Abstract
The first embryonic division represents a starting point for the development of a new individual. In many species, tight control over the first embryonic division ensures its accuracy. However, the first division in humans is often erroneous and can impair embryo development. To delineate the spatiotemporal organization of the first mitotic division typical for normal human embryo development, we systematically analyzed a unique timelapse dataset of 300 IVF embryos that developed into healthy newborns. The zygotic division pattern of these best-quality embryos was compared to their siblings that failed to implant or arrested during cleavage stage. We show that division at the right angle to the juxtaposed pronuclei is preferential and supports faithful zygotic division. Alternative configurations of the first mitosis are associated with reduced clustering of nucleoli and multinucleation at the 2-cell stage, which are more common in women of advanced age. Collectively, these data imply that orientation of the first division predisposes human embryos to genetic (in)stability and may contribute to aneuploidy and age-related infertility.
Similar content being viewed by others
Introduction
Upon fertilization, the oocyte undergoes a sequence of events, which result in the completion of meiotic division and a combination of parental genomes. Genetic material from both parents is first enclosed in spatially separated pronuclei (PNs), which appose in preparation for the first mitotic division. Orientation of the first mitosis is instructive for embryonic differentiation and establishing a three-dimensional (3D) body plan in animals such as Drosophila melanogaster, Caenorhabditis elegans, or Xenopus laevis1,2,3. In these species, the orientation of the first mitosis biases arising cells to differentiation trajectories by distributing cytoplasmic and membrane-bound factors. Whereas the geometry of egg-to-embryo transition in lower species is well documented, the impact of spatiotemporal control over the first mitosis in mammals remains largely unexplained4,5. Several studies on mice postulated that the position of the polar body (PB) and sperm entry site define the axis for the first embryonic mitosis6,7. Therefore, the existence of spatial clues might explain the high fidelity of the first mitosis in mouse embryos, which display little to no errors in chromosome segregation during their preimplantation development.
Unlike in mice, the first mitosis in human embryos is highly susceptible to chromosome segregation errors. Genetic imbalances adversely affect embryo development and constitute a major limitation to human reproduction8,9,10. Depending on the degree of chromosome missegregation, abnormal cell divisions result in micro- or multinucleation that are linked to aneuploidy9,11. The frequency of chromosomal abnormalities in human embryos is known to further increase with female reproductive aging, leading to an age-associated fertility decline12.
While the efficiency of infertility treatment remains low, much effort is put into assessing the developmental potential of in vitro-produced embryos to prioritize high-quality embryos for transfer. Positive outcomes of in vitro fertilization (IVF) based treatments are associated with certain morphological patterns observed in human zygotes13,14. It is believed that, similarly to mouse embryos, parental PN in human zygotes should align with the PB axis to ensure the correct development15,16,17,18. It has been reported that some PN-PB configurations and patterns of nucleoli arrangement were more frequent in embryos bearing chromosomal abnormalities19. In addition, the size, number, and position of nucleoli have been proposed as a marker of embryo quality20. However, most studies that deal with the morphological assessment of human zygotes are based on observations with a low number of time points. The dynamic nature of early human development has become more accessible with the introduction of a timelapse embryo incubation system into clinical embryology practice. Continuous monitoring of fertilized oocytes revealed that migration of PN and nucleoli is highly dynamic and may reflect the ploidy status of the embryo21,22. This observation challenges the conventional static assessment of PN and nucleoli configuration and urges investigating the causative relationship between zygote morphology and chromosome segregation errors.
The discrepancy in genetic stability of mouse and human embryos led us to investigate whether spatial control over first mitosis is relevant to normal human embryo development. To this end, we examined whole-volume timelapse records of 300 IVF embryos that gave rise to live births (LB). The divisional pattern of these high-quality embryos was compared with 207 sibling embryos that failed to develop into blastocysts (hereafter termed cleavage arrest—CA group) and 111 sibling embryos that failed to implant (FI group). Based on the analysis of geometry and the dynamics of the first mitotic division in our dataset, we propose that the orientation of the first mitotic division is important for its success and the genetic stability of human embryos.
Results
The cleavage pattern of the human embryo is determined by juxtaposed pronuclei
To investigate the role of spatial clues in normal human development, we first assessed the mutual position of the PB and juxtaposed PNs at the time of nuclear envelope breakdown (NEBD) in embryos that resulted in a live birth (LB). Based on the angle between the PB and PN axis, we categorized observed patterns as “longitudinal” (0–30°), “intermediate” (30–60°), or “orthogonal” (60–90°) (Fig. 1a). Analysis showed that these patterns were equally distributed in our dataset of LB embryos (29.7%, 32.0% and 38.3% respectively). A similar distribution was recorded in sibling CA and FI embryos. This observation indicates that the pre-determined PN-PB alignment is not required for human embryo development (Fig. 1b). Instead, we noticed that developing LB embryos tended to divide at the right angle to the axis of juxtaposed PNs. Therefore, we evaluated the direction of the first mitosis with respect to the PN axis and categorized observed patterns as (1) “perpendicular” when the cleavage direction was approximately 90° to PN axis; (2) “parallel” when cleavage direction aligned with the PN axis and (3) “oblique” for remaining patterns (Fig. 1c). Detailed multiple-observer analysis of 4D records with 5-min resolution confirmed that perpendicular division was favored in LB embryos (68.7%), whereas parallel division pattern was scarce (3.7%) (Fig. 1d). To address if this divisional trend can be attributed to the inherent quality of LB embryos, we performed the same analysis in embryos which failed to produce pregnancy. We noted that the first mitosis orientation in FI embryos resembled the pattern observed in LB embryos. On the contrary, in the group of low-quality CA embryos, the incidence of parallel division was almost three times higher than in LB embryos (14.9% vs 3.7%, respectively; Fig. 1d). These data suggest that the direction of zygotic cleavage is spatially controlled and linked to the development to the blastocyst stage.
a Orientation of pronuclei (PN) to the polar body (PB) in human IVF embryos; yellow dashed line marks borders of pronuclei. b Quantification of PN-PB alignment in human embryos resulting in a live birth (LB) and their failed-to-implant (FI) and cleavage-arrested (CA) siblings. c Stills from timelapse movies of preimplantation development of three zygotes and corresponding 2-cell embryos representative of different divisional patterns. d Quantification of CA, FI, and LB embryos dividing parallel, oblique and perpendicular direction to the PN axis. The number of embryos in each group is specified in italics. Chi-square test: “ns” (non-significant difference) p > 0.05; ***p < 0.001. Source data are provided as a Source Data file.
Next, we asked if dissimilar cleavage patterns differ in the onset and duration of first mitosis. Our data showed that the first mitosis in LB embryos typically started at 22.36 h after ICSI and lasted, on average, 2.68 h. No significant difference in the onset and duration of the cell division between individual patterns of the first mitosis was observed (Supplementary Fig. 1a, b). Next, we assessed the length of the first cell cycle, which was defined as an interval between the extrusion of the second PB and the 2-cell stage. On average, the first cell cycle duration was 22.34 h, but again, no significant difference between individual division patterns was found (Supplementary Fig. 1c).
Taken together, our analysis of clinical timelapse records revealed that the first mitotic division in human embryos occurs predominantly at the right angle to the axis of juxtaposed PNs, irrespective of the PB position. Because adopting the alternative configuration did not extend the duration of cell division, the timing of cell division appears to be independent of its orientation.
The pattern of the first mitotic cleavage is associated with the clustering of nucleoli
The geometry of the spindle apparatus defines the orientation of cell division. We hypothesized that forming the spindle that ensures division perpendicularly to the PN direction would require positioning duplicated centrosomes close to the PN interface. Recent evidence indicates that centrosomes are also implicated in microtubule-dependent chromatin and nucleoli clustering at the pronuclear interface in bovine and human zygotes23. Therefore, we sought to determine if the first mitotic division’s geometry, presumably dependent on centrosome position, is related to nucleolar clustering in human embryos.
The distribution of nucleoli was assessed 5 min before NEBD, and nucleoli were considered clustered if they all aligned along the pronuclear interface in both PNs (Fig. 2a). We have found that the overall incidence of incomplete nucleolar clustering was remarkably high in our dataset (48.3%). The unclustered nucleolar phenotype was particularly pronounced in non-developing CA embryos (63.7%) and FI embryos (63.0%). However, we noticed that also 39.4% of LB zygotes failed to cluster nucleoli before mitosis onset (Fig. 2a). This observation challenges the predictive value of nucleolar configuration scoring for embryo quality assessment, as the presence of unclustered nucleoli shortly before NEBD was compatible with normal embryo development.
a Representative images of nucleolar clustering patterns observed in human embryos and their incidence in cleavage arrested (CA), failed to implant (FI) and cleavage arrested (CA) embryos; yellow dashed line marks borders of pronuclei, red dots indicate the position of individual nucleoli within each pronucleus. b Quantification of observed clustering patterns with respect to the orientation of the first mitotic division. The number of embryos is specified in italics. Chi-square test: *p < 0.05; ***p < 0.001. Source data are provided as a Source Data file.
Next, we investigated the inefficiency of nucleolar clustering with respect to the orientation of the first mitosis in our dataset. We observed that, regardless of their developmental outcome, zygotes dividing in a non-perpendicular (parallel or oblique) direction were significantly less successful in nucleoli clustering than perpendicularly dividing embryos from the same subgroup (Fig. 2b). This trend was particularly pronounced in non-developing CA group, where 69.6% of embryos that divided with a non-perpendicular phenotype failed to cluster nucleoli before the onset of mitosis (Fig. 2b). These data suggest that the nucleolar clustering and division pattern of the first cleavage are interlinked.
Division orientation and nucleolar clustering support accurate first zygotic division
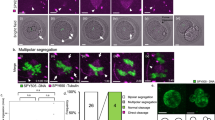
We reasoned that the non-perpendicular division geometry may be attributed to atypical centrosome positioning, which could disrupt first mitotic division either through insufficient chromatin clustering or a failure to capture condensed chromosomes. Errors in chromosome segregation during zygotic division may manifest in the nucleolar status of resulting blastomeres8 or completely arrest embryo development24. Here, we assessed the incidence of multinucleation (MuN), defined as the presence of multiple nuclear variants (visible under the conventional light microscope) in at least one blastomere of the 2-cell stage embryos (Fig. 3a). We noted that the incidence of MuN embryos was remarkably high in the group of poor-quality CA embryos (82.9%) and lower in FI embryos (57.6%). But importantly, more than half (50.7%) of the LB embryos capable of producing healthy pregnancy also presented more than one nucleus in one or both blastomeres (Fig. 3b). This finding implies that MuN at the 2-cell stage does not impede successful embryo development and thus should not be regarded as a deselection marker in clinical practice.
a Representative brightfield images of monunucleated (MoN) 2-cell stage embryos displaying a single nucleus in each blastomere and different degrees of multinucleation (MuN). b Bar charts showing the percentage of MoN and MuN embryos at the 2-cell in cleavage arrested (CA), failed to implant (FI) and cleavage arrested (CA) embryos. c percentage of MuN and MoN embryos resulting from perpendicular and non-perpendicular division. d The proportion of MoN/MuN 2-cell stage CA/FI/LB embryos entering mitosis with (un)clustered nucleoli. The number of embryos is specified in italics. Chi-square test: “ns” (non-significant difference) p > 0.05; *p < 0.05; **p < 0.01; ***p < 0.001. Source data are provided as a Source Data file.
Next, we set out to explore if the nuclear status of blastomeres at the 2-cell stage is related to the orientation of zygotic division. In cases where the first mitosis resulted in two mononucleated (MoN) blastomeres, perpendicular mitosis was seen more often than alternative division patterns. This was true for LB embryos as well as their FI and CA siblings (66.2%, 61.7%, and 75.7%, respectively). However, non-perpendicular mitosis could only account for 28.9% of multinucleations in embryos that resulted in full-term pregnancies (Fig. 3c). This data indicates that successful zygotic division producing two MoN blastomeres requires the involvement of additional mechanisms other than spindle geometry.
Our previous results showed that the perpendicular pattern of mitosis is linked to nucleoli clustering (Fig. 2b). Therefore, we sought to determine if clustered nucleoli configuration prevented MuN. When we compared LB embryos exhibiting MoN at the 2-cell stage with their non-developing siblings, we observed that high-quality embryos were significantly more likely to enter mitosis with clustered nucleoli. On the contrary, the majority of CA embryos, which presented MuN at the 2-cell stage, failed to cluster nucleoli before NEBD (Fig. 3d).
Collectively, these data imply that both the geometry of the first mitosis and efficient nucleolar clustering determine the nuclear status of blastomeres at the 2-cell stage.
Spatial organization of the first mitosis declines with maternal age
The negative effects of reproductive aging on female fertility and outcomes of IVF treatments are well documented12. Therefore, we asked whether maternal age affects the fidelity of the first mitosis in our LB embryos. By plotting the ratio of MuN embryos at the 2-cell stage and female egg age, we observed a positive correlation between these two variables (Fig. 4a). In women older than 35 years, 75.0% of embryos exhibited multiple nuclei in at least one blastomere. However, the association between maternal age and the presence of MuN was not observed in sibling cleavage arrested (CA) and failed-to-implant (FI) embryos (Supplementary Fig. 2).
a Correlation between egg age and multinucleation (MuN) incidence at the 2-cell stage. Red dots mark embryos derived from young donors’ eggs (aged 18, 20, and 22 years). b, c Quantification of MuN in 2-cell and 4-cell stage live birth embryos in individual age groups. The number of embryos is specified in italics. d Correlation between egg age and duration of the first cell cycle (white) and mitosis (gray) in individual age groups. e Correlation between egg age and incidence of perpendicular cell division. f Correlation between egg age and incidence of unclustered nucleoli before the NEBD onset. Simple linear regression was used to establish the correlation between variables. Source data are provided as a Source Data file.
We further traced the nuclear status of blastomeres after the second mitotic division. The total incidence of MuN in LB embryos markedly decreased after the next round of cell division (50.7% at the 2-cell stage vs. 8.3% at the 4-cell stage, respectively, Fig. 4b, c). This observation is in line with previous reports proposing the existence of a molecular mechanism by which embryos ensure self-correction of chromosome segregation errors11.
We observed that the proportion of MuN-blastomeres remained higher in 4-cell stage LB embryos derived from oocytes of >35 years of age compared to younger age subgroups (Fig. 4b, c). Of note is that LB embryos that developed from very young eggs, aged 18, 20 and 22 years, also exhibited a high incidence of MuN at the 2-cell stage (Fig. 3a; marked in red). This raises the question if this phenotype might be underlined by similar mechanisms contributing to the high propensity of very young eggs to chromosome non-disjunction12.
The timing of the first cell cycle has been associated with the developmental potential of the human embryos25,26,27. Therefore, we analyzed the durations of the first cell cycle and mitosis in individual age groups to investigate the temporal regulation of these events in humans. The first cell cycle took, on average, 22.14 ± 2.45 h in the <30 years age group, whereas in the >35 years age group, it was 23.19 ± 3.01 h. We reasoned that a longer time to reach the 2-cell stage might result from longer mitosis in the >35 years age group and explain the high incidence of MuN embryos. The first mitosis was slightly longer in the >35 years group, but this difference was not statistically significant (Fig. 4d, Supplementary Fig. 1d–f).
Our previous results indicate that the first mitosis in humans is controlled by spatial rather than temporal cues (Supplementary Fig. 1a–c). Therefore, we further investigated how age impacts spatial control over the first mitosis and analyzed patterns of the zygotic division across different age groups. We observed that embryos in the >35 years age group were less likely to divide perpendicularly to the PN axis (55.8%) than embryos from younger age groups (69.6% in <30 and 71.2% in the 30–35 age groups). However, the correlation between egg age and the incidence of perpendicular cell division did not reach significance in our analysis (Fig. 4e).
Failure to achieve single nuclear status after cell division was accompanied by an increased ratio of embryos with unclustered nucleoli in the >35-year-old group (Fig. 4f). Altogether, these findings suggest that spatial control over the first mitotic division may decline with reproductive aging.
Discussion
Early human development is highly error-prone. Incorrect kinetochore-microtubule attachment, manifested as chromosome lagging during anaphase, can lead to chromosome missegregation and embryonic aneuploidy28,29. Gross segregation defects like tripolar cell division can completely arrest human development24,30.
The study of genetic instability during the earliest stages of human development is hindered by restrictions on producing human embryos for research. Nevertheless, introducing continuous embryo monitoring into clinical IVF practice now provides unique insight into the initial stages of the developmental process. In a present study, we retrospectively analyzed a unique collection of 300 timelapse records covering the preimplantation development of human IVF embryos that gave rise to healthy live births. The complementary analysis involved 207 sibling cleavage-arrested embryos and 111 embryos that developed to the blastocyst stage but failed to implant and establish viable pregnancies. We observed that human embryos tend to divide at the right angle to the axis of juxtaposed PNs, irrespectively to the PB position. This observation contrasts with the textbook view derived from experiments on mouse embryos, in which the position of the PB and sperm entry site heavily influences the orientation of the first cell division7.
One of the possible explanations for the different spatial organization of cell division is a distinct composition of spindle apparatus in rodent and non-rodent zygotes. In mice, the zygotic spindle is established by numerous MTOCs surrounding parental pronuclei, whereas in humans, it is crucial to correctly position paternally inherited centrosomes to ensure effective chromosome capture. Based on clinical timelapse data from IVF embryos, we hypothesize that in humans, nascent centrosomes organize the first mitotic spindle at the right angle to juxtaposed PNs (Fig. 5). The ultimate proof of the hypothesized role of centrosomes in defining the first mitotic spindle geometry and zygotic division orientation could be brought by visualizing centrosome markers using live-cell imaging. However, we could not adopt such an experimental design due to ethical restrictions. Nevertheless, our assumption is supported by previous reports showing that centrosomes drive spindle assembly in discarded 3PN zygotes using fixed or live-cell imaging31.
Schematics recapitulating key developmental events that determine the outcome of the first mitosis. The top row shows the apposition of pronuclei with subsequent clustering of nucleoli and chromatin by a centrosome-driven microtubule-dependent mechanism; centrosomes organize mitotic spindle assembly perpendicularly to the juxtaposed pronuclei, thus promoting the correct segregation of chromosomes. The bottom row depicts the deregulation of the spatial control over the first mitosis seen in embryos from reproductively aged eggs.
Our data from the large dataset of high-quality IVF embryos support recent experimental evidence revealing the role of centrosomes in microtubule-dependent clustering of chromatin and nucleoli at the PN interface bovine embryos23. We propose that the position of centrosomes close to the pronuclear interface results in optimal nucleolar clustering that, together with the perpendicular pattern of zygotic mitosis, leads to the emergence of a single nucleus per daughter blastomere. Importantly, the timing and duration of the first mitosis were not affected by different division patterns, indicating that temporal and spatial control over the first zygotic mitosis is not coupled. Therefore, even when mitotic machinery is not properly established, deviation from perpendicular cleavage geometry alone does not prevent the completion of zygotic division.
In this study, we confirmed previous reports that MuN is particularly frequent in human embryos at the 2-cell stage8,32. A study by Currie et al. has demonstrated that chromosome segregation errors during first mitosis result in altered nuclear status of daughter blastomeres8. During the revision of this paper, the preprint published by Ono et al.33 monitored the spindle dynamics in human zygotes using fluorescent live-cell imaging. Their data showed that defocusing of spindle poles is strongly associated with multinucleation at the 2-cell stage. This supports our hypothesis that non-perpendicular division is caused by alteration of spindle geometry.
The presence of multiple nuclei after the cell division was previously associated with chromosomal abnormalities in human embryos34,35,36. However, the causes and consequences of this phenotype are under debate. A study by McCoy et al.37 performed a large analysis of the genetic background of embryos developed to blastocyst or arrested in cleavage state. In their study, the authors show that the incidence of meiotic but not mitotic errors is affected by maternal age. Here, we found that spatial control over the first mitosis becomes altered with female reproductive aging, leading to an increase of MuN in LB embryos (summarized in Fig. 5). Yet, the incidence of MuN in sibling cleavage arrested embryos remained high regardless of maternal age (Supplementary Fig. 2). Additionally, we observed that multiple nuclei phenotype may disappear in the next cell divisions, as reported previously32,33. Collectively, this evidence argues against the use of MuN as a marker of embryo quality. However, more studies are needed to understand the link between nuclear status and the chromosomal composition of daughter cells.
The molecular basis of age-associated decline in spatial control over the first mitosis observed in this study awaits further research. As reported by Fishman et al., sperm-derived centrosome recruits maternal proteins to gain its function in the fertilized egg38. Hence, the age-related decline of maternal mRNAs, which is important for centrosome function, may explain the decreased developmental capacity of eggs derived from women of advanced reproductive age. Recent evidence confirms that egg transcriptome changes with reproductive aging39. An in-depth analysis is needed to answer the question of whether age-related decay of centrosomal mRNAs may explain the increased incidence of multinucleation in older patients.
In conclusion, we analyzed a unique clinical dataset of human embryos and revealed previously undescribed spatial cues affecting the outcome of the first mitotic division. Future mechanistic studies exploring how the positioning of nascent centrosomes affects zygotic cleavage direction are necessary to refine our understanding of enigmatic preimplantation development, which is particularly important in the context of assisted reproduction. A deeper knowledge of the molecular composition of sperm-derived centrosomes will bring an understanding of the causes of fertilization failure and unexplained infertility.
Methods
Study design
A collection of timelapse videos covering the preimplantation development of 618 human embryos was retrospectively analyzed in this study. All embryos were produced for the purpose of fertility treatment; standard clinical and laboratory procedures were used. Embryos that resulted in a live birth after the single embryo transfer were assigned to the LB group, whereas the same cycle “sibling” embryos that arrested in cleavage stage or failed to implant and establish pregnancy were allocated to CA and FI groups, respectively. The average age of female participants was 27.5 years; 71.67% of embryos were derived from eggs of young women with no reproductive issues who were enrolled in an egg donation program. No vitrified-thawed eggs were used; severe andrological factor was present in reported in 8% of cases. Participation in the research was voluntary and unpaid and did not affect the treatment regime. Informed consent with analysis of anonymized timelapse records, and laboratory and clinical data was obtained before the cycle commenced. The study was undertaken under ethical approvals issued by the Institutional Review Boards of the collaborating academic and clinical unit (MUNI MED 81/2022 and RF 3/2018).
Timelapse imaging of human embryos
Timelapse recording was performed in the humidified GERI timelapse incubator (Genea BIOMEDX, Australia) and started no later than 20 min after intracytoplasmic sperm injection. Embryos were cultured in individual microwells in a CSCC medium (Irvine Scientific) covered with mineral oil at 37 °C, 5% O2, 6% CO2, and under humid conditions. Transmitted light images have been acquired automatically with a 5-min frequency in 11 focal planes covering the whole embryo volume. Data from each embryo have been analyzed by at least three clinical embryologists and two basic scientists to avoid bias.
Analysis of axes orientation
The position of the first polar body defined the polar body (PB) axis. The alignment of the PB and the pronuclear (PN) axis was analyzed one timeframe before the time point when the nuclear contour disappeared at nuclear envelope breakdown (NEBD) and was categorized as either longitudinal, intermediate, or orthogonal. The relationship between the PN axis and the cell division axis has been analyzed after the successful completion of the first cell division at the first frame when sharp boundaries between cells were observed. The onset of the mitotic division was defined as an interval between the timeframe when the PB was visibly extruded and the timeframe when NEBD was observed. The duration of the mitotic division was defined as an interval between the timeframe when NEBD and the first timeframe when two separate blastomeres could be distinguished.
Data and statistical analysis
A Chi-square test was used to confirm that the actual frequency differed significantly from the expected frequency, debunking the expectation of no correlation between the groups and the phenomena. Student’s t-test was used to analyze differences in the onset and duration of the first mitosis and the duration of the first cell cycle. A simple linear regression model was applied to assess the correlation between maternal egg age as the independent variable and the incidence of the selected phenotype. All analyses were performed in GraphPad Prism v9.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Timelapse records were annotated directly in the commercially available GERI software (version 2.0) and are accessible to authorized personnel only. The embryo monitoring data are part of medical documentation and can not be made public due to legal and security reasons. Selected anonymized data can be exported upon request to the corresponding author (Z.H.) and approval of the collaborating clinical unit. Source data are provided with this paper.
References
Cha, B.-J., Serbus, L. R., Koppetsch, B. S. & Theurkauf, W. E. Kinesin I-dependent cortical exclusion restricts pole plasm to the oocyte posterior. Nat. Cell Biol. 4, 592–598 (2002).
Goldstein, B. & Hird, S. N. Specification of the anteroposterior axis in Caenorhabditis elegans. Development 122, 1467–1474 (1996).
Kikkawa, M., Takano, K. & Shinagawa, A. Location and behavior of dorsal determinants during first cell cycle in Xenopus eggs. Development 122, 3687–3696 (1996).
Hiiragi, T. & Solter, D. First cleavage plane of the mouse egg is not predetermined but defined by the topology of the two apposing pronuclei. Nature 430, 360–364 (2004).
Louvet-Vallée, S., Vinot, S. & Maro, B. Mitotic spindles and cleavage planes are oriented randomly in the two-cell mouse embryo. Curr. Biol. 15, 464–469 (2005).
Plusa, B., Grabarek, J. B., Piotrowska, K., Glover, D. M. & Zernicka-Goetz, M. Site of the previous meiotic division defines cleavage orientation in the mouse embryo. Nat. Cell Biol. 4, 811–815 (2002).
Plusa, B., Piotrowska, K. & Zernicka-Goetz, M. Sperm entry position provides a surface marker for the first cleavage plane of the mouse zygote. Genesis 32, 193–198 (2002).
Currie, C. E. et al. The first mitotic division of human embryos is highly error prone. Nat. Commun. 13, 6755 (2022).
Hardarson, T., Hanson, C., Sjögren, A. & Lundin, K. Human embryos with unevenly sized blastomeres have lower pregnancy and implantation rates: indications for aneuploidy and multinucleation. Hum. Reprod. 16, 313–318 (2001).
Wartosch, L. et al. Origins and mechanisms leading to aneuploidy in human eggs. Prenat. Diagn. 41, 620–630 (2021).
Daughtry, B. L. et al. Single-cell sequencing of primate preimplantation embryos reveals chromosome elimination via cellular fragmentation and blastomere exclusion. Genome Res. 29, 367–382 (2019).
Gruhn, J. R. et al. Chromosome errors in human eggs shape natural fertility over reproductive life span. Science 365, 1466–1469 (2019).
Scott, L. Pronuclear scoring as a predictor of embryo development. Reprod. Biomed. Online 6, 201–214 (2003).
Gámiz, P. et al. The effect of pronuclear morphology on early development and chromosomal abnormalities in cleavage‐stage embryos. Hum. Reprod. 18, 2413–2419 (2003).
Gardner, R. L. Specification of embryonic axes begins before cleavage in normal mouse development. Development 128, 839–847 (2001).
Gardner, R. L. & Davies, T. J. An investigation of the origin and significance of bilateral symmetry of the pronuclear zygote in the mouse. Hum. Reprod. 21, 492–502 (2006).
Garello, C. et al. Pronuclear orientation, polar body placement, and embryo quality after intracytoplasmic sperm injection and in-vitro fertilization: further evidence for polarity in human oocytes? Hum. Reprod. 14, 2588–2595 (1999).
Payne, D., Flaherty, S. P., Barry, M. F. & Matthews, C. D. Preliminary observations on polar body extrusion and pronuclear formation in human oocytes using time-lapse video cinematography. Hum. Reprod. 12, 532–541 (1997).
Gianaroli, L., Magli, M. C., Ferraretti, A. P., Fortini, D. & Grieco, N. Pronuclear morphology and chromosomal abnormalities as scoring criteria for embryo selection. Fertil. Steril. 80, 341–349 (2003).
Tesarik, J. & Greco, E. The probability of abnormal preimplantation development can be predicted by a single static observation on pronuclear stage morphology. Hum. Reprod. 14, 1318–1323 (1999).
Coticchio, G., Borini, A., Zacà, C., Makrakis, E. & Sfontouris, I. Fertilization signatures as biomarkers of embryo quality. Hum. Reprod. 37, deac123 (2022).
Inoue, T. et al. The migration speed of nucleolar precursor bodies in pronuclei affects in vitro fertilization-derived human embryo ploidy status and live birth. Reprod. Med. Biol. 22, e12497 (2023).
Cavazza, T. et al. Parental genome unification is highly error-prone in mammalian embryos. Cell 184, 2860–2877.e22 (2021).
Ottolini, C. S. et al. Tripolar mitosis and partitioning of the genome arrests human preimplantation development in vitro. Sci. Rep. 7, 9744 (2017).
Fenwick, J. Time from insemination to first cleavage predicts developmental competence of human preimplantation embryos in vitro. Hum. Reprod. 17, 407–412 (2002).
Lundin, K., Bergh, C. & Hardarson, T. Early embryo cleavage is a strong indicator of embryo quality in human IVF. Hum. Reprod. 16, 2652–2657 (2001).
Sakkas, D., Shoukir, Y., Chardonnens, D., Bianchi, P. G. & Campana, A. Early cleavage of human embryos to the two-cell stage after intracytoplasmic sperm injection as an indicator of embryo viability. Hum. Reprod. 13, 182–187 (1998).
Vázquez-Diez, C., Paim, L. M. G. & FitzHarris, G. Cell-size-independent spindle checkpoint failure underlies chromosome segregation error in mouse embryos. Curr. Biol. 29, 865–873.e3 (2019).
Paim, L. M. G. & FitzHarris, G. Tetraploidy causes chromosomal instability in acentriolar mouse embryos. Nat. Commun. 10, 4834 (2019).
McCollin, A., Swann, R. L., Summers, M. C., Handyside, A. H. & Ottolini, C. S. Abnormal cleavage and developmental arrest of human preimplantation embryos in vitro. Eur. J. Med. Genet. 63, 103651 (2020).
Kai, Y., Kawano, H. & Yamashita, N. First mitotic spindle formation is led by sperm centrosome-dependent MTOCs in humans. Reproduction 161, V19–V22 (2021).
Hashimoto, S. et al. Multinucleation per se is not always sufficient as a marker of abnormality to decide against transferring human embryos. Fertil. Steril. 106, 133–139.e6 (2016).
Ono, Y. et al. Shape of the first mitotic spindles impacts nucleation in human embryos. Nat. Commun. 15, 5381 (2024).
Daphnis, D. D. et al. Detailed FISH analysis of day 5 human embryos reveals the mechanisms leading to mosaic aneuploidy. Hum. Reprod. 20, 129–137 (2005).
Kligman, I., Benadiva, C., Alikani, M. & Munné, S. Fertilization and early embryology: the presence of multinucleated blastomerés in human embryos is correlated with chromosomal abnormalities. Hum. Reprod. 11, 1492–1498 (1996).
Staessen, C. & Van Steirteghem, A. The genetic constitution of multinuclear blastomeres and their derivative daughter blastomeres. Hum. Reprod. 13, 1625–1631 (1998).
McCoy, R. C. et al. Meiotic and mitotic aneuploidies drive arrest of in vitro fertilized human preimplantation embryos. Genome Med. 15, 77 (2023).
Fishman, E. L. et al. A novel atypical sperm centriole is functional during human fertilization. Nat. Commun. 9, 2210 (2018).
Alvarez, N. S., Brachova, P. & Christenson, L. K. Codon composition in human oocytes reveals age-associated defects in mRNA decay. Preprint at bioRxiv https://doi.org/10.1101/2021.01.05.425501 (2021).
Acknowledgements
We thank Reprofit International for enabling this research by providing access to clinical timelapse records. We would like to express our gratitude to all patients who participated in this study and the IVF clinic staff for cooperation, administration of relevant documentation and long-term follow-up of clinical outcomes. We also thank Prof Mary Herbert and Dr. Anna MacGilavry-Danylevska for valuable discussions and critical reading of the manuscript. We thank Tami Bockova for her input on statistical analysis. V.P. and Z.H. are supported by funds from Masaryk University (MUNI/A/1301/2022; MUNI/A/1598/2023; MUNI/IGA/1127/2021). V.P. was supported by Boehringer Ingelheim Fond PhD Fellowship. We are also thankful for continuous support from the FutureLife consortium and Brno City Municipality–Brno PhD talent programme.
Author information
Authors and Affiliations
Contributions
V.P.: Data analysis, and interpretation, manuscript writing, and figure design; D.K.: Annotation of timelapse movies, clinical outcome follow-up; M.M. and T.K.: Annotation of timelapse movies; P.O. and S.K.: Patient’s recruitment, clinical data curation, relevant documentation management and manuscript critical reading; Z.H.: Project design, organization and supervision, data analysis and interpretation, manuscript writing. All authors discussed the results and viewed the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Porokh, V., Kyjovská, D., Martonová, M. et al. Zygotic spindle orientation defines cleavage pattern and nuclear status of human embryos. Nat Commun 15, 6369 (2024). https://doi.org/10.1038/s41467-024-50732-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50732-z
- Springer Nature Limited