Abstract
Clonal hematopoiesis of indeterminate potential (CHIP) is linked to diverse aging-related diseases, including hematologic malignancy and atherosclerotic cardiovascular disease (ASCVD). While CHIP is common among older adults, the underlying factors driving its development are largely unknown. To address this, we performed whole-exome sequencing on 8,374 blood DNA samples collected from 4,187 Atherosclerosis Risk in Communities Study (ARIC) participants over a median follow-up of 21 years. During this period, 735 participants developed incident CHIP. Splicing factor genes (SF3B1, SRSF2, U2AF1, and ZRSR2) and TET2 CHIP grow significantly faster than DNMT3A non-R882 clones. We find that age at baseline and sex significantly influence the incidence of CHIP, while ASCVD and other traditional ASCVD risk factors do not exhibit such associations. Additionally, baseline synonymous passenger mutations are strongly associated with CHIP status and are predictive of new CHIP clone acquisition and clonal growth over extended follow-up, providing valuable insights into clonal dynamics of aging hematopoietic stem and progenitor cells. This study also reveals associations between germline genetic variants and incident CHIP. Our comprehensive longitudinal assessment yields insights into cell-intrinsic and -extrinsic factors contributing to the development and progression of CHIP clones in older adults.
Similar content being viewed by others
Introduction
Clonal hematopoiesis (CH) is a common aging-related phenomenon whereby blood cells are predominantly derived from a few hematopoietic stem and progenitor cells (HSPC) with acquired somatic mutation(s) in known leukemia driver genes that foster clonal expansion. CH with a variant allele fraction (VAF) ≥ 2% is termed clonal hematopoiesis of indeterminate potential (CHIP)1,2. The most frequently mutated genes in CHIP include epigenetic regulators DNMT3A, TET2, and ASXL1, splicing factor genes SF3B1, SRSF2, and U2AF1, and DNA damage response genes TP53 and PPM1D, and JAK23. CHIP is associated with many age-related conditions, including hematologic malignancy4,5, atherosclerotic cardiovascular disease (ASCVD)6, stroke7, chronic liver disease8, and heart failure9.
Clinical consequences of CHIP differ depending on driver genes, types of mutations, growth rate, and the size of the clones10,11,12,13. Cell-intrinsic factors such as driver gene, mutation type, genetic background, and cell-extrinsic factors such as chronological age and several putative exposures and disease states across the life course may influence clonal expansion. Additionally, genetic3,14,15,16,17 and environmental factors18,19,20,21,22,23 are associated with increased odds of CHIP in cross-sectional analyses24, but their roles in the emergence or expansion of CHIP clones remain elusive. Small studies have evaluated determinants of incident CHIP or progression of CHIP clones—defined as the emergence or expansion of clones (VAF ≥ 2%)—through short-term serial sequencing12,25,26, a comprehensive understanding of the risk factors is yet to be established firmly.
In this work, we profile incident CHIP in 4187 middle-aged participants from the Atherosclerosis Risk in Communities Study (ARIC) over a median follow-up of 21 years via serial blood-based whole-exome sequencing to identify the determinants of incidence and progression of CHIP clones in older adults. We show that both cell-intrinsic factors, such as driver gene and genetic background, and cell-extrinsic factors, such as chronological age and (self-reported) sex, contribute to the incidence and progression of CHIP clones in older adults, providing valuable insight into clonal dynamics of aging HSPC.
Results
CHIP at baseline visit
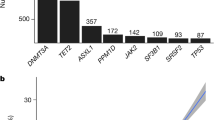
We investigated CHIP in a cohort of 10,871 participants from the ARIC Study baseline visits using whole-exome sequencing (WES) with the HiSeq 2000 platform (Supplementary Table 1). After excluding individuals with prevalent hematologic malignancy and those without WES data at a follow-up visit (Supplementary Fig. 1), our analysis focused on 4187 study participants. Table 1 and Supplementary Table 1 presents the characteristics of these participants, among whom 2478 (59.2%) were female, 951 (22.7%) were African American, and the mean (SD) age was 55.5 (5.5) years at the time of enrollment. A total of 576 CHIP clones with mutations in 51 driver genes were detected among 457 participants (Supplementary Data 1). Baseline CHIP prevalence was 10.9% (457/4187) at VAF ≥ 2% and 3.8% (161/4,187) at VAF ≥ 10% (Fig. 1a). Notably, in all age categories, most of the participants with CHIP (383/457; 83.8%) carried a single mutated clone (Fig. 1b). The frequently mutated genes included DNMT3A, TET2, and ASXL1, with median VAF ranging from 6.6 to 15.3% (Fig. 1c, d). Our study provides comprehensive insights into the prevalence and characteristics of CHIP in middle-aged participants.
Distributions of clonal hematopoiesis of indeterminate potential (CHIP) at baseline (a–d) and follow-up (e–h) visits. CHIP prevalence increases with age, reaching approximately 30% when individuals reach 80 years (a, e). Error bands in (a, e) show a 95% confidence interval around the fitted logistic regression line. Most individuals with CHIP typically carry a single clone (b, f). In the earlier decades of life (under 70 years), many individuals carry mutations in DNMT3A or other less commonly mutated CHIP genes (c, g). However, individuals aged 70 and older show a higher incidence of CHIP involving genes such as TET2, ASXL1, and splicing factors (c, g), and the size of these clones tends to be relatively larger in later years (d, h). Box plots display median (center line), 25th and 75th percentiles (box edges), and whiskers extend to ±1.5 * interquartile range. VAF variant allele fraction.
CHIP at the follow-up visit
We further ascertained CHIP at a later visit, with a median follow-up duration of 21 years (range = 5–27; mean = 20.3; SD = 2.03). The mean (SD) age of participants at follow-up blood draw was 75.8 (5.2) years. For this analysis, we performed WES on 4187 samples using the NovaSeq 6000 platform and identified 1302 CHIP clones at VAF ≥ 2% in 50 driver genes among 1047 participants (Supplementary Data 2). The prevalence of CHIP showed an age-dependent increase, reaching an overall prevalence of 25.0% (1047/4187) and 11.8% (496/4187) at VAF ≥ 2% and ≥10%, respectively (Fig. 1e, f). With advancing age, we observed a shift in the proportions of individuals carrying specific CHIP subtypes. While the prevalence of DNMT3A CHIP and mutations in other less-frequently mutated genes decreased, mutations in TET2, ASXL1, and splicing factor genes increased (Fig. 1g). Additionally, clone sizes tended to increase with advancing age, with a median VAF ranging from 7.7 to 10.2% (Fig. 1h). The shifting patterns and increasing clone sizes of CHIP subtypes in older adults show a dynamic nature of CHIP over time. However, a total of 269 mutations detected at baseline at VAF ≥ 2% were lost during follow-up (Supplementary Fig. 2), whereas 972 clones newly emerged at the follow-up visit (Supplementary Fig. 3). We detected 352 clones at both visits at a VAF ≥ 2%. Among these clones, 233 (66%) were growing, 33 (9%) were shrinking, and 86 (24%) remained static during the follow-up period (Fig. 2).
Concordance of CHIP calls from the HiSeq vs NovaSeq sequencing platforms
Here, baseline and follow-up visits were sequenced using two different sequencing platforms, HiSeq 2000 and NovaSeq 6000. We re-sequenced 786 samples from the same baseline visit using NovaSeq to assess the systematic difference in estimated VAF and the concordance of CHIP ascertainment between the two sequencing platforms. Concordance estimates for CHIP clones detected “yes/no” by the two sequencing platforms were 83% (654/786) and 93% (731/786) at VAF ≥ 2% and VAF ≥ 10%, respectively. We also observed a strong correlation (Pearson’s r = 0.80) between the VAF estimates from these two platforms (Supplementary Fig. 4). As the VAF did not correlate perfectly, our primary analysis was on determinants of incident CHIP, defined as a CHIP clone at VAF ≥ 2% detected at the follow-up visit only without any prevalent CHIP clone at baseline (i.e. VAF < 2% at baseline). In a secondary analysis, we considered factors affecting clonal growth rate.
Incident CHIP in the ARIC Study
We identified 3730 participants without prevalent CHIP, of which 59.7% (2226/3730) were female, 23.2% (865/3730) were African American, and 53.7% (2004/3730) had a history of smoking (Table 1). A total of 735 (19.7%) participants developed incident CHIP (VAF ≥ 2%) during the follow-up, of which 37% (272/735) had large clones (VAF ≥ 10%). Individuals with incident CHIP were relatively older at the baseline visit (median age of 56 vs 54 years; Wilcoxon rank sum test P = 2.4E−6). Here, 876 incident clones were detected in 735 participants, where the majority (615/735; ~84%) acquired a single clone during follow-up (Fig. 3a). Most incident CHIP mutations occurred in DTA (DNMT3A, TET2, or ASXL1), followed by SF (SF3B1, SRSF2, U2AF1, or ZRSR2) and DDR (PPM1D or TP53) genes, and large clones (VAF ≥ 10%) in ASXL1, SF3B1, JAK2, ZNF318, U2AF1, and ZRSR2 (Fig. 3b, c). CHIP incidence increased with advancing age, where >23% of participants older than 75 years acquired incident CHIP (Fig. 3d).
a The vast majority (over 83%) of individuals with incident CHIP carry a single clone, b with approximately 37% of the clones exhibiting an expanded state (VAF > = 10%). b DNMT3A and TET2 show higher proportions of smaller incident clones (VAF between 2% and 10%), while ASXL1, JAK2, U2AF1, and ZRSR2 display expanded clones. c The median clone size exceeds 10% for ASXL1, SF3B1, U2AF1, and ZRSR2 CHIP. Box plots display median (center line), 25th and 75th percentiles (box edges), and whiskers extend to ±1.5 * interquartile range. d Like CHIP prevalence (Fig. 1), CHIP incidence increases with age. Error bands show a 95% confidence interval around the fitted logistic regression line. VAF variant allele fraction.
Clinical predictors of incident CHIP
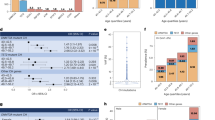
First, we performed univariable logistic regression to examine the associations of baseline risk factors such as age, sex, race, body mass index (BMI), high-density lipoprotein cholesterol (HDL-C), non-HDL-C, history of smoking, hypertension, ASCVD, and T2D with incident CHIP vs. non-CHIP (binary outcome) (Supplementary Fig. 5). In univariable analyses, we observed significant associations (P < 0.0025, considering 20 independent tests at a 5% level of significance) of age at baseline, male sex, and European with incident CHIP categories (Supplementary Fig. 5). Age was significantly associated with a higher incidence of overall CHIP, TET2, and SF mutations (1.03≤ odds ratio (OR) ≤ 1.09; 5.4E−06 ≤ P ≤ 3.2E−4), and nominally associated (0.0025 < P < 0.05) with incidence of ASXL1 and DDR mutations (Supplementary Fig. 5). However, no association was found between age and incident DNMT3A mutations. Male sex was significantly associated with a higher incidence of overall CHIP, ASXL1, and SF (1.32 ≤ OR ≤ 2.82; 6.9E−5 ≤ P ≤ 9E−4) and nominally associated with higher incidence of DDR mutations (Supplementary Fig. 5). European American was significantly associated with lower incidence of DNMT3A (OR = 0.67; 95% confidence interval (CI) = 0.52–0.86; P = 1.8E−3), and nominally associated with higher incidence of SF mutations (Supplementary Fig. 5). Additionally, there were nominal non-significant (0.0025 < P < 0.05) associations between smoking status (never vs. ever) and incident SF, BMI (inverse rank normalized) and incident DNMT3A, history of ASCVD and incident DNMT3A, and non-HDL-C level (inverse rank normalized) and incident TET2 (Supplementary Fig. 5).
Next, we performed multivariable-adjusted logistic regression analyses of incident CHIP categories (CHIP vs. non-CHIP) vs. baseline risk factors, including age, sex, race, BMI, HDL-C, non-HDL-C, history of smoking, hypertension, ASCVD, and T2D (Fig. 4, and Supplementary Fig. 6). Fully adjusted regression models included baseline risk factors and covariates such as age, sex, race, BMI, HDL-C, non-HDL-C, cholesterol-lowering medication use, history of smoking, hypertension, ASCVD, T2D, baseline visits, and visit center. Age was independently associated with a higher incidence of overall CHIP, as well as gene-specific CHIP subtypes (1.04 ≤ OR ≤ 1.10; 2.0E−7 ≤ P ≤ 7.4E−5), with higher odds for splicing factor genes (Fig. 4 and Supplementary Fig. 6). Male sex was nominally associated with a higher incidence of overall and DDR CHIP, and significantly associated with a higher incidence of ASXL1 and SF CHIP (1.30 ≤ OR ≤ 2.79; 7.5E−4 ≤ P < 0.05). European ancestry was nominally associated with a lower incidence of CHIP in DNMT3A and a higher incidence of CHIP in DDR. Interestingly, no significant association was observed between BMI, HDL-C, non-HDL-C, history of smoking, hypertension, T2D, and ASCVD, and incident CHIP categories (Fig. 4 and Supplementary Fig. 6). However, we observed a nominal non-significant association of higher BMI with reduced incident DNMT3A, and history of ASCVD with increased incident DNMT3A but reduced incident TET2 (Fig. 4). We performed sensitivity analyses with stringent incident CHIP case-control definition; findings were robust to the case-control status (“Methods” section; Supplementary Data 3 and 4).
Forest plot showing odds ratio (OR) with a 95% confidence interval (CI) from multivariable logistic regression analyses examining the association between incident overall CHIP (N = 3614; 709 cases and 2905 controls), incident DNMT3A CHIP (N = 3614; 302 cases and 3312 controls,) and incident TET2 CHIP (N = 3614; 160 cases and 3454 controls), and baseline cardiovascular risk factors. Uncorrected P-values are from two-sided Z-tests. The adjusted model accounted for age, sex, race, body mass index (BMI), high-density lipoprotein cholesterol (HDL-C), non-HDL-C, cholesterol medication usage, smoking history, hypertension, atherosclerotic cardiovascular disease (ASCVD, including coronary heart disease and/or ischemic stroke), type 2 diabetes (T2D), and batch effects. Inverse rank normalization was performed before the analysis to account for potential variations in BMI, HDL-C, and non-HDL-C distribution. The results indicate that age is independently and significantly associated with a higher incidence of CHIP. Additionally, nominal non-significant associations are observed for male sex, European American, BMI, and history of ASCVD with incident CHIP categories. ***P < 0.0025 (0.05/20); **P < 0.01; *P < 0.05.
In secondary analyses, we tested the associations of smoking categories (never vs. former or current smokers), BMI categories (BMI < 25 vs. 25–30 or >30 kg/m2), triglyceride to HDL-C (TG/HDL-C) ratio, dyslipidemia, male sex by smoking status (never vs. ever) with incident CHIP categories in fully adjusted models (Supplementary Figs. 7 and 8). There were no significant associations between smoking status (never vs. former or current smoker) or BMI categories and incident CHIP categories (Supplementary Fig. 7). However, we observed a nominal non-significant association between current smoking status and incident CHIP in splicing factor genes, and between BMI > 30 kg/m2 and incident ASXL1 (Supplementary Fig. 7).
No significant associations were observed between TG/HDL-C ratio and incident CHIP (Supplementary Fig. 8a), although dyslipidemia did significantly associate with increased odds of incident TET2 (OR = 2.51; P = 2.5E−3; Supplementary Fig. 8b). A nominal association was observed between dyslipidemia and increased odds of incident ASXL1 (Supplementary Fig. 8b). We also tested interactions between sex and smoking history vs. incident CHIP categories in exploratory analyses. Fully adjusted models did not reveal significant interactions between sex and smoking history on incident CHIP categories. Nevertheless, there was a nominal interaction between sex and smoking history: we observed lower odds of incident DNMT3A in the male sex by ever-smoker status (OR = 0.48; P = 0.0036) (Supplementary Fig. 8c).
Shared genetic predisposition in prevalent and incident CHIP
We separately assessed the association of an independently derived prevalent CHIP polygenic risk score (PRS), consisting of 21 conditionally independent and genome-wide significant (P < 5E−8; Supplementary Data 5) variants15, with incident CHIP categories in African American (AA) and European American (EA) participants, followed by inverse-variance weighted fixed-effect meta-analysis (Fig. 5a, b). We found that per SD increase in genetic liability to prevalent CHIP was significantly associated with incident CHIP (OR = 1.23; 95% CI 1.12–1.35; P = 8.9E−6). Furthermore, genetic liability to prevalent CHIP was associated with incident DNMT3A (OR = 1.28; 95% CI 1.12–1.46), TET2 (OR = 1.30; 95% CI = 1.09–1.55), and ASXL1 (OR = 1.66; 95% CI = 1.26–2.17) CHIP (Fig. 5b).
Association of prevalent clonal hematopoiesis of indeterminate potential (CHIP) with incident CHIP: a, b polygenic risk score (PRS) and c germline variants analysis. a Distribution of PRS in ARIC AA and EA participants. b Ancestry-stratified PRS was calculated using 21 independent variants (P < 5E−8; Supplementary Data 5) from Kessler et al.15. Logistic regression was performed, adjusting for age, sex, smoking status, top five principal components of ancestry, and batch effect. Data are presented as odds ratio (OR) with a 95% confidence interval (CI) and uncorrected P-value from a two-sided Z-test. The results show a strong association between prevalent CHIP PRS and incident CHIP. c Forest plot of odds ratio with 95% CI from inverse variant-weighted fixed-effect meta-analyses of single-variant associations for incident overall CHIP, incident DNMT3A CHIP, and incident TET2 CHIP in ARIC AA (n = 637) and ARIC EA (n = 2378) participants. Uncorrected P-values are from two-sided Z-tests. Single-variant association adjusted for age, age2, sex, top ten principal components of ancestry, and batch effect, followed by multi-ancestry meta-analysis, was conducted to investigate the association of genome-wide significant (P < 5E−8) prevalent CHIP-associated variants15 and incident CHIP at a significant level of P < 0.05. This figure presents the lead variant(s) from each locus, and the full list is available in Supplementary Data 6–8. Several germline variants previously associated with prevalent CHIP were found to be associated with incident CHIP, indicating a shared genetic basis between prevalent and incident CHIP. ARIC AA the Atherosclerosis Risk in Communities Study African American; ARIC EA European American; EA effect allele; EAF effect allele frequency; SNV single nucleotide variant.
Next, to test the associations of genome-wide significant (P < 5E−8) variants for prevalent CHIP with risk of incident CHIP, we performed targeted single-variant associations in AA and EA participants from ARIC. We tested known loci associated with prevalent CHIP derived in the UK Biobank by Kessler et al.15. For matching variants with minor allele frequency ≥1%, we performed ancestry-stratified single-variant associations for incident CHIP, DNMT3A, and TET2 in the ARIC study, followed by a multi-ancestry inverse-variance weighted fixed-effect meta-analysis. A P threshold of <0.05 was considered statistically significant. At this threshold, we reported several prevalent CHIP loci associated with incident CHIP (Fig. 5c and Supplementary Data 6–8). Notably, the risk alleles in SMC4, TERT, HBS1L, ZNF318, GSDMC, ATM, TCL1A, and SETBP1 loci were associated with a higher incidence of overall CHIP (1.16 ≤ OR ≤ 1.41; 0.0011 ≤ P ≤ 0.044; Fig. 5c and Supplementary Data 6). Risk alleles in SMC4, TERT, CD164, HBS1L, GSDMC, SETBP1, and BCL2L1 loci were associated with higher incidence of DNMT3A CHIP (1.24 ≤ OR ≤ 1.61; 0.0017 ≤ P ≤ 0.043; Fig. 5c and Supplementary Data 7). The risk alleles in SMC4, TERT, CD164, HBS1L, H2AFV, GSDMC, TCL1A, DNAH2, SETBP1, and RUNX1 loci were associated with higher incidence of TET2 CHIP (1.35 ≤ OR ≤ 1.71; 0.0013 ≤ P ≤ 0.048; Fig. 5c and Supplementary Data 8). Here, rs2887399-T, the non-DNMT3A CHIP-associated protective variants in TCL1A promoter8, were nominally associated with incident overall CHIP and incident TET2 CHIP (P < 0.2) with similar effect estimate for TET2 (Supplementary Table 2). Importantly, variants in the TCL1A locus at discovery P < 0.05 were in strong linkage equilibrium with rs2887399-T (Supplementary Table 3).
Determinants of clonal growth
To identify determinants of clonal growth rate—calculated as log(VAFFollowup/VAFBaseline)/ (AgeFollowup – AgeBaseline) when dVAF >0, we examined cell-intrinsic factors such as type of mutation and CHIP driver gene, and cell-extrinsic factors such as age at baseline, self-reported sex (female vs. male), self-reported race (African American vs. European American), smoking status (never vs ever), prevalent disease status, HDL-C, non-HDL-C, and BMI. In aggregate clonal growth rate was strongly associated with driver gene, but not with mutation type (Table 2 and Supplementary Table 4). Compared to DNMT3A non-R882 clones, splicing factor genes that included SF3B1, SRSF2, U2AF1, and ZRSR2 clones, and TET2 clones grew significantly (P < 0.0025) faster, followed by nominal associations for DNA damage response genes (PPM1D and TP53), other gene category, and ASXL1 clones. However, we did not observe a significant difference in the growth rate of DNMT3A non-R882 and DNMT3A R882 clones. Age was positively associated with clonal growth rate, though there was no other significant association between growth rate and traditional ASCVD risk factors, including sex, race, smoking, BMI, hypertension, HDL-C, or non-HDL-C level (Table 2 and Supplementary Table 4).
Inferring CHIP occurrences from synonymous passenger mutations
HSPCs acquire somatic mutations with aging, the majority of which are neutral and remain below the detection threshold or stochastically disappear from the cell population27. Neutral somatic mutations cooccurring with a (detected or undetected) driver can be positively selected and reach a higher VAF simply by hitchhiking—these are known as passenger mutations28. It is known from previous studies that passenger mutations can be used to identify new driver genes29,30,31 and estimate the fitness effect of known drivers8. However, the utility of synonymous passenger mutations detected in the WES of healthy individuals for predicting CHIP occurrences is not known. Here, we analyzed 4187 WES from ARIC baseline visits with longitudinal CHIP ascertainment and identified 6789 synonymous passenger mutations (1% ≤ VAF ≤ 25%) in 1018 ARIC participants (Supplementary Data 9 and Supplementary Fig. 9).
Approximately half of the detected clones are within the VAF range of 5–10%, and 81% of clones are within the range of 1% ≤ VAF ≤ 10%. The mutational spectrum of nonsynonymous CHIP mutations and synonymous passenger mutations is presented in Supplementary Fig. 10a–d. C>T transitions are the most frequent nonsynonymous mutation in CHIP, followed by T>C transitions (Supplementary Fig. 10a, b), whereas T>C transitions are the most frequent synonymous mutation, followed by C>T transitions (Supplementary Fig. 10c, d). To assess the utility of synonymous passenger counts, we performed multivariable regression analyses between CHIP outcomes and passenger counts (“Methods” section) and found that passenger counts were strongly associated with the presence of CHIP (1.1E−30 ≤ P < 0.05) and were predictive of both prevalent and incident CHIP (Fig. 6a). Additionally, passenger counts were strongly associated with the number of CHIP clones in a multivariable multinomial logistic regression (P < 5.6E−9; Fig. 6b). Furthermore, passenger counts were significantly associated with clonal growth (dVAF>0) in a multivariable logistic regression (P = 5.0E−4; Fig. 6c). Associations between CHIP categories and synonymous passenger counts in Fig. 6a–c were independent of demographic and other clinical exposures including enrollment age, age2, sex, race, smoking status, BMI, HDL-C, non-HDL-C, and prevalent disease status for hypertension, ASCVD, and T2D. Additionally, the clone size of synonymous passenger mutations (i.e., VAF) was positively correlated (Pearson’s r > 0.10, P < 0.05; Supplementary Table 5) and strongly associated with the growth rate of CHIP clones (P < 0.05; Supplementary Table 6).
Association of synonymous passenger mutation counts with a presence of CHIP, b number of CHIP clones, and c growth of CHIP clones. Forest plot of odds ratio (OR) with a 95% confidence interval (CI) showing the effects of (inverse rank normalized) synonymous passenger counts on CHIP-related outcomes (listed below in a–c)—models accounted for age at baseline, age2, self-reported sex, self-reported race, body mass index (BMI), high-density lipoprotein cholesterol (HDL-C), non-HDL-C, cholesterol medication usage, smoking history, hypertension, atherosclerotic cardiovascular disease (ASCVD, including coronary heart disease and/or ischemic stroke), type 2 diabetes (T2D), and batch effects (including ARIC baseline visit and visit center). Uncorrected P-values are from two-sided Z-tests. a Model 1: four separate multivariable logistic regression analyses were performed for outcomes (i) no CHIP (n = 2905) vs. presence of CHIP at either visit (n = 1147), (ii) no CHIP at baseline (n = 3614) vs. CHIP detected at baseline (i.e. prevalent CHIP, n = 438), (iii) no CHIP at follow-up visit (n = 3043) vs. CHIP detected at the follow-up visit (n = 1009), and (iv) no CHIP (n = 2905) vs. incident CHIP (n = 709). b Model 2: multivariable multinomial logistic regression was performed for the number of CHIP clones as outcomes, i.e., no CHIP clone (n = 2905) vs. one clone (n = 877), 2 clones (n = 206), 3 clones (n = 43), or ≥4 clones (n = 21). c Multivariable logistic regression was performed for no CHIP clone (or no change in clone size, dVAF = 0; n = 2909) vs. growing CHIP clone (i.e. dVAF > 0; n = 936). BMI, HDL-C, and non-HDL-C values were inverse rank normalized. CHIP: clonal hematopoiesis of indeterminate potential; dVAF = VAFfollow-up − VAFbaseline.
Discussion
We conducted one of the largest long-term longitudinal whole-exome sequencing studies on clonal hematopoiesis involving 4,187 healthy participants from the ARIC study with a median follow-up of 21 years. We identified CHIP (VAF ≥ 2%) in 11% of the participants at baseline and 25% at the follow-up visits. Consistent with previous studies3,6, we showed that the prevalence of CHIP increases with advancing age.
We observed incident CHIP in approximately 17% of participants below 70 years, and this proportion increased to around 30% in individuals above 85 years of age among those without CHIP in middle age. These results reinforce the substantial impact of age on the acquisition of CHIP. Furthermore, our study revealed an interesting pattern of decreasing diversity in mutated CHIP genes with advancing age. Specifically, we noted an increase in incident clones in genes such as TET2, ASXL1, and splicing factor genes (SF3B1, U2AF1, SRSF2, and ZRSR2), as well as PPM1D and TP53. In multivariable linear regression, we further show that splicing factor genes and TET2 clones expanded significantly faster than DNMT3A non-R882 clones over a median 21-year follow-up. The clonal growth rate after CHIP was manifest was not associated with traditional ASCVD risk factors. These findings corroborate and extend recent reports highlighting reduced clonal diversity (VAF ≥ 2%), faster expansion, and the acquisition of new mutations in these genes as individuals age12,32,33.
Previous cross-sectional studies reported associations of age, smoking, T2D, and BMI with prevalence of CHIP4,5,18,34. Interestingly, the impact of age on CHIP subtypes varied, with stronger associations observed for splicing factor genes, TET2, and ASXL1 CHIP, but not for DNMT3A CHIP. Surprisingly, besides age, we did not find any significant association between certain traditional risk factors for atherosclerosis, such as a history of smoking, hypertension, T2D, BMI, and LDL-C, and the risk for incident CHIP, contrary to previous cross-sectional observations related to prevalent CHIP. Our findings aligned with a recent longitudinal study demonstrating no association between smoking, being overweight, and acquiring new CHIP mutations33. Another study reported an inverse association between HDL-C and clonal expansion35. While we observed a nominal association between non-HDL-C and incident TET2 CHIP in univariable analysis, this association did not hold in a fully adjusted logistic regression model.
Our genetic analyses provide compelling evidence for a shared genetic basis between prevalent and incident CHIP. For the first time, we demonstrated strong associations between prevalent CHIP PRS and increased odds of incident CHIP, underscoring the predictive value of CHIP PRS in identifying individuals at risk of developing CHIP. Furthermore, the genetic variants previously linked to prevalent CHIP15 also showed associations with incident CHIP in our study using the ARIC dataset. Specifically, we observed significant associations for SMC4, TERT, ATM, TCL1A, and SETBP1 loci for incident CHIP. These findings further support the notion of a shared genetic underpinning between prevalent and incident CHIP, emphasizing the relevance of these specific loci in the development and progression of clonal hematopoiesis. Importantly, these results provide evidence for the causality of key genes in the development of CHIP itself.
However, it is worth noting that only a quarter of the prevalent CHIP-associated loci were associated with incident CHIP. Notably, certain loci, including PARP1, LY75, SENP7, TET2, CD164, and ITPR2, showed no association with incident CHIP. This could be due to lower statistical power, poor imputation quality, or true biological differences between prevalent and incident CHIP. The CD164 locus is an example of the latter, where this locus is strongly associated with prevalent CHIP15,16,17 but lacks any association with incident CHIP. The opposite effect directions observed at the CD164 locus for incident DNMT3A and TET2 CHIP could explain this null association in overall CHIP. Previously, we and others reported opposite associations at the TCL1A locus with prevalent DNMT3A and TET2 CHIP, leading to a null association with overall CHIP3,15,16,17. This study found an association at the TCL1A locus with incident overall CHIP and TET2 CHIP but not with incident DNMT3A CHIP. While our study was insufficiently powered to identify the lead variant TCL1A rs2887399-T, reported in Weinstock et al.8, we observed an identical effect estimate for incident TET2 CHIP, reaching statistical significance (P~0.05) (Supplementary Table 2). Notably, the association of incident DNMT3A tended toward null, with P approaching 1.0. Furthermore, it is crucial to highlight that incident TET2 and overall CHIP-associated variants identified in this study exhibit strong linkage disequilibrium with TCL1A rs2887399-T (Supplementary Table 3). Our findings align with recent research demonstrating the involvement of TCL1A in the expansion of various non-DNMT3A CHIP subtypes, including TET28.
The ongoing debate surrounding the relationship between CHIP and the development of ASCVD has led to investigations exploring the role of CHIP if it triggers inflammation and contributes to the development of ASCVD6,36, or if ASCVD itself or clinical risk factors for ASCVD promote the development of CHIP37, as well as a potential bidirectional link between CHIP and inflammation38. While baseline ASCVD status showed a nominal association with incident DNMT3A and TET2 CHIP, the associations were not statistically significant and were in opposite directions. Secondary analyses revealed a strong link between dyslipidemia and incident TET2 and potentially ASXL1 CHIP, while no such associations were found for DNMT3A or overall CHIP. These findings partially support the hypothesis that dyslipidemia may influence CHIP development37.
Nonetheless, explanations based solely on ASCVD (or associated risk factors) are insufficient in understanding the development of CHIP among older adults. The results from our clinical and genetic analyses point to intricate relationships between environmental and genetic risk factors in the development of clonal hematopoiesis, providing strong evidence for a bidirectional association between CHIP and inflammation. Notably, studies in mice and zebrafish support a positive feedback loop between CHIP expansion and inflammation, where CHIP promotes inflammation, and the relative fitness and resistance to inflammation of CHIP clones further fuel clonal expansion, creating a vicious cycle of inflammation and expansion38,39,40. These findings highlight the complexity of the interactions between exposome, inherited, and somatic genomes, shedding light on the pathogenesis of CHIP in the context of (inflamm-)aging41,42,43. Further research is crucial to fully unravel these intricate relationships’ underlying mechanisms and potential therapeutic implications.
Finally, we assessed the utility of synonymous passenger mutations in understanding clonal dynamics in aging HSPCs. We show that baseline synonymous passenger counts are strongly associated with CHIP status and are predictive of acquiring new CHIP clones (VAF ≥ 2%) and clonal growth that were ascertained after a long median follow-up interval of 21 years. Furthermore, passenger clone size was moderately correlated with the growth rate of CHIP clones. Our findings suggest that synonymous passengers detected in WES are a good proxy for clonal growth and are predictive of CHIP occurrences, providing valuable insights into the dynamics of somatic mutations in aging HSPCs.
Although our study demonstrates associations between environmental and genetic determinants, important limitations should be considered. First, variability in CHIP ascertainment: CHIP was determined using two different sequencing platforms. The technical differences, albeit with good correlation, precluded the ability to compare clonal trajectories and growth rates and investigate factors associated with VAF changes but were suitable for incident CHIP analyses. Second, homogeneity in follow-up time: the study was constrained by a lack of heterogeneity in the follow-up time among participants as well as a single follow-up sampling. This limited the ability to perform time-to-event analyses between the baseline exposures and the incidence of CHIP. Longer follow-up durations and varying time intervals could provide a more comprehensive understanding of the relationship between exposures and the development of CHIP. Third, although the long duration of almost two decades between time points was a strength in regards to examining the impact of age, a substantial number of individuals died during this time period. Participants without follow-up WES had more clinical ASCVD, T2D, hypertension, cholesterol medication usage, increased BMI, and unfavorable lipid profiles (as shown in Supplementary Table 1). Thus, individuals with the most severe ASCVD and perhaps the greatest baseline inflammation were more likely to die before the follow-up visit (ARIC visit-5), limiting our power and biasing our results towards the null. Fourth, the study may have suffered from reduced statistical power, which could have influenced the observed associations between exposures and outcomes. Larger sample sizes would enhance the study’s statistical power and provide more reliable conclusions. Fifth, WES data limited our passenger mutation search to only coding regions; focusing on only synonymous mutations further reduced the pool of putative passenger mutations in the population. This could limit the generalizability of the findings from this study to studies that include a more diverse set of mutation types detected across the genome for passenger mutation. Therefore, these limitations should be considered when interpreting the findings, and future studies should aim to address these issues to further advance our understanding of the relationships between environmental and genetic determinants and their impact on clonal hematopoiesis.
Our comprehensive long-term longitudinal assessment provides new insights into the factors that promote incident CHIP and clonal growth rate in older adults. We find that age at baseline, sex, and dyslipidemia are significant predictors of incident CHIP, while ASCVD and traditional risk factors for ASCVD are not. We also find that the factors driving clinically relevant clonal expansion (VAF ≥ 2%) may vary, to some extent, among different CHIP driver genes. Additionally, our research demonstrates a shared genetic basis between prevalent and incident CHIP, further supporting the notion that CHIP can evolve independently of prevalent ASCVD and traditional ASCVD risk factors. Taken together, these results support a bidirectional relationship between CHIP and inflammation. Early identification of incident CHIP presents an opportunity for timely intervention and preventive measures. These findings underscore the importance of further research to understand the mechanisms underlying incident CHIP better and to develop strategies for its early detection and effective management.
Methods
Informed consent was obtained from all study participants, and the study design and methods were approved by the respective institutional review boards at each of the collaborating institutions: University of Mississippi Medical Center Institutional Review Board (ARIC: Jackson Field Center); Wake Forest University Health Sciences Institutional Review Board (ARIC: Forsyth County Field Center); University of Minnesota Institutional Review Board (ARIC: Minnesota Field Center); and Johns Hopkins University School of Public Health Institutional Review Board (ARIC: Washington County Field Center). Each study received institutional certification before depositing sequencing data into dbGaP, ensuring approval by all relevant institutional ethics committees and compliance with relevant ethical regulations. The secondary use of genomic data was approved by the Mass General Brigham Institutional Review Board (protocol 2016P002395 and 2016P001308).
Study samples
There were 10,881 ARIC participants with WES data at baseline sequenced using the HiSeq 2000 platform (Illumina, Inc., CA) (ARIC sub-study phs000668 CHARGE-S44 https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000668.v6.p2). Baseline samples were from five visits with age ranges 44–84 years (Mean = 57.3; Median = 57.0; SD = 6.06). WES was also generated using the NovaSeq 6000 platform (Illumina, Inc., CA) from 4233 longitudinal visit samples (ARIC Visit 05). Among the longitudinal visit samples, 4,187 without hematologic malignancy had baseline WES data (detailed study design in Supplementary Fig. 1).
Detection of clonal hematopoiesis of indeterminate potential (CHIP)
Somatic mutations were identified from WES using Mutect2 software45 in the Terra platform (https://portal.firecloud.org/?return=terra#methods/gatk/mutect2-gatk4/21), annotated using ANNOVAR software46, and CHIP was detected using a publicly available pipeline (https://app.terra.bio/#workspaces/terra-outreach/CHIP-Detection-Mutect2/). To minimize potential artifacts in the Mutect2 calls, a panel-of-normal (PON) was created from 100 random HiSeq WES from the youngest participants, while 1000 Genomes PON was used for the NovaSeq WES. Besides, the Genome Aggregation Database (gnomAD) was used to limit germline variants in the somatic mutation call. Mutect2 calls were further filtered, and variants were kept if (i) total depth of coverage (DP) ≥ 20, (ii) number of reads supporting the alternate allele (AD_Alt) ≥ 3, (iii) ≥1 read in both forward and reverse direction supporting the alternate allele (F1R2_Alt and F2R1_Alt), (iv) variant allele fraction ≥2%, (v) gnomAD allele frequency ≤0.001 (not hotspot mutations). CHIP variants that passed sequence-based filtering underwent additional curation, wherein Mutect2 annotation criteria encompassed “PASS,” “weak_evidence,” “multiallelic,” or “germline.” Variants with frequencies exceeding those of DNMT3A R882 were excluded. Moreover, variants were eliminated if their VAF was less than 10% in the majority of clones, specifically those annotated with “weak_evidence,” and exhibited AD_Alt < 5. To remove potential oxoG artifacts, G>T and C>A substitutions with F1R2_Alt = 1 or F2R1_Alt = 1 were excluded from the analysis. Indel variants positioned at the ends of sequence reads, denoted by Mutect2 annotation “MPOS” ≤10 or >45, were also excluded. Additionally, indels in proximity to homopolymer regions were excluded if AD_Alt < 5 and the median VAF was less than 10%. A subset of the identified CHIP variants underwent manual inspection using the Integrative Genomics Viewer. In instances where CHIP was detected in Mutect2 during one visit compared to another, the sequence read pileup for the corresponding genomic coordinates was extracted from the bam or cram file. Pathogenic variants were queried in 69 genes known to drive clonal hematopoiesis and myeloid malignancies6,47 to identify CH. The detailed CHIP calling pipeline was previously reported by Bick et al. 3 and Uddin et al. 17 (https://app.terra.bio/#workspaces/terra-outreach/CHIP-Detection-Mutect).
Hotspot mutations in U2AF1
A special approach was used to identify somatic variants in U2AF1 since an erroneous segmental duplication in the hg38 reference genome resulted in a mapping score of zero for this gene during the sequence alignment from FASTQ to BAM/CRAM. We used a custom script (https://github.com/MMesbahU/U2AF1_pileup) to recover hotspot mutations: S34F, S34Y, R156H, Q157P, and Q157R. A minimum of 5 supporting reads for alternate alleles was required to include a somatic mutation in U2AF1.
HiSeq vs. NovaSeq WES
To analyze the concordance between the two platforms used for WES, we re-sequenced 786 baseline samples using the NovaSeq 6000 platform (Illumina, Inc., CA). CHIP was detected using the same pipeline described above. We compared CHIP status “yes/no” at VAF ≥ 2% and VAF ≥ 10%. We also performed Pearson’s correlation between the VAF estimates when the same clone was detected from WES generated by the two platforms.
Associations between baseline risk factors and incident CHIP
Incident CHIP was defined as the presence of a clone with a VAF of ≥2% at the follow-up visit in individuals who did not have a prevalent CHIP clone (or CHIP at VAF < 2%) at baseline. The CHIP mutations were considered if they met the following criteria: a read depth ≥20, a minimum of 3 supporting reads for the alternative allele, and at least one read from both the forward and reverse directions.
Univariable and multivariable analyses were performed using a logistic regression model to examine the associations between incident overall CHIP vs. non-CHIP and driver gene-specific incident CHIP categories (incident DNMT3A, incident TET2, incident ASXL1, incident SF, and incident DDR) vs. non-CHIP or other CHIP categories, and baseline risk factors. The tested risk factors included age, self-reported sex, self-reported race, smoking status, body mass index (BMI), high-density lipoprotein cholesterol (HDL-C), non-HDL-C, disease status for type 2 diabetes (T2D), atherosclerotic cardiovascular disease (ASCVD, including ischemic stroke and coronary heart disease), and hypertension. Additional covariates considered in the analysis included cholesterol medication usage, baseline visits, and visit centers.
Secondary analyses were performed to test the association between exposures such as smoking categories (never vs former or current smoker), BMI categories (BMI < 25 vs. 25 ≤ BMI ≤ 30 or BMI > 30 kg/m2), triglyceride to HDL-C (TG/HDL-C) ratio and dyslipidemia, and incident CHIP. Interactions between sex and smoking status were also tested in a fully adjusted model for association with incident CHIP.
Here, dyslipidemia was defined as a binary variable (“yes/no”) based on the following criteria: individuals with total cholesterol ≥240, triglyceride ≥200, LDL-C ≥ 160, HDL-C < 40 in men or HDL-C < 50 mg/dl in women, and/or individuals on statin therapy. Inverse rank normalization was performed before the analysis to account for potential variations in the distribution of BMI, HDL-C, non-HDL-C, and TG/HDL-C values. To account for multiple comparisons in the analysis, a significance threshold of <0.0025 was employed, considering 20 independent tests at a 5% significance level.
We also performed sensitivity analysis where CHIP was detected at the follow-up visit with baseline sequence coverage for corresponding positions ≥20 with all nonmutant alleles or clones with VAF < 0.1% for the mutation. Here, we applied stringent filtering where all variants newly detected at the follow-up visit required ≥5 supporting reads where 2 reads each needed from both forward and reverse directions. The findings were consistent with our primary analysis (Supplementary Data 3 and 4).
Single-variant association analysis
Imputed genotype data from the ARIC sub-study GENEVA (dbGaP accession phs000090 “phg000248” https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000090.v8.p2) was used for genetic analyses. From dbGaP, we downloaded genotype data (Affymetrix 6.0 SNP array) imputed to the whole-genome sequence using the 1000 Genomes reference panel (www.1000genomes.org, June 2011 release)48 using IMPUTE2 software49 (details in dbGaP phs000090/phg000248/). Imputed GWAS data and WES were available for 637 AA participants and 2378 EA participants. Variants with a minor allele frequency >1%, imputation accuracy (INFO score) ≥0.30, and significant association (P < 5.0E−8) with prevalent CHIP in the UK Biobank15 were considered for association analysis. We performed ancestry-stratified single-variant associations, adjusted for age, age2, sex, top ten principal components of ancestry, and batch effect, followed by multi-ancestry inverse-variance weighted fixed-effect meta-analysis for incident overall CHIP, DNMT3A, and TET2 CHIP. REGENIE software50 was used for single-variant associations, and GWAMA software51 for multi-ancestry metanalysis (scripts available in the “Code Availability” section). Variants with an association P < 0.05 were considered significant for these analyses.
Prevalent CHIP polygenic risk score (PRS)
Prevalent CHIP PRS were derived for 637 AA and 2376 EA participants using weights of 21 single nucleotide variants that were conditionally independent and significantly associated with the prevalence of CHIP in the UK Biobank at P < 5E−815. Effects for the risk-increasing allele were considered and the PLINK (version v2.00a3LM) “score” function was used to derive prevalent CHIP PRS. PRS was standardized to have a mean of zero and SD of 1. We performed associations for prevent CHIP PRS with incident CHIP categories in ARIC AA and EA participants using multivariable logistic regressions adjusted for age, sex, smoking status (never vs. ever), top five principal components of ancestry, and batch effect, followed by inverse-variance weighted fixed-effect multi-ancestry meta-analyses (scripts available in the “Code Availability” section).
Clonal trajectories
We classified CHIP clones observed at both visits (VAF ≥ 2% in one of the visits) in three different trajectories: “growing” when growth rate>0, with percent VAF change≥10% and dVAF≥0.02 (red), “shrinking” when growth rate<0, percent VAF change ≤ −10% and dVAF ≤ −0.02 (blue), otherwise “static” (black). Here, dVAF = VAFfollowup - VAFbaseline.
Determinants of clonal growth rate
A single clone per individual at dVAF > 0 was considered for growth rate analysis. Where multiple clones were detected at the follow-up visit at VAF > 2%, only a clone with max dVAF>0 was considered. If no clone was detected at baseline with coverage ≥20, we imputed baseline VAF = 0.1%. The growth rate was calculated as log(VAFFollowup/VAFBaseline)/(AgeFollowup − AgeBaseline). Multivariable linear regression was performed to model inverse rank normalized growth rate (outcome) with age, driver gene, mutation type, self-reported sex, self-reported race, BMI, smoking, HDL-C, Non-HDL-C, hypertension, T2D, ASCVD as exposures of interest, with additional covariates such as batch effect that included log(baseline sequencing coverage), baseline visit (i.e. ARIC visit 02 vs. others [visit 01, visit 03, visit 04 or MRI visit]), baseline visit center, baseline mutation detection method [MUTECT vs. manually detected in IGV or from read pileup], estimated baseline VAF vs. imputed VAF. The fully adjusted multivariable model included all these variables. Sensitivity analysis was performed where only variants at VAF ≥ 2% detected at follow-up with ≥5 supporting reads with ≥2 forward and reverse reads for the mutant allele were considered.
Detection of synonymous passenger mutations and association analysis
We analyzed ANNOVAR annotated Mutect2 variant call format files from 4187 baseline WES with longitudinal visit samples for synonymous passenger mutations (Supplementary Fig. 1). We identified synonymous single nucleotide variants (synonymousSNV) from autosomes in the VAF range 1%–25% with sequence coverage 20–400× with a minimum of three supporting reads for the alternative allele with ≥1 read each from forward and reverse directions. Variants were excluded if present in gnomAD, Mutect2 filter annotation other than “PASS”, median base quality (MBQ) < 30, median mapping quality (MMQ) < 60, median distance from the end of supporting reads (45 ≤ MPOS ≤ 10), number of events in the haplotype (ECNT) > 1, and variant observed in more than one sample (i.e. only singletons were considered for analysis). synonymousSNV that remained after filtering were considered for analysis and per-sample passenger counts were calculated. Individuals without a synonymousSNV detected in baseline WES were assigned a “0” for passenger count. Inverse rank normalized synonymous passenger counts (INTsynonymousSNV) were used as an exposure of interest for regression analyses.
To assess the association between the presence of CHIP (outcome) and (inverse rank normalized) synonymous passenger counts (exposure, INTsynonymousSNV), four separate multivariable logistic regressions were performed with varying outcome definitions, such as (i) no CHIP vs. presence of CHIP at either baseline and/or follow-up visit, (ii) no CHIP at baseline vs. prevalent CHIP (i.e. CHIP at VAF ≥ 2% detected at baseline visit), (iii) no CHIP at follow-up visit vs. CHIP detected at the follow-up visit, and (iv) no CHIP vs. incident CHIP (i.e. CHIP at VAF ≥ 2% only detected at the follow-up visit in individuals without a prevalent clone). Using a multivariable multinomial logistic regression, we further assessed the association between the number of CHIP clones (no CHIP clone vs. one clone/ 2 clones/ 3 clones or ≥4 clones) and INTsynonymousSNV. Finally, we used multivariable logistic regression to assess the association between clonal growth (no = 0/yes = 1) and INTsynonymousSNV. Here, binary outcome clonal growth was defined as no CHIP clone or no change in clone size (i.e., dVAF = 0) as “0” vs. growing CHIP clone (i.e., dVAF > 0) as “1”. All multivariable analyses accounted for additional covariates such as age at baseline, age2, self-reported sex, self-reported race, BMI, HDL-C, non-HDL-C, cholesterol medication usage, smoking history, hypertension, ASCVD, T2D, and batch effects (including ARIC baseline visit and visit center).
Use of large language models
Advanced language models like Grammarly, Inc., Bard (https://bard.google.com/), and ChatGPT (version May 24; https://chat.openai.com/) were used to enhance grammatical accuracy and improve sentence structure and clarity.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
To protect the privacy of research participants and the confidentiality of their data while ensuring that these data are available for appropriate use by researchers, all raw data used in this study are available via controlled access. Individual-level phenotypes and whole-exome sequencing data from the ARIC baseline visits participants (used in this study) are available in dbGaP (https://www.ncbi.nlm.nih.gov/gap/) accession code phs000668 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000668.v6.p2). Affymetrix 6.0 genome-wide association array dataset is available in dbGaP accession phs000557.v7.p2 (https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000557.v7.p2). Imputed genotype data from the ARIC sub-study GENEVA is available in dbGaP (accession phs000090 “phg000248” https://www.ncbi.nlm.nih.gov/projects/gap/cgi-bin/study.cgi?study_id=phs000090.v8.p2). ARIC Visit-5 WES (generated in this study) and individual-level data are available via controlled access ancillary study proposal (https://aric.cscc.unc.edu/aric9/researchers/Obtain_Submit_Data). The timeline for the approval process ranges from 3–6 weeks for ARIC ancillary studies, with specific criteria and proposal forms available at https://sites.cscc.unc.edu/aric/ancillary-studies-pfg. All other data supporting the findings described in this manuscript are available in the article and its Supplementary Information files.
Code availability
The complete scripts used in this study are available at https://github.com/MMesbahU/longitudinal-profiling-of-clonal-hematopoiesis52; CHIP calling pipeline is available at https://app.terra.bio/#workspaces/terra-outreach/CHIP-Detection-Mutect, and the custom script for mutations in U2AF1 is available at https://github.com/MMesbahU/U2AF1_pileup.
References
Khoury, J. D. et al. The 5th edition of the World Health Organization classification of haematolymphoid tumours: myeloid and histiocytic/dendritic neoplasms. Leukemia 36, 1703–1719 (2022).
Arber, D. A. et al. International consensus classification of myeloid neoplasms and acute leukemias: integrating morphologic, clinical, and genomic data. Blood 140, 1200–1228 (2022).
Bick, A. G. et al. Inherited causes of clonal haematopoiesis in 97,691 whole genomes. Nature 586, 763–768 (2020).
Genovese, G. et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 371, 2477–2487 (2014).
Jaiswal, S. et al. Age-related clonal hematopoiesis associated with adverse outcomes. N. Engl. J. Med. 371, 2488–2498 (2014).
Jaiswal, S. et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N. Engl. J. Med. 377, 111–121 (2017).
Bhattacharya, R. et al. Clonal hematopoiesis is associated with higher risk of stroke. Stroke 53, 788–797 (2022).
Weinstock, J. S. et al. Aberrant activation of TCL1A promotes stem cell expansion in clonal haematopoiesis. Nature 616, 755–763 (2023).
Yu, B. et al. Association of clonal hematopoiesis with incident heart failure. J. Am. Coll. Cardiol. 78, 42–52 (2021).
Niroula, A. et al. Distinction of lymphoid and myeloid clonal hematopoiesis. Nat. Med. 27, 1921–1927 (2021).
Desai, P. et al. Somatic mutations precede acute myeloid leukemia years before diagnosis. Nat. Med. 24, 1015–1023 (2018).
Fabre, M. A. et al. The longitudinal dynamics and natural history of clonal haematopoiesis. Nature 606, 335–342 (2022).
van der Werf, I. et al. Splicing factor gene mutations in acute myeloid leukemia offer additive value if incorporated in current risk classification. Blood Adv. 5, 3254–3265 (2021).
Zink, F. et al. Clonal hematopoiesis, with and without candidate driver mutations, is common in the elderly. Blood 130, 742–752 (2017).
Kessler, M. D. et al. Common and rare variant associations with clonal haematopoiesis phenotypes. Nature 612, 301–309 (2022).
Kar, S. P. et al. Genome-wide analyses of 200,453 individuals yield new insights into the causes and consequences of clonal hematopoiesis. Nat. Genet. 54, 1155–1166 (2022).
Uddin, M. M. et al. Germline genomic and phenomic landscape of clonal hematopoiesis in 323,112 individuals. Preprint at medRxiv https://doi.org/10.1101/2022.07.29.22278015 (2022).
Coombs, C. C. et al. Therapy-related clonal hematopoiesis in patients with non-hematologic cancers is common and associated with adverse clinical outcomes. Cell Stem Cell 21, 374–382.e374 (2017).
Bolton, K. L. et al. Cancer therapy shapes the fitness landscape of clonal hematopoiesis. Nat. Genet. 52, 1219–1226 (2020).
Dawoud, A. A. Z., Tapper, W. J. & Cross, N. C. P. Clonal myelopoiesis in the UK Biobank cohort: ASXL1 mutations are strongly associated with smoking. Leukemia 34, 2660–2672 (2020).
Bhattacharya, R. et al. Association of diet quality with prevalence of clonal hematopoiesis and adverse cardiovascular events. JAMA Cardiol. 6, 1069–1077 (2021).
Jasra, S. et al. High burden of clonal hematopoiesis in first responders exposed to the World Trade Center disaster. Nat. Med. 28, 468–471 (2022).
Mencia-Trinchant, N. et al. Clonal hematopoiesis before, during, and after human spaceflight. Cell Rep. 33, 108458 (2020).
Jakubek, Y. A., Reiner, A. P. & Honigberg, M. C. Risk factors for clonal hematopoiesis of indeterminate potential and mosaic chromosomal alterations. Transl. Res. 255, 171–180 (2023).
Robertson, N. A. et al. Longitudinal dynamics of clonal hematopoiesis identifies gene-specific fitness effects. Nat. Med. 28, 1439–1446 (2022).
Uddin, M. M. et al. Longitudinal profiling of clonal hematopoiesis provides insight into clonal dynamics. Immun. Ageing 19, 23 (2022).
Fabre, M. A. & Vassiliou, G. S. The lifelong natural history of clonal hematopoiesis and its links to myeloid malignancy. Blood 143, 573–581 (2023).
Poon, G. Y. P., Watson, C. J., Fisher, D. S. & Blundell, J. R. Synonymous mutations reveal genome-wide levels of positive selection in healthy tissues. Nat. Genet. 53, 1597–1605 (2021).
Dietlein, F. et al. Identification of cancer driver genes based on nucleotide context. Nat. Genet. 52, 208–218 (2020).
Stacey,,S. N. et al. Genetics and epidemiology of mutational barcode-defined clonal hematopoiesis. Nat. Genet. 55, 2149–2159 (2023).
Hess, J. M. et al. Passenger hotspot mutations in cancer. Cancer Cell 36, 288–301.e214 (2019).
Mitchell, E. et al. Clonal dynamics of haematopoiesis across the human lifespan. Nature 606, 343–350 (2022).
van Zeventer, I. A. et al. Evolutionary landscape of clonal hematopoiesis in 3,359 individuals from the general population. Cancer Cell 41, 1017–1031.e4 (2023).
Haring, B. et al. Healthy lifestyle and clonal hematopoiesis of indeterminate potential: results from the women’s health initiative. J. Am. Heart Assoc. 10, e018789 (2021).
Andersson-Assarsson, J. C. et al. Evolution of age-related mutation-driven clonal haematopoiesis over 20 years is associated with metabolic dysfunction in obesity. EBioMedicine 92, 104621 (2023).
Fuster, J. J. et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science 355, 842–847 (2017).
Heyde, A. et al. Increased stem cell proliferation in atherosclerosis accelerates clonal hematopoiesis. Cell 184, 1348–1361 e1322 (2021).
Rauh, M. J. Breaking the CH inflammation-expansion cycle. Blood 141, 815–816 (2023).
Avagyan, S. et al. Resistance to inflammation underlies enhanced fitness in clonal hematopoiesis. Science 374, 768–772 (2021).
Caiado, F. et al. Aging drives Tet2+/- clonal hematopoiesis via IL-1 signaling. Blood 141, 886–903 (2023).
Ferrucci, L. & Fabbri, E. Inflammageing: chronic inflammation in ageing, cardiovascular disease, and frailty. Nat. Rev. Cardiol. 15, 505–522 (2018).
Belizaire, R., Wong, W. J., Robinette, M. L. & Ebert, B. L. Clonal haematopoiesis and dysregulation of the immune system. Nat. Rev. Immunol. 23, 595–610 (2023).
Liberale, L., Montecucco, F., Tardif, J. C., Libby, P. & Camici, G. G. Inflamm-ageing: the role of inflammation in age-dependent cardiovascular disease. Eur. Heart J. 41, 2974–2982 (2020).
Psaty, B. M. et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet. 2, 73–80 (2009).
Benjamin, D. et al. Calling Somatic SNVs and Indels with Mutect2. Preprint at bioRxiv https://doi.org/10.1101/861054 (2019).
Wang, K., Li, M. & Hakonarson, H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 38, e164 (2010).
Gibson, C. J. et al. Clonal hematopoiesis associated with adverse outcomes after autologous stem-cell transplantation for lymphoma. J. Clin. Oncol. 35, 1598–1605 (2017).
Genomes Project Consortium. et al. A map of human genome variation from population-scale sequencing. Nature 467, 1061–1073 (2010). 1000.
Howie, B. N., Donnelly, P. & Marchini, J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 5, e1000529 (2009).
Mbatchou, J. et al. Computationally efficient whole-genome regression for quantitative and binary traits. Nat. Genet. 53, 1097–1103 (2021).
Magi, R. & Morris, A. P. GWAMA: software for genome-wide association meta-analysis. BMC Bioinform. 11, 288 (2010).
Uddin, M. M. et al. Long-term longitudinal analysis of 4,187 participants reveals insights into determinants of clonal hematopoiesis. Zenodo (2024). Determinants of longitudinal CHIP v1.0.0.
Acknowledgements
The Atherosclerosis Risk in Communities study has been funded in whole or in part with Federal funds from the National Heart, Lung, and Blood Institute, National Institutes of Health, Department of Health and Human Services, under Contract nos. (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700004I, HHSN268201700005I). Funding was also supported by R01HL087641, R01HL059367, and R01HL086694; National Human Genome Research Institute contract U01HG004402; and National Institutes of Health contract HHSN268200625226C. Infrastructure was partly supported by Grant Number UL1RR025005, a component of the National Institutes of Health and NIH Roadmap for Medical Research. Funding support for “Building on GWAS for NHLBI-diseases: the U.S. CHARGE consortium” was provided by the NIH through the American Recovery and Reinvestment Act of 2009 (ARRA) (5RC2HL102419). CHARGE sequencing was carried out at the Baylor College of Medicine Human Genome Sequencing Center (U54 HG003273 and R01HL086694). Funding for GO ESP was provided by NHLBI grants RC2 HL-103010 (HeartGO) and exome sequencing was performed through NHLBI grants RC2 HL-102925 (BroadGO) and RC2 HL-102926 (SeattleGO). The authors thank the staff and participants of the ARIC study for their important contributions. The authors also thank Mrs. Leslie Gaffney from the Broad Research Communication Lab for her valuable assistance in improving the display items. S.S. is supported by NIH/NHLBI T32 HL139425. A.N. was supported by funds from the Knut and Alice Wallenberg Foundation (KAW2017.0436). M.C.H. is supported by the U.S. National Heart, Lung, and Blood Institute (K08HL166687) and the American Heart Association (940166, 979465). A.G.B. is supported by a Burroughs Wellcome Foundation Career Award for Medical Scientists, the NIH Director’s Early Independence Award (DP5-OD029586), and the Pew-Stewart Scholar for Cancer Research Award, supported by the Pew Charitable Trusts and the Alexander and Margaret Stewart Trust. P.L. receives funding support from the National Heart, Lung, and Blood Institute (1R01HL134892, 1R01HL163099-01, R01AG063839, R01HL151627, R01HL157073, R01HL166538), the RRM Charitable Fund, and the Simard Fund. P.N. is supported by a Hassenfeld Scholar Award and the Paul & Phyllis Fireman Endowed Chair in Vascular Medicine from the Massachusetts General Hospital, and grants from the National Heart, Lung, and Blood Institute (R01HL148050). P.N. and B.L.E. are supported by a grant from the Fondation Leducq (TNE-18CVD04). B.L.E. is also supported by the NIH (R01HL082945, P01CA108631, and P50CA206963) and the Howard Hughes Medical Institute.
Author information
Authors and Affiliations
Contributions
M.M.U., S.S., C.M.B., and P.N. conceived and designed the study. M.M.U., A.N., A.B., B.L.E., and P.N. generated somatic mutation calls. S.S., B.Y., and C.M.B. prepared ARIC phenotypes. M.M.U. and S.S. performed bioinformatic and statistical analysis with inputs from W.E.H., S.G., K.L., A.S., M.C.H., and P.L. P.N. and C.M.B. supervised this study. M.M.U. drafted the manuscript with critical input from P.N. All authors read and provided critical revision of the manuscript.
Corresponding authors
Ethics declarations
Competing interests
M.C.H. reports research grants from Genentech, advisory board service for Miga Health, and consulting fees from Comanche Biopharma, all unrelated to the present work. P.L. is an unpaid consultant to/or involved in clinical trials for Amgen, Baim Institute, Beren Therapeutics, Esperion Therapeutics, Genentech, Kancera, Kowa Pharmaceuticals, Novo Nordisk, Novartis, Sanofi-Regeneron. P.L. is a member of the scientific advisory board for Amgen, Caristo Diagnostics, CSL Behring, DalCor Pharmaceuticals, Dewpoint Therapeutics, Eulicid Bioimaging, Kancera, Kowa Pharmaceuticals, Olatec Therapeutics, MedImmune, Novartis, PlaqueTec, Polygon Therapeutics, TenSixteen Bio, Soley Therapeutics, and XBiotech, Inc. P.L.’s laboratory has received research funding in the last 2 years from Novartis, Novo Nordisk, and Genentech. P.L. is on the Board of Directors of XBiotech, Inc. P.L. has a financial interest in Xbiotech, a company developing therapeutic human antibodies, in TenSixteen Bio, a company targeting somatic mosaicism and clonal hematopoiesis of indeterminate potential (CHIP) to discover and develop novel therapeutics to treat age-related diseases, and in Soley Therapeutics, a biotechnology company that is combining artificial intelligence with molecular and cellular response detection for discovering and developing new drugs, currently focusing on cancer therapeutics. P.L.’s interests were reviewed and are managed by Brigham and Women’s Hospital and Mass General Brigham in accordance with their conflict-of-interest policies. B.L.E. has received research funding from Novartis and Calico. He has received consulting fees from Abbvie. He is a member of the scientific advisory board and shareholder for Neomorph Inc., TenSixteen Bio, Skyhawk Therapeutics, and Exo Therapeutics, all distinct from the present work. P.N., A.G.B., and B.L.E. are scientific co-founders of TenSixteen Bio. P.N. reports research grants from Allelica, Amgen, Apple, Boston Scientific, Genentech / Roche, and Novartis, personal fees from Allelica, Apple, Astra Zeneca, Blackstone Life Sciences, Creative Education Concepts, CRISPR Therapeutics, Eli Lilly & Co, Esperion Therapeutics, Foresite Labs, Genentech / Roche, GV, HeartFlow, Magnet Biomedicine, Merck, Novartis, TenSixteen Bio, and Tourmaline Bio, equity in MyOme, Preciseli, and TenSixteen Bio, and spousal employment at Vertex Pharmaceuticals, all unrelated to the present work. C.M.B. reports grant/research support from Abbott Diagnostic, Akcea, Amgen, Arrowhead, Esperion, Ionis, Merck, New Amsterdam, Novartis, Novo Nordisk, Regeneron, Roche Diagnostic, NIH, AHA, ADA, consultation fees from Abbott Diagnostics, Alnylam Pharmaceuticals, Althera, Amarin, Amgen, Arrowhead, Astra Zeneca, Denka Seiken, Esperion, Genentech, Gilead, Illumina, Ionis, Matinas BioPharma Inc, Merck, New Amsterdam, Novartis, Novo Nordisk, Pfizer, Regeneron, Roche Diagnostic, TenSixteen Bio. The remaining authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks Tamir Chandra and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Uddin, M.M., Saadatagah, S., Niroula, A. et al. Long-term longitudinal analysis of 4,187 participants reveals insights into determinants of clonal hematopoiesis. Nat Commun 15, 7858 (2024). https://doi.org/10.1038/s41467-024-52302-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52302-9
- Springer Nature Limited