progressive disease.
Plasma PrPC level is elevated in EGFR-mutant NSCLC patients and its evolution mirrors that of the disease
We next sought to assess whether monitoring the levels of circulating PrPC may have potential clinical value, as in colon cancer [16]. PrPC plasma levels were quantified in 29 patients with EGFR-mutated NSCLC treated with first line EGFR-TKI in the metastatic setting. The overall characteristics of patients is summarized in Table S3. At diagnosis (T0), plasma PrPC levels were higher in patients with EGFR mutated NSCLC as compared with sex- and age-matched healthy controls (Fig. 7A, B). In patients previously treated with chemotherapy for localized disease, there was a tendency towards higher levels of PrPC at metastatic relapse (T0) (Supplementary Fig. S9).
A Boxplot showing the mean levels of circulating PrPC in the plasma of healthy subjects or patients with EGFR-mutated NSCLC at the time of pre-treatment with first line EGFR-TKI in the metastatic disease. B Summary of plasmatic PrPC values according to demographic information in healthy subjects or patients. C Evolution of plasma PrPC levels in paired samples from EGFR-TKI treated patients from cohort 1 between pre-treatment (T0) and evaluation (T1) with T1 corresponding to progression. D Table summarizing clinical data of patients from (C) at T0 and T1. E Evolution of plasma PrPC levels in paired samples from EGFR-TKI treated patients from cohort 1 between pre-treatment (T0) and evaluation (T1) with T1 corresponding to a clinical event other than progression. F Table summarizing clinical data of patients from (E) at T0 and T1. G Evolution of plasma PrPC levels in paired samples from EGFR-TKI patients from cohort 2 between pre-treatment (T0) and evaluation (T1). H Table summarizing clinical data of patients from (G) at T0 and T1. I Kinetics of evolution of plasma PrPC levels in samples from EGFR-TKI patients from cohort 2 according to disease history. J Table summarizing clinical data of patients from (I) at T0, T1 and T2.
Paired samples were available for 6 PLAPOU patients at T0 and T1 corresponding to TKI treatment start (first generation) and first progression event, respectively. Four patients had no change (#1.3) or increased PrPC levels (#1.1, #1.2 and #1.4) (Fig. 7C) and progression was clinically observed (Fig. 7D). There was a strong decrease in PrPC levels between T0 and T1 in patient #1.5, whose treatment was changed at T1 due to toxicity and a more moderate decrease in the case of patient #1.6, whose disease was controlled except at the cerebral level (Fig. 7E, F).
In the Onco-HEGP series, follow-up was available for 9 patients treated in first line with osimertinib. Five had paired pre-treatment (T0) and first progression event (T1) samples (Fig. 7G). PrPC levels were increased in all but one patient that had a confirmed stable disease. (Fig. 7G, H). The last series was composed of matched first line pre-treatment (T0), first evaluation (T1) and first progression event (T2). Quite remarkably, PrPC levels were consistently decreased at T1 and raised backed at T2 (Fig. 7I, J). Detailed information on patients #2.1 and #2.6 are available as supplementary information as examples of disease monitoring (Supplementary Fig. S10A, B). As a whole, these data argue that monitoring circulating PrPC levels along disease kinetics may have clinical value to objectify evolution.
Discussion
The cellular and molecular mechanisms sustaining resistance to TKI in EGFR-mutated LUAD remain imperfectly understood. Among non-genetic processes features EMT [3], which raises the question of how EMT is induced. The present work assessed PrPC as a promoter of EMT in EGFR-mutated LUAD. PrPC, a ubiquitous protein intensely studied for its implication in neurodegenerative diseases [45], is raising interest in the field of cancer, notably for its pro-migratory and pro-invasive role, as well as its contribution to chemo- and radio-resistance [15].
Our conclusion that PrPC fosters EMT is supported by a corpus of computational and experimental data. By leveraging the CCLE cell panel and four different patient datasets, including our own Onco-HEGP cohort, we systematically found prominent correlations between PRNP levels and EMT. In cell-based assays, we demonstrated that PrPC is necessary for the acquisition (upon exposure to TGFβ or EGF) or the maintenance of mesenchymal traits. From a mechanistic point of view, our data suggest that, rather than exerting its action through the regulation of soluble TGFβ levels, as we showed in the context of colon cancer [16], PrPC primes NSCLC cancer cells for TGFβ responsiveness. By controlling an ILK-RBPJ axis, PrPC appears to put cells in a permissive state, where they respond to TGFβ by the expression of EMT TF such as ZEB1. Our results are fully consistent with (i) our previous demonstration that ILK is a proximal effector of PrPC [32], (ii) the finding that ILK features among the PrPC-interacting partners in melanoma, (iii) the very high correlation between PrPC and ILK levels in LUAD, as in colon cancer (our unpublished observations) and (iv) fits in with the reported poor prognosis associated with high ILK levels in EGFR-mutated NSCLC [46]. Regarding RBPJ, our data recall the control exerted by PrPC on the Notch pathway in the context of neuronal development [26] or in pancreatic cancer cells [27]. The robustness of our findings is sustained by the converging observations gained with NSCLC cell lines and the PC3 prostate cancer cell line. Because ZEB1 has been reported to positively regulate the expression of NOTCH1 [47] and JAG1 [48] through the suppression of miR-200, we may assume that PrPC, ZEB1 and the NOTCH pathway are intricately linked within a gene regulatory network that instigates EMT. This model also aligns well with the higher expression of PRNP mRNA found in Onco-HEGP patients with a low miR-200 signature (Supplementary Fig. S1G).
A second major finding of our study is the reciprocal regulation of PrPC and EGFR. Indeed, we first gained evidence that PrPC is a transcriptional target of EGFR signalling, which is notably reflected by the increased levels of PRNP mRNA in EGFR-mutated LUAD. This observation provides some answer as to how PrPC is induced in LUAD, although other possible mechanisms may also operate. These may include positive feedback regulatory loops, as we described for TGFβ [16], ILK [32], NOTCH [26] and more recently Wnt and glucorticoid signalling [49].
We may thus envision that once induced, the expression of PrPC is self-entertained by its own downstream effectors. This hypothesis would for instance accommodate the observation that PRNP transcripts are induced upon long-term (Supplementary Fig. S3) but not short-term (Supplementary Fig. S2F,G) TGFβ exposure. Our data further highlight a PrPC-dependent regulation of EGFR expression and signalling, recalling that previously described in neuroblastoma cells [50] and neural and dental pulp stem cells [26, 51, 52]. Because PrPC and EGFR co-localize in LUAD cells (Fig. 5I), as well as in other cancerous and non-cancerous cells [40, 50], we speculate that PrPC modulates the dynamics of EGFR signalling, as already suggested over a decade ago in the study by Solis et al. [53]. Further experiments should help further delineate the intricate interplay between PrPC and EGFR.
Finally, our data firmly establish a link between PrPC expression and resistance to EGFR-TKI. This conclusion is based upon (i) the increase in PRNP mRNAs in various in vitro models of EGFR-TKI resistance (Fig. 6A-C), (ii) the reduced cell viability of H1975 cells exposed to osimertinib when PrPC is silenced (Fig. 6D), (iii) the increased PRNP levels in cancer cells of EGFR-mutated LUAD patients having acquired drug resistance (progression) as compared to their corresponding stable (residual) disease state (Fig. 6E). Most importantly, our results provide the proof of principle that the longitudinal quantification of circulating PrPC may have clinical value for the follow-up of EGFR-mutated LUAD patients under TKI treatment. In line with our data obtained in patients with metastatic colon cancer [16], we found higher levels of plasma PrPC in two independent cohorts of EGFR-mutated LUAD patients. Of note, the availability of serial samples for several patients of each cohort allowed us to highlight increases in plasma PrPC that precede clinical or biological evidence for disease progression. Thus, although our data cannot discriminate whether circulating PrPC originates from tumour cells themselves or other cell types, they support the notion that an elevation of its level reflects a worsening of the disease.
As a conclusion, our work introduces PrPC as a missing link between EMT and EGFR-TKI resistance in LUAD and suggest that monitoring plasma PrPC levels may represent a valuable non-invasive strategy for patient follow-up. Given our previous proof of concept of a beneficial effect of PrPC neutralization in cancer cells [54], this study also argues that PrPC may represent a valuable target for the development of new therapeutic strategies for the management of EGFR-mutated LUAD patients.
Materials and methods
Reagents
References and sources of primary antibodies used for western blots are available in Table S1. QLT0267 was provided by Dr. Shoukat Dedhar. The appropriate QLT concentration was determined through dose-response analyses in previous studies [32]. Human recombinant TGFβ1 was purchased from R&D systems (Minneapolis, MN, USA) (reference 240-B) and reconstituted following supplier instructions with HCl 4 mM, BSA at a concentration of 1 mg/ml. HCC827 cells were exposed 24 h to TGFβ or control (HCl, BSA) at a final concentration of 10 ng/ml. Human recombinant EGF was purchased from R&D systems (reference 236-EG-200) and reconstituted following supplier instructions with PBS. H1650 cells were exposed 24 h to EGF or control (PBS) at a final concentration of 25 ng/ml.
Osimertinib (AZD-9291/mereletinib, 5 mg) was purchased from MedChemExpress (Princeton, NJ, USA) and diluted in dimethylsulfoxide (DMSO).
Cell culture
The human non-small cell lung carcinomas cell lines H1650, HCC827 and H1975 were purchased from the American Type Culture Collection. These cell lines were grown in RPMI-1640. The human colorectal cancer MDST8 and the human prostate PC3 cell lines were purchased from Sigma. MDST8 cells were grown in DMEM and PC3 cells were grown in RPMI-1640 medium. All culture media were supplemented with 10% (v/v) fetal bovine serum (FBS) (Gibco, France) and 1% (v/v) penicillin-streptomycin (Gibco, France). All cell lines were grown at 37 °C and 5% CO2 in a humidified incubator and regularly tested for mycoplasma contamination. The mutational status of cell lines was validated by NGS (AmpliSeq custom panel WG_IAD196383V2, that covers 119 amplicons in 23 cancer genes, Life Technologies-Thermo Fisher Scientific). For transient siRNA-mediated silencing, cells were transfected with siRNA sequences (50 nM) using the Lipofectamine 2000 reagent according to the manufacturer’s instructions (Invitrogen). Specific siRNA sequences used were 5′-CAGUACAGCAACCAGAACA-3′ (sense siPRNP) 5′-AACGAUGACACGAACACAC-3′ (sense scramble). Cellular mRNAs or protein extract were collected 72 h after PrPC silencing. siRNA against JAG1 were from Thermo Fischer Scientific (Silencer® select Assay ID 146914).
Preparation of protein extracts and western blot analyses
Cells were washed in PBS and incubated for 30 min at 4 °C in NaDOC lysis buffer [50 mM Tris·HCl (pH 7.4)/150 mM NaCl/5 mM EDTA/0.5% Triton X-100/0.5% sodium deoxycholate] and a mixture of phosphatase (Thermo Fischer Scientific, Waltham, MA, USA) and protease (Roche, Mannheim, Germany) inhibitors. Extracts were centrifuged at 14,000 × g for 15 min. Protein concentrations in the supernatant were measured by using the bicinchonic acid method (Pierce, Rockford, IL, USA). Protein extracts (15 µg) were resolved by 4–12% SDS-PAGE (Invitrogen) and transferred to nitrocellulose membranes (iBlot, Invitrogen). Membranes were blocked with SEA BLOCK blocking buffer (Thermo Fischer Scientific) for 1 h at room temperature and then incubated 1 h at room temperature with primary antibody. Bound antibody was revealed by infrared detection using a secondary antibody coupled to IRDye fluorophores (Li-Cor biosciences, Lincoln, NE, USA). Western blot read out was performed with the Odyssey Infrared Imaging System (Li-Cor Bsiosciences).
Isolation of total RNA and RT-PCR analysis
RNA was isolated by using the RNeasy extraction kit (Qiagen, Limburg, Netherlands), as recommended by the manufacturer’s instructions. For reverse transcriptase-polymerase chain reaction (RT-PCR) analysis, first-strand cDNA was synthesized with oligo(dT) primer and random 6mers, using the SensiFAST cDNA Synthesis Kit (Meridian, Memphis, TN, USA) according to the manufacturer’s protocol. Real-time PCR was performed using ABsolute QPCR SYBR Green ROX Mix (Thermo-Scientific, Waltham, MA, USA) on QuantStudio 12 K Flex (Applied Biosystems, Life Technologies Corporation, Carlsbad, CA, USA). Real-time PCR analyses were performed with the SDS software 2.3 (Applied biosystems). Primer sequences are available in Table S2. Results are expressed as a relative quantification of a target gene transcript normalized to the RPL13A housekeeping gene using the ∆∆Ct method.
Immunofluorescence and confocal microscopy
H1975 cells were cultured on glass coverslips, fixed with 4% paraformaldehyde in PBS, and blocked with PBS containing 1% BSA and 20 mM glycine. The cells were also permeabilized with 0.1% Triton in PBS containing 20 mM glycine for 15 min. The primary antibodies anti-PrPC (12F10, Bertin Pharma, Montigny Le Bretonneux, France) and anti-EGFR (#4267, Cell Signaling, Danvers, MA, USA), were used at 1/20 and 1/50 respectively in PBS with 1% BSA and 0.1% Tween for 1 h. Alexa Fluor 488 and 555 secondary antibodies were incubated for 1 h alongside TRITC-phalloidin for F-actin labelling. Nuclei were stained with DAPI. Images were recorded using a Zeiss Axio Observer Z1 for immunofluorescence and with a Zeiss LSM 710 for confocal microscopy. Images were processed with FIJI software.
Cell sensitivity to osimertinib
Twenty four hours after plating, H1975 cells were serum starved (1% FBS) and submitted to PRNP silencing after another 24 h. 24 h after siRNA transfection, cells were exposed to different concentrations of osimertinib or control (DMSO) for 72 h. Cell number, diameter and volume were determined using the CASY TT cell counter (Schärfe System GmbH, Reutlingen, Germany).
Real-time cell migration and invasion
Migration and invasion assays were performed using CIM-Plate 16 (Agilent, Santa Clara, CA, USA) and monitored on the xCELLigence Real-Time Cell Analyzer (RTCA) Dual Purpose instrument (Agilent) according to the manufacturer’s instructions. H1975 cells were seeded at a density of 20,000 cells/well in 3% FBS in the upper chamber and forced to move towards the lower chamber containing 10% FBS. For invasion, the upper chamber was pre-coated with 30 µl of Cultrex (1:15 in medium, Biotechne, Minneapolis, MN, USA). The impedance was recorded every 15 min for up to 48 h. The experiments were performed in triplicate.
3′RNA‑seq
PolyA-RNAseq libraries were prepared using the Quant-Seq 3′mRNA-Seq Kit FWD for Illumina (Lexogen™) according to the manufacturer’s instructions. Libraries were sequenced on a NovaSeq6000. Fastq RNAseq files were analysed using a standard bioinformatical pipeline, with adaptations related to polyA sequencing. Briefly, reads were mapped by STAR (v2.7.9a) [55]. All genes with a HGNC symbol were kept. Analyses were carried using the DESeq2 package version 1.38.3 with R studio 4.2.2.
Gene expression analyses
The following datasets were retrieved from public sources, exclusively on cells and patients with lung adenocarcinoma (LUAD): Cancer Cell Line Encyclopaedia (CCLE,) (n = 45) [18], TCGA (n = 511) [56], Onco-HEGP cohort (n = 107) [19], the patients from the proteogenomic studies by Chen et al. (n = 90) [20] and Lehtiö et al. (n = 90) [21]. Other datasets include GSE49644 [25], GSE121634 [13], GSE131594 [42], GSE193258 [43], GSE17373 [37] and PRJNA591860 [44]. GSEA (Gene Set Enrichment Analysis) was performed using the Broad Institute platform (http://www.broadinstitute.org/gsea/index.jsp; Version 2.0.14).
Collection of blood samples and analysis of plasma PrPC
Analyses were carried on a collection of plasmas from a set of 29 patients with EGFR-mutated metastatic NSCLC treated by EGFR-TKI as first line treatment. All methods were performed in accordance with the relevant guidelines and regulations. Seventeen patients were from the cohort “PLAPOU”, and twelve patients from Onco-HEGP. PLAPOU patients were prospectively included between June 2013 and November 2015 at the Hôpital Européen Georges Pompidou (HEGP) and have been previously described [57]. Blood sample collection was approved by the Ethics Committee for the Protection of Persons Ile-de France II (CPP Ile-de-France II n°2013-06-21 SC), and informed written consent had been obtained from patients. The second group of patients has been selected in a retrospective cohort of healthcare plasmas from Onco-HEGP. Patients have given their consent and collection of material was approved by the Ethics Committee for the Protection of Persons Ile-de France II (CPP Ile-de-France II n°2013- A01283-42), and informed written consent had been obtained from patients. The biological samples have been provided by the Biological Resources Center and Tumor Bank Platform (BB-0033-00063). Blood samples from 71 age-matched healthy individuals without any indication of malignancy were collected as a control group [16]. The overall characteristics of patients are summarized in Table S3. All plasma samples were frozen at −70 °C until analysis. The levels of PrPC were quantified in plasmas by DELFIA as in [16]. Experiments were all carried out under blinded conditions. ctDNA analyses in patients from Onco-HEGP cohort were carried out as in [57].
Statistical analyses
All statistical analyses were performed in R studio (version 4.2.2) using the stat_compare_means function from the ggpubr package. The results from experimental data in cell lines are reported as the means ± standard errors of the means (s.e.m.) with graphs generated using GraphPad PRISM version 9.4.1. Analyses involving two groups were carried out using the Shapiro test followed by Student’s t-test or Mann-Whitney rank-sum test according to normality. Results from analysis in public datasets or patient cohorts are expressed as median and interquartile range with graphs generated with ggplot2 in R studio. Statistical analysis was performed using the Mann-Whitney rank-sum test for two groups or one-way ANOVA followed by Wilcoxon rank-sum tests with Holm’s correction for multiple comparisons for >2 groups.
Data availability
Data supporting the observations of this study, including the methodology, are available upon reasonable request from the corresponding authors.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49.
Yang C-Y, Yang JC-H, Yang P-C. Precision management of advanced non-small cell lung cancer. Annu Rev Med. 2020;71:117–36.
Rotow J, Bivona TG. Understanding and targeting resistance mechanisms in NSCLC. Nat Rev Cancer. 2017;17:637–58.
Passaro A, Jänne PA, Mok T, Peters S. Overcoming therapy resistance in EGFR-mutant lung cancer. Nat Cancer. 2021;2:377–91.
Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26.
Zhang Z, Lee JC, Lin L, Olivas V, Au V, LaFramboise T, et al. Activation of the AXL kinase causes resistance to EGFR-targeted therapy in lung cancer. Nat Genet. 2012;44:852–60.
Weng C-H, Chen L-Y, Lin Y-C, Shih J-Y, Lin Y-C, Tseng R-Y, et al. Epithelial-mesenchymal transition (EMT) beyond EGFR mutations per se is a common mechanism for acquired resistance to EGFR TKI. Oncogene. 2019;38:455–68.
Aissa AF, Islam ABMMK, Ariss MM, Go CC, Rader AE, Conrardy RD, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat Commun. 2021;12:1628.
Bronte G, Bravaccini S, Bronte E, Burgio MA, Rolfo C, Delmonte A, et al. Epithelial-to-mesenchymal transition in the context of epidermal growth factor receptor inhibition in non-small-cell lung cancer. Biol Rev Camb Philos Soc. 2018;93:1735–46.
Zhu X, Chen L, Liu L, Niu X. EMT-mediated acquired EGFR-TKI resistance in NSCLC: mechanisms and strategies. Front Oncol. 2019;9:1044.
Raoof S, Mulford IJ, Frisco-Cabanos H, Nangia V, Timonina D, Labrot E, et al. Targeting FGFR overcomes EMT-mediated resistance in EGFR mutant non-small cell lung cancer. Oncogene. 2019;38:6399–413.
Yochum ZA, Cades J, Wang H, Chatterjee S, Simons BW, O’Brien JP, et al. Targeting the EMT transcription factor TWIST1 overcomes resistance to EGFR inhibitors in EGFR-mutant non-small-cell lung cancer. Oncogene 2019;38:656–70.
Nilsson MB, Sun H, Robichaux J, Pfeifer M, McDermott U, Travers J, et al. A YAP/FOXM1 axis mediates EMT-associated EGFR inhibitor resistance and increased expression of spindle assembly checkpoint components. Sci Transl Med. 2020;12:eaaz4589.
Tulchinsky E, Demidov O, Kriajevska M, Barlev NA, Imyanitov E. EMT: A mechanism for escape from EGFR-targeted therapy in lung cancer. Biochim Biophys Acta Rev Cancer. 2019;1871:29–39.
Mouillet-Richard S, Ghazi A, Laurent-Puig P. The cellular prion protein and the hallmarks of cancer. Cancers. 2021;13:5032.
Le Corre D, Ghazi A, Balogoun R, Pilati C, Aparicio T, Martin-Lannerée S, et al. The cellular prion protein controls the mesenchymal-like molecular subtype and predicts disease outcome in colorectal cancer. EBioMedicine. 2019;46:94–104.
Lin S-C, Lin C-H, Shih N-C, Liu H-L, Wang W-C, Lin K-Y, et al. Cellular prion protein transcriptionally regulated by NFIL3 enhances lung cancer cell lamellipodium formation and migration through JNK signaling. Oncogene. 2020;39:385–98.
Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483:603–7.
Garinet S, Didelot A, Denize T, Perrier A, Beinse G, Leclere J-B, et al. Clinical assessment of the miR-34, miR-200, ZEB1 and SNAIL EMT regulation hub underlines the differential prognostic value of EMT miRs to drive mesenchymal transition and prognosis in resected NSCLC. Br J Cancer. 2021;125:1544–51.
Chen Y-J, Roumeliotis TI, Chang Y-H, Chen C-T, Han C-L, Lin M-H, et al. Proteogenomics of non-smoking lung cancer in East Asia delineates molecular signatures of pathogenesis and progression. Cell. 2020;182:226–44.e17.
Lehtiö J, Arslan T, Siavelis I, Pan Y, Socciarelli F, Berkovska O, et al. Proteogenomics of non-small cell lung cancer reveals molecular subtypes associated with specific therapeutic targets and immune evasion mechanisms. Nat Cancer. 2021;2:1224–42.
Mak MP, Tong P, Diao L, Cardnell RJ, Gibbons DL, William WN, et al. A patient-derived, pan-cancer EMT signature identifies global molecular alterations and immune target enrichment following epithelial-to-mesenchymal transition. Clin Cancer Res J Am Assoc Cancer Res. 2016;22:609–20.
de Reyniès A, Jaurand M-C, Renier A, Couchy G, Hysi I, Elarouci N, et al. Molecular classification of malignant pleural mesothelioma: identification of a poor prognosis subgroup linked to the epithelial-to-mesenchymal transition. Clin Cancer Res. 2014;20:1323–34.
Garinet S, Didelot A, Marisa L, Beinse G, Sroussi M, Le Pimpec-Barthes F, et al. A novel Chr1-miR-200 driven whole transcriptome signature shapes tumor immune microenvironment and predicts relapse in early-stage lung adenocarcinoma. J Transl Med. 2023;21:324.
Sun Y, Daemen A, Hatzivassiliou G, Arnott D, Wilson C, Zhuang G, et al. Metabolic and transcriptional profiling reveals pyruvate dehydrogenase kinase 4 as a mediator of epithelial-mesenchymal transition and drug resistance in tumor cells. Cancer Metab. 2014;2:20.
Martin-Lannerée S, Halliez S, Hirsch TZ, Hernandez-Rapp J, Passet B, Tomkiewicz C, et al. The cellular prion protein controls notch signaling in neural stem/progenitor cells. Stem Cells. 2017;35:754–65.
Wang Y, Yu S, Huang D, Cui M, Hu H, Zhang L, et al. Cellular prion protein mediates pancreatic cancer cell survival and invasion through association with and enhanced signaling of Notch1. Am J Pathol. 2016;186:2945–56.
Derynck R, Muthusamy BP, Saeteurn KY. Signaling pathway cooperation in TGF-β-induced epithelial-mesenchymal transition. Curr Opin Cell Biol. 2014;31:56–66.
Hori K, Sen A, Artavanis-Tsakonas S. Notch signaling at a glance. J Cell Sci. 2013;126:2135–40.
Sethi N, Dai X, Winter CG, Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205.
Manderfield LJ, High FA, Engleka KA, Liu F, Li L, Rentschler S, et al. Notch activation of Jagged1 contributes to the assembly of the arterial wall. Circulation. 2012;125:314–23.
Ghazi A, Le Corre D, Pilati C, Taieb J, Aparicio T, Didelot A, et al. Prognostic value of the PrPC-ILK-IDO1 axis in the mesenchymal colorectal cancer subtype. Oncoimmunology. 2021;10:1940674.
Li H, Zhang J, Ke J-R, Yu Z, Shi R, Gao S-S, et al. Pro-prion, as a membrane adaptor protein for E3 ligase c-Cbl, facilitates the ubiquitination of IGF-1R, promoting melanoma metastasis. Cell Rep. 2022;41:111834.
Troussard AA, McDonald PC, Wederell ED, Mawji NM, Filipenko NR, Gelmon KA, et al. Preferential dependence of breast cancer cells versus normal cells on integrin-linked kinase for protein kinase B/Akt activation and cell survival. Cancer Res. 2006;66:393–403.
Jiang H, Zhou C, Zhang Z, Wang Q, Wei H, Shi W, et al. Jagged1-Notch1-deployed tumor perivascular niche promotes breast cancer stem cell phenotype through Zeb1. Nat Commun. 2020;11:5129.
Maciaczyk D, Picard D, Zhao L, Koch K, Herrera-Rios D, Li G, et al. CBF1 is clinically prognostic and serves as a target to block cellular invasion and chemoresistance of EMT-like glioblastoma cells. Br J Cancer. 2017;117:102–12.
Regales L, Gong Y, Shen R, de Stanchina E, Vivanco I, Goel A, et al. Dual targeting of EGFR can overcome a major drug resistance mutation in mouse models of EGFR mutant lung cancer. J Clin Investig. 2009;119:3000–10.
Pallier K, Cessot A, Côté J-F, Just P-A, Cazes A, Fabre E, et al. TWIST1 a new determinant of epithelial to mesenchymal transition in EGFR mutated lung adenocarcinoma. PLoS ONE. 2012;7:e29954.
Andreu-Agullo C, Morante-Redolat JM, Delgado AC, Farinas I. Vascular niche factor PEDF modulates Notch-dependent stemness in the adult subependymal zone. Nat Neurosci. 2009;12:1514–23.
Atkinson CJ, Kawamata F, Liu C, Ham S, Győrffy B, Munn AL, et al. EGFR and Prion protein promote signaling via FOXO3a-KLF5 resulting in clinical resistance to platinum agents in colorectal cancer. Mol Oncol. 2019;13:725–37.
Zhang X, Maity TK, Ross KE, Qi Y, Cultraro CM, Bahta M, et al. Alterations in the global proteome and phosphoproteome in third generation EGFR TKI resistance reveal drug targets to circumvent resistance. Cancer Res. 2021;81:3051–66.
Kurppa KJ, Liu Y, To C, Zhang T, Fan M, Vajdi A, et al. Treatment-induced tumor dormancy through YAP-mediated transcriptional reprogramming of the apoptotic pathway. Cancer Cell. 2020;37:104–22.e12.
Gogleva A, Polychronopoulos D, Pfeifer M, Poroshin V, Ughetto M, Martin MJ, et al. Knowledge graph-based recommendation framework identifies drivers of resistance in EGFR mutant non-small cell lung cancer. Nat Commun. 2022;13:1667.
Maynard A, McCoach CE, Rotow JK, Harris L, Haderk F, Kerr DL, et al. Therapy-induced evolution of human lung cancer revealed by single-cell RNA sequencing. Cell. 2020;182:1232–51.e22.
Aguzzi A, Baumann F, Bremer J. The prion’s elusive reason for being. Annu Rev Neurosci. 2008;31:439–77.
Karachaliou N, Cardona AF, Bracht JWP, Aldeguer E, Drozdowskyj A, Fernandez-Bruno M, et al. Integrin-linked kinase (ILK) and src homology 2 domain-containing phosphatase 2 (SHP2): novel targets in EGFR-mutation positive non-small cell lung cancer (NSCLC). EBioMedicine. 2019;39:207–14.
Zhang T, Guo L, Creighton CJ, Lu Q, Gibbons DL, Yi ES, et al. A genetic cell context-dependent role for ZEB1 in lung cancer. Nat Commun. 2016;7:12231.
Brabletz S, Bajdak K, Meidhof S, Burk U, Niedermann G, Firat E, et al. The ZEB1/miR-200 feedback loop controls Notch signalling in cancer cells. EMBO J. 2011;30:770–82.
Mouillet-Richard S, Gougelet A, Passet B, Brochard C, Le Corre D, Pitasi CL, et al. Wnt, glucocorticoid and cellular prion protein cooperate to drive a mesenchymal phenotype with poor prognosis in colon cancer. J Transl Med. 2024;22:337.
Llorens F, Carulla P, Villa A, Torres JM, Fortes P, Ferrer I, et al. PrP(C) regulates epidermal growth factor receptor function and cell shape dynamics in Neuro2a cells. J Neurochem. 2013;127:124–38.
Martellucci S, Manganelli V, Santacroce C, Santilli F, Piccoli L, Sorice M, et al. Role of Prion protein-EGFR multimolecular complex during neuronal differentiation of human dental pulp-derived stem cells. Prion. 2018;12:117–26.
Groveman BR, Schwarz B, Bohrnsen E, Foliaki ST, Carroll JA, Wood AR, et al. A PrP EGFR signaling axis controls neural stem cell senescence through modulating cellular energy pathways. J Biol Chem. 2023;299:105319.
Solis GP, Schrock Y, Hulsbusch N, Wiechers M, Plattner H, Stuermer CA. Reggies/flotillins regulate E-cadherin-mediated cell contact formation by affecting EGFR trafficking. Mol Biol Cell. 2012;23:1812–25.
Mouillet-Richard S, Martin-Lannerée S, Le Corre D, Hirsch TZ, Ghazi A, Sroussi M, et al. A proof of concept for targeting the PrPC - Amyloid β peptide interaction in basal prostate cancer and mesenchymal colon cancer. Oncogene. 2022;41:4397–404.
Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21.
Cancer Genome Atlas Research Network. Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50.
Pécuchet N, Zonta E, Didelot A, Combe P, Thibault C, Gibault L, et al. Base-position error rate analysis of next-generation sequencing applied to circulating tumor DNA in non-small cell lung cancer: a prospective study. PLoS Med. 2016;13:e1002199.
Acknowledgements
The authors wish to thank N. Pierron for expert technical assistance, T. Balde for helping with dataset retrieval and the Biological Resources Center and Tumor Bank Platform (BB-0033-00063) for providing plasma samples. CL was supported by grants from Association pour la Recherche contre le Cancer and Labex Immuno-oncology. HBerthou was supported by grants from Alliance pour la Recherche contre le Cancer (APREC). This work was supported by Funding grants from Labex ImmunoOnco, Association pour la Recherche sur le Cancer as well as INSERM.
Author information
Authors and Affiliations
Contributions
CL: data acquisition; data analysis and interpretation; data processing and presentation; management of biological samples and clinical annotations. AD: data acquisition; data analysis and interpretation; management of biological samples and clinical annotations. SG: performed transcriptomic analysis of in house patient cohort. HBerthou, SM-L, VP, FD, AP: data acquisition. MS and AdR: support for bioinformatics analyses. SD: provided material. EF, FLP-B and AM-L: provided and characterized patient samples. J-ML: performed biological analyses. PL-P: data analysis and interpretation; securing funding. HBlons: study concept and design; data analysis and interpretation; management of biological samples and clinical annotations; general study supervision; securing funding. SM-R: study concept and design; data analysis and interpretation; general study supervision; manuscript drafting; securing funding. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Lailler, C., Didelot, A., Garinet, S. et al. PrPC controls epithelial-to-mesenchymal transition in EGFR-mutated NSCLC: implications for TKI resistance and patient follow-up. Oncogene 43, 2781–2794 (2024). https://doi.org/10.1038/s41388-024-03130-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/s41388-024-03130-0
- Springer Nature Limited