Abstract
Environmental stimuli not only alter gene expression profiles but also induce structural changes in cells. How distinct nuclear bodies respond to cellular stress is poorly understood. Here, we identify a subnuclear organelle named the nucleolar stress body (NoSB), the formation of which is induced by the inhibition of rRNA transcription or inactivation of rRNA processing and maturation in C. elegans. NoSB does not colocalize with other previously described subnuclear organelles. We conduct forward genetic screening and identify a bZIP transcription factor, named nucleolar stress response-1 (NOSR-1), that is required for NoSB formation. The inhibition of rRNA transcription or inactivation of rRNA processing and maturation increases nosr-1 expression. By using transcriptome analysis of wild-type animals subjected to different nucleolar stress conditions and nosr-1 mutants, we identify that the SR-like protein NUMR-1 (nuclear localized metal responsive) is the target of NOSR-1. Interestingly, NUMR-1 is a component of NoSB and itself per se is required for the formation of NoSB. We conclude that the NOSR-1/NUMR-1 axis likely responds to nucleolar stress and mediates downstream stress-responsive transcription programs and subnuclear morphology alterations in C. elegans.
Similar content being viewed by others
Introduction
The nucleus contains several different dynamic bodies that are variously composed of proteins or specific RNA molecules1. Intriguingly, nuclear bodies are not surrounded by membranes yet are retained as separate and distinct entities within the nucleoplasm, formed by a process termed liquid-liquid phase separation (LLPS)2,3. It is well established that LLPS-associated proteins that contain folded domains, intrinsically disordered regions (IDRs), and low complexity (LC) regions play vital roles in the process of nucleation4. For instance, paraspeckles require NONO, PCPC1, and PSF5,6. Cajal bodies are disrupted or disappear in the absence of COILIN, SMN, FAM118B, or WRAP537,8, PML bodies are nucleated by PML9, and SON and SRRM2 are essential for nuclear speckle formation10.
Nuclear body formation is generally dynamic, especially in response to a variety of stress conditions11,12,13. For example, heat shock, UV radiation, and chemical toxicity trigger the formation of nuclear stress bodies (nSBs), which may further influence the reprogramming of gene expression14. Mitochondrial stress leads to altered morphology and numbers of nuclear paraspeckles15. Polycomb Group (PcG) bodies were shown to repress damaged DNA upon UV irradiation16. However, how these nuclear bodies respond to cellular stresses and their physiological roles are still poorly understood.
The nucleolus is the most prominent subnuclear structure, where the main processes of rRNA and ribosome biogenesis occur. In the mammalian nucleolus, rDNA genes are transcribed by RNA polymerase I into 47S pre-rRNA, which is subsequently processed and modified to generate 28S, 18S, and 5.8S rRNAs. Genes encoding 5S rRNA are transcribed by Pol III17,18. These rRNAs are assembled with ribosomal proteins (RPs) to form small and large preribosome subunits, which are exported to the cytoplasm and eventually form the mature 40S and 60S ribosome subunits19.
Ribosome assembly factors maintain their greatest steady-state concentrations within nucleoli under normal growth conditions20, and blocking ribosome biosynthesis stimulates nucleolar stress21. For instance, actinomycin D inhibits Pol I transcription elongation and causes nucleolar segregation22. Starvation or rapamycin treatment downregulates mTOR signaling in yeast and mammalian cells, leading to a significant nucleolar size reduction and inhibition of rRNA transcription23. Mutations in ribosome assembly factors or ribosomal proteins (r-proteins) lead to nucleolar stress and induce a number of human disorders24.
In mammals, nucleolar stress signaling pathways are classified into two types: p53-dependent and p53-independent pathways20. p53 is a crucial transcription factor that responds to many cellular stresses. Nucleolar stress releases p53 from inhibition by MDM2 and activates a set of target genes25,26. The mechanism of the p53-independent pathway remains poorly understood. Nucleolar stress may induce cell proliferation or cell cycle arrest via the regulation of c-Myc, E2F-1, p21 Waf1/Cip1, or p27Kip1, independent of p5327,28,29,30. Yeast and plants do not express p53 or MDM2 and, therefore, use mechanisms independent of the p53 pathway. In plant cells, the plant-specific transcription factor ANAC082 may play critical roles in the response to nucleolar stress31.
C. elegans expresses p53/CEP-1 but lacks MDM2. Active p53/CEP-1 is regulated at the translational level32,33. Mutation in the conserved nucleolar protein NOL-6 enhances resistance to bacterial infection by activating p53/CEP-1 and its target gene, sym-134. The depletion of a nucleolar protein, WDR-46, activates xenobiotic detoxification genes via Nrf/SKN-1 and p53/CEP-135. Interestingly, the transcription factor FoxA/PHA-4, instead of p53/CEP-1, acts as a nucleolar stress sensor to induce the expression of lipogenic genes and lipid accumulation36, suggesting additional mechanisms for the nucleolar stress response.
Previously, we reported that nucleolar stress could induce the accumulation of erroneous rRNAs that lead to the recruitment of RdRPs to synthesize antisense ribosomal siRNAs (risiRNAs), which subsequently turn on the nuclear RNAi-mediated gene silencing pathway to inhibit pre-rRNA expression37,38,39. In addition, EXOS-10 exhibits nucleoplasmic translocation upon cold-worm shock in C. elegans40 .Third, abnormal accumulation of 27SA2 rRNA intermediates upon the depletion of class I RPLs reshaped spherical nucleoli to a ring-shaped nucleolar structure with an enlarged nucleolar vacuole (NoV) in C. elegans41. Therefore, it is very likely that a number of cellular events may occur concurrently to respond to nucleolar stress.
Here, we identified that nucleolar stress induced the formation of a subnuclear organelle, named the nucleolar stress body (NoSB), in C. elegans. The inhibition of rRNA transcription by actinomycin D, inactivation of rRNA processing and maturation, or mutation in the exosome ribonuclease complex, which degrades erroneous rRNAs, results in NoSB formation. To search for factors required for the formation of nucleolar stress-induced NoSB, we conducted forward genetic screening and identified a bZIP-containing protein, nucleolar stress required-1 (NOSR-1). In the absence of NOSR-1, nucleolar stress failed to induce NoSB formation. Interestingly, NOSR-1 acts as a sensor of nucleolar stress to activate the expression of the SR-like protein NUMR-1, consequently promoting NoSB formation. Therefore, our work revealed a NOSR-1/NUMR-1-involved pathway that responds to nucleolar stress and mediates nuclear substructure alterations in C. elegans.
Results
Deficiency in rRNA processing and maturation induced the formation of a distinct nucleolar stress body
Our recent work showed that the depletion of class I RPL proteins and a number of 26S rRNA processing factors led to nucleolar reshaping, in which the 27SA2 rRNA intermediate accumulated, nucleolar rings formed, and nucleolar vacuoles enlarged (Fig. 1A)41. To further investigate the nucleolar stress-induced substructure alterations in the nucleus, we generated an mCherry::DIS-3 strain and visualized the structure of the nucleus and nucleolus in hypodermal cells upon a number of nucleolar stresses. DIS-3 (also known as Rrp44 or EXOSC11) is a subunit of the 3’ to 5’ exoribonuclease complex, which degrades erroneous rRNAs in the nucleus and accumulates in the nucleoplasm (Fig. 1B)38,42. The single-copy mCherry::DIS-3 transgene was integrated into the C. elegans’ chromosome III of the strain EG8080 by MosSCI technology38. The transgene was driven by its own promoter, did not affect the brood size, but rescued dis-3(ust56) dysfunction in risiRNA biogenesis, suggesting that the transgene recapitulated the function of endogenous DIS-338.
A Schematic diagram of the formation of nucleolar rings, enlarged nucleolar vacuoles, and nucleolar stress bodies (NoSBs) in C. elegans. B DIC and fluorescence microscopy images of nuclei in hypodermal cells after knockdown of the indicated genes in the indicated L4 stage animals (scale bar, 5 μm). The nucleoplasm is marked by mCherry::DIS-3, the yellow arrows indicate NoSB. The images were taken by the Leica THUNDER imaging System. C Quantification of NoSB and ring-shaped nucleoli after RNAi knockdown of the indicated genes. The percentage of animals containing at least three cells out of hypodermal cells that have NoSB or ring-shaped nucleoli structure was shown (20 independent animals were tested).
We performed feeding RNAi experiments and knocked down approximately 100 genes involved in rRNA processing and modification in mCherry::DIS-3 animals (Supplementary Data 1). Then, we visualized the hypodermal cells using Nomarski (DIC) and fluorescence microscopy. Consistent with previous work, we observed the formation of nucleolar ring structures and enlarged nucleolar vacuoles upon knocking down a number of genes involved in pre-60S ribosome maturation (Fig. 1B, C)41. The nucleolar vacuole (NoV) is an evolutionarily conserved nucleolar subcompartment, present in the nucleoli of various plants and animals43,44,45,46. Our recent work showed that faithful rRNA processing is essential to maintain the structure of nucleolus41. When the rRNA precursor 27SA2 rRNA was inappropriately accumulated, the spherical nucleolus transformed into a ring-shaped nucleolus, in which a NoV appears. In the circumstance, the rRNA transcription and processing machineries were enriched in the ring, while many usual nucleoplasmic proteins accumulated in the NoV41. NoVs may have important roles in germline development or mRNA metabolism and may be involved in the transport of nucleolar substances from nucleolus to nucleoplasm and temporary storage of certain materials.
Interestingly, we noticed the appearance of an additional vacuole per cell in the nucleoplasm in hypodermal cells (Fig. 1B, C). The vacuole excludes DIS-3 and is also visible by DIC microscopy. We named this vacuole the nucleolar stress body (NoSB) (see below).
Ribosomal proteins of the small subunit (RPS) fall into two main categories according to their functions in pre-rRNA processing47. i-RPSs are strictly required for initiating the processing of sequences flanking the 18S rRNA. p-RPS proteins are required for subsequent nuclear and cytoplasmic maturation steps. Knocking down both i-RPS and p-RPS proteins induced the formation of NoSB but not the formation of nucleolar rings (Fig. 1B, C and Supplementary Fig. 1A, C)41.
RPLs are proteins of the 60S ribosome large subunit that are involved in pre-rRNA processing, including 27SA2, 27SA3, 27SB pre-rRNAs, and 7S pre-rRNA maturation48. C. elegans’ genome encodes 48 large subunit (rpl) genes. Our recent work classified RPL proteins into two main functional categories according to whether they are required for the processing of 27SA2 pre-rRNAs and the suppression of ring-shaped nucleoli and enlarged nucleolar vacuoles (NoV)41. The Class I RPLs are required for 27SA2 pre-rRNA processing, and the knockdown of this group significantly increased the proportion of vacuole-contained nucleoli and the accumulation of 27SA2 pre-rRNA41. However, Class II RPLs were likely involved in the processing of other pre-rRNA intermediates, and the knockdown of Class II RPLs did not lead to the formation of NoV41. The depletion of both classes of RPL proteins induced NoSB formation, yet only class I RPLs triggered the formation of ring-shaped nucleoli (Supplementary Fig. 1B, C).
Taken together, these results suggested that defective rRNA biogenesis and ribosome assembly could induce alterations in subnuclear structures in C. elegans.
Mutation of exosome ribonucleases induced NoSB formation
To investigate the mechanism of NoSB formation, we first conducted forward genetic screening to search for mutants that reveal NoSB formation. We mutagenized the mCherry::DIS-3 strain with ethyl methanesulfonate (EMS), followed by clonal screening with DIC and fluorescence microscopy (Fig. 2A). From the 2000 haploid genome, we isolated two mutants, ust242 and ust243. Using whole-genome resequencing, we identified two point mutations in the coding regions of exos-4.2 (ust243, G54R) and exos-10 (ust242, D368N) (Fig. 2B). Both EXOS-4.2 and EXOS-10 are core factors of the exosome 3’-5’-exoribonuclease complex38,42,49. exos-4.2 encodes a ribonuclease (RNase) pleckstrin homology (PH)-like protein, and exos-10 encodes a 3′-5′-exoribonuclease.
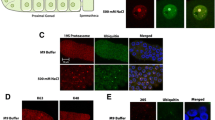
A Schematic diagram of forward genetic screening to search for factors suppressing NoSB formation in C. elegans nuclei. B Schematic of the gene structure of exosome genes. exos-4.2 (ust243) and exos-10 (ust242) are the mutants obtained from the forward genetic screen; exos-1 (ust57) and exos-10 (ok2269) are obtained from the National Bioresource Project and the CGC; dis-3 (ust56), a conserved amino acid Arg363, was mutated to cysteine38,40. C DIC and fluorescence microscopy images of nuclei in hypodermal cells of respective L4 stage animals expressing the mCherry::DIS-3 (scale bar, 5 μm). The yellow dashed line represents the nucleus outlines, and the yellow arrows indicate NoSB (see also in Fig. 2F). D Quantification of NoSB in hypodermic cells in exosome mutants. Red dots indicate the percentage of hypodermis cells containing NoSB in each worm. n indicates the number of independent animals tested. For box plots, the horizontal line represents the median value, the lower and upper quartiles represent the 25th and 75th percentile, and the whiskers show the maximum and minimum values; a two-tailed t-test was performed to determine statistical significance (see also in Fig. 2E). E The size of NoSB in exosome mutants. Blue dots indicate the area of NoSB. n of nucleoli from 20 independent animals were measured. F DIC and fluorescence microscopy images of C. elegans nuclei in hypodermal cells after actinomycin D treatment for 48 h. The nucleoplasm is marked by mCherry::DIS-3 (magenta), and the nucleolus is marked by FIB-1::GFP (green). Three times were repeated independently with similar results. All images were taken by the Leica THUNDER imaging System. Source data are provided as a Source Data file.
To confirm that the exosome complex suppresses NoSB formation, we generated additional alleles of exos-4.2 and exos-10 by CRISPR/Cas9 technology and obtained mutants of other exosome subunits. The mutation of exos-1, exos-4.2, and exos-10 resulted in NoSB formation (Fig. 2C–E). Consistently, feeding mCherry::DIS-3 animals dsRNAs targeting exos-2, exos-3, exos-4.1, exos-5, exos-7, exos-8, or exos-9 induced the formation of NoSB (Supplementary Fig. 2A).
We measured the size of NoSB and nucleoli in the exosome mutants. The size of NoSB is ~10% of that of nucleoli, and the exosome mutants exhibited modestly enlarged nucleoli (Fig. 2E and Supplementary Fig. 2B). Similarly, in S. cerevisiae and D. melanogaster, the depletion of ribosomal proteins also led to enlarged nucleoli50.
LMN-1 is an ortholog of human lamin A/C and localizes to the inner side of the nuclear membrane. NoSB localized inside the nuclei, as shown by LMN-1::mCherry (Supplementary Fig. 2C). MTR-4 and NRDE-3 are two nucleoplasm-localized proteins and are involved in nuclear and nucleolar RNAi38. Our previous work showed that deficient rRNA processing and maturation led to the accumulation of erroneous rRNAs and induced the production of a class of antisense ribosomal siRNA (risiRNA) which further translocated the Argonaute protein NRDE-3 from the cytoplasm to nucleoli37,38,39. Similarly, the disruption of RNA exosomes also resulted in the production of risiRNAs and triggered the nucleolar accumulation of NRDE-338. To test whether nucleolar stress could also induce the production of risiRNAs and enrich NRDE-3 in NoSBs, we visualized the subcellular localization of NRDE-3 and MTR-4 upon exos-9 RNAi. However, both proteins were excluded from the exos-9 depletion-induced NoSB (Supplementary Fig. 2D), suggesting that it is unlikely risiRNA could enrich in NoSB.
Therefore, we concluded that a deficient exosome ribonuclease complex could induce NoSB formation.
The inhibition of RNA polymerase I induced NoSB formation
Previous work showed that actinomycin D inhibits rRNA transcription and induces nucleolar stress41,51,52. We generated an mCherry::DIS-3;FIB-1::GFP strain and visualized the structure of the nucleolus in hypodermal cells in the presence of actinomycin D. FIB-1 encodes the C. elegans ortholog of human fibrillarin and Saccharomyces cerevisiae Nop1p53 and localizes to the nucleolus.
At a concentration of 5 µg/ml actinomycin D, we did not observe pronounced nuclear and nucleolar substructure changes in hypodermal cells (Fig. 2F and Supplementary Fig. 3A). However, at a higher concentration of 20 to 30 µg/ml actinomycin D, FIB-1 likely condensed and detached from some unidentified nucleolar border. The NoSB excludes DIS-3 in the nucleoplasm, which is also visible by DIC and fluorescence microscopy. FIB-1 did not accumulate in NoSB, suggesting that the vacuole is distinct from nucleoli. The sizes of NoSB increased at a higher concentration of actinomycin D (Supplementary Fig. 3B). LMN-1::mCherry confirmed that actinomycin D-induced NoSB localized in the nucleus (Supplementary Fig. 3C). MTR-4 and NRDE-3 were also excluded from actinomycin D-induced NoSB (Supplementary Fig. 3D). Notably, actinomycin D treatment has been shown to inhibit the formation of nucleolar ring and nucleolar vacuole41, which is oppositive to actinomycin D-induced NoSB formation.
We tested whether other kinds of environmental conditions could induce the formation of NoSB. However, growing animals at 15 or 25 °C, heat shock at 37 °C or stravation failed to induce the formation of NoSB (Supplementary Fig. 3E–G, also see Supplementary Fig. 18A, B). In addition, the ectopic expression of mcherry::DIS-3 did not affect the efficacy of NoSB formation (Supplementary Fig. 4A, B).
Taken together, these data suggested that the inhibition of RNA polymerase I by actinomycin D induced the formation of NoSBs.
NoSB is not a portion of nucleoli that blebbed off upon nucleolar stress
In mammalian cells, the factors involved in rRNA transcription and processing and ribosome assembly usually localize to the FC, DFC, and GC nucleolar subcompartments. Recent work showed that C. elegans nucleoli contain two phase-separated subcompartments, with FIB-1 and GARR-1 in the FC and NUCL-1 and LPD-6 in the GC region43.
To test the possibility that NoSB may be a portion of the nucleolus that has blebbed off upon nucleolar stress, we generated a number of single-copy transgenes of nucleoli-localized proteins by CRISPR-Cas9 technology (Fig. 3A–C and Supplementary Fig. 5A). Among them, FIB-1 is a methyltransferase for pre-rRNA processing and modification. RBD-1 is an ortholog of human RBM19, which is required for 90S preribosome maturation. RRP-8 is a rRNA processing factor involved in N1-methyladenosine (m1A) modification of 26S rRNAs. DAO-5 is an rRNA transcription factor. T06E6.1 is an rRNA processing factor. GARR-1 is involved in snoRNA-guided rRNA pseudouridine synthesis. RPOA-1, RPOA-2, and RPAC-19 are subunits of the RNA polymerase I complex. We observed that upon exos-9 RNAi, all these proteins were excluded from the NoSB (Fig. 3B, C, Supplementary Fig. 5A, and summarized in Supplementary Fig. 5B).
A List of marker genes of subnuclear organelles and their C. elegans homologs. B–D DIC and fluorescence microscopy images of hypodermic cells of L4 animals expressing TagRFP::DAO-5, RBD-1::mCherry, RRP-8::GFP and HSF-1::GFP after exos-9 knockdown (Scale bar, 5 μm). Yellow arrows indicate NoSB. Three times were repeated independently with similar results. E Schematic diagram of the domain structure and predicted intrinsically disordered regions of NUCL-1 and RRP-8. F DIC and fluorescence microscopy images of nuclei with the indicated transgenes after exos-9 knockdown (scale bar, 5 μm). Three times were repeated independently with similar results. All images were taken by the Leica THUNDER imaging System.
To further test that NoSB may be a reservoir for partially constructed ribosomes, we generated single-copy transgenes of ribosomal subunits by CRISPR-Cas9 technology and then examined their localization upon knocking down exos-9 by RNAi (Supplementary Fig. 5A, B). RPS-4 and RPS-19 are small ribosomal subunit proteins and are involved in ribosomal small subunit biogenesis. RPL-7 and RPL-14 are large ribosomal subunit proteins and are involved in ribosomal large subunit biogenesis. RPS-4 is enriched in nucleolus; RPS-19 accumulates in the nucleolus and nucleoplasm during the larval stage; RPL-7 is enriched in nucleolus, and RPL-14 is likely evenly distributed in the cell. Upon knocking down exos-9 by RNAi, however, RPS-4, RPS-19, RPL-7, and RPL-14 were all excluded from NoSB (Supplementary Fig. 5A, B).
Therefore, we concluded that NoSB is not a portion of the nucleolus that has blebbed off upon nucleolar stress, or the reservoir for partially constructed ribosomes.
NoSB is a distinct subnuclear organelle
To test whether NoSB is a distinct subnuclear organelle or colocalizes with other known subnuclear structures, we generated a number of fluorescence-labeled protein markers to reveal the Cajal body, paraspeckle, PcG body, and nuclear stress body (Fig. 3A)54. The exos-9 depletion-induced NoSB did not colocalize with any of these known subnuclear organelles (Fig. 3D and Supplementary Fig. 6A–D). The heat shock protein HSF-1 responds to heat stress by forming subnuclear structures termed nuclear stress granules in C. elegans. However, we did not find colocalization of NoSB with HSF-1::GFP (Fig. 3D).
We tested whether NoSB is a nuclear lipid droplet. Recent work reported that lipid droplets, which form at the nuclear envelope, may enter the nucleoplasm by penetrating the nuclear lamina and associated peripheral heterochromatin to form nuclear lipid drops (nLDs)55,56. Actinomycin D-triggered nucleolar stress could induce excessive lipid accumulation in C. elegans36. To test whether NoSB is a kind of nLD, we used Oil Red O to stain neutral lipids of postfixed animals with or without exos-9 RNAi but failed to observe lipid accumulation in NoSBs (Supplementary Fig. 6E). Therefore, NoSB is unlikely to form nuclear neutral lipid droplets.
An interesting observation is NUCL-1. NUCL-1 is located in the GC region and is required for nucleolar vacuole formation41. NUCL-1 encodes an evolutionarily conserved protein exhibiting extensive homology to yeast and human nucleolin and has been shown to interact with exosome proteins57. The N-terminal domain of NUCL-1 is a long IDR containing a GAR/RGG domain that is required for subnucleolar compartmentalization43 (Fig. 3E). IDRs are likely one of the driving forces of phase separation and have been identified in many proteins capable of phase separation. RRP-8 encodes an evolutionarily conserved protein exhibiting extensive homology to yeast and human RRP-8. The N-terminal domain of RRP-8 is also a long IDR (Fig. 3E). We constructed GFP- or mCherry-tagged NUCL-1, NUCL-1ΔRGG, RRP-8, and RRP-8ΔIDR transgenic animals. The depletion of RNA exosome did not translocate NUCL-1 or NUCL-1(ΔRGG) into NoSB in live worms (Supplementary Fig. 7B, C). However, further treatment with NaN3 following actinomycin D or exos-9 RNAi could induce an accumulation of NUCL-1 or NUCL-1(ΔRGG) to NoSB (Fig. 3F and Supplementary Fig. 7A–C). Yet nucl-1 is not required for the formation of NoSB (Supplementary Fig. 7D). Double treatment of nucleolar stress followed by NaN3 could recruit NUCL-1 to NoSB, yet the mechanism is unclear. Neither RRP-8 nor RRP-8(ΔIDR) translocated to NoSB upon nucleolar stress followed by NaN3 treatment (Fig. 3F).
In conclusion, we speculated that NoSB is a distinct subnuclear structure induced by nucleolar stress.
Forward genetic screening identified a bZIP transcription factor NOSR-1 required for NoSB formation
To further understand the mechanism and regulation of NoSB, we performed a second forward genetic screening to search for factors that were required for NoSB formation. We chemically mutagenized mCherry::DIS-3;exos-10(ust242) animals followed by clonal screening with DIC and fluorescence microscopy and isolated mutants in which NoSB was depleted (Fig. 4A). From approximately 2000 haploid genomes, we isolated one mutant, ust303, in which NoSB was depleted in mCherry::DIS-3;exos-10(ust242) animals (Fig. 4B). We named the gene nucleolar stress response-1 (nosr-1).
A Schematic diagram of the forward genetic screening to search for factors required for NoSB formation in C. elegans nuclei. B DIC and fluorescence microscopy images of nuclei in the indicated L4 stage animals expressing the mCherry::DIS-3 (scale bar, 5 μm). The yellow dashed line represents the nucleus, and the yellow arrows indicate NoSB (see also in Fig. 4E). C Schematic of the nosr-1 exon structure. nosr-1 (ust303) is the mutant obtained from the forward genetic screen. The deletion alleles ust307, ust308, and ust309 were constructed by CRISPR/Cas9 technology (see Methods). D Quantification of NoSB in hypodermic cells in the indicated animals. Red dots indicate the percentage of hypodermis cells containing NoSB in each L4 stage worm. n indicates the number of independent animals tested. A two-tailed t-test was performed to determine statistical significance. For box plots, the horizontal line represents the median value, the lower and upper quartiles represent the 25th and 75th percentile, and the whiskers show the maximum and minimum values (see also in Fig. 4F). E Top: Schematic diagram of the formation of nucleolar rings, enlarged nucleolar vacuoles and nucleolar stress bodies (NoSBs) in rpl-14(RNAi) animals. Bottom: Images of mCherry::DIS-3 transgene in nosr-1(-);rpl-14(RNAi) animals (scale bar, 5 μm). The green arrows indicate the nucleolar ring. F Quantification of nucleolar rings and NoSB in hypodermic cells in the indicated animals. Every dot indicates the percentage of hypodermis cells containing NoSB or nucleolar rings in each worm. Red dots indicate NoSB and green dots indicate nucleolar rings. n indicates the number of independent animals tested. A two-tailed t-test was performed to determine statistical significance. All images were taken by the Leica THUNDER imaging System. Source data are provided as a Source Data file.
We deep-sequenced the nosr-1(ust303) genome and identified Y17G7B.20 (Fig. 4C), in which the amino acid threonine242 was mutated to a proline (Fig. 4C). Y17G7B.20 was predicted to have a basic leucine zipper (bZIP) domain that can bind to DNA and mediate protein dimerization58,59. To further confirm that y17g7b.20 is nosr-1, we generated three additional deletion alleles of y17g7b.20 by CRISPR/Cas9 technology with two single-guide RNAs (sgRNAs)60 and examined NoSB formation in exos-10(ust242) animals. All three alleles, ust307, ust308, and ust309, deleted most of the gene sequence and caused a frame shift; therefore, they are likely null alleles. In the three y17g7b.20 mutants, exos-10(ust242) mutation or RNAi knockdown of exos-9 failed to induce NoSB formation (Fig. 4B, D and Supplementary Fig. 8A, B). In addition, the nosr-1 mutation blocked actinomycin D-induced NoSB formation (Supplementary Fig. 8C). Therefore, y17g7b.20 is nosr-1.
To test whether NOSR-1 is the key player in NoSB formation from all types of defects in ribosome synthesis, we observed that (1) nosr-1 is required for both exos-9 RNAi- and actinomycin D-induced NoSB formation (Supplementary Fig. 8A–C); (2) after knocking down the components of snoRNP, pre-40S and pre-60S particles, and RPSs and RPLs, ~50% hypodermal cells in each worm revealed NoSB structure which also depended on nosr-1 (Supplementary Figs. 9A, B, 10A, B). These data suggested that NOSR-1 likely suppresses the formation of NoSB induced by all types of defects in ribosome biogenesis.
Interestingly, the depletion of nosr-1 inhibited rpl-14 knockdown-induced NoSB formation but did not prohibit the formation of ring-shaped nucleoli (Fig. 4E, F), suggesting that the formation of NoSB and ring-shaped nucleoli involved two distinct mechanisms.
NOSR-1 promotes animal fertility and lifespan under nucleolar stress conditions
The bZIP transcription factor family is a gene family conserved from yeast to humans and has roles in a wide variety of processes61. C. elegans underwent a relatively recent expansion of bZIP genes, some of which do not appear to have direct homologs in mammals, likely including NOSR-158,61.
We investigated the role of NOSR-1 in C. elegans. nosr-1 mutants revealed brood sizes similar to those of wild-type N2 animals at 20 °C but strongly reduced fertility at 25 °C (Fig. 5A, B). In addition, nosr-1 mutation reduced the brood size of exos-10 animals (Fig. 5C, D).
A–D Brood size of the indicated animals at 20 and 25 °C. Bleached embryos were hatched and grown at 20 or 25 °C. Then, L3 worms were transferred individually onto fresh NGM plates. n of progeny worms was scored. A two-tailed t-test was performed to determine statistical significance. E Survival curves of the indicated animals at 20 °C. Three biological replicates were performed. F Histogram displaying the average lifespan of the indicated animals. Mean ± SD of three independent experiments. n, the number of animals tested. A two-tailed t-test was performed to determine statistical significance. Source data are provided as a Source Data file.
We failed to detect a significant change in the lifespan of nosr-1 animals compared to that of N2 animals (Supplementary Fig. 11A, B). However, the nosr-1 mutation shortened the lifespan of exos-10 animals (Fig. 5E, F). The depletion of nosr-1 did not significantly change the oxidative stress resistance or heat stress resistance in N2 background worms (Supplementary Fig. 11C, D).
Therefore, we concluded that nosr-1 is required for fertility and lifespan regulation in animals under nucleolar stress conditions.
Nucleolar stress activates nosr-1 expression
To investigate the mechanism of nosr-1 in NoSB regulation, we tested whether nosr-1 responds to nucleolar stresses. We compared the transcriptomes of animals with or without nucleolar stress using mRNA deep-sequencing. The nosr-1 mRNA levels were upregulated upon actinomycin D treatment, exos-10 mutation, or RNAi knockdown of exos-9 and rpl-14 (Fig. 6A).
A Volcano plot comparing the transcriptomes of the indicated animals. Volcano plot showing log2 (fold change) of gene product-annotated transcript abundances in stressed vs control-animals (x axis) and −log10 (P value) (y axis), calculated using an exact test in edgeR. The dashed line represents the significance threshold. The expression of nosr-1 is highlighted. B Fluorescence microscopy images of nosr-1p::NLS::GFP(ustIS285) after actinomycin D (abbreviated as Act D) treatment. C Fluorescence microscope images of nosr-1p::NLS::GFP(ustIS285) in the indicated animals. D Fluorescence microscope images of nosr-1p::NLS::GFP after knocking down the indicated genes by RNAi (scale bar, 20 μm). All worms were imaged at the L4 stage and all images are representative of more than ten animals (B–D). All images were taken by the Leica THUNDER imaging System. E, F Relative mRNA levels of nosr-1 measured by quantitative real-time PCR in the indicated animals. Data were presented as the means ± SD of three and four biological replicates, respectively. A two-tailed t-test was performed to determine statistical significance. Source data are provided as a Source Data file.
Then, we constructed a GFP-fused transcription fusion nosr-1P::NLS::GFP and inserted it at the ttTi4348 site on chromosome I using CRISPR/Cas9 gene editing technology to monitor nosr-1 expression60. Actinomycin D treatment significantly increased the expression of nosr-1P::NLS::GFP (Fig. 6B and Supplementary Fig. 12A). Knocking down exos-9 by RNAi or mutations in exosome genes significantly increased the expression of nosr-1P::NLS::GFP (Fig. 6C, D and Supplementary Fig. 12B, C). We measured the mRNA levels of endogenous nosr-1 mRNA by quantitative real-time PCR and identified an increase in the mRNA levels of nosr-1 (Fig. 6E, F). Similarly, knocking down rRNA biogenesis and ribosome assembly factors, the mutation of which induced NoSB formation, increased the expression of nosr-1P::NLS::GFP as well (Supplementary Fig. 12D–G).
Taken together, these results suggested that nucleolar stress could activate the expression of nosr-1.
nosr-1 may engage in an uncharted nucleolar stress response pathway
We tested whether other known stress response pathways are involved in the formation of nucleolar stress-induced NoSB. CEP-1 is an ortholog of mammalian p53 in C. elegans and has an essential role in nucleolar stress34. SKN-1 senses nucleolar stress to activate xenobiotic detoxification genes35. PHA-4 is a sensor of nucleolar stress and transactivates the expression of lipogenic genes36; DAF-16 is a FOXO transcription factor involved in lifespan regulation as well as in oxidative stress and heat stress resistance62,63. However, none of these genes are required for exos-10(ust242) mutation-induced NoSB formation (Supplementary Fig. 13A, B). Therefore, we speculated that nosr-1 may engage in an uncharted nucleolar stress response pathway.
Transcriptome analysis identified numr-1 as the NOSR-1 target
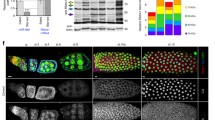
We performed mRNA-seq to explore the NOSR-1-targeted genes involved in NoSB formation. We first compared transcriptomes of wild-type and nosr-1(-) mutant animals under different stress conditions (Fig. 7A and Supplementary Fig. 14A–C) and identified 11 genes that were consistently upregulated by nucleolar stress (Fig. 7B), which depend on the presence of functional NOSR-1 (Fig. 7C–E)
A Schematic of the animal with or without nucleolar stress. B Venn diagrams reveal the overlapping target genes that are upregulated or downregulated under three nucleolar stress conditions. C–E Heatmap of the standardized fragments per kilobase of transcript per million mapped reads (FPKM) for the 11 coupregulated target genes in the indicated animals. The expression levels are indicated by the color bar. F DIC and fluorescence microscopy images of hypodermal nuclei in exos-10(ust242);mCherry::DIS-3 worms after knockdown of the indicated genes by RNAi (scale bar, 5 μm). The yellow dashed line represents the nucleus, and the yellow arrows indicate NoSB. Three times were repeated independently with similar results. Images were taken by the Leica THUNDER imaging System.
We then tested whether nucleolar stress-induced NoSB depends on the 11 genes. We performed feeding RNAi experiments and knocked down the 11 genes in mCherry::DIS-3;exos-10(ust242) animals (Fig. 7F). RNAi knockdown of numr-1 results in the prohibition of NoSB formation, suggesting that nucleolar stress-induced NoSB requires the presence of NUMR-1.
Nucleolar stress induced the accumulation of NUMR-1 in NoSB
NUMR-1 (nuclear-localized metal responsive) was previously identified as an SR-like protein that promotes cadmium tolerance (Fig. 8A)64,65,66. NUMR-1 contains an RNA-recognition motif (RRM), serine- and arginine-rich regions that are common in SR proteins, and a C-terminal tail rich in histidine and glycine, suggesting that NUMR-1 might function in RNA metabolism67,68,69. Interestingly, the SR family of proteins is strongly associated with liquid‒liquid phase separation (LLPS) and is enriched in nuclear bodies, especially in nuclear speckles70. These features suggest that NUMR-1 might function in RNA metabolism and promote the formation of various biomolecular condensates.
A Schematic of the NUMR-1 protein structure. B DIC and fluorescence microscopy images of hypodermal nuclei after actinomycin D treatment and RNAi knockdown of exos-9 and rpl-14 in the indicated animals expressing the GFP::NUMR-1 (scale bar, 5 μm). The yellow arrows indicate NoSB. The images were taken by the Leica THUNDER imaging System. C Quantification of NoSB in indicated tissues with indicated treatment. Every dot indicates the percentage of cells containing NoSB in hypodermis cells (red), intestine cells (blue), germline cells (black), and oocyte cells (purple). n, the number of independent animals tested, see also Supplementary Fig. 16A–D. The horizontal line represents the median value, the lower and upper quartiles represent the 25th and 75th percentile, and the whiskers show the maximum and minimum values. Source data are provided as a Source Data file.
To study the function of NUMR-1 in NoSB formation, we constructed a 3xFLAG–GFP–tagged NUMR-1 transgene (abbreviated GFP::NUMR-1) using CRISPR/Cas9-directed in situ gene editing technology. In wild-type animals cultured under normal laboratory conditions, we did not observe explicit GFP::NUMR-1 expression (Fig. 8B and Supplementary Fig. 15A). Strikingly, upon nucleolar stress by the inhibition of rRNA transcription using actinomycin D or RNAi knockdown of exos-9 or rpl-14, GFP::NUMR-1 expression was activated, and the protein accumulated in distinct nuclear foci under fluorescence microscopy (Fig. 8B and Supplementary Fig. 15A). DIC microscopy indicated that NUMR-1 foci colocalized with NoSB (Fig. 8B). In addition, RPOA-1 is the core subunit of Pol I and contributes to the polymerase activity. Feeding GFP::NUMR-1 animals with dsRNAs targeting rpoa-1 induced the formation of NoSB (Supplementary Fig. 15B, C), which is consistent with the result that inhibiting Pol I activity by actinomycin D elicited NoSB formation. As expected, the nosr-1 mutation inhibited GFP::NUMR-1 expression (Fig. 8B and Supplementary Fig. 15A).
We also quantified the percentage of NoSB formation in different tissues (Fig. 8C and Supplementary Fig. 16A–D). NoSB was mostly found in the hypodermal and intestinal cells, but rarely noticeable in germline and oocyte cells upon nucleolar stress.
To test whether NUMR-1 alone is sufficient to direct NoSB formation, we generated a fib-1p::NUMR-1::GFP::fib-1utr transgene to constitutively express NUMR-1 in all tissues. The ectopically expressed fib-1p::NUMR-1::GFP evenly diffused in the nucleoplasm without condensation into NoSB in the absence of nucleolar stress (Supplementary Fig. 17A, B). The nucleolar stress treatment via actinomycin D or exos-9 RNAi induced the NoSB accumulation of fib-1p::NUMR-1::GFP, similar to the endogenous numr-1 promoter-driven NUMR-1::GFP. Interestingly, this constitutively expressed fib-1p::NUMR-1::GFP did not rescue nosr-1 mutation and failed to direct NoSB formation upon nucleolar stress. In the nosr-1(-); fib-1p::NUMR-1::GFP::fib-1utr animal, we did not observe noticeable condensation of NUMR-1::GFP and formation of NoSB. We speculated that there are additional NOSR-1-dependent factors besides NUMR-1 to synergistically mediate NoSB formation, therefore the constant expression of NUMR-1 only is not sufficient to rescue all nosr-1(-) defects. The identification of the components, both RNAs and proteins, in NoSB will greatly help to clarify the function and regulation of NoSB.
Together, these data suggest that NUMR-1 is a downstream target of NOSR-1 and is a crucial component of NoSB. Additional factors, besides NUMR-1, could act downstream of NOSR-1 and mediate NoSB formation, as NUMR-1::GFP failed to replace NOSR-1 functionality in terms of forming NoSB.
NoSB may not obviously contain liquid-liquid phase separation (LLPS) properties
To test whether NUMR-1 can undergo phase separation, we first visualized the fine structure of NoSB by collecting Z-stack images of GFP::NUMR-1 in NoSB (Fig. 9A). GFP::NUMR-1 revealed even distribution throughout NoSB, and did not display noticeable subcompartments.
A Z-stack images of the exos-10(ust242); GFP::NUMR-1 worms with a step size of 0.5 μm. B, C (Top) FRAP assay of GFP::NUMR-1 transgene in indicated regions after actinomycin D treatment or in exos-10(ust242) mutants. (Bottom) Quantification of FRAP data. Mean ± SD, n = 3 independent animals. Scale bar, 2 μm. All images were taken by the Zeiss LSM980 confocal microscope. Source data are provided as a Source Data file.
Then, we induced NoSB formation via actinomycin D treatment or exos-10 depletion and performed fluorescence recovery after photobleaching (FRAP) assays (Fig. 9B, C). However, we failed to observe the recovery of GFP::NUMR-1 foci after photobleaching. These data suggested that NoSB may not obviously contain liquid-liquid phase separation (LLPS) properties.
To explore the recovery dynamics of NoSB from nucleolar stress, we treated worms with actinomycin D for 48 hours, then transferred the stressed worms to normal growth media without actinomycin D, and quantified the percentage of NoSB thereafter (Supplementary Fig. 18A–C). As expected, the percentage of NoSB, as well as the expression of GFP::NUMR-1 gradually decreased upon the removal of actinomycin D (Supplementary Fig. 18B, C), suggesting that constant nucleolar stress is required for the maintenance of NoSB condensation.
Using GFP::NUMR-1 as the marker, we tested again whether other kinds of environmental conditions besides nucleolar stress could induce the formation of NoSB. However, growing animals at 15 or 25 °C, heat shock at 37 °C, or stravation failed to induce the formation of NoSB (Supplementary Fig. 19A, B, also see Supplementary Fig. 3E–G). Yet occasionally, we observed one or two small GFP::NUMR-1 foci in a few animals at normal growing conditions as well as under certain environmental stresses (Supplementary Fig. 19A, B), which were hardly detectable by DIC.
Cadmium stress-induced NUMR-1 expression and nosr-1-independent NoSB formation
NUMR-1 (nuclear-localized metal responsive) was previously identified as an SR-like protein that promotes cadmium tolerance64,65,66. Cadmium was shown to increase numr-1/-2 mRNA expression in pharyngeal and intestinal cells. Cadmium arrests larval development and decreases body size, and these effects were exacerbated by numr-1/-2 RNAi64. We then tested whether cadmium exposure could translocate NUMR-1 to NoSB.
As expected, the treatment with cadmium increased the expression of GFP::NUMR-1 (Supplementary Fig. 20A). In addition, cadmium induced the aggregation of GFP::NUMR-1 in the nucleus in intestine cells, but not in the hypodermis (Fig. 10A, B). Interestingly, the cadmium-induced NUMR-1 expression and condensation did not dependent on nosr-1. In nosr-1 mutants, cadmium could still stimulate NUMR-1 expression and aggregation (Fig. 10A, B and Supplementary Fig. 20A), suggesting a nosr-1-independent pathway is involved in the formation of cadmium-induced NUMR-1 condensation.
A DIC and fluorescence microscopy images of hypodermal and intestine nuclei after cadmium treatment in the indicated animals expressing the GFP::NUMR-1. Arrows indicate NoSB (scale bar, 5 μm). All images were taken by the Leica THUNDER imaging System. B Quantification of NoSB in hypodermic cells and intestine cells from (A). Every dot indicates the percentage of hypodermis or intestine cells containing NoSB in each worm. Red dots indicate wild-type animals, blue dots nosr-1(ust303) mutants, green dots nosr-1(ust308) mutants. n, the number of animals tested. The horizontal line represents the median value, the lower and upper quartiles represent the 25th and 75th percentile, and the whiskers show the maximum and minimum values. A two-tailed t-test was performed to determine statistical significance. C A working model of NoSB formation under nucleolar stress in C. elegans. Source data are provided as a Source Data file.
Discussion
Environmental stresses not only alter gene expression but also trigger structural changes in cells. However, how distinct subcellular structures respond to stresses remains largely mysterious. Here, we showed that nucleolar stress induced the formation of a distinct subnuclear organelle, nucleolar stress body (NoSB), which is regulated by a bZIP transcription factor, NOSR-1. Further experiments identified that NUMR-1 is the critical component of NoSB, is induced by NOSR-1 and is per se required for NoSB formation (Fig. 10C), suggesting that NUMR-1 may be involved in building up the scaffold and recruiting NoSB components.
Failures in ribosome biogenesis or functions result in a condition termed nucleolar stress, which ultimately leads to disruptions in cell homeostasis. The RNA exosome is a conserved, multi-subunit ribonucleolytic complex that contributes to the 3’ to 5’ processing and degradation of RNAs in eukaryotic cells71. All RNA exosome components are required during the 3’ processing of the 5.8S rRNA72. The disruption of RNA exosomes resulted in the deficiency of rRNA processing, which may ultimately lead to nucleolar stress.
In mammals, the tumor suppressor protein p53/CEP-1 is considered an essential factor in monitoring nucleolar stress and the integrity of ribosome biogenesis20. Additional pathways have also been identified to respond to nucleolar stresses in the absence of functional p5373,74. The ubiquitin E3 ligase MDM2 negatively regulates p53 by marking it for ubiquitin-mediated proteasomal degradation. In plant cells, nucleolar stress activates the expression of ANAC082 by releasing ribosome stalling at the upstream open reading frame (uORF)31. These multiple layers of regulation enable transcription factors to execute cellular responses to specific nucleolar stress conditions. Here, we found that the disruption of RNA exosomes or nucleolar stress activates the expression of NOSR-1 and identified an uncharted NOSR-1/NUMR-1-dependent pathway that links nucleolar stress with the formation of NoSB. However, how the NOSR-1/NUMR-1 axis senses perturbed ribosome biogenesis and nucleolar disorders is unclear. Further investigation is required for searching the factors that activated the NOSR-1/NUMR-1 pathway upon the disruption of RNA exosome or nucleolar stress.
NUMR-1 is upregulated by NOSR-1 and is required for NoSB formation upon nucleolar stress, suggesting that NUMR-1 functions downstream of NOSR-1. We also identified a diverse set of mRNAs that are upregulated by NOSR-1 but do not participate in NoSB formation, indicating that NOSR-1 may be involved in a variety of additional cellular processes. Further experiments, for example, chromatin immunoprecipitation followed by deep sequencing (ChIP-seq), are required to identify the direct targets of NOSR-1.
NUMR-1 encodes RRM and serine- and arginine-rich regions that are found in canonical SR proteins64. Many SR proteins have reported functions in protein/RNA nucleation. For example, the lncRNA metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) localizes to nuclear speckles where it interacts with SR proteins75. SRSF1 and SRSF9 assemble nSBs when cells are exposed to high temperatures76. Some SR proteins (SRSF1, SRSF2, SRSF3, SRSF7, and SRSF10) are recruited to SGs along with nontranslated mRNAs77. Whether and how NUMR-1 uses a similar mechanism to promote NoSB formation requires further investigation.
RNAs, especially noncoding RNA (ncRNA), may act as the molecular scaffold during the formation of phase-separated subcellular organelles, provide multivalent sites to bind one or more RNA binding proteins (RBPs), and drive the occurrence of phase separation. For example, our recent work showed that 27SA2 rRNA intermediate could promote nucleolar reshaping41. Ribosome deficiency may lead to the accumulation of erroneous RNAs and abnormal rRNA intermediate accumulation. The RRM regions in NUMR-1 implies its potential to interact with these RNA molecules and, therefore, elicit the formation of NoSB.
Yet whether or how ncRNAs or specific types of ncRNAs are involved in NoSB formation is unclear. Since the Argonaute protein NRDE-3, which binds endogenous 22 G RNAs, did not accumulate in NoSB (Supplementary Fig. 2D), we guess that it is very unlikely that siRNAs or their targeted mRNAs could enrich in NoSB. In addition, the ribosomal subunit proteins also did not enrich in NoSB (Supplementary Fig. 5A, B), implying that rRNAs may not enrich in NoSB either. In the future, we will try to identify the types of RNAs in NoSB by utilizing ncRNA-seq and smFISH, and explore their roles in NoSB.
The components of NoSB are very intriguing. Till now, we found that (1) treating worms with actinomycin D or inactivation of RNA exosome did not directly translocate NUCL-1 or NUCL-1(ΔRGG) into NoSB. However, further treatment with NaN3 after actinomycin D or inactivation of RNA exosome could induce an accumulation of NUCL-1 or NUCL-1(ΔRGG) to NoSB. (2) An SR-like protein NUMR-1 is specifically enriched in NoSB after nucleolar stress via actinomycin D, inactivation of RNA exosome, or cadmium treatment.
NoSB and GFP::NUMR-1 are rarely detectable in steady-state cells; however, their expression drastically increases in the hypodermal cells and intestinal cells upon nucleolar stress. We speculated that the reason for tissue-specificity of NoSB formation upon nucleolar stress may be: (1) NOSR-1/NUMR-1 axis mostly takes place in somatic cells, yet the pathway is not activated in germ cells upon nucleolar stress; (2) other soma-specific factors, including RNAs or proteins, may cooperate with NUMR-1 to engage in the formation of NoSBs.
The NoSB also disassembles following stress removal. Therefore, we speculated that NUMR-1 might recruit stress-induced proteins and RNAs to NoSB upon nucleolar stress. How NoSB interact with stress responsive proteins and RNAs is unknown. Further investigation to identify the protein and RNA components in NoSB is crucial to elucidate the functions of NoSB and the underlying mechanisms.
NaN3 can quickly immobilize nematodes, which makes it an ideal tool to paralyze worms and is widely used in imaging experiments78,79. Treating C. elegans with NaN3 induces hypoxia and eventually leads to cell death80. For all the proteins tested in the work, including markers for nucleolus and other subnuclear organelles, we only observed that NUCL-1 and NUCL-1(ΔRGG) aggregated to NoSB after the treatment of NaN3 following the nucleolar stress. NUCL-1 is not required for NoSB formation. The reason is not clear yet. Nucleolin likely functions in rRNA transcription and processing. In C. elegans, NUCL-1 is located in the GC region and is required for nucleolar vacuole formation41. We did observe that double treatment of nucleolar stress followed by NaN3 could recruit NUCL-1 to NoSB. The exact role of NaN3 in NUCL-1 recruitment to NoSB is unclear. We guess that NaN3 treatment or hypoxia stress might change the biochemical properties of NUCL-1 or induce other stress reactions and import NUCL-1 into NoSB. In the future, we will focus on how and why NUCL-1 is used in NoSBs.
Membraneless organelles (MLOs) possess a continuum of dynamicity, ranging from liquid to solid, or even amyloid-like aggregates. Many MOLs possess liquid-like properties, and molecules diffuse in and out of MLOs and exchange with the surrounding environment. Some RNA granules including stress granules and processing bodies (P bodies) are highly dynamic and behave like liquid droplets. Yet other MLOs, for example, centrosomes, nuclear pores, and amyloid-like bodies, revealed less dynamic properties. NUMR-1 did not exhibit fluid-like dynamics by FRAP assay, suggesting that NoSB might be a relatively stable structure with a slow turnover in stressed cells.
NUMR-1 was previously identified as an SR-like protein that promotes cadmium tolerance64,65,66. Cadmium arrests larval development and decreases body size, and these effects were exacerbated by numr-1/2 RNAi64. Cadmium was shown to increase numr-1/2 mRNA expression. Therefore, we speculated that NUMR-1/2 and NoSB may involve in protecting animals from adverse environmental conditions. The identification of the components, both RNAs and proteins, in NoSB will greatly help to clarify the function and regulation of NoSB.
Methods
Strains
Bristol strain N2 was used as the standard wild-type strain. All strains were grown at 20 °C unless otherwise specified. The strains used in this study are listed in Supplementary Data 2.
Construction of transgenic strains
For ectopic transgene expression of nosr-1p::NLS::GFP, the NOSR-1 promoter and UTR were amplified from N2 genomic DNA. The EGL-3 NLS sequence was amplified from N2 genomic DNA. A GFP::3xFLAG sequence was PCR amplified from SHG326 genomic DNA. The vector fragment was PCR amplified from the plasmid pSG274. These fragments were joined together by Gibson assembly to form the repair plasmid with the ClonExpress MultiS One Step Cloning Kit (Vazyme Biotech, Nanjing, China, Cat. No. C113-01/02). The transgene was integrated into C. elegans chromosome I via a modified counterselection (cs)-CRISPR method60. The sequences of the primers are listed in Supplementary Data 3.
For in situ knock-in transgenes, the 3xFLAG::GFP sequence was PCR amplified from YY178 genomic DNA. The GFP::3xFLAG sequence was PCR amplified from SHG326 genomic DNA. The tagRFP coding sequence was PCR amplified from YY1446 genomic DNA. Homologous left and right arms (1.5 kb) were PCR amplified from N2 genomic DNA. The backbone was PCR amplified from the plasmid pCFJ151. All these fragments were joined together by Gibson assembly to form the repair plasmid with the ClonExpress MultiS One Step Cloning Kit. The plasmid was coinjected into N2 animals with three sgRNA expression vectors, 5 ng/µl pCFJ90, and the Cas9 II-expressing plasmid. The sequences of the primers are listed in Supplementary Data 4.
Construction of deletion mutants
For gene deletions, triple sgRNA-guided chromosome deletion was conducted60. To construct sgRNA expression vectors, the 20 bp unc-119 sgRNA guide sequence in the pU6::unc-119 sgRNA(F + E) vector was replaced with different sgRNA guide sequences. Addgene plasmid #47549 was used to express Cas9 II protein. Plasmid mixtures containing 30 ng/µl of each of the three or four sgRNA expression vectors, 50 ng/µl Cas9 II-expressing plasmid, and 5 ng/µl pCFJ90 were coinjected into N2 animals. Deletion mutants were screened by PCR amplification and confirmed by sequencing. The sgRNA sequences are listed in Supplementary Data 5.
Imaging and quantification
Images were collected on a Leica DM4 B microscope. The nucleolar area and NoSB area were quantified manually with the freehand tool using Fiji software.
Actinomycin D treatment
Actinomycin D (MedChemExpress no. HY-17559, CAS: 50-76–0) was prepared at 20 mg/ml in DMSO as a stock solution. The actinomycin D stock solution was diluted to 5 to 30 μg/ml with concentrated OP50. NGM plates were prepared and placed at room temperature overnight before use. Synchronized L1 worms were placed onto the seeded plates and grown for 48 h before imaging.
RNAi
RNAi experiments were performed at 20 °C by placing synchronized embryos on feeding plates81. HT115 bacteria expressing the empty vector L4440 (a gift from A. Fire) were used as controls. Bacterial clones expressing dsRNAs were obtained from the Ahringer RNAi library and sequenced to verify their identity. Some bacterial clones were constructed in this work, which are listed in Supplementary Data 1. Images were collected using a Leica DM4 B microscope.
Forward genetic screening
To identify factors prohibiting the formation of NoSB, we chemically mutagenized the mCherry::DIS-3 strain by ethyl methanesulfonate (EMS), followed by a clonal screen. The F2 progeny worms were visualized under a fluorescence microscope at the L4 stage. Two mutants that exhibited NoSB formation were isolated from two thousand haploid genomes. exso-10 (ust242) and exos-4.2 (ust243) were identified by genome resequencing.
To identify factors required for the formation of NoSB, we chemically mutagenized the exso-10(ust242);mCherry::DIS-3 strain by ethyl methanesulfonate (EMS), followed by a clonal screen. The F2 progeny worms were visualized under a fluorescence microscope at the L4 stage. One mutant that disrupted NoSB formation was isolated from two thousand haploid genomes. nosr-1(ust303) was identified by genome resequencing.
Postfix Oil Red O staining and bright field imaging
Briefly, L4 stage animals were fixed in 1% paraformaldehyde/PBS for 30 min with rocking. Samples were then frozen on dry ice/ethanol and thawed with running tap water three times. Samples were washed three times with 1× PBS, dehydrated in 60% isopropanol for 2 min, and stained with 0.5 ml 60% Oil Red-O (#A6000395-50G, BBI) working solution for 30 min with rocking. Oil Red O stock solution was dissolved in isopropanol at a concentration of 0.5 g/100 ml and equilibrated for several days. The Oil Red-O working solution was prepared fresh by mixing 60% volume of stock with 40% volume of water. The mix was equilibrated for 10 min and then filtered with a 0.22-μm syringe filter. Stained samples were then washed three times with 1× PBS, rehydrated in 1× PBS, and mounted in 1× PBS onto agarose-padded slides for imaging.
RNA isolation
Synchronized L4 worms were incubated with TRIzol (Invitrogen) reagent followed by five quick liquid nitrogen freeze-thaw cycles. RNA was precipitated by isopropanol followed by DNaseI digestion (Thermo Fisher).
mRNA deep-sequencing
The Illumina-generated raw reads were first filtered to remove adapters, low-quality tags, and contaminants to obtain clean reads at Novogene. The clean reads were mapped to the reference genome of WBcel235 via HISAT2 software (version 2.1.0)82. Differential expression analysis was performed using custom R scripts. A twofold-change cutoff was applied when filtering for differentially expressed genes. All plots were drawn using custom R scripts.
qRT–PCR
cDNAs were generated from RNA using HiScript III RT SuperMix for qPCR (Vazyme), which includes a random primer/oligo(dT)20VN primer mix for reverse transcription. qPCR was performed using a MyIQ2 real-time PCR system (Bio-Rad) with AceQ SYBR Green Master mix (Vazyme). The primers used in qRT‒PCR are listed in Supplementary Data 6.
Brood size
L4 hermaphrodites were singled onto NGM plates and transferred daily as adults until embryo production ceased, and the progeny numbers were scored.
Lifespan assay
Worm populations were synchronized by placing young adult worms on NGM plates for 4–6 h and then removing them. The hatching day was counted as day 1 for all lifespan measurements. Worms were transferred every other day to new plates to eliminate confounding progeny. Animals were scored as alive or dead every day. Worms were scored as dead if they did not respond to repeated prods with a platinum pick. Worms were censored if they crawled off the plates or died from vulval bursting and bagging. For each lifespan assay, >50 worms were used.
Hydrogen peroxide assay
Ten synchronized worms on day 1 of adulthood were transferred to each well, which contained 1 ml of worm S-basal buffer with various concentrations of H2O2 in a 12-well plate at 20 °C. Four hours later, 100 μl of 1 mg/ml catalase (Sigma, C9322) was added to neutralize H2O2, and the mortality of worms was scored.
Heat-shock assay
Ten synchronized hermaphrodites on day 1 of adulthood were transferred to new NGM plates and incubated at 35 °C, and their mortality was checked every 1 h until there were no living worms.
Starvation assay
L4 worms were transferred to empty NGM plates without any bacterial lawn for 12 h, and then the animals were imaged.
Cadmium treatment
Synchronized L1 worms were fed with OP50 on NGM agar plates containing 0, 100, or 300 μM of cadmium chloride for 48 h before imaging.
Fluorescence recovery after photobleaching (FRAP)
FRAP experiments were performed using a Zeiss LSM980 laser scanning confocal microscope at room temperature. Worms were anesthetized with 2 mM levamisole. A region of interest was bleached with 70% laser power for 3–4 s, and the fluorescence intensities in these regions were collected every 5 s and normalized to the initial intensity before bleaching. For analysis, image intensity was measured by mean and further analyzed by GraphPad Prism 9.0 software.
Z-stack imaging experiment
Z-stack imaging experiments were performed using a Zeiss LSM980 laser scanning confocal microscope. Worms were anesthetized with 2 mM levamisole. By capturing the filtered emissions within the 495–590 nm range, with steps of 0.5 μm, the images per section in a z-stack were acquired.
Statistics and reproducibility
For each microscopy result in Figs. 2F, 3B, D, F, 6B, 7F and Supplementary Figs. 2A, C, D, 3C–G, 5A, 6A–E, 7A–D, independent experiments were performed three times.
The mean and standard deviation of the results are presented in bar graphs with error bars. All experiments were conducted with independent C. elegans animals for the indicated number (N) of times. Statistical analysis was performed with the two-tailed Student’s t-test as indicated. GraphPad Prism 9 or R scripts were used for statistical analysis.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that the data supporting the findings of this study are available within the paper and its supplementary information files, and that all additional data are publicly available. The high throughput data reported in this paper have been deposited in the Genome Sequence Archive in the National Genomics Data Center (China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences) under accession codes CRA012745. Source data are provided with this paper.
References
Stanek, D. & Fox, A. H. Nuclear bodies: news insights into structure and function. Curr. Opin. Cell Biol. 46, 94–101 (2017).
Banani, S. F., Lee, H. O., Hyman, A. A. & Rosen, M. K. Biomolecular condensates: organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 18, 285–298 (2017).
Strom, A. R. & Brangwynne, C. P. The liquid nucleome - phase transitions in the nucleus at a glance. J. Cell Sci. 132, jcs235093 (2019).
Harmon, T. S., Holehouse, A. S. & Pappu, R. V. Differential solvation of intrinsically disordered linkers drives the formation of spatially organized droplets in ternary systems of linear multivalent proteins. New J. Phys. 20, 045002 (2018).
Chen, L. L. & Carmichael, G. G. Altered nuclear retention of mRNAs containing inverted repeats in human embryonic stem cells: functional role of a nuclear noncoding RNA. Mol. Cell 35, 467–478 (2009).
Fox, A. H. & Lamond, A. I. Paraspeckles. Cold Spring Harb. Perspect. Biol. 2, a000687 (2010).
Mahmoudi, S. et al. WRAP53 is essential for Cajal body formation and for targeting the survival of motor neuron complex to Cajal bodies. PLos Biol. 8, e1000521 (2010).
Li, Y. J. et al. Fam118B, a newly identified component of Cajal bodies, is required for Cajal body formation, snRNP biogenesis and cell viability. J. Cell Sci. 127, 2029–2039 (2014).
Lallemand-Breitenbach, V. & de The, H. PML nuclear bodies. Cold Spring Harb. Perspect. Biol. 2, a000661 (2010).
Ilik, I. A. et al. SON and SRRM2 are essential for nuclear speckle formation. Elife 9, e60579 (2020).
Krafft, C., Knetschke, T., Funk, R. H. W. & Salzer, R. Studies on stress-induced changes at the subcellular level by Raman microspectroscopic mapping. Anal. Chem. 78, 4424–4429 (2006).
Sampuda, K. M., Riley, M. & Boyd, L. Stress induced nuclear granules form in response to accumulation of misfolded proteins in Caenorhabditis elegans. BMC Cell Biol. 18, 18 (2017).
Boulon, S., Westman, B. J., Hutten, S., Boisvert, F. M. & Lamond, A. I. The nucleolus under stress. Mol. Cell 40, 216–227 (2010).
Biamonti, G. & Vourc’h, C. Nuclear Stress Bodies. Cold Spring Harb. Perspect. Biol. 2, a000695 (2010).
Wang, Y. et al. Genome-wide screening of NEAT1 regulators reveals cross-regulation between paraspeckles and mitochondria. Nat. Cell Biol. 20, 1145 (2018).
Sustackova, G. et al. Acetylation-dependent nuclear arrangement and recruitment of BMI1 protein to UV-damaged chromatin. J. Cell Physiol. 227, 1838–1850 (2012).
Klinge, S. & Woolford, J. L. Ribosome assembly coming into focus. Nat. Rev. Mol. Cell Biol. 20, 116–131 (2019).
Woolford, J. L. & Baserga, S. J. Ribosome biogenesis in the yeast Saccharomyces cerevisiae. Genetics 195, 643–681 (2013).
Boisvert, F. M., van Koningsbruggen, S., Navascues, J. & Lamond, A. I. The multifunctional nucleolus. Nat. Rev. Mol. Cell Biol. 8, 574–585 (2007).
James, A., Wang, Y. B., Raje, H., Rosby, R. & DiMario, P. Nucleolar stress with and without p53. Nucleus 5, 402–426 (2014).
Yang, K., Yang, J. & Yi, J. Nucleolar stress: hallmarks, sensing mechanism and diseases. Cell Stress 2, 125–140 (2018).
Goldblatt, P. J., Verbin, R. S. & Sullivan, R. J. Induction of nucleolar segregation by actinomycin D following inhibition of protein synthesis with cycloheximide. Exp. Cell Res. 63, 117–123 (1970).
Tsang, C. K., Bertram, P. G., Ai, W. D., Drenan, R. & Zheng, X. F. S. Chromatin-mediated regulation of nucleolar structure and RNA Pol I localization by TOR. EMBO J. 22, 6045–6056 (2003).
Kang, J. et al. Ribosomal proteins and human diseases: molecular mechanisms and targeted therapy. Signal Transduct. Target Ther. 6, 323 (2021).
Fumagalli, S. et al. Absence of nucleolar disruption after impairment of 40s ribosome biogenesis reveals an rpL11-translation-dependent mechanism of p53 induction. Nat. Cell Biol. 11, 501–U350 (2009).
Zhang, Y. P. & Lu, H. Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 (2009).
Dai, M. S., Arnold, H., Sun, X. X., Sears, R. & Lu, H. Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 26, 3332–3345 (2007).
Donati, G. et al. Selective inhibition of rRNA transcription downregulates E2F-1: a new p53-independent mechanism linking cell growth to cell proliferation. J. Cell Sci. 124, 3017–3028 (2011).
Russo, A. et al. Human rpL3 induces G(1)/S arrest or apoptosis by modulating p21 (waf1/cip1) levels in a p53-independent manner. Cell Cycle 12, 76–87 (2013).
Zhou, X., Hao, Q., Liao, J. M., Liao, P. & Lu, H. Ribosomal protein S14 negatively regulates c-Myc activity. J. Biol. Chem. 288, 21793–21801 (2013).
Ohbayashi, I. et al. Evidence for a role of ANAC082 as a ribosomal stress response mediator leading to growth defects and developmental alterations in Arabidopsis. Plant Cell 29, 2644–2660 (2017).
Schumacher, B. et al. Translational repression of C. elegans p53 by GLD-1 regulates DNA damage-induced apoptosis. Cell 122, 145–145 (2005).
Gao, M. X. et al. The SCFFSN-1 ubiquitin ligase controls germline apoptosis through CEP-1/p53 in C. elegans. Cell Death Differ. 15, 1054–1062 (2008).
Fuhrman, L. E., Goel, A. K., Smith, J. Shianna, K. V. & Aballay A. Nucleolar proteins suppress Caenorhabditis elegans innate immunity by inhibiting p53/CEP-1. PLos Genet. 5, e1000657 (2009).
Leung, C. K., Empinado, H. & Choe, K. P. Depletion of a nucleolar protein activates xenobiotic detoxification genes in Caenorhabditis elegans via Nrf/SKN-1 and p53/CEP-1. Free Radic. Biol. Med. 52, 937–950 (2012).
Wu, J. et al. PHA-4/FoxA senses nucleolar stress to regulate lipid accumulation in Caenorhabditis elegans. Nat. Commun. 9, 1195 (2018).
Zhou X, et al. RdRP-synthesized antisense ribosomal siRNAs silence pre-rRNA via the nuclear RNAi pathway. Nat Struct Mol Biol 24, 258–269 (2017).
Liao, S. et al. Antisense ribosomal siRNAs inhibit RNA polymerase I-directed transcription in C. elegans. Nucleic Acids Res. 49, 9194–9210 (2021).
Zhu, C. et al. Erroneous ribosomal RNAs promote the generation of antisense ribosomal siRNA. Proc. Natl Acad. Sci. USA 115, 10082–10087 (2018).
Xu, T. et al. A ZTF-7/RPS-2 complex mediates the cold-warm response in C. elegans. PLos Genet. 19, e1010628 (2023).
Xu, D. et al. rRNA intermediates coordinate the formation of nucleolar vacuoles in C. elegans. Cell Rep. 42, 112915 (2023).
Kilchert, C., Wittmann, S. & Vasiljeva, L. The regulation and functions of the nuclear RNA exosome complex. Nat. Rev. Mol. Cell Biol. 17, 227–239 (2016).
Spaulding, E. L., Feidler, A. M., Cook, L. A. & Updike, D. L. RG/RGG repeats in the C. elegans homologs of nucleolin and GAR1 contribute to sub-nucleolar phase separation. Nat. Commun. 13, 6585 (2022).
Shaw, P. J. & Brown, J. W. Plant nuclear bodies. Curr. Opin. Plant Biol. 7, 614–620 (2004).
Johnson, J. M. & Jones, L. E. Behavior of nucleoli and contracting nucleolar vacuoles in tobacco cells growing in microculture. Am. J. Bot. 54, 189–198 (1967).
Rose, R. J., Setterfield, G. & Fowke, L. C. Activation of nucleoli in tuber slices and the function of nucleolar vacuoles. Exp. Cell Res. 71, 1–16 (1972).
O’Donohue, M. F., Choesmel, V., Faubladier, M., Fichant, G. & Gleizes, P. E. Functional dichotomy of ribosomal proteins during the synthesis of mammalian 40S ribosomal subunits. J. Cell Biol. 190, 853–866 (2010).
Gamalinda, M. et al. A hierarchical model for assembly of eukaryotic 60S ribosomal subunit domains. Genes Dev. 28, 198–210 (2014).
Morton, D. J. et al. The RNA exosome and RNA exosome-linked disease. RNA 24, 127–142 (2018).
Neumuller, R. A. et al. Conserved regulators of nucleolar size revealed by global phenotypic analyses. Sci. Signal 6, ra70 (2013).
Perry, R. P. & Kelley, D. E. Persistent transcription of 5s RNA genes when synthesis of 18s and 28s ribosomal RNA is inhibited by low doses of actinomycin D. J. Cell Biol. 39, A103 (1968).
Perry, R. P. & Kelley, D. E. Inhibition of RNA synthesis by actinomycin D: characteristic dose-response of different RNA species. J. Cell Physiol. 76, 127–139 (1970).
Lee, L. W., Lee, C. C., Huang, C. R. & Lo, S. J. The nucleolus of Caenorhabditis elegans. J. Biomed. Biotechnol. 2012, 601274 (2012).
Pham, K., Masoudi, N., Leyva-Diaz, E. & Hobert, O. A nervous system-specific subnuclear organelle in Caenorhabditis elegans. Genetics 217, 1–17 (2021).
Ohsaki, Y. et al. PML isoform II plays a critical role in nuclear lipid droplet formation. J. Cell Biol. 212, 29–38 (2016).
Romanauska, A. & Kohler, A. The inner nuclear membrane is a metabolically active territory that generates nuclear lipid droplets. Cell 174, 700 (2018).
Salvetti, A. et al. Nuclear functions of nucleolin through global proteomics and interactomic approaches. J. Proteome Res. 15, 1659–1669 (2016).
Reinke, A. W., Baek, J., Ashenberg, O. & Keating, A. E. Networks of bZIP protein-protein interactions diversified over a billion years of evolution. Science 340, 730–734 (2013).
Reinke, V., Krause, M. & Okkema, P. Transcriptional regulation of gene expression in C. elegans. WormBook 1–34 (2013).
Chen, X. Y. et al. Dual sgRNA-directed gene knockout using CRISPR/Cas9 technology in Caenorhabditis elegans. Sci. Rep. 4, 7581 (2014).
Amoutzias, G. D. et al. One billion years of bZIP transcription factor evolution: conservation and change in dimerization and DNA-binding site specificity. Mol. Biol. Evol. 24, 827–835 (2007).
Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. daf-16: An HNF-3/forkhead family member that can function to double the life-span of Caenorhabditis elegans. Science 278, 1319–1322 (1997).
Ogg, S. et al. The Fork head transcription factor DAF-16 transduces insulin-like metabolic and longevity signals in C-elegans. Nature 389, 994–999 (1997).
Wu, C. W. et al. RNA processing errors triggered by cadmium and integrator complex disruption are signals for environmental stress. BMC Biol. 17, 56 (2019).
Shepard, P. J. & Hertel, K. J. The SR protein family. Genome Biol. 10, 242 (2009).
Tvermoes, B. E., Boyd, W. A. & Freedman, J. H. Molecular characterization of numr-1 and numr-2: genes that increase both resistance to metal-induced stress and lifespan in Caenorhabditis elegans. J. Cell Sci. 123, 2123–2133 (2010).
Jeong, S. SR proteins: binders, regulators, and connectors of RNA. Mol. Cells 40, 1–9 (2017).
Nagai, K., Oubridge, C., Jessen, T. H., Li, J. & Evans, P. R. Crystal-structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein-A. Nature 348, 515–520 (1990).
Perez-Canadillas, J. M. & Varani, G. Recent advances in RNA-protein recognition. Curr. Opin. Struc. Biol. 11, 53–58 (2001).
Orti, F., Navarro, A. M., Rabinovich, A., Wodak, S. J. & Marino-Buslje, C. Insight into membraneless organelles and their associated proteins: drivers, clients and regulators. Comput. Struct. Biotechnol. 19, 3964–3977 (2021).
Puno, M. R., Weick, E. M., Das, M. & Lima, C. D. SnapShot: the RNA exosome. Cell 179, 282–282.e281 (2019).
Allmang, C. et al. Functions of the exosome in rRNA, snoRNA and snRNA synthesis. EMBO J. 18, 5399–5410 (1999).
Lindstrom MS, Jurada D, Bursac S, Orsolic I, Bartek J, Volarevic S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 37, 2351–2366 (2018).
Holmberg Olausson K, Nister M, Lindstrom MS. p53 -Dependent and -Independent Nucleolar Stress Responses. Cells 1, 774–798 (2012).
Wagner, R. E. & Frye, M. Noncanonical functions of the serine-arginine-rich splicing factor (SR) family of proteins in development and disease. Bioessays 43, e2000242 (2021).
Ninomiya, K. et al. LncRNA-dependent nuclear stress bodies promote intron retention through SR protein phosphorylation. EMBO J. 39, e102729 (2020).
Twyffels, L., Gueydan, C. & Kruys, V. Shuttling SR proteins: more than splicing factors. FEBS J. 278, 3246–3255 (2011).
Greene, J. S. et al. Balancing selection shapes density-dependent foraging behaviour. Nature 539, 254–258 (2016).
Chen, W. W. et al. Spectroscopic coherent Raman imaging of Caenorhabditis elegans reveals lipid particle diversity. Nat. Chem. Biol. 16, 1087–1095 (2020).
Anyanful, A. et al. Paralysis and killing of Caenorhabditis elegans by enteropathogenic Escherichia coli requires the bacterial tryptophanase gene. Mol. Microbiol. 57, 988–1007 (2005).
Timmons, L., Court, D. L. & Fire, A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112 (2001).
Kim, D., Paggi, J. M., Park, C., Bennett, C. & Salzberg, S. L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 37, 907 (2019).
Acknowledgements
We are grateful to the members of the Guang lab for their comments. We are grateful to the International C. elegans Gene Knockout Consortium and the National Bioresource Project for providing the strains. Some strains were provided by the CGC, which is funded by the NIH Office of Research Infrastructure Programs (P40 OD010440). This work was supported by grants from the National Key R&D Program of China (2022YFA1302700 to S.G., and 2019YFA0802600 awarded to X.F.) and the National Natural Science Foundation of China (32230016 awarded to S.G., 32270583 awarded to C. Z., 32070619 awarded to X.F., 2023M733425 awarded to X.H., and 32300438 awarded to X.H.), and the Strategic Priority Research Program of the Chinese Academy of Sciences (XDB39010600 awarded to S.G.), the Research Funds of Center for Advanced Interdisciplinary Science and Biomedicine of IHM (QYPY20230021 awarded to S.G.) and the Fundamental Research Funds for the Central Universities awarded to S.G.
Author information
Authors and Affiliations
Contributions
M.H.: Conceptualization, data curation, formal analysis, visualization, investigation, methodology, and writing—original draft. X.Z.: Conceptualization, data curation, visualization, investigation, methodology, and resources. C.Zeng.: Conceptualization, investigation, validation, data curation, and methodology. D.X.: Software, formal analysis, visualization. T.X.: Investigation, methodology, and resources. S.L.: Conceptualization and validation. K.W.: Formal analysis and validation. C.Zhu.: Conceptualization and methodology. G.S.: Conceptualization. X.H.: Conceptualization, supervision, and project administration. X.C.: Conceptualization, supervision, and project administration. X.F.: Conceptualization, supervision, project administration, and writing—review & editing. S.G.: Conceptualization, supervision, project administration, and writing - review & editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Hong, M., Zhou, X., Zeng, C. et al. Nucleolar stress induces nucleolar stress body formation via the NOSR-1/NUMR-1 axis in Caenorhabditis elegans. Nat Commun 15, 7256 (2024). https://doi.org/10.1038/s41467-024-51693-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-51693-z
- Springer Nature Limited