Abstract
During bacterial cytokinesis, polymers of the bacterial tubulin FtsZ coalesce into the Z ring to orchestrate divisome assembly and septal cell wall synthesis. Previous studies have found that Z ring condensation and stability is critical for successful cell division. However, how FtsZ filaments condense into a Z ring remains enigmatic and whether septal cell wall synthesis can feedback to the Z ring has not been investigated. Here, we show that FtsZ-associated proteins (Zaps) play important roles in Z ring condensation and stability, and discover septal cell wall synthesis as a novel player for Z ring condensation and stabilization in Escherichia coli and Caulobacter crescentus. Moreover, we find that the interaction between the Z ring membrane anchor, FtsA, and components of the septal cell wall synthetic complex are critical for septal cell wall synthesis-mediated Z ring condensation. Altogether, these findings suggest that the divisome is a self-enhancing machine in these two gram-negative bacteria, where the Z ring and the septal cell wall synthetic complex communicate with and reinforce each other to ensure robustness of cell division.
Similar content being viewed by others
Introduction
Bacterial cell division is mediated by a large protein complex called the divisome1. At the heart of this machine is the Z ring, a discontinuous and highly dynamic ring-like structure formed by polymers of the bacterial tubulin FtsZ1,2,3,4. Previous studies showed that FtsZ filaments are first tethered to the membrane by FtsZ membrane anchors and then are pushed or attracted to the midcell region as loosely organized spiral structures by spatial regulators, which eventually condense into a coherent Z ring4,5,6,7,8,9,10,11. Treadmilling dynamics and lateral interaction of FtsZ filaments, as well as FtsZ-associated proteins (Zaps) that can crosslink FtsZ filaments, have been suggested to be important for Z ring condensation7,8,12, but the exact mechanism for Z ring condensation remains elusive.
Once a condensed Z ring is formed, it functions as a scaffold for assembly and activation of the complete division apparatus and as a guide for septal peptidoglycan (sPG) synthesis (Fig. 1a), which is an essential part of the cell wall and critical for cell constriction. Activation of the sPG synthetic complex FtsQLBWI in E. coli depends upon FtsN, the last essential division protein to arrive at the Z ring13,14,15,16,17. FtsN allosterically activates the sPG synthase FtsWI by interacting with the Z ring membrane anchor FtsA in the cytoplasm via its cytoplasmic domain (FtsNcyto) and with the FtsQLBWI complex in the periplasm through its essential domain (FtsNE)15,16,18,19,20,21,22,23,24,25,26,27. Newly synthesized sPG is then processed to denuded glycan strands by the FtsEX-EnvC-amidase cell wall cleavage system28,29,30,31,32, leading to the separation of the two daughter cells. Meanwhile, the denuded glycan strands function as landmarks to recruit more FtsN molecules to activate more FtsQLBWI complexes through the C-terminal SPOR domain of FtsN (FtsNSPOR)14,33,34,35, generating a positive feedback loop of sPG synthesis (sPG loop) to facilitate cytokinesis14,36. Intriguingly, the requirement of FtsN for sPG synthesis could be bypassed by superfission mutations in FtsA and the FtsQLBWI complex that likely result in a constitutively active FtsWI complex15,16,18,19. Recently, treadmilling FtsZ filaments were proposed to distribute inactive FtsQLBWI complexes at the septum which are activated by FtsN to synthesize sPG37,38. Moreover, the organization and dynamics of the Z ring were found to strongly affect sPG synthesis7. However, whether sPG synthesis can in turn feedback to the Z ring to affect its organization and dynamics has not been studied.
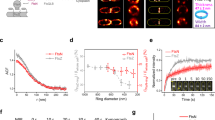
a A diagram for E. coli cytokinesis. Midcell accumulated FtsZ filaments are condensed into a Z ring with the aid of Zap proteins. The Z ring recruits the remaining cell division proteins to form the divisome and guides septal peptidoglycan (sPG) synthesis, leading to cell constriction. Once sPG synthesis is started, it is propagated via a positive feedback loop termed sPG loop (indicated by a blue arrow). b Representative images of Z rings in WT and Δzap cells labeled with FtsZ-GFP. Strains were grown at 30 °C to exponential phase and FtsZ-GFP was induced with 40 μM IPTG for 2 h before imaging. Cephalexin was added at 20 μg/mL and treated for 1 h. c Z ring width distribution in WT and Δzap strains labeled with FtsZ-GFP. Z ring width was determined by intensity projections of FtsZ-GFP in WT and ∆zap cells as described in Methods. d Percentage of condensed and decondensed Z rings in WT and Δzap strains labeled with FtsZ-GFP, ZipA-mCherry or FtsEX-GFP. Z rings with a width > 350 nm were considered decondensed for FtsZ-GFP and FtsEX-GFP, while >430 nm was set for ZipA-mCherry. Number of Z rings analyzed (FtsZ-GFP: WT = 427, Δzap = 525; FtsEX-GFP: WT = 516, Δzap = 525; ZipA-mCherry: WT = 420, Δzap = 527). e–g Representative time-lapse series of Z ring dynamics in WT (e) and Δzap cells (f), or in the presence of cephalexin (g). Strains were grown as in (b), samples were spotted onto LB-agarose pads and FtsZ-GFP was imaged for 10 min with an interval of 1 min. Three representative cells are shown for Δzap cells. h-i Representative images of Z rings labeled with FtsEX-GFP or ZipA-mCherry (h) and Z ring width distribution (i) in wild-type and Δzap cells. ZipA-mCherry was expressed from the native promoter of zipA, whereas FtsEX-GFP was induced with 15 μM IPTG for 2 h prior to imaging. All results presented in b–i are representatives of or derived from results of 3 biological replicates. Number of rings analyzed was indicated in c and i. Scale bars, b, g and h, 5 μm; e–f, 2 μm. Source data are provided as a Source Data file.
In this study, we investigate the mechanism of Z ring condensation in E. coli and show that Zap proteins (ZapA, ZapB, ZapC, and ZapD), collectively, are important for Z ring condensation and stability as well as subsequent steps in division. More importantly, we discover that sPG synthesis in turn promotes Z ring condensation and stability via interactions between the Z ring membrane anchor FtsA and components of the sPG synthetic complex, FtsN and FtsW. Such a mechanism is also conserved in Caulobacter crescentus. Thus, our study uncovers a novel mechanism for Z ring condensation and a positive feedback loop from septal cell wall synthesis to the Z ring. Based on these findings, we propose that the divisome is a self-enhancing machine in these organisms, where the Z ring and the sPG synthetic complexes enhance each other to promote division.
Results
Zap proteins work together to promote Z ring condensation and stability in E. coli
It is well documented that Zap proteins can promote Z ring formation in E. coli by crosslinking FtsZ filaments (Fig. 1a)12. Although previous studies have shown that individual Zap proteins influence Z ring organization39,40,41,42, it remains unclear how these proteins work together to facilitate Z ring condensation. As a result, the collective significance of Zap proteins on Z ring condensation is not well defined. To investigate this problem, we constructed and characterized a series of deletion strains lacking one to three of the zapA, zapB, zapC, and zapD genes. Among the single deletion strains, the ΔzapA and ΔzapB mutants had the strongest division defect, followed by the ΔzapC mutant, whereas the ΔzapD mutant had no noticeable defect (Supplementary Fig. 1a, b and Data 1), suggesting that ZapA and ZapC play more important roles in cell division than ZapD. The average cell lengths of the ΔzapA, ΔzapB, or ΔzapAB strains were similar to each other, consistent with previous findings that they are components of the Ter linkage that coordinates Z ring formation and chromosome segregation39,43,44. Deletion of zapC in the ΔzapA or ΔzapD strain significantly increased the cell length, but a triple deletion strain lacking ZapA, ZapC and ZapD (named Δzap hereafter) behaved similarly to the ΔzapAC strain (Supplementary Fig. 1a, b and Data 1). Overall, these results are in line with previous analysis of the Zap proteins: the absence of an individual Zap protein had a minor or modest division defect, but the lack of two or three Zap proteins simultaneously caused a more pronounced division problem12,41,45
To monitor Z rings, a cassette containing ftsZ-gfp under an IPTG-inducible promoter was transduced into the chromosome of the strains by P1-mediated transduction (integrated at the lambda attachment site)46. FtsZ-GFP was expressed at about 50% of the chromosomal level, which did not cause any phenotypic changes in comparison to wild-type and Δzap strains, and did not suppress the division defect of Δzap cells (Supplementary Fig. 1c–e and Data 1). As shown in Supplementary Fig. 2a, the absence of any one of the tested Zap proteins resulted in the formation of aberrant Z ring structures, such as spirals, double bands or clusters of filaments. Moreover, the Z ring abnormality became more severe in the double or triple deletion mutants, in agreement with the cell length analysis. The average width of Z rings in wild-type cells was 318 ± 46 nm, whereas it was increased to 559 ± 380 nm in the Δzap mutant (Fig. 1b–d, Supplementary Fig. 2b, Data 2 and “Methods” section). Using a width of 350 nm as a cutoff for condensed and decondensed Z rings, we found that around 80.7% of the Z rings in wild-type cells could be classified as condensed Z rings, whereas only 33.7% of the Z rings in the Δzap strain fell into this category (Fig. 1d and Supplementary Data 3).
Tracking the localization dynamics of FtsZ-GFP by time-lapse imaging revealed that Z rings appeared stable in wild-type cells, whereas they were unstable in Δzap cells, and normal ring-like structures often transitioned to mis-organized spirals or vice versa (Fig. 1e, f and Supplementary Movie 1). Moreover, the FtsZ spirals oscillated within a broad region (Supplementary Movie 1), likely due to the action of the oscillatory Min system, which pushes FtsZ filaments to the midcell6. This severe Z ring condensation defect in Δzap cells was not due to cell filamentation, because FtsZ-GFP still localized as condensed and stable Z rings in filamentous wild-type cells caused by cephalexin (Figs. 1b, g, Supplementary Moive 1 and Data 2, 3), which blocks cell division by inhibiting the transpeptidase activity of FtsI47,48. This defect was also not caused by an alteration of FtsZ treadmilling dynamics since the FtsZ’s treadmilling speeds in wild-type and Δzap cells were comparable (Supplementary Fig. 2c). Similar Z ring decondensation and instability in Δzap cells were also observed when ZipA-mCherry or FtsEX-GFP was used as proxy for Z rings (Fig. 1d, h, i, Supplementary Fig. 1d, e, 2d–g, Data 2, 3 and Movie 2), indicating that the phenotypes were independent of the fluorescent division protein fusion used.
To test if the unstable Z rings in Δzap cells eventually stabilized resulting in cell division, we tracked FtsZ-GFP and division events for 120 min with 10 min intervals. As shown in Supplementary Fig. 3a, most wild-type cells contained a condensed Z ring at midcell which gradually contracted, leading to a productive division. Also, new Z rings quickly assembled at midcell in the two daughter cells and initiated another round of division. In contrast, most Z rings in Δzap cells were unstable except for those associated with constrictions (Supplementary Fig. 3b–e). In Δzap cells of normal cell length, we often observed condensed Z rings led to successful divisions. However, formation of Z rings in the two daughter cells varied (Supplementary Fig. 3b). Frequently, FtsZ spirals coalesced into a condensed Z ring in one daughter cell but failed to do so in the other (Supplementary Fig. 3b, c). As a consequence, one daughter cell divided, while in the other division was delayed and the cell became elongated. Also, a small fraction of constricting Z rings in Δzap cells disassembled before the completion of constriction, leading to aborted divisions (Supplementary Fig. 3d). In filamentous Δzap cells, some of the condensed Z rings contracted and led to successful cell division, however, a significant fraction of them exhibited dynamic behaviors, transitioning intermittently back and forth between spirals and condensed Z rings or disappearing (Supplementary Fig. 3e). Based on these observations, we concluded that Zap proteins are critical for Z ring condensation and stability in E. coli. In their absence, FtsZ filaments tend to accumulate at potential division sites but have difficulty in condensing into sharp Z rings, and even if they do, these Z rings are unstable unless constriction begins.
Z ring condensation and stability are critical for divisome assembly and sPG synthesis in E. coli
To test how Z ring decondensation led to a severe division defect, we checked if divisome assembly and sPG synthesis were affected. To do this, we scored the co-localization of ZipA-mCherry (a proxy for the Z ring) with GFP-FtsI or GFP-FtsN, the last two essential components of the divisome, and co-localization of ZipA-mCherry with HADA, a D-alanine analog for tracking nascent PG49. As shown in Supplementary Fig. 4a–d and Data 4, GFP-FtsI and GFP-FtsN co-localized with ZipA-mCherry rings at 46.1% and 76.1%, respectively, in wild-type cells. However, co-localization of these proteins in Δzap cells decreased to 13.7% and 22.6%, respectively. The higher co-localization of GFP-FtsN with ZipA-mCherry is likely due to the ectopic expression of the functional GFP-FtsN, which enhances FtsN interaction with FtsA, resulting in its early recruitment. Moreover, these late proteins were rarely found to associate with aberrant structures of ZipA-mCherry (Supplementary Fig. 4a, c). Meanwhile, the co-localization of ZipA-mCherry with HADA decreased from 50.6% in wild-type cells to 16.7% in Δzap cells (Supplementary Fig. 4e, f and Data 4). Additionally, the HADA signal was not present at aberrant structures of ZipA-mCherry, suggesting a condensed and persistent Z ring is also required for sPG synthesis in E. coli.
Time-lapse imaging of the localization dynamics of ZipA-mCherry and GFP-FtsI showed that once GFP-FtsI was recruited to a ZipA-mCherry ring in wild-type cells, it persisted over the time course of the experiment (Supplementary Fig. 5a, arrow). However, in Δzap cells, localized GFP-FtsI quickly delocalized when the ZipA-mCherry rings became decondensed (Supplementary Fig. 5b, arrow). Moreover, GFP-FtsI was not recruited to decondensed Z rings (Supplementary Fig. 5c), indicating that Z ring condensation and stability are required for the recruitment and maintenance of the downstream division proteins at potential division sites. The absence of multiple FtsZ binding proteins in Bacillus subtilis also resulted in defects of divisome assembly and sPG synthesis7, indicating that the requirement of Z ring condensation for cytokinesis is broadly conserved.
Overexpression of FtsN facilitates Z ring condensation and division in Δzap cells
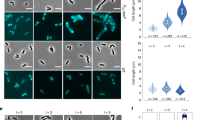
In the above experiments, we noticed that ectopic expression of a functional GFP-FtsN partially suppressed the division defect of Δzap cells (Supplementary Fig. 4c and Data 1). Given the role of FtsN in the activation of sPG synthesis (Fig. 2a), we speculated that expression of GFP-FtsN in Δzap cells enhanced sPG synthesis in the periplasm, which somehow facilitated Z ring condensation and stability in the cytoplasm. Consistent with this hypothesis, overexpression of FtsN greatly reduced the average cell length of Δzap cells (Fig. 2b and Supplementary Data 1). More importantly, it reduced the average width of Z rings of Δzap cells from 411 ± 153 nm to 312 ± 60 nm and greatly increased the percentage of condensed Z rings (Fig. 2c–e and Supplementary Data 2, 3). Tracking the dynamics of FtsZ-GFP revealed that Z rings were relatively stable in Δzap cells overexpressing FtsN and most of them led to successful divisions (Supplementary Fig. 6a–d and Movie 1). In contrast, overexpression of FtsND5N or FtsNWY/AA, which are deficient in interaction with FtsA or interaction with the FtsQLBWI complex15,50, respectively, was less effective in restoring division and Z ring condensation in Δzap cells (Fig. 2b–e and Supplementary Data 1–3). However, overexpression of FtsNΔSPOR, which is defective in binding denuded glycan strands, significantly improved cell division and restored condensed Z rings to Δzap cells. Western blot of a 6×His tagged FtsN and its variants showed that they were expressed at a similar level (Supplementary Fig. 6e), suggesting that the different effect was not due to a difference in protein level. However, it should be noted that in this experiment chromosomal FtsN was present in the Δzap cells in addition to the overexpressed FtsNΔSPOR, so the sPG loop likely still existed in these cells. Consistent with this, when we deleted ftsN from the chromosome in Δzap cells, overexpression of FtsNΔSPOR, was not able to rescue division as effectively as wild-type FtsN (Supplementary Fig. 6f, g). Thus, both FtsN’s interactions with FtsA and FtsQLBWI (sPG synthesis activation), and its ability to maintain the sPG loop, were important for it to facilitate Z ring condensation in the Δzap cells.
a A diagram showing the activation pathway of sPG synthesis in E. coli. The cytoplasmic domain of FtsN (FtsNCyto) interacts with FtsA, while the E-domain of FtsN (FtsNE) interacts with FtsQLBWI. These interactions initiate a signaling cascade to stimulate sPG synthesis by FtsWI. The SPOR domain of FtsN (FtsNSPOR) binds to denuded glycan strands generated during constriction, enhancing FtsN recruitment and generating the sPG loop. Cyto: cytoplasm; IM: Inner membrane; Peri: periplasm; *: superfission mutations. b Cell length distribution of WT and Δzap strains overexpressing FtsN or its variants (results from 3 biological replicates were pooled together for the violin plot analysis). Strains were grown at 30 °C in exponential phase and FtsN was constitutively expressed from a multicopy plasmid, number of cells analyzed (WT/control: 250; WT/FtsN: 250; WT/FtsND5N: 238; WT/FtsNWY/AA: 216; WT/FtsNΔSPOR: 238; Δzap/control: 213; Δzap/FtsN: 226; Δzap/FtsND5N: 204; Δzap/FtsNWY/AA: 211; Δzap/FtsNΔSPOR: 228). Data are presented as mean values +/- s.d.. ****P < 0.0001; ns, not significant (P > 0.05), two-tailed Student’s t test. c Representative images of Z rings in Δzap cells overexpressing FtsN or its variants from 3 biological replicates. Strains were grown at 30 °C in exponential phase, FtsN was constitutively expressed from a multicopy plasmid and FtsZ-GFP was induced with 40 μM IPTG for 2 h before imaging. Scale bar 5 μm. d, e Z ring width distribution (d) and percentage of condensed and decondensed Z rings (e) in WT and Δzap strains overexpressing FtsN or its variants (3 biological replicates). Number of Z rings analyzed in d-e (WT/control: 550; Δzap/control: 539, Δzap/FtsN: 459, Δzap/FtsND5N: 474, Δzap/FtsNWY/AA: 499, Δzap/FtsNΔSPOR: 470). The box plots in (e) show the 25th and 75th percentiles as the box limits, with the mean at the center, whiskers extend from -s.d. to +s.d. of the mean. Source data are provided as a Source Data file.
Superfission mutations promote Z ring condensation and stabilization in Δzap cells
To further examine the role of sPG synthesis in Z ring condensation and stability, we introduced superfission mutations [ftsA* (R286W), ftsL*(G92D), ftsI*(K211I) and ftsW*(E289G)]15,18,19,51 into Δzap cells and checked if Z ring condensation and cell division were restored. As shown in Fig. 3a, b and Supplementary Data 1, Δzap cells carrying a superfission mutation were much shorter than their counterparts without the mutation. Moreover, the average width of Z rings/structures in Δzap cells was also reduced from 559 ± 380 nm to ~287 ± 36 nm to 310 ± 79 nm by the superfission mutations (Fig. 3c and Supplementary Data 2). The percentage of condensed Z rings in Δzap cells was restored to the wild-type level in the presence of superfission mutations (Fig. 3d and Supplementary Data 3). Concomitantly, co-localization of Z ring and GFP-FtsI, or HADA signal, in Δzap cells was markedly increased by the superfission mutations (Fig. 3e, f, Supplementary Fig. 7a, b and Data 4). Importantly, although spiral Z rings and aberrant FtsZ structures were still observed in some Δzap cells carrying superfission mutations (ftsA* or ftsL*), most of them condensed into stable Z rings at midcell resulting in division (Supplementary Fig. 8 and Movie 3). Thus, superfission mutations promoted Z ring condensation and stability in Δzap cells and ultimately improved divisome assembly and sPG synthesis. Among the superfission mutations, ftsA* had the strongest impact on Z ring condensation (Fig. 3, Supplementary Fig. 7, 8 and Data 1–3), probably because FtsA* not only recruits and activates the sPG synthase better, but also promotes FtsZ filament bundling (also see “Discussion” section)52,53.
a Cell length distribution of WT and Δzap strains with or without superfission mutations from 3 biological replicates. Strains were grown at 30 °C in exponential phase, number of cells analyzed (WT: 234; Δzap: 213; Δzap ftsA*: 274; Δzap ftsI*: 211; Δzap ftsL*: 234, Δzap ftsW*: 235). b Representative images of Z rings in Δzap cells with or without superfission mutations from 3 biological replicates. Strains were grown at 30 °C in exponential phase. Scale bar 5 μm. c-d Z ring width distribution (c) and percentage of condensed and decondensed Z rings (d) in WT and Δzap strains with or without superfission mutations from 3 biological replicates. Number of Z rings analyzed in c, d (WT: 427; Δzap: 525, Δzap ftsA*: 499, Δzap ftsI*: 455, Δzap ftsL*: 420, Δzap ftsW*: 523). The box plots (c) show the 25th and 75th percentiles as the box limits, with the mean at the center, whiskers extend from -s.d. to +s.d. of the mean. e Percentage of co-localization (Methods) of ZipA-mCherry with GFP-FtsI in WT and Δzap strains with or without superfission mutations (2 biological replicates). Strains were grown at 30 °C in exponential phase, ZipA-mCherry was constitutively expressed from its chromosomal locus while GFP-FtsI was induced with 5 μM IPTG for 2 h before imaging. Number of ZipA-mCherry/GFP-FtsI rings analyzed (WT: 970/489; Δzap: 965/157; Δzap ftsA*: 1064/478; Δzap ftsI*: 907/288; Δzap ftsL*: 810/320; Δzap ftsW*: 775/265). f Percentage of co-localization of FtsZ-GFP with HADA in WT and Δzap strains with or without superfission mutations (2 biological replicates). Strains were grown at 30 °C in exponential phase, FtsZ-GFP was induced with 40 μM IPTG for 2 h before imaging, HADA was added to a final concentration of 0.25 mM and incubated for 1 min. Number of FtsZ-GFP/HADA rings analyzed (WT: 1318/567; Δzap: 848/154; Δzap ftsA*:759/365; Δzap ftsI*:952/446; Δzap ftsL*:730/327; Δzap ftsW*:711/315). Data in a, e and f are presented as mean values ±s.d.. **P < 0.01, ***P < 0.001, ****P < 0.0001, two-tailed Student’s t test. Source data are provided as a Source Data file.
Ongoing sPG synthesis is necessary for superfission mutations to promote Z ring condensation and stability
So far, the results clearly demonstrate that FtsN overproduction and superfission mutations promote Z ring condensation and stability in Δzap cells. Such mutations, as well as FtsN overproduction, short circuit regulation of sPG synthesis leading to premature FtsWI activation. However, it was not clear if active sPG synthesis was required for their effect. To test this, we examined if cephalexin treatment, which inhibits the activity of FtsI, would eliminate the impact of superfission mutations on Z ring condensation. As expected, cephalexin treatment greatly reduced the ability of superfission mutations (except ftsA*) to restore Z ring condensation in Δzap cells (Fig. 4a, b and Supplementary Data 2). In a second approach, we inhibited the glycosyltransferase activity of FtsW by employing the ftsWI302C allele and treating the cells with MTSES (2-sulfonatoethyl methanethiosulfonate), which reacts with FtsWI302C37,54,55. MTSES had no effect on cell division in the absence of the ftsWI302C mutation, but it blocked division in its presence, suggesting no other unexpected effects (Supplementary Fig. 9). The addition of MTSES to Δzap cells harboring the ftsWI302C allele and superfission mutations resulted in FtsZ-GFP mainly forming spiral structures (Fig. 4c and Supplementary Movies 4, 5), and the width of Z rings in these cells was similar to that of Δzap cells without the mutations (Fig. 4d and Supplementary Data 2). Thus, inhibiting the activity of FtsI or FtsW eliminates the ability of superfission mutations to promote Z ring condensation in Δzap cells. Notably, blocking sPG synthesis, either by inhibiting the activity of FtsI or FtsW, also resulted in modest Z ring decondensation in wild-type cells (Fig. 4a, c, e and Supplementary Movie 4), indicating that ongoing sPG synthesis also enhances Z ring condensation in wild-type cells.
a Representative images of Z rings in WT and Δzap cells with or without superfission mutations and with or without cephalexin treatment (3 biological replicates). Strains were grown at 30 °C in exponential phase, FtsZ-GFP was induced with 40 μM IPTG for 2 h before imaging. Cephalexin was added at 20 μg/mL and treated for 1 h. Scale bar 5 μm. b Z ring width distribution in WT and Δzap strains with or without superfission mutations upon cephalexin treatment (3 biological replicates). Number of Z rings analyzed (WT: 669; Δzap: 360; Δzap ftsA*: 501; Δzap ftsI*: 310; Δzap ftsL*: 422; Δzap ftsW*: 428). c Representative images of Z rings/structures in WT and Δzap cells with or without superfission mutations and with or without MTSES treatment to block FtsW activity (3 biological replicates). Strains were grown at 30 °C in exponential phase. MTSES was added to a final concentration of 1 mM and treated for 1.5 h. Scale bar 5 μm. d Z ring width distribution in WT and Δzap strains with or without superfission mutations upon MTSES treatment. Number of Z rings analyzed (WT: 435; Δzap: 548; Δzap ftsA*: 536; Δzap ftsI*: 394; Δzap ftsL*: 695). e Z ring width distribution in WT or ftsWI302C cells with or without cephalexin or MTSES treatment. Number of Z rings analyzed (WT-no cephalexin: 427; WT-with cephalexin: 669; ftsWI302C-no MTSES: 426; ftsWI302C-with MTSES: 435). The box plots in (b, d, e) show the 25th and 75th percentiles as the box limits, with the mean at the center. Whiskers extend from -s.d. to +s.d. of the mean. Source data are provided as a Source Data file.
Interplay between the Z ring and the sPG synthetic complex is required for sPG synthesis to promote Z ring condensation and stabilization
The results above demonstrated that enhancing sPG synthesis overcomes the Z ring condensation and stability problem of Δzap cells, however, the underlying mechanism was not clear. We hypothesized that the superfission mutations (and increasing FtsN) facilitate the activation of sPG synthesis, which locks the sPG synthetic complex in place, creating a beacon to attract FtsZ filaments or prevent them from spiraling away from the division site via an interaction between the sPG synthetic complex and FtsA (Fig. 5a). The retention of FtsZ filaments at the septum might promote their condensation into coherent Z rings, which in turn, may enhance recruitment and activation of the sPG synthetic complex. Over time, this positive feedback loop between the Z ring and sPG synthesis may drive cell division in the absence of the Zap proteins. Following this logic, the interaction between FtsA and the sPG synthetic complex, as well as maintaining the sPG loop, would be critical for sPG synthesis to promote Z ring condensation and stability.
a A diagram for testing the importance of FtsA-FtsN interaction and sPG loop on sPG synthesis-mediated Z ring condensation. Cyto: cytoplasm; IM: Inner membrane; Peri: periplasm. Red cross means disruption of the interaction. b Cell length distribution of ΔzapΔftsN cells with or without superfission mutations complemented with FtsN or its variants (3 biological replicates). Strains were grown at 30 °C in exponential phase. Number of cells analyzed (ΔftsN/FtsN: 256; ΔftsN/FtsND5N: 256; ΔftsN/FtsNΔSPOR: 256; Δzap ΔftsN/FtsN: 249; Δzap ΔftsN/FtsND5N: 237; Δzap ΔftsN/FtsNΔSPOR: 244; Δzap ΔftsN ftsA*/FtsN: 256; Δzap ΔftsN ftsA*/FtsND5N: 238; Δzap ΔftsN ftsA*/FtsNΔSPOR: 256; Δzap ΔftsN ftsI*/FtsN: 256; Δzap ΔftsN ftsI*/FtsND5N: 256; ΔzapΔftsN ftsI*/FtsNΔSPOR: 256; Δzap ΔftsN ftsL*/FtsN: 254; Δzap ΔftsN ftsL*/FtsND5N: 241; Δzap ΔftsN ftsL*/FtsNΔSPOR: 256; Δzap ΔftsN ftsW*/FtsN: 256; Δzap ΔftsN ftsW*/FtsND5N: 241; Δzap ΔftsN ftsW*/FtsNΔSPOR: 256). Data are presented as mean values +/- s.d. *P < 0.05, **P < 0.01, ****P < 0.0001, two-tailed Student’s t-test. c-d Z ring width distribution (c) and percentage of condensed and decondensed Z rings (d) in ΔzapΔftsN cells with or without superfission mutations and complemented with FtsN or its variants (3 biological replicates). Number of Z rings analyzed in c-d (ΔftsN/FtsN: 429; ΔftsN/FtsND5N: 456; ΔftsN/FtsNΔSPOR: 430; Δzap ΔftsN/FtsN: 365; Δzap ΔftsN/FtsND5N: 356; Δzap ΔftsN/FtsNΔSPOR: 432; Δzap ΔftsN ftsA*/FtsN: 347; Δzap ΔftsN ftsA*/FtsND5N: 407; Δzap ΔftsN ftsA*/FtsNΔSPOR: 364; Δzap ΔftsN ftsI*/FtsN: 384; Δzap ΔftsN ftsI*/FtsND5N: 302; Δzap ΔftsN ftsI*/FtsNΔSPOR: 339; Δzap ΔftsN ftsL*/FtsN: 384; Δzap ΔftsN ftsL*/FtsND5N: 349; Δzap ΔftsN ftsL*/FtsNΔSPOR: 372; Δzap ΔftsN ftsW*/FtsN: 308; Δzap ΔftsN ftsW*/FtsND5N: 268; Δzap ΔftsN ftsW*/FtsNΔSPOR: 315). The box plots in (c) show the 25th and 75th percentiles as the box limits, with the mean at the center, whiskers extend from -s.d. to +s.d. of the mean. Source data are provided as a Source Data file.
To test the above model, we deleted ftsN from the chromosome and complemented the strain with FtsN or variants that are deficient in interaction with FtsA (FtsND5N) or the ability to maintain the sPG loop (FtsNΔSPOR)15,50, and examined FtsZ-GFP localization. Although the average cell lengths of ΔftsN cells complemented with FtsN variants were longer than that of ΔftsN cells complemented with wild-type FtsN, the width of Z rings and the percentage of condensed Z rings were similar in the three strains (Fig. 5b–d), suggesting that in the presence of the Zap proteins, the interaction between FtsA and FtsN and the sPG loop were not that critical for Z ring condensation. However, in ΔzapΔftsN cells, disruption of the FtsN-FtsA interaction (complemented with FtsND5N), and to a lesser extent disruption of the sPG loop (complemented with FtsNΔSPOR), greatly reduced the ability of the superfission mutations, except for ftsA*, to restore Z ring condensation and cell division (Fig. 5b–d, Supplementary Fig. 10a, and Data 1–3). Thus, the interaction between FtsN and FtsA, and the sPG loop, are necessary for the superfission mutations to facilitate Z ring condensation and division in ΔzapΔftsN cells. In this experiment, FtsNΔSPOR was the only form of FtsN inside the cells, whereas in Fig. 2 wild-type FtsN was still expressed from the chromosome. Thus, the importance of the SPOR domain of FtsN differed in these two experiments.
In addition to FtsN, FtsW also interacts with FtsA and this interaction appears important for the relay of the activation signal from FtsN through FtsA to the FtsQLBWI complex (Fig. 6a)22. To test if the interaction between FtsA and FtsW was important for the superfission mutations to suppress the defects of Δzap cells, we deleted ftsW from the chromosome and complemented the strain with FtsW or a variant defective in the interaction (FtsWR172D/E289G: R172D eliminates the interaction with FtsA and is lethal, but it is rescued by the superfission mutation E289G)22. As expected, the superfission mutations facilitated Z ring condensation and division in ΔzapΔftsW cells complemented with wild-type FtsW, but not in cells complemented with FtsWR172D/E289G (Fig. 6b–d and Supplementary Fig. 10b). The average width of Z rings in ΔzapΔftsW cells harboring superfission mutations and complemented with FtsWR172D/E289G was comparable to that of Δzap cells (Fig. 6c and Supplementary Data 2). Thus, the interaction between FtsA and FtsW is also critical for sPG synthesis to promote Z ring condensation.
a A diagram for testing the importance of FtsA-FtsW interaction on sPG synthesis-mediated Z ring condensation. Cyto: cytoplasm; IM: Inner membrane; Peri: periplasm. Red cross means disruption of the interaction. b Cell length distribution of Δzap ΔftsW cells with or without superfission mutations complemented with FtsW or its variant (3 biological replicates). Strains were grown at 30 °C in exponential phase. Number of cells analyzed (ΔftsW/FtsW: 249; ΔftsW/FtsNR172D/E289G: 251; Δzap ΔftsW/FtsW: 252; Δzap ΔftsW/FtsNR172D/E289G: 132; Δzap ΔftsW ftsA*/FtsW: 204; Δzap ΔftsW ftsA*/FtsNR172D/E289G: 122; Δzap ΔftsW ftsI*/FtsW: 255; Δzap ΔftsW ftsI*/FtsNR172D/E289G: 122; Δzap ΔftsW ftsL*/FtsW: 245; Δzap ΔftsW ftsL*/FtsNR172D/E289G: 138). Data are presented as mean values +/- s.d.. ****P < 0.0001, ns: not significant (P > 0.05), two-tailed Student’s t-test. c, d Z ring width distribution (c) and percentage of condensed and decondensed Z rings (d) in Δzap ΔftsW cells with or without superfission mutations and complemented with FtsW or its variant (3 biological replicates). Strains were grown at 30 °C in exponential phase. Number of Z ring analyzed in c, d (ΔftsW/FtsW: 329; ΔftsW/FtsNR172D/E289G: 358; Δzap ΔftsW/FtsW: 327; Δzap ΔftsW/FtsNR172D/E289G: 370; Δzap ΔftsW ftsA*/FtsW: 247; Δzap ΔftsW ftsA*/FtsNR172D/E289G: 317; Δzap ΔftsW ftsI*/FtsW: 275; Δzap ΔftsW ftsI*/FtsNR172D/E289G: 315; Δzap ΔftsW ftsL*/FtsW: 289; Δzap ΔftsW ftsL*/FtsNR172D/E289G: 302). The box plots in (c) show the 25th and 75th percentiles as the box limits, with the mean at the center, whiskers extend from -s.d. to +s.d. of the mean. RE/DG stands for the double mutation R172D/E289G in d. Source data are provided as a Source Data file.
Enhancing sPG synthesis promotes Z ring condensation and stability in C. crescentus
Having established the positive impact of sPG synthesis on Z ring condensation and stability in E. coli, we tested if this phenomenon was conserved in another model bacterium C. crescentus, where FtsN is required for activation of sPG synthesis and superfission mutations in FtsW and FtsI have been isolated56,57,58. As shown in Fig. 7a–d and Supplementary Data 1–3, deletion of zapA increased cell length by 30.4% and the width of Z rings (labeled with ectopically expressed FtsZ-mNeonGreen) by 29% when compared to those of wild-type cells (369 ± 86 vs 476 ± 171 nm). However, these increases were largely suppressed by the presence of the ftsW* (ftsWA246T) or ftsW**ftsI* (ftsWF145L, A246T ftsII45V) mutations (Fig. 7a–d). Time-lapse imaging of FtsZ-mNeonGreen revealed that deletion of zapA resulted in formation of dynamic aberrant Z rings throughout the cells, but the addition of superfission mutations largely restored Z ring condensation and stability (Fig. 7e and Supplementary Movie 6). Therefore, enhancing sPG synthesis also promotes Z ring condensation and persistence in C. crescentus.
a Cell length distribution of WT and ΔzapA C. crescentus cells with or without superfission mutations (3 biological replicates). Strains were grown in PYE medium at 30 °C in exponential phase, Number of cells analyzed (WT: 638; ΔzapA: 627; ΔzapA ftsW*: 770; ΔzapA ftsW** ftsI*: 606). Data are presented as mean values +/- s.d.. ****P < 0.0001, two-tailed Student’s t test. b Representative images of Z rings in WT and ΔzapA C. crescentus cells with or without superfission mutations (3 biological replicates). Strains were grown in PYE medium at 30 °C with xylose to induce expression of FtsZ-mNeonGreen. Scale bar 2 μm. c-d Z ring width distribution (c) and percentage of condensed and decondensed Z rings (d) in WT and ΔzapA C. crescentus cells with or without superfission mutations (3 biological replicates). Number of rings analyzed (WT: 478; ΔzapA: 519; ΔzapA ftsW*: 482; ΔzapA ftsW** ftsI*: 364). The box plots in (c) show the 25th and 75th percentiles as the box limits, with the mean at the center, whiskers extend from -s.d. to +s.d. of the mean. e Representative time-lapse series of Z ring dynamics in WT and Δzap C. crescentus cells from 3 biological replicates. Samples were spotted onto PYE-agarose pad and imaged for 10 min with an interval of 1 min. Scale bar 2 μm. Source data are provided as a Source Data file.
Discussion
Coalescence of FtsZ polymers into the Z ring at the future division site is the first step of bacterial cytokinesis. Time-lapse imaging of FtsZ fluorescent protein fusions revealed that FtsZ filaments first accumulate as loosely organized spiral structures at midcell and then rapidly coalesce into condensed Z rings5,7,8,59,60. The accumulation of FtsZ filaments in the midcell region is likely due to the regulation by spatial regulators6. However, what mediates the condensation of FtsZ filaments into a mature Z ring has not been elucidated. In this study, we show that the absence of any one of the tested Zap proteins, a group of FtsZ crosslinking proteins, resulted in less condensed Z rings in E. coli. Moreover, the combined absence of ZapA, ZapC, and ZapD resulted in unstable FtsZ structures that often failed to condense into sharp Z rings (Fig. 1). In cases where condensed Z rings were formed, they often transitioned back and forth between condensed Z rings and spiral structures. As a consequence, late division proteins were not efficiently recruited to the division site and sPG synthesis did not occur (Supplementary Fig. 4). These findings echo a recent report in B. subtilis where the absence of multiple FtsZ-binding proteins resulted in Z ring decondensation and defects in divisome assembly and sPG synthesis7. Therefore, FtsZ crosslinking proteins, although not essential individually, together play a critical role in Z ring condensation and stability in diverse organisms.
Although E. coli cells lacking the three Zap proteins have severe Z ring condensation and stability defects, condensed Z rings were still assembled in some cells, indicating the existence of additional mechanism(s) for Z ring condensation and stability. Indeed, we showed that enhancing sPG synthesis, either by FtsN overproduction or superfission mutations, promoted Z ring condensation and stability in Δzap cells (Figs. 2 and 3). This positive effect of sPG synthesis on Z ring condensation required interaction between FtsA, a membrane anchor for FtsZ filaments, and FtsN and FtsW, two components of the sPG synthetic complex FtsQLBWIN (Figs. 4–6). In addition, the ability of FtsN to maintain the sPG loop was important. Thus, a picture emerging from these findings is that sPG synthesis, once initiated, becomes a positioning signal for the Z ring and an enhancer for its stabilization, which requires communication between the Z ring and the sPG synthetic complex (Fig. 8). This impact is readily apparent in Δzap cells, but also occurs in wild type cells as inhibition of sPG synthesis resulted in less condensed Z rings. Additionally, sPG synthesis promotes Z ring condensation and stability in C. crescentus (Fig. 7), suggesting that the role of sPG synthesis in Z ring condensation and stability is at least conserved in organisms relying on FtsN for activation of sPG synthesis. A recent study showed that depletion of FtsL in B. subtilis, a critical component of the sPG synthetic complex (DivIB-FtsL-DivIC-FtsWI, equivalent to FtsQLBWI), results in Z ring decondensation61, implying that sPG synthesis may also have a positive impact on Z ring condensation in Gram-positive bacteria.
In newborn cells, FtsZ filaments anchored to the membrane by FtsA (and ZipA, not shown) accumulate at midcell as a loosely organized structure (nascent Z ring). Zap proteins promote Z ring condensation by crosslinking FtsZ filaments. Once a mature Z ring is formed, the sPG cleavage complex FtsEX-EnvC, the sPG synthase complex FtsQLBWI, and FtsN are recruited sequentially to form the complete divisome. FtsN activates sPG synthesis by switching FtsA and the FtsQLBWI complex to the active state. Newly synthesized sPG is processed by the FtsEX-EnvC-amidase system to denuded glycan strands, which recruit additional FtsN molecules via the SPOR domain to activate more FtsQLBWI complexes, forming a positive feedback loop of sPG synthesis (sPG loop). Meanwhile, sPG serves as a beacon for FtsZ filaments via the interaction between the FtsQLBWIN complex on the sPG track and FtsA, which anchors FtsZ filaments to the membrane, thus promoting Z ring condensation and divisome stability. In the absence of Zap proteins, FtsZ filaments cannot condense into Z ring effectively, resulting in a division delay. However, superfission mutations facilitate Z ring condensation and stability by prematurely activating and increasing sPG synthesis, thus compensating for the loss of Zap proteins. PG: peptidoglycan; sPG: septal peptidoglycan; IM: inner membrane. ZapA is depicted as a dark-red dumbbell, ZapC as a red ball and ZapD as pink half-overlay ovals. For simplicity, many division proteins are not shown.
Interestingly, a recent study reported that Z rings were less condensed in E. coli mutant cells lacking the amidase activator EnvC62, which is necessary for sPG degradation and cell separation63,64. It was proposed that sPG degradation plays an important role in Z ring condensation and divisome stability during cell constriction. Moreover, this effect was postulated to be mediated by proteins containing the SPOR domain (FtsN and DedD), which bind simultaneously denuded glycan strands in the periplasm and the FtsZ membrane anchor FtsA in the cytoplasm. This report corroborates our findings and suggests that sPG remodeling (synthesis and degradation) in the periplasm can alter the organization and stability of the Z ring in the cytoplasm. However, unlike the Δzap mutant, introduction of a superfission mutation, ftsL*(FtsLE88K), into the ΔenvC strain did not restore normal Z ring condensation62. Why is there a difference? We suspect that this discrepancy may be due to the pleiotropic effect of the envC deletion. In the current study, severe Z ring decondensation and instability stemmed from the absence of Zap proteins which directly interact with FtsZ in the cytoplasm, whereas in ΔenvC cells the cause of the Z ring condensation defect was indirect62. EnvC forms a tight complex with the ABC-binding cassette transporter-like complex FtsEX and amidases to coordinate sPG synthesis and degradation in E. coli18,32. It is not only required for ATP hydrolysis by the FtsEX complex, which is important for sPG synthesis activation, but also critical for the stimulation of amidase activity. In its absence, FtsEX cannot hydrolyze ATP effectively such that sPG synthesis is compromised, meanwhile, the amidases are not activated to process the newly synthesized sPG to generate denuded glycan strands18,30,32,65. As a consequence, less FtsN molecules will be recruited to the division site, leading to the disruption of the sPG loop. Moreover, timely sPG degradation by the FtsEX-EnvC-amidase system is also required for invagination of the outer membrane (OM). Thus, the lack of EnvC not only causes a loss of sPG degradation, but also a reduction of sPG synthesis and a disruption of the coordinated constriction of the PG layer and the OM. Thus, it is not surprising that the superfission mutations almost completely suppressed the Z ring condensation defect of Δzap cells but not that of ΔenvC cells.
In addition to Zap proteins and sPG remodeling, the lateral interaction of FtsZ filaments likely contributes to Z ring condensation and stability. Previous studies showed that an FtsZ mutant (FtsZL169R) displaying increased bundling in vitro, or FtsA*, which promotes FtsZ filament bundling on lipid monolayers as well as hyperactivating sPG synthesis, suppressed the division deficiency of ΔzapAC cells15,45,52,53,66. Although it was not examined in the previous studies, the division deficiency of ΔzapAC cells is due to a defect in Z ring condensation and stability as we shown in this study. Thus, the ability of FtsZL169R to rescue division of ΔzapAC cells may be due to increased FtsZ filament lateral interaction of the mutant protein, whereas FtsA* does it by enhancing FtsZ filament lateral interaction and sPG synthesis simultaneously45,52. The ability of FtsA* to promote FtsZ filament lateral interaction makes it unique among the superfission mutations and explains why blocking sPG synthesis or disruption of the interplay between the Z ring and the sPG synthetic complex had the least impact on the ability of FtsA* to rescue Z ring condensation in Δzap cells.
A question raised by this study is why E. coli evolved so many strategies to promote and maintain Z ring condensation? We speculate that Z ring condensation is required for multiple reasons. First, a condensed and stable Z ring is necessary for the efficient recruitment and maintenance of the other division proteins at the division site to form the complete divisome. The hierarchical recruitment pathway determined by localization dependency experiments depicts a one-way street for the assembly of the divisome complex67. However, it is likely that the so-called ‘late’ divisome components are already present early on the premature Z rings, at least transiently. Yet, because the interactions between the Z ring and the late division proteins are relatively weak and transient, only when the local concentration of FtsZ filaments in the Z ring is high enough, are they stably retained to form the complete divisome complex. It has been reported that superfission mutations (e.g. ftsLE88K) shortened both the constriction time and the division period16, indicating that Z-ring condensation is accelerated by a higher number of active sPG synthases in nascent Z rings. This is also consistent with the ability of the superfission mutations to restore Z ring condensation in the Δzap cells. Also in agreement with this, we showed that FtsI/FtsN was recruited and maintained only at condensed Z rings. Second, Z ring condensation is likely required to concentrate the activity of the sPG synthetic complex in a narrow region to promote cell constriction7,8. Third, a condensed Z ring is probably important for the generation of smooth septa. Recent studies showed that once cell constriction is initiated in B. subtilis and Staphylococcus aureus, or proceeds to a late stage as in E. coli (diameter of the Z ring <300 nm), Z ring dynamics or even the Z ring is no longer necessary for completion of cytokinesis8,68,69. However, the septa generated in these cells were often aberrant. This is also exemplified by the appearance of abnormal septa in ΔzapA cells and in cells with FtsZ mutants deficient in GTPase activity39,70.
Since Z ring condensation and stability are critical for divisome assembly and efficient sPG synthesis, the finding that sPG remodeling in turn can promote Z ring condensation and stability indicates that the bacterial divisome is a self-enhancing machine (Fig. 8). In this model, FtsZ filaments present around midcell of newborn cells are condensed into mature Z rings, primarily by Zap proteins. Once a condensed Z ring is formed, the sPG synthetic complex FtsQLBWI is recruited and sPG synthesis is initiated by the arrival of FtsN. Newly synthesized sPG is processed by the FtsEX-EnvC-amidase system to form denuded glycan strands which recruit additional FtsN molecules via their SPOR domains. Accumulated FtsN molecules capture and activate additional FtsQLBWI complexes, producing more sPG. Meanwhile, these FtsN molecules and FtsQLBWI complexes also interact with FtsA which can attract FtsZ filaments or prevent them from migrating away from the developing septum, thereby increasing the local concentration of FtsZ filaments and maintaining Z ring condensation. In turn, condensed Z rings recruit late divisome proteins and guide sPG synthesis more efficiently. Thus, once sPG synthesis is initiated, the divisome complex is ‘locked’ in place and becomes more stable until it is time for its disassembly. The absence of Zap proteins severely compromises Z ring condensation and leads to pleiotropic effects on downstream division steps. However, superfission mutations likely initiate sPG synthesis even before the Z ring is sufficiently condensed since the sPG synthetic complexes are constitutively active and require little or no FtsN. As a result, once a complex is recruited to the division site, sPG is produced immediately, which propagates via the sPG loop to further stabilize FtsZ filaments at the division site, leading to Z ring condensation in Δzap cells.
The self-enhancing property of the bacterial divisome offers an explanation for many intriguing observations. For example, overexpression of FtsQAZ suppresses division deficiencies resulting from the lack of essential division proteins, such as FtsEX or FtsK, or from temperature-sensitive mutations in many division genes50,51,71,72, but why is not clear. It is plausible that increasing the level of FtsQAZ results in more FtsZ filaments in Z rings that increase the recruitment efficiency of the sPG synthetic complex so that the deficiency is rescued. Another interesting observation is the resistance of constricting Z rings to the Min oscillation when it switches from pole-to-pole in non-dividing cells to pole-to-midcell in late constricting cells73. In the early stage of constriction, the positive forces for Z ring condensation provided by the Zap proteins and sPG remodeling might outcompete the inhibitory activity of the Min system so that the Z ring persists. However, as constriction proceeds, the local concentration of Min proteins gradually increases. The balance between the positive and negative forces for Z ring condensation leans towards the negative side, leading to the timely disassembly of the Z ring. In other words, the dynamic balance between the positive and negative regulators of the Z rings during the cell cycle governs the timely assembly and disassembly of the Z ring. Additionally, the impairment of self-enhancing ability due to the absence of EnvC or the SPOR domain of FtsN might be responsible for their synthetic lethal/sick phenotype with the Min system14,74.
In summary, we show that Zap proteins play critical roles in Z ring condensation and stability in E. coli and C. crescentus, and provide compelling evidence that sPG synthesis feedbacks to the Z ring to facilitate its condensation and stability. These findings suggest that the divisome is a self-enhancing apparatus that senses and integrates signals from inside (the Z ring) and outside (the cell wall) of the cytoplasmic membrane to execute cell division with high fidelity and robustness. Such a self-enhancing property may be a conserved feature of the division machineries across the tree of life.
Methods
Culture conditions
All strains and plasmids used in this study were listed in Supplementary Data 5–7. E. coli cells were grown in Luria–Bertani (LB) medium (1% tryptone, 0.5% yeast extract, 0.5% NaCl, and pH was adjusted to 7.5 by adding 1 M NaOH solution) at indicated temperatures. C. crescentus cells were grown in PYE medium (0.2% peptone, 0.1% yeast extract, 0.8 mM MgSO4, 0.5 mM CaCl2) at 30 °C. Antibiotics were used at the following concentrations when they were needed: ampicillin = 100 μg/mL; spectinomycin = 25 μg/mL; kanamycin = 25 μg/mL; tetracycline = 12.5 μg/mL; chloramphenicol = 15 μg/mL; cephalexin = 20 μg/mL; and MTSES = 1 mM. Arabinose, xylose or IPTG was added at appropriate concentrations to induce the expression of specific proteins. Mutant alleles were moved between strains by phage P1-mediated transduction, and the antibiotic cassette was removed using FLP recombinase expressed from pCP2075.
Strain construction
Construction of zap gene deletion strains
All deletion strains of the zap genes were constructed by λ-Red recombination system in E. coli75. Double and triple deletion strains were created by successively deletion of the respective genes by P1-mediated transduction76. For instance, for the deletion of zapA, a DNA fragment containing the kanamycin resistance gene kan and 60bp-long homologous arms upstream and downstream of zapA was amplified from the pKD13 plasmid using primer pair: 5-zapA-up and 3-zapA-down. The fragment was transferred into W3110/pKD46 (a temperature-sensitive plasmid expressing recombinase induced with 0.05% arabinosse) by electroporation (1.8 kV, 200Ω). Transformants were selected on LB plates with 25 μg/mL of kanamycin and incubated at 37 °C overnight to cure the plasmid pKD46. The correct transformants were confirmed by colony PCR and sequencing, saved as SH1 (W3110, ΔzapA::kanR). The antibiotic marker was removed by FLP recombinase expressed from pCP2075, generating strain SH19 (W3110, ΔzapA < >frt). Deletion of zapC or zapD was carried out similarly. The zapC::kanR and zapD::kanR cassettes were transduced into strain SH19 by P1-mediated transduction and FLP-mediated removal of the antibiotic marker, resulting in strain SH23 (W3110, ΔzapACD::kanR) and SH68 (W3110, ΔzapACD).
Introduction of superfission mutations into the chromosome
Superfission mutations were introduced into the Δzap strain SH23 (W3110, ΔzapACD::kanR) by P1-mediated transduction. P1 phages grown on PS2343 (W3110, leu::Tn10 ftsAR286W), LYA8 (W3110, leu::Tn10 ftsIK211I), SD246 (W3110, leu::Tn10 ftsLG92D), or SD488 (W3110, leu::Tn10 ftsWE289G) were used to infect strain SH23 following the procedure as described previously19,76,77. Transductants were selected on LB plates with 12.5 μg/mL tetracycline, 25 μg/mL kanamycin, and 8 mM sodium citrate at 30 °C overnight. Since the leu::Tn10 marker has roughly a 50% co-transduction frequency with division genes in the dcw gene cluster, 8–10 transductants were restreaked on selective plates and the transfer of the superfission mutations were verified by sequencing. The correct strains were named SH24 (W3110, ΔzapACD::kanR, leu::Tn10 ftsIK211I), SH25 (W3110, ΔzapACD::kanR, leu::Tn10 ftsLG92D), SH28 (W3110, ΔzapACD::kanR, leu::Tn10 ftsAR286W), and SH29 (W3110, ΔzapACD::kanR, leu::Tn10 ftsWE289G), respectively.
Construction of ftsN and ftsW chromosomal deletion strains
Strain SH74 (W3110, ΔftsN::kanR/pBH21[pBAD33-ftsN]) was created by λ-Red recombination as previously described14. The ftsN::kanR cassette was transferred into strain WGZ (W3110, ftsZ-gfp-ampR) carrying a plasmid expressing wild-type FtsN (pGH98[pSC101-ftsN]) or its variants (pGH97[pSC101-ftsNΔSPOR], or pGH99 [pSC101-ftsND5N]) by P1-mediated transduction. Transductants were selected on LB plates with 25 μg/mL of kanamycin and incubated at 30 °C. The correct transductants were confirmed by PCR and sequencing. To obtain chromosomal ftsW deletion strains, SD237 (W3110, ΔftsW::kanR/pDSW406[pBAD33-ftsW]) was created as previously described78. P1 grown on SD237 was used to transduce strain WGZ carrying a plasmid expressing wild-type FtsW (pGH100 [pSC101-ftsW]) or its variant FtsWR172D/E289G (pGH101 [pSC101-ftsWR172D/E289G]). Transductants were selected on LB plates with 25 μg/mL of kanamycin and incubated at 30 °C for overnight. The correct transductants were confirmed by PCR and sequencing. Similarly, ftsN and ftsW were also knocked out in Δzap strain and its derivatives carrying different superfission mutations via the same way.
Introduction of zipA-mCherry into chromosome
Strain CYA13 (TB28, zipA-mCherry-spcR) was constructed by replacing the chromosomal zipA with a cassette expressing zipA-mCherry under its own promoter using λ-Red recombination. Strain SH31 (W3110, zipA-mCherry-spcR) and SH32 (W3110, ΔzapACD::kanR, zipA-mCherry-spcR) were created by transduction with P1 grown on CYA13 into W3110 and SH23, respectively. Transductants were selected on LB plates containing 25 μg/mL spectinomycin and 8 mM sodium citrate at 30 °C for overnight. The correct transductants were verified by colony PCR and the presence of red fluorescence. The cassette containing zipA-mCherry was introduced into other Δzap strains carrying different superfission mutations similarly. These strains were named SH33 (W3110, ΔzapACD::kanR, leu::Tn10 ftsIK211I, zipA-mCherry-spcR), SH34 (W3110, ΔzapACD::kanR, leu::Tn10 ftsLG92D, zipA-mCherry-spcR), SH36 (W3110, ΔzapACD::kanR, leu::Tn10 ftsAR286W, zipA-mCherry-spcR), and SH37 (W3110, ΔzapACD::kanR, leu::Tn10 ftsWE289G, zipA-mCherry-spcR), respectively.
Introduction of ftsZ-gfp into the chromosome
Strains expressing FtsZ-GFP were created by transduction with P1 grown on WGZ (W3110, ftsZ-gfp-ampR) into respective strains46,79. Transductants were selected on LB plates containing 25 μg/mL ampicillin, and 8 mM sodium citrate at 30 °C for overnight. The correct transductants were verified by colony PCR and the presence green fluorescence. These correctly constructed strains were named SH172 (W3110, ΔzapACD::kanR, leu::Tn10 ftsAR286W, ftsZ-gfp-ampR), SH173 (W3110, ΔzapACD::kanR, leu::Tn10 ftsIK211I, ftsZ-gfp-ampR), SH174 (W3110, ΔzapACD::kanR, leu::Tn10 ftsLG92D, ftsZ-gfp-ampR), and SH176 (W3110, ΔzapACD::kanR, leu::Tn10 ftsWE289G, ftsZ-gfp-ampR), respectively.
Introduction of gfp-ftsI into chromosome
Strain SH116 (W3110, zipA-mcherry-spcR, gfp-ftsI-ampR) and SH117 (W3110, ΔzapACD::kanR, zipA-mcherry-spcR, gfp-ftsI-ampR) was created by transduction with P1 grown on EC436 (gfp-ftsI-ampR) into strain SH31 and SH32, respectively46,80. Transductants were selected on LB plates containing 25 μg/mL ampicillin and 8 mM sodium citrate at 30 °C for overnight. The correct transductants were verified by colony PCR and the presence of green and red fluorescence. Other strains carrying gfp-ftsI were constructed similarly. These correctly constructed strains were named SH118 (W3110, ΔzapACD::kanR, leu::Tn10 ftsIK211I, zipA-mcherry-spcR, gfp-ftsI-ampR), SH119 (W3110, ΔzapACD::kanR, leu::Tn10 ftsLG92D, zipA-mcherry-spcR, gfp-ftsI-ampR), SH121 (W3110, ΔzapACD::kanR, leu::Tn10 ftsAR286W, zipA-mcherry-spcR, gfp-ftsI-ampR), and SH122 (W3110, ΔzapACD::kanR, leu::Tn10 ftsWE289G, zipA-mcherry-spcR), respectively.
Construction of strains in C. crescentus
SH245 (EG865, ftsW*, ΔzapA) and SH246 (EG865, ftsW** ftsI*, ΔzapA) were created by homologous recombination in C. crescentus as previously described80. To delete zapA, the suicide plasmid pEG562 which contains homologous arms upstream and downstream of zapA and is resistant to kanamycin but sensitive to sucrose was transferred into strain EG1556 (EG865, ftsW*) and EG1557 (EG865, ftsW** ftsI*), respectively, by electroporation (2.4 kV, 200Ω). Transformants (Integrants) were selected on PYE plates containing 25 μg/mL kanamycin at 30 °C and confirmed by colony PCR. Correct transformants were grown in PYE medium until OD600 ~ 0.6, then diluted and plated on PYE plates with 0.2% sucrose at 30 °C for double exchange screening. Finally, the correct ΔzapA strains were confirmed by PCR and sequencing. To create strains for Z ring observation, the integrated plasmid pEG1365 which expresses FtsZ-mNeonGreen and is resistant to kanamycin was transformed into strain EG865 (WT), EG1080 (ΔzapA), EG1556 (ftsW*), EG1557 (ftsW**ftsI*), SH245 (ftsW*, ΔzapA) and SH246(ftsW** ftsI*, ΔzapA), respectively, by electroporation. The transformants were selected on PYE plates containing 25 μg/mL kanamycin at 30 °C and successful integration of the plasmid in the chromosome was verified by the presence of fluorescence.
Plasmids construction
Plasmid pBH20 which expresses FtsN under the control of the PBAD promoter was constructed by inserting a SalI and HindIII digested fragment containing ftsN into the pBAD33 plasmid digested by SalI and HindIII. The fragment was amplified from W3110 chromosomal DNA using primer pairs: 5-up-ftsN-SalI and 3-HindIII-ftsN.
Plasmid pGH20 which expresses FtsN fused to GFP under the control of an IPTG-inducible promoter was constructed by inserting an EcoRI and SalI digested fragment into the pDSW209 plasmid digested by the same restriction enzymes. The fragment was amplified from W3110 chromosomal DNA using primer pairs: 5-ftsN-EcoRI and 3-ftsN-SalI.
Plasmid pGH85 was created by replacing the bla coding sequence of pBR322 with spcR amplified from plasmid pGB2 using primer pairs: 5-PstI-spcR and 3-ScaI-spcR.The amplified fragment was digested with PstI and ScaI and ligated to pBR322 digested with the same enzymes.
Plasmid pGH87, which contains ftsN as well as its upstream 822 bp and downstream 351 bp, was constructed by inserting a BamHI and SalI digested fragment into pGH85. The fragment was amplified from W3110 chromosomal DNA using primer pair 5-BamHI-ftsN-up and 3-SalI-ftsN-down.
Plasmid pGH97, which expresses ftsNΔSPOR was constructed by inserting a BamHI and SalI digested fragment into the pGB2 plasmid. The fragment was amplified from W3110 chromosomal DNA using primer pair: U-SalI-ftsN-F and U-BamHI-ftsNΔSPOR-R.
Plasmid pGH98, which expresses ftsN was constructed by inserting a BamHI and SalI digested fragment into the pGB2 plasmid. The fragment was amplified from W3110 chromosomal DNA using primer pair: U-SalI-ftsN-F and U-BamHI-ftsN-R.
Plasmid pGH99, which expresses ftsND5N was constructed by inserting a BamHI and SalI digested fragment into the pGB2 plasmid. The fragment was amplified from plasmid pGH87-D5N using primer pair: U-SalI-ftsN-F and U-BamHI-ftsN-R.
Plasmid pGH100, which expresses ftsW was constructed by inserting a BamHI and ScaI digested fragment containing ftsW into the pGB2 plasmid digested with the same enzymes. The fragment was amplified from W3110 chromosomal DNA using primer pair 5-up-ftsW-SalI and 3-HindIII-ftsW.
Plasmid pGH101, which expresses ftsWR172D/E289G, was constructed by inserting a BamHI and ScaI digested fragment into the pGB2 plasmid digested with the same enzymes. The fragment was amplified from pSEB429-R172D/E289G using primer pair: 5-up-ftsW-SalI and 3-HindIII-ftsW.
Mutagenesis
Mutagenesis was carried out by site-directed mutagenesis using the Quickchange II kit (Agilent) following the protocol provided with the kit or by overlap PCR. The oligonucleotides used for mutagenesis are provided in Supplementary Data 8. All mutations were confirmed by sequencing. Plasmids carrying a mutated gene were named after the original plasmid with the substitution, such as pBH20-D5N, which expresses ftsND5N.
Microscopy: sample preparation
Cells to be observed were inoculated in 5 mL LB medium and grown at 37 °C overnight. Overnight cultures were diluted 1:100 in 5 mL fresh LB medium with appropriate antibiotics, grown at 30 °C for 3 h. Then the cultures were diluted 1:10 in 20 mL fresh LB medium again with appropriate antibiotics and inducers, grown at 30 °C for 2 h until OD600 reached 0.4–0.6. 1 mL of cells were collected by centrifugation (10,000×g for 1 min) and resuspended with appropriate fresh LB. 2 μL of concentrated cells were pipetted onto 2% LB-agarose pads for imaging at room temperature.
Microscopy: phase contrast, epifluorescence
Phase contrast and epifluorescence images were collected on an Olympus BX53 upright microscope with a Retiga R1 camera from QImaging, a CoolLED pE-4000 light source and a U-Plan X Apochromat phase contrast objective lens (100×, 1.45 numerical aperture [NA], oil immersion) using VisiView software. Fluorescence images of GFP, mCherry, and HADA were acquired using the Chroma EGFP filter EGFP/49002, mCherry/Texas Red filter mCherry/49008, and the DAPI filter DAPI/49000, respectively. For FtsZ-GFP and FtsEX-GFP, the exposure time for wild-type strain was 100 ms, but for Δzap strains, the exposure time was set at 200 ms. For ZipA-mCherry, the exposure time was set at 500 ms and 800 ms for wild-type and Δzap strains, respectively. For HADA, the exposure time was 500 ms.
Microscopy: observation of Z ring/structure in E. coli
Overnight cultures of wild-type and zap gene deletion strains expressing different label for the Z ring were diluted 1:100 in fresh LB medium with antibiotics, grown at 30 °C for 3 h. Then the cultures were diluted 1:10 in fresh LB medium again and IPTG was added to 40 μM or 15 μM to induce the expression of FtsZ-GFP and FtsEX-GFP, respectively. ZipA-mCherry was expressed from its native promoter from the chromosome. The strains were grown at 30 °C for 2 h when OD600 reached 0.4–0.6. Cells were concentrated by centrifugation and immobilized on 2% LB-agarose pads before imaging.
To check the impact of cephalexin treatment on Z ring condensation and stability, wild-type and Δzap strains expressing FtsZ-GFP were cultured as above. 1 h post the addition of IPTG to induce the expression of FtsZ-GFP, the cultures were split into two halves, cephalexin was added to one at a final concentration of 20 μg/mL, while the other was kept as a control. 1 h after the cephalexin treatment, samples were harvested and immobilized on 2% LB-agarose pads for imaging.
To determine the impact of inhibition of FtsW activity on Z ring condensation, MTSES (2-sulfonatoethyl methanethiosulfonate) was used to inhibit the glycosyltransferase activity of FtsW as previously described by Yang, et al. 37. Wild-type and Δzap strains carrying ftsWI302C and expressing FtsZ-GFP were grown as above. 1 h post the addition of IPTG to induce the expression of FtsZ-GFP, the cultures were split into two-halves, MTSES was added to a final concentration of 1 mM in one halves of the cultures, while the other halves were kept as controls. 1.5 h after the MTSES treatment, samples were harvested and immobilized on 2% LB-agarose pads for imaging.
To verify the impact of FtsN overexpression on Z ring condensation and stability, plasmid pGH87 and its derivatives expressing FtsN variants were transformed into strain WGZ and SH171, respectively. The strains carrying the plasmids were cultured as above and FtsZ-GFP was induced with 40 μM IPTG. After adding IPTG for 2 h, cells were concentrated and immobilized on 2% LB-agarose pads for imaging.
Microscopy: co-localization of Z ring and downstream division protein FtsI or FtsN
To check the co-localization of ZipA-mCherry and GFP-FtsI or GFP-FtsN, overnight cultures of wild-type and Δzap strains expressing these fluorescent proteins were diluted 1:100 in fresh LB medium with antibiotics, grown at 30 °C for 3 h. The cultures were then diluted 1:10 in fresh LB medium containing 5 μM or 20 μM IPTG for induction of GFP-FtsI or GFP-FtsN, respectively. GFP-FtsI was expressed from a cassette integrated in the chromosome, whereas GFP-FtsN was expressed from the plasmid pGH20 (pDSW209-ftsN). When the OD600 of cultures reached 0.4–0.6, cells were concentrated and immobilized on 2% LB-agarose pads for imaging.
Microscopy: tracking sPG synthesis by HADA staining
Wild-type and Δzap strains expressing FtsZ-GFP or ZipA-mCherry were grown as above in the method for “Observation of Z ring/structure in E. coli” and HADA (7-hydroxycoumarin-3-carboxylic acid-D-alanine) was used to label nascent PG as previously reported with slight modifications19,49. 200 μL of cells were mixed with 0.25 mM HADA and incubated at 30 °C for 1 min. Then cells were fixed with 2.6% polyformaldehyde and 0.04% glutaraldehyde, and incubated on ice for 15 min. Treated cells were collected via centrifugation (10,000×g for 1 min) and washed 5 times with equal volume of PBS (Phosphate Buffered Saline) and resuspended in 50 μL PBS. Cells were then immobilized on 2% LB-agarose pads for imaging.
Microscopy: observation of Z ring/structure in C. crescentus
To check the Z ring or FtsZ structures in wild-type and ΔzapA C. crescentus strains, the integrated plasmid pEG1365 expressing ftsZ-mNeonGreen was transformed into wild-type and ΔzapA strains by electroporation (2.5 kV,200 Ω). Overnight cultures of wild-type and ΔzapA strains carrying ftsZ-mNeonGreen were diluted 1:100 in fresh PYE medium with antibiotics, grown at 30 °C for 4 h. The cultures were then diluted 1:10 in fresh PYE medium again with 0.03% xylose to induce ftsZ-mNeonGreen. 4 h post the induction, the OD600 of the cultures reached 0.4–0.6, cells were harvested and immobilized on 2% water-agarose pads for imaging.
Microscopy: time-lapse imaging
To follow the dynamics of Z ring/structures, WGZ (ftsZ-gfp), SH171 (ΔzapACD::kanR, ftsZ-gfp) and its derivatives (SH172-176) carrying different superfission mutations were grown as above, FtsZ-GFP localization was tracked in real time for 10 min (1 min interval) or 2 h (10 min interval) to determine its dynamics in short and long term, respectively. To track the dynamics of ZipA-mCherry, strain SH31 (zipA-mCherry) and SH32 (ΔzapACD::kanR, zipA-mCherry) were cultured as described above and observed via fluorescence microscopy for 10 min (1 min interval). To track the dynamics of FtsEX-GFP, strain W3110/pSD242(ftsEX-gfp) and SH23/pSD242 were grown as above and induced with 15 μM IPTG for 2 h prior imaging for 10 min (1 min interval).
To check the localization dynamics of GFP-FtsI, SH116 (zipA-mCherry, gfp-ftsI), SH117 (ΔzapACD::kanR, zipA-mCherry, gfp-ftsI) were cultured as above. The cells was immobilized on 2% LB-agarose pads, and localization of ZipA-mCherry and GFP-FtsI were tracked by time-lapse imaging for 15 min (3 min interval) at 30 °C.
To follow the dynamics of Z ring/structures more closely, cells were cultured as described above and observed on a High Intelligent and Sensitive SIM (HIS-SIM) P-104WT microscope81. An Apo 100×/1.5 Oil objective was used for cell imaging. For E. coli, WGZ (ftsZ-gfp), SH171 (ΔzapACD::kanR, ftsZ-gfp) and other strains carrying FtsZ-GFP or FtsEX-GFP were grown to log phase with the addition of inducers, FtsZ-GFP/FtsEX-GFP localization in real time were acquired at 10 s frame rates for 10 min from a 488 nm laser. For observation of FtsZ-GFP and FtsEX-GFP, the exposure time and illumination intensity for wild-type strain were 30 ms and 0.1 (W/cm2), but for Δzap strains they were set at 100 ms and 0.2 (W/cm2), respectively. For C. crescentus, images were acquired at 10 s frame rates for 10 min with 200 ms exposure time and illumination intensity of 0.3 W/cm2. Samples were enclosed in a confocal glass bottom dish. Images were processed and used for production of movies in Fiji82.
Microscopy: treadmilling speed of FtsZ filaments
Images for determining FtsZ treadmilling velocity was captured using a Nikon inverted microscope equipped with a TELEDYNE CMOS (Prime BSI Express) camera and a Nikon 100× NA 1.49 TIRF objective, similar to our previous setup83. FtsZ-GFP was excited using a 488-nm laser under total internal reflection fluorescence (TIRF) mode. The images were acquired in 1-second intervals with an exposure time of 50 milliseconds for 150 frames.
Western blot
For FtsZ immunoblotting, 1 mL of W3110, WGZ (ftsZ-gfp), SH23 (Δzap), and SH171 (Δzap, ftsZ-gfp) cells cultured to log phase were harvested by centrifugation at 14,500 × g for 1 min, resuspended in 50 μL 1× loading buffer and boiled at 95 °C for 10 min. Samples were loaded onto SDS-polyacrylamide gel for electrophoresis and then transferred onto a nitrocellulose membrane. The membrane was blocked with 20 mL of TPBS containing 5% non-fat milk for 1 h at room temperature and then incubated with 20 mL of TPBS containing 5% non-fat milk and rabbit serum raised against FtsZ (1:10,000) for 1 h. The nitrocellulose membranes were washed with fresh TBST for three times (10 min/time) and then incubated with 20 mL TPBS containing 5% milk and goat-anti-rabbit secondary antibody (1:10000; ABclonal, AS014) for 1 h. As the anti-FtsZ serum recognized both untagged FtsZ and FtsZ-GFP, we compared the level of FtsZ and FtsZ-GFP by the density of the corresponding bands in Fiji. A blot for FtsA using the same samples and anti-FtsA serum was carried out as a control.
Quantification and statistical analysis
Statistical analysis was performed using Prism software (GraphPad, v.9.1.1) or OriginLab 2021b. The statistical details of analysis applied in this paper are provided alongside in the result section, Supplementary Data and Figure Legends.
Cell length measurements
Cell lengths were measured by ImageJ/Fiji. Briefly, phase contrast images were imported into ImageJ/Fiji, converted to 8-bit versions with scale bar embedded. Each cell was outlined and a central line was added along the cell body and measured. Cells exhibiting partial view were excluded. Collected cell length data were processed using the Graphpad Prism (v.9.9.1) software. More than 200 cells were analyzed for each strain. Strains with division defects showed larger average cell length and larger standard deviation.
Z ring dynamic change across the cell cycle
Time-lapse images of Z rings were acquired as described above in “Microscopy” section. Images were analyzed by using Fiji for cell segmentation and movie production. Representative images were cropped to small regions of interest (ROIs) in Fiji. Each image stack was aligned using the HyperstackReg plugin to eliminate the drifting and generate the time-series and movies.
Z-ring identification and width measurement
Phase-contrast images of cells were segmented using Cellpose 2.084, a versatile algorithm for cellular segmentation. For cells exhibiting different morphologies, such as the Δzap strains, we re-segmented 100–200 cells and trained a new high-quality neural network model based on the pre-trained model cyto2 using the embedded function of Cellpose 2.0. Manual segmentation was employed for some excessively long filamentous cells that were not properly segmented by the model.
The analysis of Z-ring width was modified from a previously described method7. Cell outlines segmented by Cellpose 2.0 were converted to binary masks using the Mask from roi(s) function in Fiji82. The midlines along the long axis of each cell were determined using Microbe J85, which provided the average fluorescence intensity profile of FtsZ along the long axis of each cell. Using a custom MATLAB code, the uneven fluorescence background of the cytoplasm was removed by applying a high-pass filter on the intensity profile. Z-ring center positions were detected, and their widths were calculated as the full width at half maximum (FWHM). The resulting width distributions were constructed and plotted in OriginLab 2021b. Details of the code for Z ring width calculation can be achieved at https://github.com/BacImgLab/RingWidthCalc.
Quantification of condensed and decondensed Z rings
To quantify the percentage of condensed Z rings, strains carrying FtsZ-GFP, FtsEX-GFP or ZipA-mCherry were grown as described above. Cells were imaged in phase contrast and epifluorescence mode as described above. Fluorescent images were imported into Fiji for further analyzed. In E. coli, Z-rings with a width below 350 nm were identified as condensed Z-rings, whereas Z-rings with a width above 350 nm were considered as decondensed Z rings. Note that the cutoff for condensed and decondensed Z rings labeled with ZipA-mCherry was set at 430 nm. In C. crescentus, the width threshold for condensed Z-rings was set at 450 nm. With these cutoffs, condensed Z rings appeared as bright, tight and sharp fluorescent bands in cells, while decondensed Z rings largely appeared as spiral-like structures, multiple bands/helices within a distance of 1 μm, or fluorescent bands surrounded with spotted fluorescence. Fluorescent images from at least two repeated experiments were selected for quantification, and more than 300 Z rings were calculated for each strain.
FtsZ filament treadmilling speed analysis
Raw time-lapse fluorescence images were first denoised using the PureDenoise plugin (Florian Luisier at the Biomedical Imaging Group (BIG), EPFL, Switzerland) in Fiji85 with a moving average over a 3-frame window for 10 iterations, employing global estimation. The images were then enlarged to a pixel size of 21.7 nm using the bicubic interpolation method in Fiji85. Additionally, drift correction was applied using the StackReg plugin in Fiji. Kymographs were generated using the kymograph plugin (https://github.com/remiberthoz/imagej-live-kymographer) with a 13-pixel wide (~280 nm) line along the Z ring. The polymerization or depolymerization speeds of the FtsZ clusters were calculated by manually measuring the slopes of the leading or trailing edges of the fluorescence zigzags in the kymographs. The box diagram plotted in OriginLab software was based on data from three independent experiments.
Quantification of co-localization of Z ring and FtsI and FtsN
Images of Z rings and GFP-FtsI or GFP-FtsN were acquired as described above in “Microscopy” section and processed in Fiji. Total number of Z rings and GFP-FtsI or GFP-FtsN rings was determined by the fluorescence intensity at midcell or perspective division sites. Co-localization was scored if the two different fluorescent rings were clearly overlapped in the merged images. Number of rings analyzed was indicated in the figure legends.
Quantification of co-localization of Z ring and HADA
Images of Z rings and HADA staining were acquired as described above in “Microscopy” section and processed in Fiji. Total number of Z rings and HADA rings was determined by the fluorescence intensity at midcell or perspective division sites. Co-localization was scored if the fluorescence for Z rings and the fluorescence for HADA were overlapped in the merged images. Number of rings analyzed for each strain was indicated in the figure legends.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All strains, plasmids, primers and reagents/chemicals/softwares used in this study are listed in Supplementary Data 5–9, respectively. All data presented in this study is deposited in Figshare https://doi.org/10.6084/m9.figshare.26567548 and is publicly available as of the data of publication. Source data are provided with this paper.
Code availability
This paper reports original code for Z ring width calculation: https://github.com/BacImgLab/RingWidthCalc.
References
Cameron, T. A. & Margolin, W. Insights into the assembly and regulation of the bacterial divisome. Nat. Rev. Microbiol. https://doi.org/10.1038/s41579-023-00942-x (2023).
Bi, E. F. & Lutkenhaus, J. FtsZ ring structure associated with division in Escherichia coli. Nature 354, 161–164 (1991).
Du, S. & Lutkenhaus, J. At the heart of bacterial cytokinesis: the Z ring. Trends Microbiol. https://doi.org/10.1016/j.tim.2019.04.011 (2019).
Erickson, H. P., Anderson, D. E. & Osawa, M. FtsZ in bacterial cytokinesis: cytoskeleton and force generator all in one. Microbiol. Mol. Biol. Rev. 74, 504–528 (2010).
Thanedar, S. & Margolin, W. FtsZ exhibits rapid movement and oscillation waves in helix-like patterns in. Curr. Biol. 14, 1167–1173 (2004).
Lutkenhaus, J. Assembly dynamics of the bacterial MinCDE system and spatial regulation of the Z ring. Annu. Rev. Biochem. 76, 539–562 (2007).
Squyres, G. R. et al. Single-molecule imaging reveals that Z-ring condensation is essential for cell division in Bacillus subtilis. Nat. Microbiol. 6, 553–562 (2021).
Whitley, K. D. et al. FtsZ treadmilling is essential for Z-ring condensation and septal constriction initiation in Bacillus subtilis cell division. Nat. Commun. 12, 2448 (2021).
Hale, C. A. & de Boer, P. A. Direct binding of FtsZ to ZipA, an essential component of the septal ring structure that mediates cell division in E. coli. Cell 88, 175–185 (1997).
Naha, A., Haeusser, D. P. & Margolin, W. Anchors: a way for FtsZ filaments to stay membrane bound. Mol. Microbiol. https://doi.org/10.1111/mmi.15067 (2023).
Pichoff, S. & Lutkenhaus, J. Unique and overlapping roles for ZipA and FtsA in septal ring assembly in Escherichia coli. EMBO J. 21, 685–693 (2002).
Huang, K. H., Durand-Heredia, J. & Janakiraman, A. FtsZ ring stability: of bundles, tubules, crosslinks, and curves. J. Bacteriol. 195, 1859–1868 (2013).
Addinall, S. G., Cao, C. & Lutkenhaus, J. FtsN, a late recruit to the septum in Escherichia coli. Mol. Microbiol. 25, 303–309 (1997).
Gerding, M. A. et al. Self-enhanced accumulation of FtsN at division sites and roles for other proteins with a SPOR domain (DamX, DedD, and RlpA) in Escherichia coli cell constriction. J. Bacteriol. 191, 7383–7401 (2009).
Liu, B., Persons, L., Lee, L. & de Boer, P. A. Roles for both FtsA and the FtsBLQ subcomplex in FtsN-stimulated cell constriction in Escherichia coli. Mol. Microbiol. 95, 945–970 (2015).
Tsang, M. J. & Bernhardt, T. G. A role for the FtsQLB complex in cytokinetic ring activation revealed by an ftsL allele that accelerates division. Mol. Microbiol. 95, 925–944 (2015).
Lutkenhaus, J. FtsN–trigger for septation. J. Bacteriol. 191, 7381–7382 (2009).
Du, S., Pichoff, S. & Lutkenhaus, J. FtsEX acts on FtsA to regulate divisome assembly and activity. Proc. Natl Acad. Sci. USA 113, E5052–E5061 (2016).
Li, Y. et al. Genetic analysis of the septal peptidoglycan synthase FtsWI complex supports a conserved activation mechanism for SEDS-bPBP complexes. PLoS Genet. 17, e1009366 (2021).
Marmont, L. S. & Bernhardt, T. G. A conserved subcomplex within the bacterial cytokinetic ring activates cell wall synthesis by the FtsW-FtsI synthase. Proc. Natl Acad. Sci. USA 117, 23879–23885 (2020).
Park, K. T., Du, S. & Lutkenhaus, J. Essential role for ftsl in activation of septal peptidoglycan synthesis. mBio https://doi.org/10.1128/mBio.03012-20 (2020).
Park, K. T., Pichoff, S., Du, S. & Lutkenhaus, J. FtsA acts through FtsW to promote cell wall synthesis during cell division in Escherichia coli. Proc Natl Acad Sci USA https://doi.org/10.1073/pnas.2107210118 (2021).
Weiss, D. S. Last but not least: new insights into how FtsN triggers constriction during Escherichia coli cell division. Mol. Microbiol. 95, 903–909 (2015).
Britton, B. M. et al. Conformational changes in the essential E. coli septal cell wall synthesis complex suggest an activation mechanism. Nat. Commun. 14, 4585 (2023).
Nierhaus, T. et al. Bacterial divisome protein FtsA forms curved antiparallel double filaments when binding to FtsN. Nat. Microbiol. 7, 1686–1701 (2022).
Radler, P. et al. In vitro reconstitution of Escherichia coli divisome activation. Nat. Commun. 13, 2635 (2022).
Taguchi, A. et al. FtsW is a peptidoglycan polymerase that is functional only in complex with its cognate penicillin-binding protein. Nat. Microbiol. 4, 587–594 (2019).
Cook, J. et al. Activator-induced conformational changes regulate division-associated peptidoglycan amidases. Proc. Natl Acad. Sci. USA 120, e2302580120 (2023).
Cook, J. et al. Insights into bacterial cell division from a structure of EnvC bound to the FtsX periplasmic domain. Proc. Natl Acad. Sci. USA 117, 28355–28365 (2020).
Du, S., Pichoff, S. & Lutkenhaus, J. Roles of ATP hydrolysis by FtsEX and iteraction with FtsA in regulation of septal peptidoglycan synthesis and hydrolysis. mBio https://doi.org/10.1128/mBio.01247-20 (2020).
Pichoff, S., Du, S. & Lutkenhaus, J. Roles of FtsEX in cell division. Res. Microbiol. 170, 374–380 (2019).
Yang, D. C. et al. An ATP-binding cassette transporter-like complex governs cell-wall hydrolysis at the bacterial cytokinetic ring. Proc. Natl Acad. Sci. USA 108, E1052–E1060 (2011).
Arends, S. J. et al. Discovery and characterization of three new Escherichia coli septal ring proteins that contain a SPOR domain: DamX, DedD, and RlpA. J. Bacteriol. 192, 242–255 (2010).
Yahashiri, A., Jorgenson, M. A. & Weiss, D. S. Bacterial SPOR domains are recruited to septal peptidoglycan by binding to glycan strands that lack stem peptides. Proc. Natl Acad. Sci. USA 112, 11347–11352 (2015).
Yahashiri, A., Jorgenson, M. A. & Weiss, D. S. The SPOR domain, a widely conserved peptidoglycan binding domain that targets proteins to the site of cell division. J. Bacteriol. https://doi.org/10.1128/JB.00118-17 (2017).
Tsang, M. J. & Bernhardt, T. G. Guiding divisome assembly and controlling its activity. Curr. Opin. Microbiol. 24, 60–65 (2015).
Yang, X. et al. A two-track model for the spatiotemporal coordination of bacterial septal cell wall synthesis revealed by single-molecule imaging of FtsW. Nat. Microbiol. 6, 584–593 (2021).
Lyu, Z. et al. FtsN maintains active septal cell wall synthesis by forming a processive complex with the septum-specific peptidoglycan synthases in E. coli. Nat. Commun. 13, 5751 (2022).
Buss, J. et al. In vivo organization of the FtsZ-ring by ZapA and ZapB revealed by quantitative super-resolution microscopy. Mol. Microbiol. 89, 1099–1120 (2013).
Hale, C. A. et al. Identification of Escherichia coli ZapC (YcbW) as a component of the division apparatus that binds and bundles FtsZ polymers. J. Bacteriol. 193, 1393–1404 (2011).
Durand-Heredia, J. M., Yu, H. H., De Carlo, S., Lesser, C. F. & Janakiraman, A. Identification and characterization of ZapC, a stabilizer of the FtsZ ring in Escherichia coli. J. Bacteriol. 193, 1405–1413 (2011).
Durand-Heredia, J., Rivkin, E., Fan, G., Morales, J. & Janakiraman, A. Identification of ZapD as a cell division factor that promotes the assembly of FtsZ in Escherichia coli. J. Bacteriol. 194, 3189–3198 (2012).
Espeli, O. et al. A MatP-divisome interaction coordinates chromosome segregation with cell division in E. coli. EMBO J. 31, 3198–3211 (2012).
Buss, J. A., Peters, N. T., Xiao, J. & Bernhardt, T. G. ZapA and ZapB form an FtsZ-independent structure at midcell. Mol. Microbiol. 104, 652–663 (2017).
Krupka, M., Sobrinos-Sanguino, M., Jimenez, M., Rivas, G. & Margolin, W. Escherichia coli ZipA organizes FtsZ polymers into dynamic ring-like protofilament structures. mBio https://doi.org/10.1128/mBio.01008-18 (2018).
Weiss, D. S., Chen, J. C., Ghigo, J. M., Boyd, D. & Beckwith, J. Localization of FtsI (PBP3) to the septal ring requires its membrane anchor, the Z ring, FtsA, FtsQ, and FtsL. J. Bacteriol. 181, 508–520 (1999).
Botta, G. A. & Park, J. T. Evidence for involvement of penicillin-binding protein 3 in murein synthesis during septation but not during cell elongation. J. Bacteriol. 145, 333–340 (1981).
Hedge, P. J. & Spratt, B. G. Resistance to beta-lactam antibiotics by re-modelling the active site of an E. coli penicillin-binding protein. Nature 318, 478–480 (1985).
Kuru, E. et al. In Situ probing of newly synthesized peptidoglycan in live bacteria with fluorescent D-amino acids. Angew. Chem. Int Ed. Engl. 51, 12519–12523 (2012).
Pichoff, S., Du, S. & Lutkenhaus, J. The bypass of ZipA by overexpression of FtsN requires a previously unknown conserved FtsN motif essential for FtsA-FtsN interaction supporting a model in which FtsA monomers recruit late cell division proteins to the Z ring. Mol. Microbiol. 95, 971–987 (2015).
Geissler, B., Elraheb, D. & Margolin, W. A gain-of-function mutation in ftsA bypasses the requirement for the essential cell division gene zipA in Escherichia coli. Proc. Natl Acad. Sci. USA 100, 4197–4202 (2003).
Krupka, M. et al. Escherichia coli FtsA forms lipid-bound minirings that antagonize lateral interactions between FtsZ protofilaments. Nat. Commun. 8, 15957 (2017).
Schoenemann, K. M. et al. Gain-of-function variants of FtsA form diverse oligomeric structures on lipids and enhance FtsZ protofilament bundling. Mol. Microbiol. 109, 676–693 (2018).
Butler, E. K., Davis, R. M., Bari, V., Nicholson, P. A. & Ruiz, N. Structure-function analysis of MurJ reveals a solvent-exposed cavity containing residues essential for peptidoglycan biogenesis in Escherichia coli. J. Bacteriol. 195, 4639–4649 (2013).
Karlin, A. & Akabas, M. H. Substituted-cysteine accessibility method. Methods Enzymol. 293, 123–145 (1998).
Modell, J. W., Hopkins, A. C. & Laub, M. T. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 25, 1328–1343 (2011).
Modell, J. W., Kambara, T. K., Perchuk, B. S. & Laub, M. T. A DNA damage-induced, SOS-independent checkpoint regulates cell division in Caulobacter crescentus. PLoS Biol. 12, e1001977 (2014).
Lariviere, P. J. et al. An essential regulator of bacterial division links FtsZ to cell wall synthase activation. Curr. Biol. 29, 1460–1470 e1464 (2019).
McQuillen, R. & Xiao, J. Insights into the structure, function, and dynamics of the bacterial cytokinetic FtsZ-ring. Annu. Rev. Biophys. 49, 309–341 (2020).
Walker, B. E., Mannik, J. & Mannik, J. Transient membrane-linked ftsz assemblies precede Z-ring formation in Escherichia coli. Curr. Biol. 30, 499–508 e496 (2020).
White, M. L., Hough-Neidig, A., Khan, S. J. & Eswara, P. J. MraZ transcriptionally controls the critical level of FtsL required for focusing Z-rings and kickstarting septation in Bacillus subtilis. J. Bacteriol. 204, e0024322 (2022).
Navarro, P. P. et al. Cell wall synthesis and remodelling dynamics determine division site architecture and cell shape in Escherichia coli. Nat. Microbiol. 7, 1621–1634 (2022).
Uehara, T., Parzych, K. R., Dinh, T. & Bernhardt, T. G. Daughter cell separation is controlled by cytokinetic ring-activated cell wall hydrolysis. EMBO J. 29, 1412–1422 (2010).
Peters, N. T., Dinh, T. & Bernhardt, T. G. A fail-safe mechanism in the septal ring assembly pathway generated by the sequential recruitment of cell separation amidases and their activators. J. Bacteriol. 193, 4973–4983 (2011).
Xu, X. et al. Mechanistic insights into the regulation of cell wall hydrolysis by FtsEX and EnvC at the bacterial division site. Proc. Natl Acad. Sci. USA 120, e2301897120 (2023).
Haeusser, D. P., Rowlett, V. W. & Margolin, W. A mutation in Escherichia coli ftsZ bypasses the requirement for the essential division gene zipA and confers resistance to FtsZ assembly inhibitors by stabilizing protofilament bundling. Mol. Microbiol. 97, 988–1005 (2015).
Buddelmeijer, N. & Beckwith, J. Assembly of cell division proteins at the E. coli cell center. Curr. Opin. Microbiol. 5, 553–557 (2002).
Monteiro, J. M. et al. Peptidoglycan synthesis drives an FtsZ-treadmilling-independent step of cytokinesis. Nature 554, 528–532 (2018).
Corbin Goodman, L. C. & Erickson, H. P. FtsZ at mid-cell is essential in Escherichia coli until the late stage of constriction. Microbiology (Reading), https://doi.org/10.1099/mic.0.001194 (2022).
Addinall, S. G. & Lutkenhaus, J. FtsZ-spirals and -arcs determine the shape of the invaginating septa in some mutants of Escherichia coli. Mol. Microbiol. 22, 231–237 (1996).
Reddy, M. Role of FtsEX in cell division of Escherichia coli: viability of ftsEX mutants is dependent on functional SufI or high osmotic strength. J. Bacteriol. 189, 98–108 (2007).
Geissler, B. & Margolin, W. Evidence for functional overlap among multiple bacterial cell division proteins: compensating for the loss of FtsK. Mol. Microbiol. 58, 596–612 (2005).
Juarez, J. R. & Margolin, W. Changes in the Min oscillation pattern before and after cell birth. J. Bacteriol. 192, 4134–4142 (2010).
Bernhardt, T. G. & de Boer, P. A. Screening for synthetic lethal mutants in Escherichia coli and identification of EnvC (YibP) as a periplasmic septal ring factor with murein hydrolase activity. Mol. Microbiol. 52, 1255–1269 (2004).
Datsenko, K. A. & Wanner, B. L. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl Acad. Sci. USA 97, 6640–6645 (2000).
Lennox, E. S. Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206 (1955).
Yanofsky, C. & Lennox, E. S. Transduction and recombination study of linkage relationships among the genes controlling tryptophan synthesis in Escherichia coli. Virology 8, 425–447 (1959).
Mercer, K. L. & Weiss, D. S. The Escherichia coli cell division protein FtsW is required to recruit its cognate transpeptidase, FtsI (PBP3), to the division site. J. Bacteriol. 184, 904–912 (2002).
Dajkovic, A., Mukherjee, A. & Lutkenhaus, J. Investigation of regulation of FtsZ assembly by SulA and development of a model for FtsZ polymerization. J. Bacteriol. 190, 2513–2526 (2008).
Woldemeskel, S. A., McQuillen, R., Hessel, A. M., Xiao, J. & Goley, E. D. A conserved coiled-coil protein pair focuses the cytokinetic Z-ring in Caulobacter crescentus. Mol. Microbiol. 105, 721–740 (2017).
Huang, X. et al. Fast, long-term, super-resolution imaging with Hessian structured illumination microscopy. Nat. Biotechnol. 36, 451–459 (2018).
Schindelin, J. et al. Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012).
Yan, D., Xue, J., Xiao, J., Lyu, Z. & Yang, X. Protocol for single-molecule labeling and tracking of bacterial cell division proteins. STAR Protoc. 5, 102766 (2024).
Pachitariu, M. & Stringer, C. Cellpose 2.0: how to train your own model. Nat. Methods 19, 1634–1641 (2022).
Ducret, A., Quardokus, E. M. & Brun, Y. V. MicrobeJ, a tool for high throughput bacterial cell detection and quantitative analysis. Nat. Microbiol. 1, 16077 (2016).
Acknowledgements
We thank members of the Du lab, Yang lab, Chen lab and Lutkenhaus lab for advice and helpful discussions to carry out this study. We thank Dr. Michael Laub at MIT and Dr. Erin Goley at Johns Hopkins University for sending us the C. crescentus strains and plasmids. We thank Dr. Georgia Squyres at Caltech for the helpful discussion on data analysis. This study was supported by National Natural Science Foundation of China (grant 32270049 and 32070032, http://www.nsfc.gov.cn/), the Fundamental Research Funds for the Central Universities (grant 2042021kf0198) and Wuhan University (https://www.whu.edu.cn/) to S.D.; D.Y., X.W., H.H., and X.Y.’s research is supported by National Natural Science Foundation of China (grant 32270035, http://www.nsfc.gov.cn/) and Anhui Provincial Natural Science Foundation (Award 2208085MC40).
Author information
Authors and Affiliations
Contributions
Conceptualization, S.D., H.G., and X.Y.; Methodology, H.G., D.Y., Y.L., Y.C., J.Y., W.Y, R.Z., Q.W., X.W., and H.H.; Investigation, H.G., D.Y., Y.L., Y.C., J.Y., W.Y, R.Z., Q.W., X.W., and H.H.; Writing – Original Draft, H.G., S.D., J.L., and X.Y.; Writing –review & editing, H.G., S.D., D.Y., J.L., and X.Y.; Funding acquisition, S.D. and X.Y.; Resources, S.D., X.C., J.L., and X.Y.; Supervision, S.D., X.C., J.L., and X.Y.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Kevin Whitley and the other, anonymous, reviewer for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Gong, H., Yan, D., Cui, Y. et al. The divisome is a self-enhancing machine in Escherichia coli and Caulobacter crescentus. Nat Commun 15, 8198 (2024). https://doi.org/10.1038/s41467-024-52217-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52217-5
- Springer Nature Limited