Abstract
Pathogens of the enterovirus genus, including poliovirus and coxsackieviruses, typically circulate in the summer months suggesting a possible positive association between warmer weather and transmission. Here we evaluate the environmental and demographic drivers of enterovirus transmission, as well as the implications of climate change for future enterovirus circulation. We leverage pre-vaccination era data on polio in the US as well as data on two enterovirus A serotypes in China and Japan that are known to cause hand, foot, and mouth disease. Using mechanistic modeling and statistical approaches, we find that enterovirus transmission appears positively correlated with temperature although demographic factors, particularly the timing of school semesters, remain important. We use temperature projections from Coupled Model Intercomparison Project Phase 6 (CMIP6) to simulate future outbreaks under late 21st-century climate change for Chinese provinces. We find that outbreak size increases with climate change on average, though results differ across climate models depending on the degree of wintertime warming. In the worst-case scenario, we project peak outbreaks in some locations could increase by up to 40%.
Similar content being viewed by others
Introduction
Understanding the implications of climate change for infectious disease is critical for public health planning and adaptation1,2,3,4,5. Many directly transmitted diseases—diseases that are transmitted directly person-to-person e.g., influenza—exhibit a sensitivity to climate factors in both experimental and population-level settings2,6,7. The transmission of several wintertime directly transmitted diseases, such as influenza and respiratory syncytial virus, has been shown to be negatively correlated with absolute humidity8,9,10, although a possible non-linear response may explain the timing of outbreaks in tropical locations11,12. In contrast, the directly-transmitted enteroviruses cause summertime disease outbreaks in temperate climates, exhibiting a correlation with warmer conditions13,14,15. Given the positive association with warmer weather, it is important to explore how climate change might impact enterovirus outbreaks in the future.
The enterovirus genus is a large group of non-enveloped RNA viruses including the polioviruses, coxsackieviruses, and other serotypes15. Perhaps the best-known enteroviruses are the polioviruses—three serotypes of enterovirus C which are responsible for poliomyelitis, a severe disease that can cause paralysis. Highly effective vaccines developed in the 1950s and 1960s have led to the local elimination of polio in almost all countries16, but global eradication has remained an elusive long-term goal17. Several enterovirus serotypes are also responsible for hand, foot, and mouth disease (HFMD)—an infection that causes a fever, mouth sores, and a rash on hands and feet, typically in young children, with neurological complications sometimes occurring in severe cases18. HFMD has been a particular concern in Asian countries since the 1990s, with high caseloads in China and Japan19, although outbreaks have occurred globally20.
Both polio and non-polio enteroviruses exhibit latitudinal gradients in the timing of epidemics, with earlier outbreaks observed in lower latitude locations13,14,15. Preliminary evidence suggests a role for climate in driving seasonal transmission: dew point temperature was found to correlate with transmission intensity for non-polio enteroviruses in the US15, whereas humidity appeared to influence the viability of the poliovirus in laboratory settings21. However, more work is needed to understand the role of climate in determining seasonal enterovirus outbreaks in other locations. In particular, locations closer to the tropics have been observed to have two enterovirus outbreaks year—a phenomenon that cannot be explained by a linear temperature relationship22.
Here we use two datasets of two enterovirus serotypes from China and Japan to explore possible climate drivers and assess the implications of climate change on epidemic peak size. Enterovirus 71 (EVA71) and coxsackievirus 16 (CVA16) are two of the leading causes of hand, foot, and mouth disease (HFMD) in China and Japan23,24. We use weekly time series from 2009 to 2013 of EVA71 and CVA16 case counts from Chinese provinces. These time series are derived from total province-level HFMD case data as well as laboratory testing for EVA71 and CVA16 serotypes for a subsample of cases, as described in previous work23. In addition to the Chinese data, we analyse national time series of weekly EVA71 and CVA16 cases in Japan spanning the period 1982–2015. We compare our results for the effect of climate on EVA71 and CVA16 with an analysis of pre-vaccination era weekly polio case data from the continental US states spanning 1940–1960 available from Project Tycho25. While we do not consider the implications of climate change for polio, comparing climate sensitivity across these pathogens allows us to explore whether a common set of environmental drivers exists across distinct enterovirus serotypes.
Results
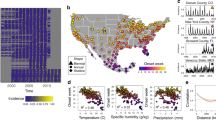
We first examine spatial patterns in the timing and intensity of EVA71, CVA16, and polio outbreaks in China, Japan, and the USA. In Fig. 1A we map the mean timing of cases (see “Methods”) of these diseases. A latitudinal gradient is observed for all three pathogens where earlier outbreaks occur in more southern locations and later outbreaks occur in more northern locations. In Fig. 1B we plot the relationship between mean temperature and mean timing of cases across locations. We find a significant negative effect of mean temperature on outbreak timing (p << 0.001, p << 0.001, p = 0.024 for polio, EVA71, and CVA16 respectively for the coefficient of an ordinary least squares (OLS) regression model) where for all three serotypes, earlier outbreaks are associated with higher mean temperatures. For EVA71 and CVA16 the relationship appears nonlinear; this is driven by the emergence of a second annual peak that is observed for both serotypes in southern locations in China (Fig. 1C). The second annual peak increases in size as latitudes decline and becomes similar in magnitude to the primary peak in tropical Hainan province. For polio, a secondary peak is not observed; however, outbreaks appear longer lasting in more southern locations. In Fig. 1C, both Nevada and Tibet appear to be outliers in terms of seasonality based on latitude. This is likely due to the sparse population numbers in these locations (Nevada was the least populous US state in 1950, apart from Alaska) making it hard to discern the seasonal trend.
A Maps showing the mean timing of cases in terms of week-of-year of polio outbreaks in the USA, EVA71 outbreaks in mainland China, CVA16 outbreaks in mainland China. B Scatter plot of mean annual temperature for US state or Chinese province against mean timing of cases for polio, EVA71, and CVA16. Local polynomial regression line is shown in black with 95% confidence intervals (CVA16, EVA71, n = 31; polio, n = 49). C Normalized average weekly cases of polio in the USA and EVA71 in mainland China and Japan. Locations are ordered by latitude. Supplementary Fig. 1 shows the result for CVA16. D Scatter plot of average temperature range (mean max.–mean. min) and epidemic intensity (see “Methods”) for polio, EVA71, and CVA16. Again, each point indicates a US state or Chinese province.
In Fig. 1D we characterize the annual distribution of cases by calculating the epidemic intensity (see “Methods”) where low-intensity implies cases are more distributed throughout the year, and high-intensity implies cases are more concentrated in time26. We find that more intense outbreaks occur in locations with a larger seasonal range of temperature values and that this pattern is significant (p << 0.001 for polio, EVA71, and CVA16 respectively, based on OLS regression of temperature range on outbreak intensity) across all three serotypes.
Estimating the effect of climate on transmission
The spatial correlations shown in Fig. 1 imply a role for climate in determining the timing of and intensity of enterovirus outbreaks. In order to further test this theory while accounting for the non-linear dynamics of the disease system, we estimate the effect of climate on enterovirus transmission. We calculate the weekly empirical transmission rate for EVA71 and CVA16 in Japan and Chinese provinces, and polio in US states, based on the discrete time version of the Susceptible-Infected-Recovered (TSIR) model, following a methodology developed in previous work8,27 (TSIR model fits are shown in Supplementary Fig. 3). The empirical transmission rate is given by \(Em{\beta }_{t}=\frac{{I}_{t+1}{N}_{t}}{{I}_{t}^{\alpha }{S}_{t}}\) where I is the infected population, S is the susceptible population, N is the total population, and α is a constant that accounts for the discretization of the continuous-time process. We fix α at 0.975 based on previous estimates24. The time step t represents the generation time of the enterovirus infection and is set at 1 week23.
We merge the dataset of transmission rates for polio, EVA71, and CVA16 with spatially and temporally resolved climate data. For EVA71 and CVA16 we use specific humidity, relative humidity, and temperature data from the ERA5 reanalysis28, and precipitation from the Climate Hazards Group InfraRed Precipitation with Station data (CHIRPS)29. Because ERA5 data is not available for the span of the 1940–1960 polio time series, we use historic temperature data from Berkeley Earth for the polio analysis30. Gridded-specific humidity, relative humidity, and precipitation data are not available for this time period. All gridded climate datasets are spatially and temporally averaged to match the resolution of the disease data (for a list of data sources see Supplementary Table 1).
We fit separate regressions (EVA71, CVA16, polio) of temperature, specific humidity, relative humidity, and precipitation on log empirical transmission rates. Prior work suggests that regressing climate factors on transmission, as opposed to incidence, provides an unbiased estimate of the climate effect31. We find a significant positive effect of temperature on transmission; the effect of specific humidity, relative humidity, and precipitation was not found to be significant (Supplementary Table 2). In order to remove possible spatial and temporal biases, we additively include location and time (year-level) dummies to the panel regression of temperature on transmission32 (Fig. 2A). The positive coefficient on temperature remains significant even with the additional controls.
A Estimated effects of temperature on transmission for polio, EVA71, and CVA16 (n = 30823, 8161, 8810). X-axis depicts the coefficient on temperature and the bar shows the 95% confidence interval on this estimate. For each pathogen, fixed effects (dummy variables) for location, year, and either schooling or week are included additively (y-axis). B Simulation results over varied seasonal temperature ranges and mean transmission rate using the EVA71 estimated temperature and schooling coefficients. The time series of observed mean weekly cases for five provinces is shown on the left-hand side. Points on surface indicate the predicted dynamics within these locations. Similar plots for CVA16 and polio are shown in (C) and (D), respectively. The ratio of the schooling peak to temperature peak for locations with two annual peaks for EVA71 is shown in (E). White region indicates there is only one peak per year.
We separately test for the effect of dew point temperature on transmission (Supplementary Table 3) which was found to be the best predictor of enterovirus seasonality in prior work on the US15. We find dew point temperature is significantly associated with transmission but is a marginally worse predictor of enterovirus transmission than temperature (Supplementary Table 3). Given the strong correlation and functional relationship between temperature and dew point temperature, and the lack of association between relative humidity and transmission, our results suggest that dew point temperature is capturing the temperature effect on transmission in the regression model. We retain temperature as our main predictor for future projections.
Seasonal aggregation of children via schooling is expected to be a key driver of transmission for several childhood infections33,34. We add a dummy variable for school timing in the CVA16 and EVA71 regressions based on estimates of school semester dates in China and Japan, and for polio we add a weekly dummy variable to account for school timing (historic data on schooling is not available). For EVA71 and CVA16 we find a significant effect of schooling on transmission in a regression model that includes both temperature and schooling (Supplementary Table 4). In addition, adding schooling to the model increased the alignment of the temperature coefficient across all the three pathogens, suggesting a possible common effect of temperature across enterovirus serotypes (Fig. 2A).
Our results imply that both temperature and schooling impact enterovirus transmission. We assess whether these factors can explain regional differences in disease dynamics by incorporating the estimated effect on transmission into our epidemiological model. We use our model to simulate disease outbreaks over a range of seasonal temperature values and mean transmission rates while keeping school semester dates fixed at the values for China. We repeat this exercise for EVA71 (Fig. 2B), CVA16 (Fig. 2C), and polio (Fig. 2D).
The image plot (right side, Fig. 2B) shows the periodicity of modeled outbreaks, while the time series plots (left side) show the observed seasonality of EVA71 in different Chinese provinces. As the seasonal range of temperature increases, modeled outbreaks shift from semi-annual i.e., two outbreaks per year (periodicity = 0.5) to annual (periodicity = 1). In the semi-annual regime, the first peak is driven by the seasonal peak of temperature (which overlaps with the spring schooling semester), and the second peak is driven by schooling alone in the fall semester. As the temperature range increases, the relative effect of schooling becomes smaller compared to the seasonal temperature forcing, eventually leading to a single annual outbreak (e.g., Heilongjiang). For locations with a large seasonal temperature range and higher mean transmission, our model predicts periodic, or even chaotic, outbreaks can occur. For instance, the model predicts that Liaoning in northern China may experience biennial outbreaks, defined as alternating years of high and low case numbers. Our observational data suggests that biennial outbreaks occur in Liaoning (Supplementary Fig. 4). Triennial outbreaks have also been observed in transient in Japan24, and other locations15,35.
Our simulations in Fig. 2 suggest that the combined effect of temperature and schooling can explain the secondary annual enterovirus peak observed in tropical locations e.g., Hainan. In more temperate regions, the strong seasonal effect of temperature dominates and the schooling effect is diminished. In Fig. 2E we show the modeled relative size of the (secondary) schooling peak to the (primary) temperature peak. In Hainan, the model predicts that the ratio of the schooling peak to temperature peak may be >1 i.e., the schooling peak size may exceed the temperature-driven peak however, for the majority of the locations, the ratio is <1, and the schooling peak disappears eventually as the temperature range increases (white region of Fig. 2E).
Implications of climate change for enterovirus outbreaks
We use our climate-driven enterovirus model for CVA16 and EVA71 to explore the implications of climate change for outbreaks of these two diseases in China. We do not consider polio in these projections as our model for polio is based on pre-vaccination era dynamics that do not reflect current (limited) circulation.
We run simulations for EVA71 and CVA16 in China taking temperature projections from individual CMIP6 climate models. We use the delta change method to bias-correct the climate projections i.e., we calculate the weekly temperature climatology under shared socioeconomic pathway 5 (SSP585) for the future period (2080–2100) and the historic period (1990–2010) for each climate model, for each Chinese province. We then run simulations using present-day climate observations from ERA5 (spanning 1990–2020), including inter-annual variability, and these same observations plus the computed temperature change to reflect the future scenario. We specifically choose to include inter-annual variability to explore the importance of shorter-term weather changes, including extremes, on the risk of larger outbreaks36.
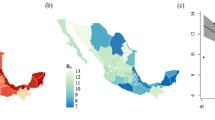
In Fig. 3 we show the impact of climate change on epidemic peak size, which is defined as the weekly maximum number infected within a year, across Chinese provinces. We show two outcomes: the impact of climate change on the average epidemic peak size across a 30-year simulation, and the impact of climate change on the maximum peak size across the 30-year simulation. Note that these two values would be the same if we had not included temperature variability within the model runs. We run simulations for each Chinese province and CMIP6 model.
On average, we find that climate change leads to an increase in average and maximum epidemic peak size in China with slightly larger increases observed for CVA16. There is substantial heterogeneity in the results, however, by both province and climate models. Some models, e.g., INM-CM4-8, predict a decline in epidemic peak size for both serotypes and the majority of provinces. This is due to the fact that INM-CM4-8 projects that wintertime temperatures in China will increase more than summertime temperatures under SSP585 (Supplementary Figs. 5, 6). In this case, the decline in the seasonal range of temperature values leads to a decline in the seasonal range of transmission values, and outbreaks become less intense. Improved understanding of the greenhouse gas-induced changes to temperature seasonality will help narrow the uncertainty of enterovirus projections.
An additional source of heterogeneity across climate models comes from the non-linear effect of temperature on transmission (Supplementary Fig. 7). In some locations, although the seasonal range in temperature declines, the seasonal range of transmission may increase due to the estimated convex temperature-transmission curve. This effect dominates for climate models, and locations, where the projected decrease in temperature change is marginal.
The majority of climate models imply an increase in outbreak size for most provinces in China. In the worst-case scenario, we find that epidemic peak size could increase by up to 40% due to climate change, depending on location and climate model. Averaging across models, we find that the change in maximum epidemic peak size varies from −5% to 24% across provinces for CVA16, and −4% to 8% across provinces for EVA71. Declines in average peak size occur in provinces such as Heilongjiang in the northernmost region of China, where several climate models again project wintertime warming exceeds summertime warming.
Accounting for temperature variability
Our climate change results also reveal the importance of variability as implied by the difference between results for maximum epidemic peak size and average epidemic peak size (Fig. 3). We find that the largest projected outbreaks occur when a relatively cooler year is followed by a warmer year. Lower-intensity outbreaks in cooler years lead to a larger pool of susceptibles in subsequent years such that if transmission increases, large outbreaks may occur. This crucial role of variability and interactions with susceptible dynamics has been underexplored in studies of climate change and disease outbreaks37.
In Fig. 4 we show the relative contribution of interannual temperature variability, the temperature coefficient, and the climate model in determining the projected epidemic peak size in Beijing under climate change. This analysis is conducted by running simulations where we sample the temperature coefficient from a normal distribution of the estimate (Fig. 2A) coupled with output from each climate model and for 30 years of weather variability: the simulation results in 12,600 annual outbreaks where we record the epidemic peak size in absolute terms. We then perform an analysis of variance (ANOVA) to analyse the relative role of these three factors in determining the outbreak size by calculating their contribution to the regression sum of squares.
We find that interannual temperature variability is the dominant driver of projected future enterovirus outbreaks for both EVA71 and CVA16 in China. Uncertainty in the temperature coefficient also plays an important role. These two factors matter substantially more than the climate model used. To build intuition behind this result: as the temperature coefficient increases, the average epidemic peak size increases across all climate models and this increase is greater than differences across models. These results suggest that improving precision of the temperature coefficient estimate could substantially reduce uncertainty in projected future outbreak size.
Discussion
Our climate change projection results and our climate-driven simulations (Fig. 2B) highlight the importance of seasonal temperature ranges in determining epidemic peak size. Seasonal temperature range determines the intensity of enterovirus outbreaks through its impact on the underlying susceptible dynamics: warmer winters lead to more year-round spread of the disease, and the supply of susceptibles limits the size of the summertime outbreak. In contrast, when climate change increases the seasonal range of temperature values, larger summertime outbreaks occur and it takes longer for the susceptible population to replenish: this can result in biennial or higher-order outbreaks. Interannual temperature variability may complicate this picture: we find that the coupled effect of climate change and temperature variability could lead to epidemic peak sizes up to 40% larger than in the present. This important role of variability should be explored for other pathogens; a better understanding of this factor could also help improve near-term infectious disease forecasts.
There are several caveats to our results. While schooling and temperature are able to predict regional differences in outbreak dynamics, these two predictors alone do not perfectly capture the shape of estimated seasonal transmission. Other factors such as seasonal mobility or national holidays may also contribute to transmission and could be explored in future work. The time series data we have from China only spans a 5-year period—this limits our ability to discern longer-term trends such as biennial outbreaks that are predicted for certain provinces. Longer time series could help further explore these dynamics as well as reduce uncertainty in the temperature coefficient (Fig. 4). A longer time series is available for Japan; however, serotype-specific data is very sparse at the prefecture level. To overcome this we use only the national-level data, but this may average over important prefecture-level differences. Although there are limitations in both the Japan and China time series, these remain globally some of the best data sources on serotype-specific enterovirus dynamics. Our projection results are limited to China, but future work should expand to other locations, depending on data availability.
There are several other aspects of global change that may influence future enterovirus outbreaks that are not considered in this study1. Demographic changes may impact future outbreaks: declining birth rates could reduce the size of the susceptible population and hence the projected epidemic peak size (although this does not impact the effect of climate change: Supplementary Fig. 8). Our main results also do not consider vaccination, although a vaccine for EVA71 was introduced in 201638. In Supplementary Fig. 9 we explore climate change simulations assuming 60% vaccination coverage starting at birth. Vaccination does not substantially alter our climate change results.
Our results suggest that a combination of climate and demographic factors determines the present-day seasonality of enterovirus circulation. Climate change implies more severe outbreaks in the future, but subtle interactions with the seasonal warming gradient complicate the picture: changes to the seasonal range of temperature may matter more than changes to mean temperature for these types of diseases. The proposed development of a multivalent vaccine may be one of the most promising approaches to mitigate possible intensifying outbreaks39. Meanwhile, nonpharmaceutical interventions have been shown to be effective at limiting enterovirus transmission14. These measures could be leveraged during periods of predicted high transmission.
We find that reducing uncertainty in the temperature coefficient could help improve precision of projections, but variability matters. The influence of variability speaks to a possible role for incorporating near-term climate forecasts into models to predict the intensity of present-day outbreaks. These early warning systems could then be used to target interventions. More broadly, the role of variability and uncertainty needs to be more carefully considered in future work on the impacts of climate change on infectious diseases.
Methods
Data
Polio case data comes from Project Tycho25; we take all data for 1940–1960 from all states. CVA16 and EVA71 data for Chinese provinces (2009–2013) were published online by Takahashi23 based on a collaboration with China CDC. CVA16 and EVA71 data for Japan, spanning 1982–2015, was taken from ref. 24. In China, the serotype-specific incidence data are calculated as the product of the weekly reports of HFMD from syndromic surveillance at the province level and the weekly prevalence of virologically-confirmed enterovirus serotypes from a subset of HFMD cases also at the province level, as derived in ref. 23. In Japan, the serotype-specific incidence data are similarly calculated as the product of the weekly reports of HFMD per sentinel site from syndromic surveillance at the national level, the number of sentinel sites at the national level, and the weekly prevalence of virologically-confirmed enterovirus serotypes from a subset of HFMD cases also at the national level, as derived in ref. 24.
Temperature data used in the polio analysis come from Berkeley Earth30 which provides historic temperature data back to 1880. The Berkeley Earth data is gridded at 1 * 1 degree latitude and longitude resolution, with the daily records. We construct spatial averages of the gridded data for each US state. For the analysis of EVA71 and CVA16 in both Japan and China we use temperature, relative humidity, and specific humidity data from ERA528 and precipitation data from CHIRPS29. ERA5 is gridded at the 0.25 * 0.25 degree latitude and longitude level, hourly. We construct spatial averages of the gridded ERA5 data for Chinese provinces and Japan. A table of climate and disease data sources is provided in Supplementary Table 1.
Birth and population data were obtained for China from the National Bureau of Statistics of China and for Japan from the Statistics Bureau of the Japanese Ministry of Internal Affairs and Communications; both datasets are also available in refs. 23,24. The timing of school holidays was estimated for China based on 2022 data from the Beijing Educational Commission40. Historic demographic data for the USA comes from the Centres for Disease Control41.
Projection data come from Coupled Model Intercomparison Project Phase 6. We use projections under Shared Socioeconomic Pathway 5, corresponding to forcing of 85 watts/meter2 (SSP585). We find 14 models have SSP585 projections. Raw grids are interpolated to a new 1 × 1 grid (i.e., lat of 89.5S–89.5N, lon of 0.5–359.5). Historic and future climatology is calculated by averaging over 20-year periods: 2080–2100 for the future, 1990–2010 for the historic period.
Epidemic intensity
Epidemic intensity is based on the inverse of the Shannon entropy as described in ref. 26. Intensity is given by \(Int={(\!-\!\! \sum {p}_{j}log({p}_{j}))}^{-1}\) where pj is the proportion of cases occurring in week j based on case numbers averaged across all years for a particular region. To make Fig. 1D, region-level intensity scores are normalized between 0 and 1 for EVA71, CVA16, and polio separately.
Mean timing of cases
We calculate the mean timing of cases by calculating the center of gravity using the “circular” package in R, following14. The center of gravity calculates the arithmetic weighted mean timing of the cases. The formula is given by:
where w is the week of year (from 1 to 52) converted to units of radians by w = week/52 ∗ 2π, and Iw is the mean cases in week w, averaged across all years for a particular location.
Maps for the mean timing of cases are produced using sub-national cartographic boundary files for China42 and the USA43.
Estimating transmission
We follow the approach developed in ref. 31 to estimate the time-varying empirical transmission rate. We estimate the empirical transmission rate using the time-series susceptible infected recovered (TSIR) model for each week in the Japan, China, and USA datasets.
It is important to note that this approach assumes that infection with a specific enterovirus serotype is immunizing. Data on EVA71 neutralization titers suggest that antibody titers remain high over time and as such the seroreversion rate is low: longitudinal serological data from Yang et al.44 estimated that the probability of EVA71 seroreversion was < 10% at 11 years of age. Furthermore, SIRS models with waning immunity fit to time series case data on EVA71, CVA16, and other non-polio enterovirus serotypes from Japan suggesting that the duration of protective immunity is long-lasting, on the order of multiple years to life-long15. However, Huang et al.45 find small rates of recurrence of HFMD infection with the same serotype: 0.05% for EVA71 and 0.04% for CVA16. Simulations generated using a SIRS model assuming a low rate of loss of immunity do not alter our main result (Supplementary Fig. 10).
The empirical transmission rate (Emβt) is given by:
where It, St, and Nt are the number of infected individuals, susceptible individuals, and total population at time period t where increments of t are fixed at the generation time, ~1 week for the enteroviruses23,24. α is a constant that accounts for the discretization of the continuous-time process and is fixed at 0.975 based on previous estimates24. It and Nt are observed in the data but St must be estimated. We estimate St by fitting the TSIR model to the time series from each location. We use the tsiR package developed by Becker et al.46 to fit the model to our data. In the TSIR model the susceptible equation is given by:
where Bt are births and ut is additive noise, with E [ut] = 0. The susceptible population can be described by \({S}_{t}=\bar{S}+{Z}_{t}\) where \(\bar{S}\) is the mean number of susceptible individuals in the population and Zt is the unknown deviation from the mean number of susceptible individuals at each time step. Equation (2) can be rewritten as:
where ρ is the reporting rate, Irk is the reported incidence and Z0 is the starting condition. From equation (3) we find that Zt can be estimated as the residuals from the linear regression of cumulative births on cumulative cases. The reporting rate ρ can be estimated as the inverse slope of the regression line. We find ρ to be ~1% for polio and 2% for EVA71 and CVA16. The expected number of infections, E [It+1], is modeled as:
which is log-linearized as:
where βt is the seasonal transmission rate estimated biweekly. The mean number of susceptible individuals, \(\bar{S}\), is estimated using marginal profile likelihoods for a range of candidate values. The \(\bar{S}\) estimates and derivation of Zt from equation (3) can be used to estimate the susceptible population and input into equation (1) to calculate the empirical transmission rate for all location-by-weeks in the dataset.
Regression model
The empirical transmission rate is used as the dependent variable in a panel regression model to estimate the effect of climate and demographic factors, specifically schooling. Panel regression models are run using location and year-fixed effects (dummy variables) and clustering standard errors at the location level to account for autocorrelation. Chinese and Japanese data are pooled together for the regression analysis and the impact of climate is explored separately for EVA71 and CVA16 (location-specific dummy variables remove mean transmission differences between Chinese provinces and Japan). Polio is analysed separately using only temperature data, as other climate variables are not available for the historic 1940–1960 time period. The polio model includes a weekly fixed effect to account for the effect of schooling.
Present-day simulations
We simulate the infectious disease model under different values for mean transmission and temperature range in Fig. 2B–E. Specifically, we calculate a weekly transmission rate as:
where βw is transmission in week w, Tr is the temperature range varied from 10 to 50 degrees Celsius reflecting the observed range in China, α is the estimated effect of temperature on transmission (from the regression model (EVA71, CVA16, and polio models used separately for Fig. B, C, and D respectively), γ is the estimated effect of schooling on transmission, Sw is a dummy variable that is equal to one if schools are in session in week w and βmean is the baseline transmission rate varied from 1 to 4 based on estimates from the regression model. Baseline transmission is estimated in the regression model as the sum of the location-specific fixed effect for a particular province, and the intercept term (which is the common baseline transmission rate across all locations). The sinusoidal term in Eq. (7) allows us to smoothly vary temperature across a range of values.
The weekly transmission term is input into the TSIR model to simulate the infected time series for a total of 100 years. We examine outbreak dynamics in the last 10 years of the model run, assuming a 90-year burn-in period to remove transient dynamics. We first assess whether there are two outbreaks within a year by examining weekly incidence for one year and then apply the “find_peaks” function in R (https://github.com/stas-g/findPeaks). This function defines a peak as a point such that m points either side of it have a lower or equal value to it where we set m to 3. We then calculate the maximum peak size for each year in the 10-year simulation. We use the number of unique peaks within 10 years to identify the outbreak periodicity i.e., two unique peaks imply biennial outbreaks.
Future simulations
We run deterministic disease simulations under both historic and future warming scenarios for individual provinces in mainland China. Simulations are run at a weekly time step for 100 years with a 70-year burn-in period to remove transient dynamics. Birth rates are fixed at the mean weekly birth rate for 2013 (the most recent year in our China dataset). Population numbers are also fixed at 2013 values.
Climate model output can be biased compared to observations due to a variety of factors such as the spatial scale of the data. In order to account for these biases we use the delta correction method i.e., we calculate the difference (delta) between future climatology (2080–2100 average) and present-day climatology (1990–2010 average) using the climate model output and then add this difference to our observational (ERA5) data. To compare present and future enterovirus trajectories we run two simulations: a present-day simulation using the observational data and a future simulation using the observational data plus the calculated difference (delta) between present and future climatologies. We also include 30 years of interannual temperature variability from the observational dataset.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data used in the study is publicly available. Polio data is available from Project Tycho25 and EVA71 and CVA16 data is published in prior work by Takahashi et al.23,24. Climate (temperature, relative humidity, and specific humidity) data are available from ERA528 and precipitation data from CHIRPS29.
Code availability
Data and code to run the main analysis are available via Zenodo at: https://doi.org/10.5281/zenodo.1276401247.
References
Baker, R. E. et al. Infectious disease in an era of global change. Nat. Rev. Microbiol. 20, 193–205 (2022).
Mahmud, A. S., Martinez, P. P., He, J. & Baker, R. E. The impact of climate change on vaccine-preventable diseases: insights from current research and new directions. Curr. Environ. Health Rep. 7, 384–391 (2020).
Mora, C. et al. Over half of known human pathogenic diseases can be aggravated by climate change. Nat. Clim. Change 12, 869–875 (2022).
Mordecai, E. A., Ryan, S. J., Caldwell, J. M., Shah, M. M. & LaBeaud, A. D. Climate change could shift disease burden from malaria to arboviruses in Africa. Lancet Planet. Health 4, e416–e423 (2020).
Ryan, S. J., Carlson, C. J., Mordecai, E. A. & Johnson, L. R. Global expansion and redistribution of Aedes-borne virus transmission risk with climate change.PLoS Negl. Trop. Dis. 13, e0007213 (2019).
Shaman, J. & Kohn, M. Absolute humidity modulates influenza survival, transmission, and seasonality. Proc. Natl Acad. Sci. 106, 3243–3248 (2009).
Viboud, C., Alonso, W. J. & Simonsen, L. Influenza in tropical regions. PLoS Med. 3, e89 (2006).
Baker, R. E. et al. Epidemic dynamics of respiratory syncytial virus in current and future climates. Nat. Commun. 10, 1–8 (2019).
Shaman, J., Pitzer, V. E., Viboud, C., Grenfell, B. T. & Lipsitch, M. Absolute humidity and the seasonal onset of influenza in the continental United States. PLoS Biol. 8, e1000316 (2010).
Yang, W., Lipsitch, M. & Shaman, J. Inference of seasonal and pandemic influenza transmission dynamics. Proc. Natl Acad. Sci. 112, 2723–2728 (2015).
Mahmud, A. S., Martinez, P. P. & Baker, R. E. The impact of current and future climates on spatiotemporal dynamics of influenza in a tropical setting. PNAS Nexus 2, pgad307 (2023).
Yuan, H., Kramer, S. C., Lau, E. H., Cowling, B. J. & Yang, W. Modeling influenza seasonality in the tropics and subtropics. PLoS Comput. Biol. 17, e1009050 (2021).
Martinez-Bakker, M., King, A. A. & Rohani, P. Unraveling the transmission ecology of polio. PLoS Biol. 13, e1002172 (2015).
Park, S. W. et al. Epidemiological dynamics of enterovirus d68 in the United States and implications for acute flaccid myelitis. Sci. Transl. Med. 13, eabd2400 (2021).
Pons-Salort, M. et al. The seasonality of nonpolio enteroviruses in the United States: patterns and drivers. Proc. Natl Acad. Sci. 115, 3078–3083 (2018).
Larson, H. J. & Ghinai, I. Lessons from polio eradication. Nat. 473, 446–447 (2011).
Metcalf, C. J. E. et al. Challenges in evaluating risks and policy options around endemic establishment or elimination of novel pathogens. Epidemics 37, 100507 (2021).
Chan, Y. F., Sam, I.-C., Wee, K.-L. & Abubakar, S. Enterovirus 71 in malaysia: a decade later. Neurol. Asia 16, 1–15 (2011).
Li, P. et al. Analysis of HFMD transmissibility among the whole population and age groups in a large city of China. Front. Public Health 10, 850369 (2022).
Zhu, P. et al. Current status of hand-foot-and-mouth disease. J. Biomed. Sci. 30, 15 (2023).
Harper, G. Airborne micro-organisms: survival tests with four viruses. Epidemiol. Infect. 59, 479–486 (1961).
Xing, W. et al. Hand, foot, and mouth disease in China, 2008–12: an epidemiological study. Lancet Infect. Dis. 14, 308–318 (2014).
Takahashi, S. et al. Hand, foot, and mouth disease in China: modeling epidemic dynamics of enterovirus serotypes and implications for vaccination. PLoS Med. 13, e1001958 (2016).
Takahashi, S. et al. Epidemic dynamics, interactions and predictability of enteroviruses associated with hand, foot and mouth disease in japan. J. R. Soc. Interface 15, 20180507 (2018).
van Panhuis, W. G., Cross, A. & Burke, D. S. Project tycho 2.0: a repository to improve the integration and reuse of data for global population health. J. Am. Med. Inform. Assoc. 25, 1608–1617 (2018).
Dalziel, B. D. et al. Urbanization and humidity shape the intensity of influenza epidemics in US cities. Sci. 362, 75–79 (2018).
Wagner, C. E. et al. Climatological, virological and sociological drivers of current and projected dengue fever outbreak dynamics in Sri Lanka. J. R. Soc. Interface 17, 20200075 (2020).
Hersbach, H. et al. The era5 global reanalysis. Q. J. R. Meteorol. Soc. 146, 1999–2049 (2020).
Funk, C. et al. The climate hazards infrared precipitation with stations—a new environmental record for monitoring extremes. Sci. Data 2, 1–21 (2015).
Rohde, R. A. & Hausfather, Z. The Berkeley earth land/ocean temperature record. Earth Syst. Sci. Data 12, 3469–3479 (2020).
Baker, R. E., Mahmud, A. S. & Metcalf, C. J. E. Dynamic response of airborne infections to climate change: predictions for varicella. Clim. Change 148, 547–560 (2018).
Dell, M., Jones, B. F. & Olken, B. A. What do we learn from the weather? the new climate-economy literature. J. Econ. Litt. 52, 740–798 (2014).
Ewing, A., Lee, E. C., Viboud, C. & Bansal, S. Contact, travel, and transmission: The impact of winter holidays on influenza dynamics in the United States. J. Infect. Dis. 215, 732–739 (2017).
Becker, A. D., Wesolowski, A., Bjørnstad, O. N. & Grenfell, B. T. Long-term dynamics of measles in London: Titrating the impact of wars, the 1918 pandemic, and vaccination.PLoS Comput. Biol. 15, e1007305 (2019).
Pons-Salort, M. & Grassly, N. C. Serotype-specific immunity explains the incidence of diseases caused by human enteroviruses. Sci. 361, 800–803 (2018).
Baker, R. E., Yang, W., Vecchi, G. A., Metcalf, C. J. E. & Grenfell, B. T. Assessing the influence of climate on wintertime sars-cov-2 outbreaks. Nat. Commun. 12, 1–7 (2021).
Hooshyar, M. et al. Cyclic epidemics and extreme outbreaks induced by hydro-climatic variability and memory. J. R. Soc. Interface 17, 20200521 (2020).
He, F. et al. Surveillance, epidemiology and impact of ev-a71 vaccination on hand, foot, and mouth disease in Nanchang, China, 2010–2019. Front. Microbiol. 12, 811553 (2022).
Deng, H. et al. Development of a multivalent enterovirus subunit vaccine based on immunoinformatic design principles for the prevention of HFMD. Vaccine 38, 3671–3681 (2020).
Beijing Municipal Education Commission. http://jw.beijing.gov.cn/.
Vital statistics, US Centers for Disease Control. https://www.cdc.gov/nchs/data/vsus/vsrates1940_60.pdf.
Ocha regional office for Asia and the Pacific (roap). https://data.humdata.org/dataset/cod-ab-chn.
US Census Bureau. https://www.census.gov/geographies/mapping-files/time-series/geo/cartographic-boundary.html.
Yang, J. et al. Seroepidemiology of enterovirus a71 infection in prospective cohort studies of children in southern China, 2013-2018. Nat. Commun. 13, 7280 (2022).
Huang, J. et al. Epidemiology of recurrent hand, foot and mouth disease, china, 2008–2015. Emerg. Infect. Dis. 24, 432 (2018).
Becker, A. D. & Grenfell, B. T. tsir: an R package for time-series susceptible-infected-recovered models of epidemics. PloS One 12, e0185528 (2017).
Baker, R. E. Increasing intensity of enterovirus outbreaks projected with climate change. Zenodo. https://doi.org/10.5281/zenodo.12764013 (2024).
Acknowledgements
This study was supported by the Cooperative Institute for Modeling the Earth System (CIMES) NOAA Grant NA18OAR4320123.
Author information
Authors and Affiliations
Contributions
Conceptualization: R.E.B., W.Y., G.A.V., and S.T.; Data curation: R.E.B., W.Y. and S.T.; Formal analysis: R.E.B.; Methodology: R.E.B.; Visualization: R.E.B., Writing—original draft: R.E.B.; Writing—reviewing and editing: R.E.B., W.Y., G.A.V., and S.T.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Baker, R.E., Yang, W., Vecchi, G.A. et al. Increasing intensity of enterovirus outbreaks projected with climate change. Nat Commun 15, 6466 (2024). https://doi.org/10.1038/s41467-024-50936-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-50936-3
- Springer Nature Limited