Abstract
Precision mapping of site-specific glycans using mass spectrometry is vital in glycoproteomics. However, the diversity of glycan compositions across species often exceeds database capacity, hindering the identification of rare glycans. Here, we introduce pGlycoNovo, a software within the pGlyco3 software environment, which employs a glycan first-based full-range Y-ion dynamic searching strategy. pGlycoNovo enables de novo identification of intact glycopeptides with rare glycans by considering all possible monosaccharide combinations, expanding the glycan search space to 16~1000 times compared to non-open search methods, while maintaining accuracy, sensitivity and speed. Reanalysis of SARS Covid-2 spike protein glycosylation data revealed 230 additional site-specific N-glycans and 30 previously unreported O-glycans. pGlycoNovo demonstrated high complementarity to six other tools and superior search speed. It enables characterization of site-specific N-glycosylation across five evolutionarily distant species, contributing to a dataset of 32,549 site-specific glycans on 4602 proteins, including 2409 site-specific rare glycans, and uncovering unexpected glycan fragments.
Similar content being viewed by others
Introduction
Protein glycosylation is one of the widespread and intricate post-translational modifications (PTMs) that involves in various biological processes and is present in nearly all life forms1,2,3. Unlike proteins and nucleic acids, which directly stem from a DNA template, glycans result from a complex biosynthetic pathway influenced by a multitude of genetic and environmental factors4,5. The template-free nature of glycan synthesis poses a considerable challenge for predicting the glycan composition within living organisms, thereby presenting a substantial obstacle to comprehensive glycoproteomic analysis6,7.
The analysis of intact glycopeptides using liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS) has emerged as an efficient approach for more comprehensive glycoproteomic analysis, encompassing peptide sequences, glycosylation sites, and the attached glycans8,9,10,11,12,13. The collisional activation of glycopeptides typically yields fragments of both peptides and glycans6,14. Modern glycoproteomics software tools commonly separate the glycopeptide search into peptide and glycan searches12,15. Based on the prioritization of search components, three primary search strategies emerge: peptide-first, glycan-removal (also regarded as the 3-step glycan-first, where glycopeptide ions are used first to narrow down the peptide search space, with peptide identification preceding the comprehensive characterization of the glycan.), and glycan-first. The peptide-first search strategy was widely adopted by several tools16,17,18,19,20,21,22,23, leveraging the advancements in modern proteomic methods to directly match peptide b and y ions in the spectrum before inferring the glycan component through the mass difference between the precursor and peptide part. To bolster the reliability of the glycan identification, some tools simultaneously consider partial B/Y ions while matching potential glycan masses in a glycan database16,17. Two recently developed tools MSFragger-Glyco17 and MetaMoroheus O-Pair16 embraced and refined this strategy by incorporating peptide fragment-ion indexing to accelerate the search process. The glycan-removal strategy utilized by tools such as MAGIC24, StrucGP25, and GlycoDecioher26 also first determines the peptide parts based on the distinctive Y0 ion, which differs from the peptide-first search. Subsequently, it proceeds to interpret glycan from the glycan database. In contrast, the glycan-first strategy, typically employed in pGlyco software series15,27,28 and other tools such as Sweet-Heart29 and GlycoMaster DB30, entails initiating the search for glycan components before delving into peptide parts. This strategy enhances the reliability of the glycan identification by prioritizing the retrieval of the glycan component to eliminate unreliable glycans. Based on this approach, pGlyco 2.0 marks the first instance of achieving comprehensive quality control at the glycan, peptide, and glycopeptide levels28. pGlyco 3 adheres to this strategy, further integrating the glycan ion-indexing approach and optimizing the utilization of Y and B ions, thereby enhancing accuracy and expediting glycopeptide matching15. Currently, the majority of intact glycopeptide identification software tools, including pGlyco315, MSFragger-Glyco17, and MetaMoroheus O-Pair16, typically identify site-specific glycans within predefined glycan libraries. The discovery of rare glycans beyond the scope of the glycan libraries is challenging11,14.
The most recent software tools, such as Glyco-Decipher26 and PEAKS GlycanFinder31, demonstrate the ability to identify rare glycans. In the analysis of a given glycopeptide spectrum, these algorithms typically start by detecting the peptide component to ascertain the peptide mass. Subsequently, the glycan part can be identified by utilizing the deduced glycan mass (precursor mass minus peptide mass) along with glycan fragments. These methods are highly effective in identifying peptides when the quality of peptide fragments is high, which in turn facilitates the straightforward identification of glycans. However, in many cases of glycopeptides, the fragmentation of peptide parts may not be optimal, potentially causing the oversight of true candidate peptides in the initial search step. For the discovery of rare glycans, it is essential that the glycans are substantiated by high-quality fragment signals, which aid in accurate glycan fragments. Therefore, the glycan-first search, which requires high-quality glycan fragment spectra, is particularly suitable for discovering rare glycans. The key challenge of this kind of analysis lies in the necessity to conduct an open search for glycans without relying on glycan libraries. Some peptide-first open search tools can identify rare glycans due to their ability to handle a wide variety of mass offsets associated with glycans17,32. However, these open search algorithms only accommodate glycan masses and require additional steps to interpret these masses and elucidate glycan compositions or structures. StrucGP25 is considered capable of de novo sequencing at the structure level, but it relies on prior knowledge of glycan structures and cannot perform de novo sequencing for arbitrarily specified monosaccharide compositions.
Here, we present pGlycoNovo, an innovative tool within the pGlyco3 software environment, featuring a full-range glycan search algorithm that enables open glycan search and glycopeptide identification without relying on glycan libraries. This strategy significantly expands the glycan diversity by allowing the de novo identification of rare glycans beyond existing libraries, using arbitrarily specified monosaccharide compositions. The glycan-first search strategy enhances global Y-ion utilization, supporting quality control through careful parameter tuning and supplemented by manual validation of rare glycans for accurate identification. We validated pGlycoNovo’s superior performance than other tools in terms of rare glycan uncovering and search speed by identifying intact N-glycopeptides and O-glycopeptides in the SARS-CoV-2 Spike protein expressed in insect cells. We highlighted pGlycoNovo’s capability for rare glycan identification by mapping site-specific N-glycans across five evolutionarily distant species, yielding a dataset of 32,549 site-specific glycans on 4602 proteins when used in conjunction with pGlyco3, among which 2409 site-specific rare glycans were exclusively identified by pGlycoNovo. The reliability of pGlycoNovo was demonstrated through the comprehensive glycopeptide 13C- and 15N-labeling strategy in yeasts and plants. Notably, the comprehensive Y-ion matching capability of pGlycoNovo revealed a significant number of unexpected glycan fragments in large-scale glycopeptide spectra across different species samples, raising concerns about the accuracy of software aimed at resolving site-specific glycan structures.
Results
Evolving the pGlycoNovo approach
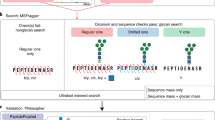
pGlycoNovo is a fire-new software package developed within the pGlyco315 platform, characterized by its highly efficient glycan library-free search capabilities. pGlycoNovo follows a glycan-first search strategy and integrates a full range Y-matching method for glycan composition de novo sequencing without knowing the peptide part (Fig. 1a). In contrast to existing tools that primarily rely on the core Y ions, pGlycoNovo utilizes the full-range Y ions and significantly expands the glycan search space for both N- and O-glycopeptide identification (Fig. 1b).
a The main strategy of full-range Y-ion dynamic searching in pGlycoNovo. b pGlycoNovo is distinguished by its utilization of a full-range of Y ions, enabling the identification of N-/O-glycopeptide across a significantly expanded glycan search space. c The schema of pGlycoNovo workflow. Detailed software algorithms and processes are provided in Online Methods. d A significant expansion of the glycan search space was achieved by pGlycoNovo in comparison to other tools. The glycan library-dependent software’s search space is limited to the number of glycan compositions recorded in its library, while the glycan library-independent software, StrucGP, is currently restricted to mammalian species due to its reliance on prior knowledge and can roughly handle up to 10,000 glycan compositions, with the largest N-glycan being HexNAc(9)Hex(10)Fuc(5)Ac(4)Gc(4). In contrast, pGlycoNovo imposes no limitations on glycan library size and glyco units, allowing flexible customization. In this study, we employed an optimized glycan search space size of 168,000, as we found that a larger search space did not significantly enhance identification results (data not shown). e Sensitivity and precision evaluation of pGlycoNovo in a yeast dataset with a precursor tolerance of 2 ppm under different glyco-gap and glycan score conditions. f pGlycoNovo’s precision and sensitivity across three datasets using the parameters adopted in our study (2 ppm for precursor tolerance, a glyco-gap of 2, and a glycan score threshold of 20). Three benchmark datasets, from budding yeast, mouse, and human species, possessing well-established glycan libraries and in-depth knowledge of glycan compositions, were used for the demonstration of pGlycoNovo’s precision and sensitivity (Supplementary Note 1). We compare pGlycoNovo’s identification results with those of pGlyco3, which relies on a known glycan library, using the formula in the figure to evaluate pGlycoNovo’s precision and sensitivity. Detailed search parameter optimization and comparisons are provided in Supplementary Note 1. g Comparison of search speed using the three benchmark datasets. The comparison includes an assessment of the pGlyco3 and pGlycoNovo algorithms (both searching the same MGF files generated by pParse56).
The schema of the whole pGlycoNovo process is shown in Fig. 1c. For a given glycopeptide MS2 spectrum, pGlycoNovo encompassed four key steps: glycan enumeration and matching of peak complementary masses (complementary mass refers to precursor mass minus peak mass); generation of Y-ion graph based on the identified Y-ions; deduction and filtration of glycan candidates from the Y-ion graph; retrieval of peptides for the identified glycans, followed by glycopeptide scoring and quality control. The detail of the pGlycoNovo algorithm is provided in the Online Methods (Methods). By employing the full-range Y-ion searching strategy, pGlycoNovo substantially expands the glycan search space, enabling de novo sequencing of ~168,000 glycan compositions. This expansion is 16 to 1000 times larger than that achieved by commonly used non-open-search approaches (Fig.1 d).
We utilized intact N-glycopeptide data from budding yeast, mouse liver, and HeLa cells as our benchmark dataset (Supplementary Note 1), as these species are recognized for possessing well-established glycan libraries. This allowed us to fine-tune the search parameters and demonstrate pGlycoNovo’s performance in terms of sensitivity, precision, and search speed. After systematically optimizing the search parameters (Supplementary Note 1, Fig. 1e), pGlycoNovo exhibited a high precision exceeding 98.5% while maintaining efficient sensitivity of over 70.9% across three datasets using the parameters adopted in our study (Fig. 1f, Supplementary Note 1), putting it on par with well-established peptide de novo techniques33,34,35. Moreover, the efficient dynamic programming implemented in pGlycoNovo ensured a high-speed search (Fig. 1g), even outpacing the proven speed leadership of pGlyco3 among various software tools15.
Assessing pGlycoNovo’s search performance on the SARS-CoV-2 spike protein
To assess the performance of pGlycoNovo in glycopeptide identification, especially in the context of rare glycan identification, we conducted a comparative analysis with six other software tools: pGlyco3, MSFragger-open, StrucGP, Byonic, GlycanFinder, and Glyco-Decipher using one of the thirty raw data from a publicly available N-glycopeptide dataset of the SARS-Cov-2 Spike protein (PXD001850636). All seven software tools employed the same protein database containing the SARS-CoV-2 Spike protein sequences for protein searches, along with their respective N-glycan databases for glycan searches. Notably, pGlycoNovo does not rely on a predefined glycan database. Instead, it employs a maximum of forty-nine monosaccharides for glycan identification. The search details are listed in the Supplementary Note 2. The comparative results shown in Fig. 2a demonstrate that in the terms of overall identification results, pGlycoNovo performed comparably to GlycanFinder and Glyco-Decipher (216 vs. 219 vs. 218), and outperformed the other four tools (Supplementary Data). Even in comparison with MSFragger-Glyco open search17, GlycanFinder31, and Glyco-Decipher26, which specialize in-depth identification, pGlycoNovo stands out for its remarkable complementary capacities for the identification. It is noteworthy that the glycopeptide data we used here originated from the SARS-CoV-2 Spike protein expressed in insects, potentially containing rare glycans. StrucGP, which relies on prior glycan knowledge25, can only identify 0.85% of glycopeptides attached with rare glycans. Other five software tools including pGlyco3 are restricted by the limitations of their glycan libraries, making them incapable of identifying glycan types beyond those present in mammalian glycan databases. In contrast, pGlycoNovo detects 12.96% of glycopeptides attached with rare glycans (Fig. 2a, Supplementary Note 2, Supplementary Data).
a Site-specific N-glycan identification comparison using different software tools on a single N-glycopeptide dataset of the SARS-CoV-2 Spike protein. b Search speed comparison using the same N-glycopeptide data (PXD001850636). All searches were initiated from RAW files, using the same protein database. Notably, pGlycoNovo performed searches in a glycan search space 16–1000 times larger than the other four tools. Detailed comparison procedures are in Supplementary Note 2. c Expanded N-glycoproteome revealed by pGlycoNovo in public SARS-CoV-2 Spike Protein MS data (PXD001850636, 30 RAW files). Results were compared with those published using Byonic for the same data (Supplementary Note 3). d Distribution of site-specific N-glycans on the SARS-CoV-2 Spike protein. We performed statistical analysis of the 523 site-specific glycans identified by combining results from pGlycoNovo and the published data. Glycans were categorized into five groups based on monosaccharide composition: oligomannose type, hybrid type, complex type, truncated glycans, and unclassified type, where “unclassified type” refers to rare glycans not included in existing glycan database-dependent search engines (Supplementary Note 4). e Analysis of site-specific O-glycans using pGlycoNovo on SARS-CoV-2 Spike protein. It is noted that this public data does not include ETD spectra, making it impossible for us to differentiate between neighboring sites. f An annotated spectrum of an intact N-glycopeptide with a rare glycan attached. g An annotated spectrum of an intact O-glycopepide attached with a rare glycan attached. Peptide sequence with “J” indicating the N-glycosylation site. The glycan symbols are as follows: green circle for Hex, blue square for HexNAc, red triangle for fucose, yellow star for xylose, and color block diamond for HexA. Here, we used publicly available datasets (PXD001850636, totally 30 RAW files). In figure a, one RAW data file was used to compare the seven software tools (Supplementary Note 2). In figure b, six RAW data files were used to compare the five software tools, and additional search time comparisons were provided in Supplementary Note 2. In figures c and d, all 30 RAW files were used to identify an expanded glycoproteome dataset with rare glycans (Supplementary Note 3).
Furthermore, we performed a runtime comparison of pGlycoNovo with four other software tools using the same dataset. All these tools processed the data from the RAW file in the same computer environment and hardware conditions (Supplementary Note 2). pGlycoNovo completed the search of 6 RAW data files in an ultra-large glycan search space in just 6.7 minutes, significantly outperforming the other tools, which searched against the restricted glycan search space inherent to their glycan libraries (Fig. 2b, Supplementary Note 2). Building on the pGlyco3 infrastructure, pGlycoNovo also employs multi-processing to search MS data, with each RAW file assigned to a separate CPU core, operating independently of other analyses. Unlike multi-threading-based searches, this approach allows multiple RAW files to be processed simultaneously, reducing the IO time required to access multiple runs (Supplementary Note 2). However, when searching a single file, only one CPU core is utilized, which results in a slower process compared to multi-threading strategies.
Then, we used pGlycoNovo to analyze all the thirty glycopeptide raw data from the SARS-Cov-2 Spike glycoprotein (PXD001850636). When comparing our results to previously published findings36, which were generated through Byonic searches (detailed methods and procedures are provided in Supplementary Note 3), pGlycoNovo demonstrated the ability to encompass approximately 71% of the published data, corresponding to 208 site-specific N-glycans out of the initially reported 293, and an additional identification of 230 site-specific N-glycans (Fig. 2c). The 230 site-specific N-glycans uniquely identified by pGlycoNovo covered all the 15 previously reported sites. These uniquely identified site-specific N-glycans show no bias in the distribution of the number of glycans at each site or in the presence of shorter glycans (Supplementary Note 3). This not only demonstrates the high matching capability of pGlycoNovo but also requires high-quality glycopeptide spectra. As a result, pGlycoNovo greatly expanded the publicly accessible dataset for the SARS-Cov-2 Spike N-glycoproteome, increasing it from 293 to 523 site-specific N-glycans. With a total of 523 site-specific N-glycans, we re-depicted the glycan compositions at the 17 N-glycosylation sites on the SARS-Cov-2 Spike glycoprotein (Fig. 2d). To convey the main glycosylation features at each site, we classified the glycans into five groups: oligomannose type, hybrid type, complex type, truncated glycans, and unclassified type, as depicted in Fig. 2d. Here, the “unclassified type” refers to rare N-glycan composition that is not included in existing glycan database-dependent search engines (Supplementary Note 4). Notably, our analysis revealed the presence of rare glycans at 12 sites, with the highest occurrence observed near the connector domain, spanning from N1074 to N1194 (Fig. 2d). The unique identifications by pGlycoNovo contributed rare N-glycans at different sites (Supplementary Note 3).
Additionally, pGlycoNovo identified 30 O-glycan compositions with indistinguishable adjacent sites in this dataset, with 12 of them being rare glycans that contain the monosaccharides of xylose and hexuronic acid (Fig. 2e). It is noted that pGlycoNovo does not support site localization of glycans on HCD data. The public data we used does not include ETD spectra, which are necessary for differentiating between neighboring sites on a glycopeptide. Two annotated spectra illustrating an intact N-glycopeptide and an intact O-glycopepide are presented in Fig. 2f and g, both featuring rare glycans. Other annotated spectra are provided in Supplementary Note 3 and Supplementary Data. It is evident that pGlycoNovo effectively utilizes the full-range Y-ion fragments, including those containing xylose and hexuronic acid. This enables extensive interpretation of intact glycopeptides especially those with rare glycans in spectra that were previously challenging to decipher.
Extensive N-glycoproteomics Across Diverse Model Species with pGlycoNovo
To further show the remarkable potential of pGlycoNovo in identifying diverse glycan compositions, we extended its application to the analysis of site-specific N-glycans across five evolutionarily distant species, with over a billion years of divergence, including plant (A. thaliana), worm (C. elegans), fly (D. melanogaster), zebrafish (D. rerio), and mouse (M. musculus). We employed the optimized LC-sceHCD-MS/MS methods in conjunction with both glycan library-independent pGlycoNovo and the glycan library-based pGlyco3, leading to the successful generation of an extensive N-glycoproteome dataset (Methods, Supplementary Note 5). This dataset comprises 32,549 site-specific N-glycans on 4,602 glycoproteins, with 643,045 glycopeptide-spectrum matches (GPSMs), all confidently identified at a 1% FDR at the intact glycopeptide level (Fig. 3a, Supplementary Data). We established the largest N-glycopeptide mass spectra data for the five species to data (Fig. 3b), and identified site-specific N-glycans in plant, worm, fly, and zebrafish for the first time on such a large scale, while also expanding the scale of site-specific N-glycans in mouse (Fig. 3c).
a Overall workflow of intact glycopeptide profiling in five species. b Number of identified glycopeptide spectra in each species. c, Number of identified glycoproteins and site-specific glycans in each species. d Contribution of glycopeptide spectra identified by pGlycoNovo and pGlyco3. e Contribution of site-specific glycans identified by pGlycoNovo and pGlyco3. f Classification of site-specific glycans in each species by glycan type. Glycans were categorized into five groups based on their monosaccharide composition: oligomannose type, hybrid type, complex type, truncated glycans, and unclassified type, where “unclassified type” refers to rare glycans not included in existing glycan database-dependent search engines (Supplementary Note 4). g Distribution of specific monosaccharide-containing glycopeptides in each species. h An annotated spectrum of a glycopeptide with four fucoses. The peptide sequence with “J” indicating the N-glycosylation site. The glycan symbols are as follows: green circle for Hex, blue square for HexNAc and red triangle for fucose. i The workflows of the comprehensive 13C/15N isotopic-labeling strategy for the FDR validation. j Validation results from the isotopically labeled fission yeast and A. thaliana. The element-level error rate (incorrect number of N or C elements) of the identified glycopeptides was tested via the 15N-/13C-labeled precursor signals. Data are presented as mean values ± SD. Each bar represents the average value across biological replicates. Yeast experiments include three biological replicates, and plant experiments include two biological replicates. Each point represents an individual measurement, with error bars indicating the SD.
The level of overlap in the identification results between pGlyco3 and pGlycoNovo is illustrated in Figs. 3d and 3e. Across the five species, both software tools exhibited an overlap in GPSMs and glycopeptide identifications ranging from approximately 54.47% to 73.74%. (Supplementary Note 5, Supplementary Data). Remarkably, about 8.21–23.08% of the identifications were exclusively reported by pGlycoNovo across different species (Fig. 3e). The species with the highest proportion of pGlycoNovo-only identifications were observed in worm, with 14.50% at the GPSMs level (Figs. 3d) and 23.08% at the site-specific glycan level (Fig. 3e). This suggests that within this dataset, a notable number of glycopeptides carry glycans not included in the glycan libraries, with C. elegans displaying the highest incidence of such rare glycans.
We then classified the site-specific glycans identified in the five species into five types, revealing varying degrees of unclassified glycans across the five species (Fig. 3f). Among these species, worms exhibit a dominant high-mannose glycan type comprising 67.32%, accompanied by the highest proportion of unclassified glycans, accounting for 18.73%. In contrast, mice demonstrate the lowest proportion of unclassified glycans at just 5.04%, while displaying a relatively higher prevalence of complex glycan types. The other three species exhibit approximately 5.98%-7.84% unclassified glycans, with zebrafish and plants having a significant presence of high-mannose and hybrid/complex glycan types, while fly has the highest proportion of high-mannose glycans. The distribution of monosaccharides in site-specific N-glycans for each species is depicted in Fig. 3g. All species exhibit a high proportion of fucose-modified glycans, with plants reaching the highest at 61.43%. Xylose-modified glycans are prevalent in plants as well, accounting for 66.29%, which aligns with existing knowledge37. It is worth mentioning that we also observed small amounts of xylose-modified N-glycans in the other four species. Sialic acid was hardly detected in plant, worm, and fly, while zebrafish primarily contained NeuAc, and mouse exhibited both sialic acid types (Fig. 3g). As demonstrated by our comparative analyses of glycopeptides identified by pGlyco3 and solely by pGlycoNovo, our understanding of glycan types in various species is enhanced by pGlycoNovo’s capability to discover rare glycans not recorded in databases across different species (Supplementary Note 5).
A series of matched Y ions within the annotated spectrum of a multi-fucose glycopeptide illustrates the precise deciphering capabilities of pGlycoNovo for glycopeptide fragments (Fig. 3h, Supplementary Note 5). To further validate the reliability of pGlycoNovo, we conducted N-glycopeptide analysis on mixed, isotope-labeled samples and performed FDR analysis using NaN ratio (Fig. 3i, Supplementary Note 6, Methods). The NaN ratio is calculated based on the MS intensity of an unlabeled glycopeptide and its 15N/13C-labeled counterpart, which has been previously shown to be an effective strategy for glycopeptide FDR validation by us15,28 and also used by other software tools38. In this study, we performed NaN ratio analyses on two different species: fission yeast (with an unlabeled/15N/13C sample ratio of 1:1:1) and plant (with an unlabeled/15N sample ratio of 1:1) (Supplementary Note 6). The results demonstrated that both pGlycoNovo and pGlyco3 reported NaN ratios below 1% for both species, confirming the reliability of pGlycoNovo identification, maintaining corresponding FDR control below 1%.
Characteristics of site-specific N-glycoproteome in Five Evolutionarily Distant Species
With this extensive N-glycoproteome dataset, we performed statistical analyses to explore the diversity of glycosylation and enhance our understanding of glycan modification characteristics across the five species. Our analysis revealed that, within each species, a majority of N-glycoproteins (exceeding 50%) predominantly feature a single glycosylation site (Fig. 4a), and more than half of the glycosylation sites undergo multiple glycan modifications (Fig. 4b). This macro- and microheterogeneity is evident across all five species (Figs. 4a, b). In mice, the number of proteins containing multiple glycosylation sites and the number of sites carrying various glycans are the largest, indicating the highest level of diversity (Fig. 4a, b). In addition, the average size of glycan chains in mice is also the largest, followed by zebrafish, which displays a glycan size distribution similar to that of mouse (Fig. 4c). In contrast, worm and fly exhibited similar glycan size distributions, with evenly distributed chains composed of 6–12 monosaccharides, as well as approximately 10% smaller glycans. Arabidopsis, on the other hand, primarily featured glycans consisting of 8–9 monosaccharides (Fig. 4c).
a Distribution of singly and multiply glycosylated proteins in each species. b Distribution of the number of glycans at each site in each species. c, Distribution of glycan size in each species. d Cellular localization of glycoproteins and the distribution of different glycan types on cellular localization in each species. e Recognition sequence motifs and the distribution of glycan types on different motifs in each species. (NXS/NXT, where N is asparagine, X is any amino acid except proline, S is serine, T is threonine; Others refer to N-X-Any motifs, where N is asparagine, X is any amino acid except proline, and Any represents any amino acid except serine and threonine.). f Secondary structure localization and the distribution of different glycan types on secondary structures in each species. In the Fig. 4d–f, the distribution of each glycan type was determined by normalizing through dividing the number of GPSMs for a glycan type within one category by the total number of GPSMs in that specific category (Supplementary Data). The secondary structure information in each species in Fig. 4f was obtained using a previously reported 3-state protein secondary structure prediction method57 (Supplementary Note 7). g Correlation of overall site-specific glycosylation in five mouse tissues. h Correlation of overall site-specific glycosylation in three plant organs. In Fig. 4g, h, the correlation of tissue/organ-specific site-specific glycosylation in five mouse tissues and three plant organs is depicted using Pearson correlation analysis of the GPSMs in each species/organ. The numerical values in the heatmap represent the degree of correlation, with values closer to 1 indicating higher inter-tissue/organ correlation, while values closer to 0 indicating lower inter-tissue/organ correlation.
The cellular localization analysis of the N-glycoproteome in five different species exhibits a degree of conservation predominantly occurring in expected extracellular regions or certain intracellular compartments like the Golgi apparatus (Fig. 4d). This observation is in agreement with the conserved molecular machinery underlying N-glycosylation across diverse eukaryotes39,40. However, the characteristics of modified glycan types within these predominant subcellular organelles vary among these distinct species (Fig. 4d). For instance, proteins with the complex glycan type in plants and mice are primarily localized on the membrane surface, while in worms, they are mainly found in organelles like the Golgi apparatus. Glycoproteins with fucosylation glycans in zebrafish are predominantly located within organelles, whereas in worms, they coexist on both the membrane and within organelles, and in plant, fly, and mouse, they are distributed across different subcellular compartments. Xylose modifications discovered in flies and mice are mainly located in the endoplasmic reticulum, while in worms, they are enriched on the membrane surface. This variability may be linked to the specific functions of proteins unique to each species.
Further analyses on the relationship between site-specific glycosylation and peptide sequence reveal that N-glycosylation adheres to consistent and stringent topological constraints across these species (Figs. 4e, f, Supplementary Data). As shown in Fig. 4e, N-glycosylation occurs more frequently with the motif of threonine than serine at the second position (Supplementary Data), and it is enriched in β-sheets while being depleted in α-helices across all organisms (Fig. 4f, Supplementary Note 7, Supplementary Data). These findings align with the previous observations derived from glycosylation site data41, confirming the presence of glycosylation canonical motifs and their structural localization characteristics. Additionally, we observe that glycan types and specific monosaccharide-containing glycans in these species also exhibit a remarkably consistent distribution pattern within these canonical motifs (Fig. 4e, Supplementary Data) and structural localizations (Fig. 4f, Supplementary Data). Our analysis not only validates the previous findings regarding the precise embellishment of proteins with N-glycosylation by the core N-glycosylation machinery, in strict concordance with the sequence motifs and topological locations of the substrates41, but also suggests that various N-glycosyltransferases, which determine glycan types, exhibit conservation in both sequence and structure throughout evolution.
Additionally, comparative analyses showed that the glycopeptides uniquely identified by pGlycoNovo exhibited similar glycosylation modification patterns across the five organisms, including the number of glycosylation sites on a protein, the number of glycans at a site, and the glycan size (Supplementary Note 5). For instance, regardless of whether identified by pGlycoNovo alone (Figure S.Note5.5–3), pGlyco3 (Figure S.Note5.5–2), or both (Fig. 4), the results indicate that mice exhibit the highest diversity level, with the largest number of proteins containing multiple glycosylation sites and the largest number of sites carrying various glycans, indicating the highest level of diversity.
Furthermore, we investigate the correlation of site-specific glycosylation modifications within different organs of the same species. We performed intact glycopeptide profiling on five mouse tissues (brain, heart, kidney, liver and lung) and three plant organs (bud, leaf and seed). Pearson correlation coefficients were calculated for each pair of organs or tissues. Consistent with our previous findings28, different mouse tissues displayed distinct glycosylation patterns. Specifically, brain tissue exhibited the most distinctive glycosylation profile, while heart and lung tissues demonstrated a higher resemblance to each other compared to the other tissues (Fig. 4g). Among the plant organs, bud and leaf showed some similarity (correlation coefficient of 0.61), whereas seed displayed distinct patterns (correlation coefficients of 0.27 and 0.22, compared to bud and leaf, respectively) in site-specific glycosylation comparisons (Fig. 4h).
Unexpected glycan fragment detection in different samples
Based on the high performance and reliability of pGlycoNovo for comprehensive glycan fragment matching and de novo glycan analysis demonstrated in the previous sections, we observed prevalent presence of unexpected glycan fragments during the analysis of intact glycopeptides across different samples and experimental sources.
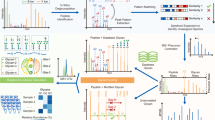
To investigate this phenomenon, we utilized both pGlyco3 and pGlycoNovo to search glycopeptide data from different sample types and mass spectrometry analyses conducted in this study and other published studies (Supplementary Note 8). Then, we annotated the matched Y ions on the co-identified set of GPSMs from both pGlycoNovo and pGlyco3, followed by a comparative analysis (Fig. 5a). pGlycoNovo could exclusively identify numerous Y ions within these GPSMs (Supplementary Data). The Y-ion analysis in Fig. 5b illustrates that, as expected, Y ions containing pentasaccharide core fragments, such as Y-H(1)N(2), Y-H(2)N(2), and Y-H(3)N(2), or those with the core-fucose attached pentasaccharide core fragments, such as Y-N(1)F(1), Y-N(2)F(1), and Y-H(3)N(2)F(1), can be co-identified by pGlyco3 and pGlycoNovo with relatively high abundance. Significantly, pGlycoNovo exhibited unique capabilities in identifying a considerable portion of glycan fragments (Figs. 5b–e), many of which constituted unexpected fragment ions. Notably, the Y ions exhibiting multiple core-fucose compositions, such as Y-H(3)N(2)F(2), were exclusively identified by pGlycoNovo in the worm, aligning with the recognized multiple core-fucosylation nature in worms42. This further demonstrated the reliability of the Y ions extracted by full-range matching and graph-based filtration. Moreover, pGlycoNovo can also detect multiple core-fucose Y ions in other species (Fig. 5b, Supplementary Data). Furthermore, a noteworthy abundance of unexpected glycan fragment ions, such as Y-H(1)N(1), Y-H(1), Y-F(1), could be identified by pGlycoNovo regardless of the sample type or mass spectrometry conditions used for analysis (Fig. 5b, Supplementary Data). Figures 5c and d illustrate the comprehensive and accurate matching of Y ions by pGlycoNovo, encompassing those unexpected glycan fragment Y ions.
a The main pipeline for the analysis of Y ions in GPSMs. b Distribution of GPSMs containing specific pGlycoNovo-matched Y ions among co-identified GPSMs in each sample. c Annotated spectrum of a glycopeptide identified in mouse brain data (PXD02585925) with low-energy HCD (HCD@20) fragmentation analysis. d Annotated spectrum of a glycopeptide identified in worm data with sceHCD (HCD@30 ± 10) fragmentation analysis. e Proportion of GPSMs containing specific Y ions in a particular glycan type. f Proportion of GPSMs containing specific pGlycoNovo-matched Y ions among co-identified GPSMs in the plant (left), and the proportion of GPSMs containing specific Y ions in a particular glycan type (right). g Annotated spectrum of a glycopeptide identified in plant data. In figure b, e, and f, blue font indicates Y-ions matched by both pGlycoNovo and pGlyco3, and fuchsias font indicates Y-ions exclusively matched by pGlycoNovo. In figure c, d, and g, spectra peaks are magnified for better visibility; unexpected fragment ions matched by pGlycoNovo are highlighted in red dashed boxes. The peptide sequence with “J” indicates the N-glycosylation site. the glycan symbols are as follows: green circle for Hex, blue square for HexNAc, red triangle for fucose, and yellow star for xylose.
It is worth noting that these unexpected glycan fragment ions consistently appeared in the spectra generated using both high and low-energy HCD fragmentation (Figs. 5b–d Supplementary Data), and Y-H(1)N(1) proportion is even higher at NCE = 20 (73.40%) than that at NCE = 33 (61.18%) (Fig. 5b). Moreover, all types of glycans, including truncated ones, were found to produce unexpected Y ions (Fig. 5e, Supplementary Data). Interestingly, the proportion of specific unexpected Y ions varies among different glycan types. For example, Y-H(1)N(1) is more prevalent in high-mannose glycan type, while Y-F(1) is more abundant in the complex glycan type. This potential pattern is related to the monosaccharide composition of the glycan types and is observed across different species (Fig. 5e, Supplementary Data). This phenomenon is widespread and is also evident in xylose-containing glycans in plant, where the proportion of Y-N(1)X(1) is 30% to the non-rearranged Y-H(1)N(2)X(1) (15.53% over 51.01%, Fig. 5f). The occurrence frequency of xylose-containing unexpected Y ions is relatively consistent across different glycan types (Fig. 5f). This may be attributed to the fact that xylose is typically attached to the first mannose in any glycan types, resulting in a uniform probability of rearrangement. Figure 5g illustrates the matching quality of the unexpected xylose fragments deciphered by pGlycoNovo.
We suspect that these unexpected fragments are possibly resulted from the glyco-rearrangement in MS. Many noteworthy discoveries regarding glycan rearrangements during collisions have been reported, such as rearrangements of hexose43, fucose44,45,46, sialic acid47, and xylose48. However, to the best of our knowledge, there are currently no convenient ways to extensively explore glycan rearrangements at intact glycopeptide level. The full-range matching capability of pGlycoNovo in this context plays a unique role in detecting a wide range of rearrangements involving different monosaccharide combinations, including the rearrangement of big pieces of fragments, such as Y-H(3)N(1) and Y-H(4)N(1) (Fig. 5c, Supplementary Data).
The wide range of glyco-rearrangement challenges the tree structure elucidation for glycans by using the fragment information, but it does not mean that the structure interpretation is impossible from MS information, especially for N-glycans of which we know the prior knowledge about the core and the possible branching structures. However, we shall always keep in mind that the frequent glyco-rearrangement may result in false tree structure assignment of rare glycans when glycan structure database is unavailable.
Discussions
Recent years have witnessed significant advancements in MS-based analytical methods and interpretation software tools, significantly expanding our toolbox for intact glycopeptide analysis. However, the identification of glycans beyond scope of the glycan libraries remains a formidable challenge. The glycans in living organisms are complex and diverse2,3,4,5. It has been estimated that only in mammals, approximately 700 proteins contribute to the full diversity of glycans, which are assembled from ten different monosaccharides4. While glycosylation sites have been extensively explored across various species41,49, the majority of known protein site-specific glycans are on mammalian proteins6,7,8,11,12,50,51,52,53, with only a limited number of site-specific glycans identified in lower organisms28,36,54. The current limitation lies in the lack of appropriate tools capable of deciphering intact glycopeptide with glycans that extend beyond established glycan libraries. Even in the case of extensively studied glycoproteins, such as the SARS-CoV-2 spike, glycopeptide identification could only be performed using the existing limited glycan database (Supplementary Table 1).
Currently, some software tools, such as StrucGP25 and PEAKS GlycanFinder31, possess glycan de novo analysis capabilities, contributing to the enhancement of sensitivity in glycan identification. However, these tools exhibit limited proficiency in identifying glycan compositions beyond existing glycan libraries. For instance, StrucGP requires prior knowledge for establishing modularized glycans and is currently applicable only to mammals, presenting constraints in its identification capabilities for species with limited information on glycan composition and structure25. Similarly, PEAKS GlycanFinder, which only supports the five common monosaccharides, faces challenges in precisely identifying intact glycopeptides in lower organisms (Supplementary Data, Fig. 2a, Supplementary Notes 2, 10). Additionally, the algorithms employed by these tools rely on peptide-first search for obtaining glycan mass (precursor mass minus peptide mass) before initiating glycan de novo analysis, which may compromise the sensitivity and accuracy of glycan identification, particularly when poor peptide fragmentation leads to errors in the initial selection of candidate peptides. Although poor peptide fragmentation can also affect the final identification accuracy in glycan-first searches, the glycan-first method can reduce the likelihood of false positives by narrowing down the correct glycan candidates early in the process, especially in spectra where glycan fragmentation is good but peptide fragmentation is suboptimal. pGlycoNovo presents an efficient glycan first-based full-range Y-ion de novo search strategy, providing a precise and convenient approach to conduct an open search for glycans without the availability of glycan libraries in intact glycopeptide identification. In contrast to existing tools that primarily rely on the core Y ions, pGlycoNovo is specifically designed to harness the full-range Y ions, significantly expanding the glycan search space for both N- and O-glycopeptide identification (Fig. 1a, b). Furthermore, the adoption of the glycan-first strategy by pGlycoNovo enhances the overall efficiency of Y-ion utilization, facilitating effective quality control and accurate identification of glycan components. Recognizing the critical role of glycan-level quality control in the development of glycopeptide search engines, as emphasized in our studies15,28 and others26,38, pGlycoNovo incorporates glycan-level quality control measures by utilizing empirical score cutoffs for glycans (Fig. 1c). However, it is important to note that these empirical cutoffs depend on careful parameter tuning and, particularly for the identification of rare glycans, often require meticulous manual determination to ensure reasonable accuracy and reliability, which reflects a limitation compared to the glycan FDR control available in pGlyco3. Nonetheless, it offers alternative quality control options. In this study, pGlycoNovo consistently used a precursor tolerance of 2 ppm. As demonstrated in the data shown in Supplementary Note 11, the precursor tolerance did not significantly impact the accuracy of pGlycoNovo under common conditions (up to 10 ppm). Thus, we suggest using a maximum tolerance of 10 ppm as a starting point for pGlycoNovo analysis. However, considering that a 10 ppm tolerance in complex samples can result in FDRs in the range of 2–4% (Supplementary Note 11), we recommend a tighter tolerance of 2 ppm for more complex samples, such as those from mouse and human. Additionally, a glyco-gap higher than 2 is recommended only for exploratory tests by experienced users; generally, a glyco-gap of 2 should be used (Supplementary Note 11). Even so, users should also keep in mind that, similar to de novo peptide sequencing, isobaric compositions would result in higher FDRs by introducing partially incorrect identifications. We have tested pGlycoNovo on different isobaric glyco compositions, and the results are shown in Supplementary Note 12.
pGlycoNovo demonstrates significant utility in situations where glycan libraries are unavailable for intact glycopeptide identification, providing enhanced flexibility and efficiency in intact glycopeptide analysis (Figs. 2, 3). As shown in the comparative analyses (Fig. 2, Supplementary Notes 2, 10), although pGlycoNovo alone does not achieve the highest number of identifications, it stands out for its remarkable complementary capabilities, particularly in identifying rare glycans. When used in combination with pGlyco3, it helps achieve identification results comparable to the best existing levels. Using the combination of pGlycoNovo and pGlyco3, we successfully generated the largest intact N-glycopeptide dataset to date across five evolutionarily distant species with over a billion years of divergence, including plant, worm, fly, zebrafish, and mouse. The site-specific N-glycoproteomic analysis across these diverse species highlights the interspecies diversity and reveals conserved relationships between the protein sequence motifs, topological locations and modified N-glycans (Fig. 4). With such data, our understanding of the extent and evolution of N-glycoproteomes could be significantly enhanced.
Glycan rearrangements, initially reported in 199755, have been the subject of research for a period43,44,45,46,47,48, but exploring these rearranged glycan fragments has been challenging due to the complexity of glycan fragments and the lack of relevant high-throughput approaches. pGlycoNovo, with its global Y-ion matching capability, offers an efficient way to explore the rearrangements across a wide range (Figs. 5b, f, Supplementary Data). Additionally, analyzing these unexpected Y ions based on the extensive glycopeptide data conducted in this study could help us observe the occurrence of rearrangements in different types of glycans (Fig. 5e).
pGlycoNovo was developed within the same software environment as pGlyco3. To enhance convenience for researchers, we have seamlessly integrated pGlycoNovo into the pGlyco3 software interface. To achieve a more comprehensive site-specific glycan analysis, we recommend users to leverage the combined functionality of pGlycoNovo and pGlyco3, especially when dealing with species that have limited glycan libraries, as we have thoroughly demonstrated in this paper. When using pGlyco3 and pGlycoNovo in combination, there are instances of inconsistent identification results, although these inconsistencies are very rare in our data—generally below 1%, and below 2% in C. elegans (Supplementary Note 13). We generally recommend relying on the results from pGlyco3, which is based on a glycan database and maintains strict quality control for glycans. For users particularly concerned about specific glycopeptides with inconsistent results between the two software tools, we suggest manually checking the oxonium ion data.
The glycan-level quality control strategy of pGlycoNovo, with empirical score cutoffs for glycans, largely ensures the reliability of pGlycoNovo identifications, as significantly validated by isotope-labeled complex glycopeptides (Fig. 3j, Supplementary Note 6). As a glycan-level de novo sequencing strategy, pGlycoNovo requires optimized searching parameters. As demonstrated in Supplementary Note 11, lower tolerance can be beneficial but not significant, whereas a smaller glyco-gap is necessary. A higher glyco-gap significantly increases false positives due to the need for optimized fragmentation for good Y ion matches. However, this is always a balance between sensitivity and accuracy. To help users achieve the best results using the pGlyco package, we have provided a detailed description of the effect of different searching parameters of pGlycoNovo (Supplementary Note 11). pGlycoNovo’s glycan-first-based full-range Y-ion de novo search strategy relies on high-quality glycopeptide fragmentation spectra and is currently limited to Orbitrap-generated data, which can be considered a current limitation.
Methods
Our research complies with all relevant ethical regulations. The study protocol was approved by the Ethics Committee at Fudan University. All procedures performed in this study involving animal participants were in accordance with the ethical standards of the institutional and/or national research committee.
Full-range glycan enumeration and matching
A table of full-range glycan is generated which enumerates all glycan compositions based on the user-defined maximal number of each glyco unit. In this study, the default maximal numbers are: HexNAc = 15, Hex = 20, Fuc = 4, NeuAc = 4, NeuGc = 4, HexA = 1, and Xyl = 1. Including zero monosaccharides, the overall number of full-range Y ions is 168,000 (\({\prod }_{i}(1+{n}_{i})\)). The mass of each glycan composition in the table are calculated and then indexed by 0.01 Da mass bin for fast (O(1) running time) mass retrieval. The glycan table is generated and indexed only once for all search spectra files in pGlycoNovo. Then, similar to pGlyco3’s glycan-first search15, for a query peak in the given MS2 spectrum, the complementary mass is calculated by precursor mass minus the peak mass to remove the peptide mass (Eq. 1), and hence only Y-complementary mass will be matched against the full-range glycan table to get the possible glycan composition for the complementary mass. If there are several compositions matched the same mass, all of them will be kept for further processing.
Building Y-complementary glycan graph
A directed graph is built that connects all matched glycan compositions for complementary masses. In the glycan table, each glycan composition is represented as a vector of glyco units. Based on the vector representation, if glycan A <glycan B and \({sum}\left({{\rm{B}}}-{{\rm{A}}}\right)\le k\) (\(k\) is the max glyco gap which is a user-defined parameter; \(k=3\) by default), then a directed edge will be generated pointing A to B. An example of the Y-complementary glycan graph is shown in Supplementary Note 9. In pGlycoNovo, we introduce the glycan zero (the zero vector) for precursor mass as the complementary mass of the precursor is always zero. The glycan zero is the source node of the directed graph. We use \((G,\, E)\) to represent this directed graph, where \(G\) is the matched Y ion set or glycan set \(\left\{{g}_{i}\right\}\) and \(E\) is the directed edge set \(\{{({g}_{i},{g}_{j})}_{{g}_{i} < {g}_{j}}\}\). If a glycan is not reachable starting from the source node, the glycan is removed to reduce the low-quality Y ion matches.
Glycan deduction and filtration from the graph
After the graph is constructed, we developed a dynamic programming algorithm to deduce the best-matched candidate glycans with their longest paths as the coarse scores. The key observation is that, the longest-matched path of a glycan must be also derived from the longest paths of its preceding nodes (glycans). This can be formulated by the optimal substructure for dynamic programming:
This dynamic programming algorithm enables to deduce the longest path scores of all glycans in only one pass on the graph. The running time is linear to the number of edges between matched glycan pairs, i.e., \(O\left(\left|E\right|\right)=O\left({N}^{k}\left|G\right|\right)\), where \(N\) is the number of glyco types (see explanation in Supple Note). For glycan \({g}_{j}\), the longest path is then normalized by dividing the glycan size \(|{g}_{j}|\) to get the coarse score:
Since Y0 ions are sometimes missing in glycopeptide spectra, we add a reducing end monosaccharide onto the deduced glycans to complete the candidate glycan list. For example, the reducing end monosaccharide, which is HexNAc for N-glycans and O-GalNAc glycans, will be added into the candidate list. Then we calculate the coarse scores in the same way. And only top-100 glycans are kept for further processing.
We then filter the candidate glycans by following the same rule in pGlyco3. For a candidate glycan, if the number of matched core Y ions are less than 2 for N-glycan or 1 for O-glycan, this glycan candidate will be removed. And if there are no pre-defined X-diagnostic B ions found for the X-containing glycan (e.g., 274 or 292 peak for X = NeuAc; 290 or 308 peak for X = NeuGc), the glycan will also be removed. The remaining glycans are then used to retrieve their peptides.
Peptide retrieval and glycopeptide scoring and quality control
For a glycan in the candidate glycan list, the peptides can be retrieved from the peptide table using the peptide mass \(({precursor\; mass}-{glycan\; mass})\). The peptide scoring schema is exactly the same as pGlyco3’s peptide score. We also use pGlyco3’s glycan scoring schema to refine the glycan’s score. To enable pGlyco3’s glycan scoring, we use the set \(\{{{g}_{j}-g}_{i}:{g}_{i}\in {G}\, {and}\, {g}_{i}\, < \, {g}_{j}\}\) as theoretical Y ions for glycan \({g}_{j}\). The glycopeptide score is the sum of the scores of glycan and peptide. Only top-1 scored glycopeptide is treated as the identity of the searched spectrum. The scoring schema is shown in Supplementary Note 14.
We then use target-decoy approach to estimate the peptide-level FDR. As there are no good methods to estimate the FDR for MS-based de novo sequencing, we use empirical score cutoff for glycan-level quality control.
Source of model organisms
D. melanogaster was bought from the Core Facility of Drosophila Resource and Technology (Shanghai Institute of Life Sciences, Chinese Academy of Sciences). C. elegans, and D. rerio were kind gifts from the Institute of Nutrition Science (Shanghai Institute of Life Sciences, Chinese Academy of Sciences) and Humangen Biotech Inc (Shanghai, China), respectively. A. thaliana, including rosette leaves, buds, and seeds, were kindly provided by Science of Plant Physiology and Ecology (Shanghai institute of life sciences, Chinese academy of sciences).
Isotopic labelling of plant samples
Arabidopsis thaliana ecotypes Col-0 was used in all experiments in this study. For stable isotope-labeling in Arabidopsis experiments, plants were grown hydroponically in one-eighth diluted Hoagland solution medium (Hoagland’s No. 2 Basal Salt Mixture, Caisson Laboratories, Inc) containing 10 mM KNO3 or 15N-KNO3 (Cambridge Isotope Laboratories Inc). The seeds harvested from these plants were grown on corresponding 14N- or 15N-medium for 5 days under constant light. The seedlings were harvested and stored at -80 °C.
Sample preparation
Washed worm pellets, whole organisms of fly, zebrafish, and plant materials were snap-frozen in liquid nitrogen, ground to a fine powder with mortar and pestle in liquid nitrogen, and stored at −80 °C until use. The grinding powder from different organisms was processed to protein extraction and digestion. The powder was dissolved in fivefold-volume lysis buffer (4% SDS, 0.1 M Tris/HCl, pH 8.0) with protease inhibitor (1 mM PMSF, 1 mM cocktail), followed by boiling at 100 °C for 10 minutes, ultra-sonication for 5 minutes and centrifugation at 12,000 g at 18 °C for 30 minutes to collect protein extracts. The protein concentration was determined by BCA method.
Protein digestion
Proteins were then reduced in 10 mM dithiothreitol at 57 °C for 30 minutes, and then alkylated in dark by 20 mM iodoacetamide at room temperature for 30 minutes. After carbamidomethylation, six volumes of acetone were added to precipitate the proteins at −20 °C for at least 3 hours. The precipitates were dissolved in a denaturing buffer (8 M urea in 50 mM NH4HCO3) following a ten-fold dilution with 50 mM NH4HCO3. Trypsin was added to a final enzyme-to-substrate ratio of 1:50 and incubated at 37 °C overnight. The reactions were terminated by adding 0.5% trifluoroacetic acid. Finally, all digested samples were centrifuged at 16,000 × g for 10 min and the supernatants were desalted using C18 column (Waters). The desalted peptides were then dried by vacuum centrifugation and used for glycopeptide enrichment.
Glycopeptide enrichment
Glycopeptides were enriched by zwitterioic hydrophilic interaction liquid chromatography (ZIC-HILIC) method. Briefly, the desalted peptides of 1 mg were resuspended in 300 μL loading buffer containing 80% acetonitrile and 1% trifluoroacetic acid and then loaded onto an in-house micro-column containing 50 mg of ZIC-HILIC particles (Merck Millipore) packed onto a C8 disk. The flow-through was collected and reloaded onto the column for additional four times. Then, the column was washed with 200 μL loading buffer for four time. Finally, the glycopeptides that have been enriched in the column were collected by eluting with 600 μL 0.1% trifluoroacetic acid and dried by vacuum centrifugation.
LC-MS/MS for glycopeptide analysis
An Orbitrap Fusion Tribrid (Thermo Scientific) coupled to an EASY-nano-LC system (Thermo Scientific) without the trap column was used. Samples were loaded onto a C18 spray tip 50 cm × 75 um i.d. column i.d. column and were separated at a flow rate of 300 nL/min. Solvent A was 0.1% formic acid in water. Solvent B was acetonitrile with 0.1% formic acid. The gradient was 360 min: 5% to 35% solvent B in 345 min, followed by an increase to 90% B in 5 min, held for another 10 min.
The parameters for glycopeptide analysis were: (1) MS: scan range (m/z) = 800–2000; resolution = 120,000; AGC target = 200,000; maximum injection time = 100 ms; included charge state = 2–6; dynamic exclusion after n times, n = 1; dynamic exclusion duration = 15 s; each selected precursor was subject to one HCD-MS/MS; (2) HCD-MS/MS: isolation window = 4; detector type = Orbitrap; resolution = 15,000; AGC target = 500,000; maximum injection time = 250 ms; collision energy = 30%; stepped collision mode on, energy difference of ±10% (10% as absolute value in the Orbitrap Fusion).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Data generated in this work, including all RAW data, Source Data, and Supplementary Data, can be downloaded from: ftp://massive.ucsd.edu, with User ID: MSV000093838_reviewer and Password: pglyconovo. The sources of other data used in this study were indicated in the Supplementary Information.
Code availability
pGlycoNovo is equipped in pGlyco3 software and is freely available on GitHub (https://github.com/pFindStudio/pGlyco3/releases).
References
Ohtsubo, K. & Marth, J. D. Glycosylation in cellular mechanisms of health and disease. Cell 126, 855–867 (2006).
Eichler, J. Extreme sweetness: protein glycosylation in archaea. Nat. Rev. Microbiol. 11, 151–156 (2013).
Nothaft, H. & Szymanski, C. M. Protein glycosylation in bacteria: sweeter than ever. Nat. Rev. Microbiol. 8, 765–778 (2010).
Moremen, K. W., Tiemeyer, M. & Nairn, A. V. Vertebrate protein glycosylation: diversity, synthesis and function. Nat. Rev. Mol. Cell Biol. 13, 448–462 (2012).
Schjoldager, K. T., Narimatsu, Y., Joshi, H. J. & Clausen, H. Global view of human protein glycosylation pathways and functions. Nat. Rev. Mol. Cell Biol. 21, 729–749 (2020).
Ruhaak, L. R., Xu, G., Li, Q., Goonatilleke, E. & Lebrilla, C. B. Mass spectrometry approaches to glycomic and glycoproteomic analyses. Chem. Rev. 118, 7886–7930 (2018).
Chau, T. H., Chernykh, A., Kawahara, R. & Thaysen-Andersen, M. Critical considerations in N-glycoproteomics. Curr. Opin. Chem. Biol. 73, 102272 (2023).
Thomas, D. R. & Scott, N. E. Glycoproteomics: growing up fast. Curr. Opin. Struct. Biol. 68, 18–25 (2021).
Marx, V. Tools to cut the sweet layer-cake that is glycoproteomics. Nat. Methods 18, 991–995 (2021).
Singh, A. Glycoproteomics. Nat. Methods 18, 28 (2021).
Cao, W. et al. Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol. Cell Proteom. 20, 100060 (2021).
Polasky, D. A. & Nesvizhskii, A. I. Recent advances in computational algorithms and software for large-scale glycoproteomics. Curr. Opin. Chem. Biol. 72, 102238 (2023).
Khoo, K. H. Glycoproteomic software solutions spotlight glycans. Nat. Methods 18, 1457–1458 (2021).
Thaysen-Andersen, M., Packer, N. H. & Schulz, B. L. Maturing Glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Mol. Cell Proteom. 15, 1773–1790 (2016).
Zeng, W. F., Cao, W. Q., Liu, M. Q., He, S. M. & Yang, P. Y. Precise, fast and comprehensive analysis of intact glycopeptides and modified glycans with pGlyco3. Nat. Methods 18, 1515–1523 (2021).
Lu, L., Riley, N. M., Shortreed, M. R., Bertozzi, C. R. & Smith, L. M. O-pair search with MetaMorpheus for O-glycopeptide characterization. Nat. Methods 17, 1133–1138 (2020).
Polasky, D. A., Yu, F., Teo, G. C. & Nesvizhskii, A. I. Fast and comprehensive N- and O-glycoproteomics analysis with MSFragger-Glyco. Nat. Methods 17, 1125–1132 (2020).
Strum, J. S. et al. Automated assignments of N- and O-site specific glycosylation with extensive glycan heterogeneity of glycoprotein mixtures. Anal. Chem. 85, 5666–5675 (2013).
Toghi Eshghi, S., Shah, P., Yang, W., Li, X. & Zhang, H. GPQuest: A Spectral library matching algorithm for site-specific assignment of Tandem Mass Spectra to intact N-glycopeptides. Anal. Chem. 87, 5181–5188 (2015).
An, Z. et al. N-linked glycopeptide identification based on open mass spectral library search. Biomed. Res. Int. 2018, 1564136 (2018).
Bern M., Kil Y. J., Becker C. Byonic: advanced peptide and protein identification software. Curr. Protoc. Bioinformatics Chapter 13, 13.20.11-13.20.14 (2012).
Pap, A., Klement, E., Hunyadi-Gulyas, E., Darula, Z. & Medzihradszky, K. F. Status report on the high-throughput characterization of complex intact O-Glycopeptide mixtures. J. Am. Soc. Mass Spectrom. 29, 1210–1220 (2018).
Xiao, K. & Tian, Z. GPSeeker enables quantitative structural N-Glycoproteomics for site- and structure-specific characterization of differentially expressed N-Glycosylation in hepatocellular carcinoma. J. Proteome Res. 18, 2885–2895 (2019).
Lynn, K. S. et al. MAGIC: an automated N-linked glycoprotein identification tool using a Y1-ion pattern matching algorithm and in silico MS² approach. Anal. Chem. 87, 2466–2473 (2015).
Shen, J. et al. StrucGP: de novo structural sequencing of site-specific N-glycan on glycoproteins using a modularization strategy. Nat. Methods 18, 921–929 (2021).
Fang, Z. et al. Glyco-Decipher enables glycan database-independent peptide matching and in-depth characterization of site-specific N-glycosylation. Nat. Commun. 13, 1900 (2022).
Zeng, W. F. et al. pGlyco: a pipeline for the identification of intact N-glycopeptides by using HCD- and CID-MS/MS and MS3. Sci. Rep. 6, 25102 (2016).
Liu, M. Q. et al. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 8, 438 (2017).
Wu, S. W., Liang, S. Y., Pu, T. H., Chang, F. Y. & Khoo, K. H. Sweet-Heart - an integrated suite of enabling computational tools for automated MS2/MS3 sequencing and identification of glycopeptides. J. Proteom. 84, 1–16 (2013).
He, L., Xin, L., Shan, B., Lajoie, G. A. & Ma, B. GlycoMaster DB: software to assist the automated identification of N-linked glycopeptides by tandem mass spectrometry. J. Proteome Res. 13, 3881–3895 (2014).
Sun, W. et al. Glycopeptide database search and de novo sequencing with PEAKS GlycanFinder enable highly sensitive glycoproteomics. Nat. Commun. 14, 4046 (2023).
Ahmad Izaham, A. R. & Scott, N. E. Open database searching enables the identification and comparison of bacterial glycoproteomes without defining glycan compositions prior to searching. Mol. Cell Proteom. 19, 1561–1574 (2020).
Yang, H., Chi, H., Zeng, W. F., Zhou, W. J. & He, S. M. pNovo 3: precise de novo peptide sequencing using a learning-to-rank framework. Bioinformatics 35, i183–i190 (2019).
Vitorino, R. et al. De novo sequencing of proteins by mass spectrometry. Expert Rev. Proteom. 17, 595–607 (2020).
Zhang, J. et al. PEAKS DB: de novo sequencing assisted database search for sensitive and accurate peptide identification. Mol. Cell Proteom. 11, M111.010587 (2012).
Zhang, Y. et al. Site-specific N-glycosylation characterization of recombinant SARS-CoV-2 spike proteins. Mol. Cell Proteom. 20, 100058 (2021).
Song, W. et al. N-glycoproteomics in plants: perspectives and challenges. J. Proteom. 74, 1463–1474 (2011).
Polasky, D. A., Geiszler, D. J., Yu, F. & Nesvizhskii, A. I. Multiattribute glycan identification and FDR control for glycoproteomics. Mol. Cell Proteom. 21, 100205 (2022).
Kelleher, D. J. & Gilmore, R. An evolving view of the eukaryotic oligosaccharyltransferase. Glycobiology 16, 47r–62r (2006).
Mohorko, E., Glockshuber, R. & Aebi, M. Oligosaccharyltransferase: the central enzyme of N-linked protein glycosylation. J. Inherit. Metab. Dis. 34, 869–878 (2011).
Zielinska, D. F., Gnad, F., Schropp, K., Wiśniewski, J. R. & Mann, M. Mapping N-glycosylation sites across seven evolutionarily distant species reveals a divergent substrate proteome despite a common core machinery. Mol. Cell 46, 542–548 (2012).
Yan, S. et al. Core richness of N-Glycans of Caenorhabditis elegans: A case study on chemical and enzymatic release. Anal. Chem. 90, 928–935 (2018).
Wuhrer, M., Koeleman, C. A. & Deelder, A. M. Hexose rearrangements upon fragmentation of N-glycopeptides and reductively aminated N-glycans. Anal. Chem. 81, 4422–4432 (2009).
Lettow, M. et al. The role of the mobile proton in fucose migration. Anal. Bioanal. Chem. 411, 4637–4645 (2019).
Lettow, M. et al. Decoding the Fucose migration product during Mass-Spectrometric analysis of Blood Group Epitopes. Angew. Chem. Int. Ed. Engl. 62, e202302883 (2023).
Ács, A., Ozohanics, O., Vékey, K., Drahos, L. & Turiák, L. Distinguishing core and antenna fucosylated glycopeptides based on low-energy Tandem mass spectra. Anal. Chem. 90, 12776–12782 (2018).
Campos, D. et al. Ghost” fragment ions in structure and site-specific glycoproteomics analysis. Anal. Chem. 95, 10145–10148 (2023).
Hecht, E. S., Loziuk, P. L. & Muddiman, D. C. Xylose migration during tandem mass spectrometry of N-linked glycans. J. Am. Soc. Mass Spectrom. 28, 729–732 (2017).
Zielinska, D. F., Gnad, F., Wiśniewski, J. R. & Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 141, 897–907 (2010).
Chen, S. Y. et al. Glycans, Glycosite, and intact Glycopeptide analysis of N-linked glycoproteins using liquid handling systems. Anal. Chem. 92, 1680–1686 (2020).
Kong, S. et al. Effective enrichment strategy using Boronic Acid-functionalized mesoporous graphene-silica composites for intact N- and O-linked glycopeptide analysis in human serum. Anal. Chem. 93, 6682–6691 (2021).
Liu, L. et al. Automated intact glycopeptide enrichment method facilitating highly reproducible analysis of serum site-specific N-Glycoproteome. Anal. Chem. 93, 7473–7480 (2021).
Stadlmann, J. et al. Comparative glycoproteomics of stem cells identifies new players in ricin toxicity. Nature 549, 538–542 (2017).
Watanabe, Y., Allen, J. D., Wrapp, D., McLellan, J. S. & Crispin, M. Site-specific glycan analysis of the SARS-CoV-2 spike. Science 369, 330–333 (2020).
Ernst, B., Müller, D. R. & Richter, W. J. False sugar sequence ions in electrospray tandem mass spectrometry of underivatized sialyl-Lewis-type oligosaccharides. Int. J. Mass Spectrom. Ion-. Process. 160, 283–290 (1997).
Yuan, Z. F. et al. pParse: a method for accurate determination of monoisotopic peaks in high-resolution mass spectra. Proteomics 12, 226–235 (2012).
Garnier, J., Gibrat, J. F. & Robson, B. GOR method for predicting protein secondary structure from amino acid sequence. Methods Enzymol. 266, 540–553 (1996).
Acknowledgements
We thank Professor Si-Min He from Institute of Computing Technology, CAS, Beijing, China for his kindly directing research, and providing valuable advices and moral support. We thank Dr. Ming-Qi Liu for his active involvement and support during his postdoctoral tenure at Fudan University. We thank the commercial version of PEAKS GlycanFinder for providing us with a free trial, which was used for comparison in this work. We thank Omicsolution Co., Ltd for their assistance with data statistics and visualization. This work was supported by grants from the National Natural Science Foundation of China Project (32271490 to W.C.). This paper is dedicated to the memory of Professor Pengyuan Yang (1949.6.12–2021.5.31) from Fudan University, who passed away during the course of this work.
Author information
Authors and Affiliations
Contributions
W.C. conducted this project, performed the experiments and data analysis, and drafted the manuscript. W.-F. Z. developed the software, analyzed the data, and revised the manuscript. G. Y. and H. Z. contribute to the MS experiments. C.L. contributes to the data analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Yi Yang, and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zeng, WF., Yan, G., Zhao, Hh. et al. Uncovering missing glycans and unexpected fragments with pGlycoNovo for site-specific glycosylation analysis across species. Nat Commun 15, 8055 (2024). https://doi.org/10.1038/s41467-024-52099-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52099-7
- Springer Nature Limited