Abstract
Usutu virus (USUV) and West Nile virus (WNV) are two closely related emerging mosquito-borne flaviviruses. Their natural hosts are wild birds, but they can also cause severe neurological disorders in humans. Both viruses are efficiently suppressed by type I interferon (IFN), which interferes with viral replication, dissemination, pathogenesis and transmission. Here, we show that the replication of USUV and WNV are inhibited through a common set of IFN–induced genes (ISGs), with the notable exception of ISG20, which USUV is resistant to. Strikingly, USUV was the only virus among all the other tested mosquito-borne flaviviruses that demonstrated resistance to the 3′–5′ exonuclease activity of ISG20. Our findings highlight that the intrinsic resistance of the USUV genome, irrespective of the presence of cellular or viral proteins or protective post-transcriptional modifications, relies on a unique sequence present in its 3′ untranslated region. Importantly, this genomic region alone can confer ISG20 resistance to a susceptible flavivirus, without compromising its infectivity, suggesting that it could be acquired by other flaviviruses. This study provides new insights into the strategy employed by emerging flaviviruses to overcome host defense mechanisms.
Similar content being viewed by others
Introduction
The Orthoflavivirus genus (hereafter referred to as flavivirus for simplicity), which belongs to the Flaviviridae family, encompasses over 70 viruses, including significant arthropod-borne human pathogens like Dengue virus (DENV), Zika virus (ZIKV), yellow fever virus (YFV), and Japanese encephalitis virus (JEV)1. Flaviviruses are small, enveloped viruses, with a capped, non-polyadenylated (polyA), single‐stranded positive sense RNA genome comprising a single open reading frame (ORF) flanked by highly structured 5′ and 3′ untranslated regions (UTR). Amidst this large viral genus, Usutu virus (USUV) poses a potential threat to animal and human health2,3,4. First isolated in South Africa in 19595, USUV has since spread across much of Europe in the last two decades and is likely to continue its geographic expansion in the future2,3,4,6,7,8. Phylogenetic analyses, based on the NS5 gene sequence, reveal eight distinct lineages: Africa 1–3 and Europe 1–58,9,10. USUV belongs to the JEV serocomplex, along with West Nile virus (WNV) and other neurotropic mosquito-borne flaviviruses11. USUV and WNV share the same transmission cycle involving wild birds as reservoirs and amplifying hosts, and Culex mosquitoes as vectors. Both viruses cause major avian epidemics, leading to mass mortality in wild birds. They can also infect mammals, including humans, who cannot transmit the virus to mosquitoes but can nevertheless develop severe neurological symptoms12. To date, there is no treatment or vaccine available for WNV or USUV infections.
Similar to other viruses, the replication of USUV and WNV is efficiently impeded by type I interferons (IFN–I)13,14,15, which are cytokines released by eukaryotic cells in response to viral infection16,17,18. These cytokines have no antiviral activity per se, but act by inducing the expression of hundreds of genes known as interferon-stimulated genes (ISGs). The products of these ISGs enable the establishment of a so-called antiviral state, rendering the cells highly resistant to any viral infection16,17,18.
Considerable efforts have been invested in identifying key ISGs responsible for the antiviral activity of IFN–I against major human pathogenic viruses17,18. This has led to the identification of several human ISGs with potent antiviral effects against flaviviruses19,20,21,22,23,24,25,26, including IFI6, SHFL, and ISG20. All these antiviral cellular factors interfere with different stages of the viral life cycle. IFI6 has robust anti-flavivirus activity through inhibiting viral factory formation in the endoplasmic reticulum23,24. SHFL (also known as C19orf66, IRAV or RyDEN) is believed to bind viral RNA and to disrupt viral translation22,27. ISG20, which belongs to the DEDDh subgroup of 3′–5′ exonucleases, contains three conserved exonuclease motifs, named Exo I, II and III28,29. ISG20 selectively degrades single-stranded RNA substrates30, and has been shown to inhibit the replication of many unrelated RNA viruses, including members of the Retroviridae31,32, Orthomyxoviridae33,34, Rhabdoviridae33,35, Picornaviridae33,36 and Flaviviridae19,36,37 families. While the conventional understanding was that ISG20′s antiviral activity primarily involved the degradation of viral RNAs, recent studies have proposed an alternative mechanism wherein ISG20 may inhibit RNA translation into viral proteins35,38.
Although ISGs have been extensively studied for their interference with widely prevalent flaviviruses including DENV, ZIKV, and WNV, no such investigations have been conducted on USUV. The identification of key anti-USUV ISGs would not only extend our knowledge of the pathophysiology of the infection but would also pinpoint potential vulnerabilities in the virus life cycle, which could then be harnessed for future therapeutic strategies.
In this work, we identify USUV as the only mosquito-transmitted flavivirus whose replication is not inhibited by ISG20, thanks to a unique sequence in its 3′–UTR that confers resistance to ISG20-mediated degradation to its genome. We tested 12 known human ISGs for their antiviral activity on USUV and report that USUV replication is sensitive to the same ISGs as WNV, with the notable exception of ISG20. Surprisingly, we found that USUV is the only mosquito-borne flavivirus tested to be resistant to ISG20-mediated restriction. We used various methods to show that the genome of USUV is intrinsically resistant to the ISG20-mediated degradation, and that this resistance is conferred by a specific domain, known as Dumbbell 2 (DB2), within its 3′–UTR. Interestingly, the replacement of WNV DB2 by that of USUV was sufficient to confer resistance to ISG20-mediated degradation, without affecting infectivity, suggesting that other flaviviruses may acquire resistance to this IFN-induced exonuclease. Altogether, our study demonstrates that the antiviral activity of ISG20 is primarily driven by its capacity to degrade viral RNA in the context of flavivirus infections and provide evidence that certain secondary structures, such as internal stem-loops, are key to resisting ISG20-mediated degradation.
Results
USUV is the only mosquito-borne flavivirus resistant to ISG20-mediated inhibition
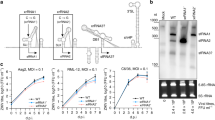
To assess the sensitivity of USUV to human ISGs, we performed a mini expression screen on 12 ISGs that are known to inhibit the replication of most flaviviruses, i.e. EIF2AK2 (also known as PKR), IFI6, IFIT1, SHFL, SLFN11, TRIM56, ZC3HAV1 (also known as ZAP), ISG20, IFITM1–3, and RSAD2 (also known as Viperin)19,20,21,22,23,24,25,39,40. The assay is based on the use of a bicistronic lentiviral vector, named SCRPSY, co-expressing an ISG and the red fluorescent protein TagRFP (Fig. 1A), as previously described20,41,42,43,44. HEK 293 T cells were first transduced with the ISG-encoding SCRPSY lentivectors at a dilution leading to around 50% of transduced cells (i.e. 1 TCID50), then challenged for 48 h with 1 TCID50 of wild-type USUV Africa 2 (AF2) or WNV lineage 2 (L2). Viral replication in TagRFP-negative and TagRFP-positive cells was assessed by flow cytometry, using a pan-flavivirus antibody (Fig. 1A). As expected, given that USUV and WNV are closely related, both viruses showed similar susceptibility to all ISGs, with the notable exception of ISG20 (Fig. 1B). Indeed, WNV replication was efficiently inhibited by ISG20, as previously described19, whereas USUV replication remained unaffected.
A Organization of the SCRPSY lentivector and graphical representation of the transient expression mini-screen and experimental procedure. B Results of the transient expression mini-screen in HEK 293 T cells, infected with WNV L2 or USUV AF2. Each dot represents one ISG. Shading highlights genes with or without observed antiviral activity. ISG20 is indicated with red dots. C Flow cytometry analysis of Vero E6, A549, HeLa and Huh7.5 cells overexpressing ISG20 and infected with WNV L2 or USUV AF2 (MOI of 1) for 48 h. An empty SCRPSY vector was used as a control. D Experimental procedure to generate stable Vero E6 and HEK 293 T cells expressing IFI6, SHFL, ISG20, and empty SCRPSY vector. E Flow cytometry analysis of TagRFP expression in stable Vero E6 cells. F Western blot analysis of IFI6, SHFL, and ISG20 expression in stable HEK 293 T cells. Molecular weight markers (in kD) are indicated. G Phylogenetic tree of the viral strains used in this study. The protein sequences of the indicated viruses were aligned with MUSCLE, refined with BMGE and tree was generated with PhyML. H Vero E6 cells stably expressing IFI6, SHFL or ISG20 were infected with four serotypes of DENV and two strains of ZIKV (MOI of 1). HEK 293 T cells stably expressing IFI6, SHFL or ISG20 were infected with two WNV lineages and six USUV lineages (MOI of 1). Infection was assessed by flow cytometry at 48 h post-infection. I Data are presented as individual biological replicates and mean ± SD, except in panel B where dots represent the mean of two biological replicates. ***, P ≤ 0.001; **, P ≤ 0.01; ns, P > 0.05 (one-way ANOVA followed by Tukey’s HSD post hoc). Exact P-values are provided in Supplementary Table 7. Abbreviations: LTR, long terminal repeat; PAC, puromycin N-acetyltransferase; TagRFP, red fluorescent protein. Source data are provided as a Source Data file.
To ensure that this was not a cell type-specific phenomenon, we compared WNV and USUV replication in four other cell types (i.e. Vero E6, A549, HeLa and Huh7.5) overexpressing ISG20 or not. In all cell types, ISG20 was able to inhibit WNV L2 replication but not that of USUV AF2 (Fig. 1C), thus demonstrating that USUV resistance to ISG20 is not cell-type specific.
Next, we sought to verify whether the observed phenotype was specific to the viruses and strains we used, or if it was a general phenotype. To this end, we generated stable cell lines expressing ISG20 from Vero E6 or HEK 293 T cells transduced with the corresponding SCRPSY lentivectors (Fig. 1D). As controls, two well-characterized ISGs, IFI6 and SHFL, known for their broad anti-flavivirus activity, were included in this experiment22,23,24,27. Transgene expression was confirmed by flow cytometry (Fig. 1E and Supplementary Fig. 1) and Western blot (Fig. 1F). We then compared the susceptibility to ISG20 of several strains of WNV and USUV along with two other phylogenetically related mosquito-borne flaviviruses: DENV and ZIKV. As we noticed that DENV and ZIKV preferably infect Vero E6 cells and already ruled out a cell-specific effect of resistance to ISG20 (Fig. 1C), these cells were used for DENV and ZIKV infections, while HEK 293 T cells were used for WNV and USUV infections.
Cells were challenged with 14 different mosquito-borne flaviviruses representative of four DENV serotypes (serotypes 1–4), the Asian lineage of ZIKV, the two lineages of WNV (lineage 1 and 2), and six different lineages of USUV (Africa 2 and 3, Europe 1–3 and 5) (Fig. 1G). As expected, all tested viruses were efficiently inhibited by IFI6 and SHFL (Fig. 1H). Strikingly, ISG20 was able to block the replication of all flaviviruses we tested, with the notable exception of USUV (Fig. 1H). Indeed, apart from USUV EU5, which corresponds to a minor lineage identified in a few birds in Germany10, all USUV strains circulating in Africa or Europe proved resistant to ISG20, including AF2, AF3, EU1, EU2 and EU38,9,10. The fact that most USUV lineages show resistance to ISG20 rules out the possibility that this is a strain-specific phenotype, therefore implying that the resistance to ISG20 is an intrinsic, distinctive and conserved property of USUV.
ISG20 is an effector of the antiviral activity of IFN–I against WNV, but not USUV
To investigate the molecular basis for the differential susceptibility of flaviviruses to ISG20, we selected a sensitive virus (WNV L2) and a resistant one (USUV AF2) for further experiments. We first evaluated the antiviral activity of the endogenous protein, following its induction by IFN–I. To select a suitable cell model for these experiments, different cell lines (i.e. HEK 293 T, A549, HT1080 and Huh7.5) were treated with IFN–I and the expression of several ISGs, namely MX1, MX2, IRF7, IFI6, SHFL and ISG20 was assessed by RT-qPCR at 8 h post-treatment (Fig. 2A). Among the treated cells, A549 cells were the most responsive to IFN–I stimulation, notably leading to a significant induction of IFI6, SHFL and ISG20, as shown by RT-qPCR (Fig. 2A), and confirmed by Western blot (Fig. 2B). Therefore, A549 cells were selected as a suitable model for further analyses.
A RT-qPCR analysis of the relative expression of ISGs in HEK 293 T, A549, HT1080 and Huh7.5 cells treated with 100 U/mL of IFN–I for 16 h. Results are presented as a heatmap with log2 of fold-change (2–ΔΔCt). B Western blot analysis of IFI6, SHFL, and ISG20 in A549 cells treated with increasing concentrations of IFN–I for 16 h. Molecular weight markers (in kD) are indicated. C Experimental procedure of the siRNA-mediated knockdown of IFI6, SHFL, and ISG20 in A549 cells followed by IFN–I treatment. D RT-qPCR validation of the siRNA-mediated knockdown of IFI6, SHFL, and ISG20 in A549 cells transfected for 24 h and treated with 100 U/mL of IFN–I for 16 h. siRNA targeting PPIB was used as a control. Data are presented as fold-change (2–ΔΔCt) compared to untreated cells. E RT-qPCR analysis of PPIDKD, IFI6KD, SHFLKD, and ISG20KD A549 cells treated with 100 U/mL of IFN–I for 16 h and infected with WNV L2 or USUV AF2 (MOI of 1) for 24 h. Data are presented as normalized fold-change (2–ΔΔCt) compared to uninfected cells. F Experimental procedure of the CRISPR/Cas9 knockout of ISG20 in A549 cells followed by IFN–I treatment and schematic representation of the resulting deletion of Exo II domain of ISG20. G Western blot analysis of ISG20 CRISPR/Cas9 knockout in A549 cells. A549 expressing non-targeting (CTR) or ISG20-specific (ISG20KO) guide RNA were treated with 100 U/mL of IFN–I for 16 h, before protein extraction. Molecular weight markers (in kD) are indicated. H Flow cytometry analysis of control and ISG20KO A549 cells treated with 100 U/mL of IFN–I for 16 h, and infected with WNV L1 or USUV AF2 (MOI of 1) for 48 h. Data are presented as individual biological replicates and mean ± SD. ***, P ≤ 0.001; ns, P > 0.05 (two-way ANOVA followed by Tukey’s HSD post hoc; data were log-transformed prior to analysis [E]). Exact P-values are provided in Supplementary Table 7. Abbreviations: NT, untreated; KD, knockdown; KO, knockout. Source data are provided as a Source Data file.
To evaluate the respective contribution of IFI6, SHFL and ISG20 in WNV and USUV restriction by IFN–I, A549 cells were transfected with irrelevant (i.e. targeting PPIB) or specific siRNAs, and treated or not with IFN–I (Fig. 2C). The knockdown (KD) efficiency was confirmed by RT-qPCR analyses showing that siRNAs targeting IFI6, SHFL, and ISG20 transcripts effectively decreased the expression of the corresponding genes upon IFN–I stimulation (Fig. 2D). Then, the cells were infected for 24 h with WNV L2 or USUV AF2 at a MOI of 1 and viral RNA yields were evaluated by RT-qPCR.
In control PPIBKD cells, IFN–I blocked the replication of both viruses (Fig. 2E), with USUV being more sensitive than WNV, as previously described15. As expected, in IFI6KD and SHFLKD cells, the antiviral effect of IFN–I was partially reversed (Fig. 2E). The KD of IFI6 led to the most efficient rescue in viral RNA quantity, demonstrating that this ISG is the main effector of the antiviral activity of IFN–I for both viruses (Fig. 2E). In ISG20KD cells, on the other hand, WNV replication was partially restored, but not USUV replication (Fig. 2E), thus demonstrating that, unlike WNV, ISG20 is not an effector of the antiviral effect of IFN–I against USUV.
This observation was further confirmed by a knockout (KO) approach, in which we generated ISG20KO A549 cells by CRISPR/Cas9, leading to the deletion of the Exo II domain of ISG20 (Fig. 2F and Supplementary Fig. 2), thus abolishing functional expression (Fig. 2G). Wild-type or ISG20KO cells were infected for 48 h with WNV L2 or USUV AF2 at a MOI of 1 and infection was evaluated by flow cytometry. We showed again that WNV and USUV equally infected control and ISG20KO cells in the absence of IFN–I, and that USUV replication was more sensitive to IFN–I than WNV (Fig. 2H). Moreover, WNV replicated significantly better in ISG20KO cells than in control cells in the presence of IFN–I, whereas USUV replication was inhibited, irrespective of the ISG20 expression status (Fig. 2H). Altogether, these results confirm that USUV is resistant to ISG20-mediated restriction, while WNV is not, regardless of whether ISG20 expression occurs by exogenous overexpression or by IFN–I endogenous induction.
The USUV genome is intrinsically resistant to ISG20-mediated degradation
Although ISG20 exonuclease activity is well characterized and its ability to degrade viral RNAs in vitro has been formally demonstrated30,31,32,33,45,46,47, it remains debated whether ISG20 exerts its antiviral activity by degrading or inhibiting translation of viral RNAs35,38.
To assess whether ISG20 was capable of degrading viral genomic RNA, HEK 293 T cells were infected for 24 h with WNV L2 or USUV AF2 at a MOI of 1. Total RNA, purified from infected cells was incubated with or without recombinant wild-type human ISG20 (rhISG20WT) for 1 h at 37 °C (Fig. 3A). As a control, a catalytically inactive32,35 mutant of ISG20 (rhISG20D94A) was also included in this experiment, in order to ensure that RNA degradation was not caused by non-specific RNase activity present in RNA or protein purifications. Following RNA incubation with or without rhISG20WT or rhISG20D94A, residual cellular (i.e. RPL13A, ACTB, and B2M housekeeping gene transcripts) and viral RNAs were quantified by RT-qPCR analyses using primers targeting the 3′–UTR of the two viruses, as this region would be the first to be degraded by ISG20. Ex cellula, ISG20 degrades all RNAs with no distinction between self and non-self transcripts30. Indeed, incubation with rhISG20WT resulted in a significant decay of both cellular and viral RNAs (Fig. 3B). In contrast, rhISG20D94A did not induce RNA degradation, thus ruling out the presence of unspecific RNase activity. Interestingly, USUV RNA was degraded with the same efficiency as housekeeping gene transcripts following incubation with rhISG20WT, whereas WNV RNA showed more extensive degradation (Fig. 3B). This greater sensitivity of WNV to ISG20-mediated degradation is better highlighted using a 2-ΔΔCt analysis, i.e. by measuring the amount of viral RNA relative to the housekeeping genes (Fig. 3C). These results suggest that USUV RNA is intrinsically more resistant to the exonuclease activity of ISG20 than that of WNV.
A Experimental procedure for in vitro ISG20 degradation (exonuclease assay) of total RNA extracts. B RT-qPCR analysis of exonuclease assay of total RNA extracted from HEK 293 T cells infected with WNV L2 or USUV AF2 (MOI of 1) for 24 h and treated with rhISG20 for 1 h. Data are presented as the percentage of residual RNA (2–ΔCt) relative to untreated samples. Catalytically inactive ISG20 mutant (D94A) was used as a control. C Specific ISG20 degradation of WNV and USUV RNA from panel B. The relative amount of RNA was normalized to the geometric mean of three housekeeping genes (RPL13A, ACTB and B2M), following the 2–ΔΔCt method. Data are presented as a percentage of the relative amount of RNA compared to untreated samples. D Positive regulation of ISG20 degradation of HBV RNA through the recruitment of YTHDF2 to m6A modifications and predicted m6A sites on the 3′–UTR of WNV L2 and USUV AF2. E Negative regulation of ISG20 degradation of HIV-1 RNA through steric hindrance of ISG20 induced by 2′-O-Me modifications and predicted 2′-O-Me sites on the 3′–UTR of WNV L2 and USUV AF2. F Experimental procedure and Western blot analysis of IS20KO A549 cells transduced with lentivectors coding for wild-type or mutant Flag-tagged ISG20 constructions. A lentivector coding for GFP was used as a control. Molecular weight markers (in kD) are indicated. G Flow cytometry analysis of control, wild-type or mutant ISG20 complemented IS20KO A549 cells infected with WNV L2 or USUV AF2 (MOI of 1) for 48 h. H Experimental procedure and Western blot analysis of HEK 293 T cells transfected with wild-type or mutant ISG20 constructions for 48 h. Molecular weight markers (in kD) are indicated. I Flow cytometry analysis of control, wild-type or mutant ISG20 expressing HEK 293 T cells infected with WNV L2 or USUV AF2 (MOI of 1) for 48 h. Data are presented as individual biological replicates and mean ± SD. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05 (two-way ANOVA followed by Tukey’s HSD post hoc [B, G, I], or one-way ANOVA followed by Tukey’s HSD post hoc [C]; data were log-transformed prior to analysis [B]). Exact P-values are provided in Supplementary Table 7. Abbreviations: rhISG20, recombinant human ISG20; NT, untreated; HBV, hepatitis B virus; HIV, human immunodeficiency virus. Source data are provided as a Source Data file.
The mechanism by which ISG20 degrades viral RNAs while sparing cellular RNAs is not fully understood. While polyA tails and polyA-binding proteins likely protect cellular mRNAs47, other features on the 3′ end of the RNA, including secondary structures like stem-loops30,46, and RNA methylations, might influence RNA susceptibility to the exonuclease activity of ISG20. In this respect, ISG20 was shown to specifically degrade N6-methyladenosine (m6A) modified Hepatitis B Virus (HBV) transcripts through recruitment by YTHDF248 (Fig. 3D), and we recently demonstrated that internal 2′-O-methylations (2′-O-Me) of the HIV-1 genome conferred resistance to ISG2032 (Fig. 3E).
Considering the role of RNA methylations in ISG20 activity and the presence of m6A and 2′-O-Me modifications in the genomes of ZIKV49 and DENV50, respectively, we investigated whether the genomes of WNV and USUV had different methylation profiles, which might explain their different sensitivity to ISG20. We focused on the 3′–UTR of the two viruses, since this is the first region to undergo degradation by ISG20. In silico analysis revealed a conserved m6A profile (Fig. 3D) but divergent 2′-O-Me sites distribution (Fig. 3E) between WNV L2 and USUV AF2 genomes, therefore making 2′-O-Me a prime candidate for USUV resistance to ISG20.
To investigate whether USUV, like HIV-1, can evade ISG20 through internal 2′-O-Me within its genome, we took advantage of an ISG20 mutant (R53A/D90A) that we recently described as being able to degrade RNA regardless of the presence of 2′-O-Me32. We also included in our experimental procedure the catalytically inactive mutant of ISG20 (D94A) as a control32,35. The antiviral activity of wild-type ISG20 and both mutants was assessed by reintroducing each protein into the previously generated ISG20KO A549 cells (Fig. 2F) using lentiviral vectors (Fig. 3F). Once ISG20 expression was confirmed by Western blot analysis (Fig. 3F), cells were infected for 48 h with WNV L2 or USUV AF2 at a MOI of 1 and infection was assessed by flow cytometry. WNV replication was efficiently inhibited by wild-type ISG20 addback, while USUV remained unaffected (Fig. 3G). As expected, the inactive D94A mutant ISG20 showed no antiviral activity (Fig. 3G). In contrast, the R53A/D90A double-mutant ISG20 inhibited WNV but not USUV (Fig. 3G), suggesting that USUV resistance to ISG20 is not due to 2′-O-Me that might be found within its 3′–UTR.
To validate these findings, we replicated the experiments in HEK 293 T cells that were transfected with plasmids expressing wild-type or mutant ISG20 (Fig. 3H) and infected for 48 h with WNV L2 or USUV AF2 at a MOI of 1. Under these conditions, we observed a similar phenotype as in ISG20KO A549 cells expressing the different mutants of ISG20, although in this new condition, USUV was not entirely resistant to wild-type ISG20 overexpression, presumably because of the higher expression of ISG20 due to transient transfection (Fig. 3I). Once again, neither virus was affected by expression of the catalytically inactive D94 A mutant, while only WNV was inhibited by the R53A/D90A double-mutant (Fig. 3I). Altogether, these results confirm that RNA methylation is unlikely to contribute to the USUV resistance to ISG20. They also provide definitive proof that the anti-flaviviral activity of ISG20 is dependent on its exonuclease activity.
The USUV 3′–UTR is intrinsically resistant to ISG20-mediated degradation
ISG20 degrades RNAs from their 3′ end28,30. Given that flavivirus genomic RNA lacks a polyA tail but have a highly structured 3′–UTR51,52,53,54,55, we sought to determine whether USUV 3′–UTR could resist ISG20-mediated degradation. To this end, we used in vitro transcription to generate unmodified 3′–UTRs of WNV L2 and USUV AF2, and evaluated their susceptibility to ISG20-mediated degradation by incubating them with varying concentrations of wild-type rhISG20WT. We then quantified the amount of residual RNA by RT-qPCR (Fig. 4A and Supplementary Fig. 3). Our findings showed that USUV 3′–UTR exhibited significantly greater stability at all rhISG20WT concentrations (<500 nM), compared to WNV 3′–UTR (Fig. 4B). This relative resistance of USUV 3′–UTR compared to that of WNV can also be observed directly on an agarose gel, using RNase A as a positive control for RNA degradation (Fig. 4C). Similarly, treatment with rhISG20WT in a time course experiment confirmed the slower decay of USUV 3′–UTR compared to WNV 3′–UTR (Fig. 4D). Again, no unspecific RNase activity was observed with the catalytically inactive rhISG20D94A (Fig. 4D). Using higher concentrations of rhISG20WT (2.5 μM), we were able to observe USUV 3′–UTR degradation beginning at 15 min of incubation. Interestingly, after 1 h, an ISG20-resistant digestion product was detected, approximately 100 nucleotides lighter than the template (Fig. 4E).
A Experimental procedure for exonuclease assay on in vitro transcribed WNV and USUV 3′–UTRs. B Exonuclease assay on 25 ng of in vitro transcribed of WNV and USUV 3′–UTRs incubated with indicated concentrations of rhISG20WT for 1 h. RNA decay was assessed by RT-qPCR. Data are presented as absolute copies of 3′–UTR. C Exonuclease assay performed on 1 µg of in vitro transcribed WNV and USUV 3′–UTRs incubated with 3 µM of rhISG20WT or 1 µg/mL RNase A. RNA decay was assessed by agarose gel electrophoresis. Molecular weight markers (nucleotides) are indicated. D Exonuclease assay performed on 1 µg of in vitro transcribed WNV and USUV 3′–UTRs incubated with 500 nM of rhISG20WT. The catalytically inactive ISG20 mutant (D94A) was used as a control. RNA decay was assessed by agarose gel electrophoresis. Molecular weight markers (nucleotides) are indicated. E Exonuclease assay performed on 1 µg of in vitro transcribed USUV 3′–UTR incubated with either 500 nM or 2.5 µM of rhISG20WT. RNA decay was assessed by agarose gel electrophoresis. Molecular weight markers (nucleotides) are indicated. F Genomic organization of WNV, 3′–WNV replicon and chimeric 3′–USUV replicon. G Immunofluorescence analysis of the expression of GFP in HEK 293 T cells infected with 3′–WNV or 3′–USUV RVPs (1 TCID50) for 48 h. Scale bar: 50 µm. H Flow cytometry analysis of GFP expression in HEK 293 T cells infected with 3′–WNV or 3′–USUV RVPs for 48 h (MOI of 1). Mean fluorescence intensity (MFI) was measured on the infected cells. I HEK 293 T cells stably expressing IFI6, SHFL or ISG20 were infected with 3′–WNV or 3′–USUV RVPs for 48 h (MOI of 1). Infection was assessed by flow cytometry. J RT-qPCR analysis of rhISG20WT exonuclease assay of total RNA extracted from HEK 293 T cells infected with 3′–WNV or 3′–USUV RVPs for 24 h and treated with rhISG20WT for 1 h. Data are presented as the percentage of residual RNA (2–ΔCt) relative to untreated samples. K Specific ISG20 degradation of 3′–WNV or 3′–USUV RVPs RNA from panel H. The relative amount of RNA was normalized to the geometric mean of three housekeeping genes (RPL13A, ACTB and B2M), following the 2–ΔΔCt method. Data are presented as a percentage of the relative amount of RNA compared to untreated samples. Data are presented as individual biological replicates and mean ± SD. ***, P ≤ 0.001; ns, P > 0.05 (two-way ANOVA followed by Tukey’s HSD post hoc [B, I, J], Two-tailed Mann-Whitney [H], one-way ANOVA followed by Tukey’s HSD post hoc [K]). Exact P-values are provided in Supplementary Table 7. Abbreviations: rhISG20, recombinant human ISG20; NT, untreated; RVP, reporter virus particle; MFI, mean fluorescence intensity. Source data are provided as a Source Data file.
These results show that USUV 3′–UTR is intrinsically resistant to ISG20-mediated degradation without the contribution of any external factors or post-transcriptional modifications. They also suggest that the ISG20-resistant domain may lie around 100 nucleotides upstream of the 3′ end of USUV genome.
The USUV 3′–UTR confers resistance to ISG20-mediated degradation
Next, we sought to evaluate whether the USUV 3′–UTR would confer ISG20 resistance to a sensitive virus. To test this hypothesis, we constructed a chimeric replicon of WNV by switching the 3′–UTR of an eGFP-encoding WNV L2 replicon with that of USUV AF2 (Fig. 4F).
The wild-type WNV L2 reporter virus particles (RVPs) and the resulting chimeras (referred to as 3′–WNV and 3′–USUV RVPs, respectively) were generated by co-transfecting HEK 293 T cells with the respective replicons and a plasmid encoding WNV structural proteins. Both the parental and chimeric 3′–USUV RVPs replicated efficiently in HEK 293 T cells, as evidenced by fluorescent microscopy (Fig. 4G) and flow cytometry (Fig. 4H). Additionally, both were potently suppressed when IFI6 or SHFL were overexpressed in HEK 293 T (Fig. 4G). However, distinct behavior was observed upon ISG20 overexpression, since the replication of 3′–WNV RVPs was inhibited by ISG20, whereas the chimeric 3′–USUV RVPs exhibited complete resistance (Fig. 4I).
This finding was further explored by assessing the sensitivity of replicon RNAs to ISG20-mediated degradation. For this, total RNAs were extracted from HEK 293 T cells infected with either the 3′–WNV or 3′–USUV RVPs and were subject to wild-type rhISG20 degradation. Once again, the chimeric 3′–USUV replicon displayed similar degradation to housekeeping gene transcripts, whereas the parental 3′–WNV replicon was more susceptible (Fig. 4J). The protective role provided by USUV 3′–UTR to the WNV reporter RNA was further supported by comparing the amount of viral RNA relative to housekeeping gene expression (Fig. 4K).
Together, our results demonstrate that USUV resistance to ISG20 is conferred by its 3′–UTR. Furthermore, and importantly, this resistance can also be transferred to an ISG20-susceptible virus.
USUV DB2 is responsible for USUV 3′–UTR resistance to ISG20
The 3′–UTR of mosquito-borne flaviviruses is highly structured51,52,53,54,55, consisting of three autonomously folded domains (Fig. 5A). Domain 1, which is located just downstream of the stop codon, is the longest and most variable, featuring simple and branched stem-loop (SL) structures53. Domain 2 is comparatively more conserved and contains one or two dumbbell (DB) structures (DB1 and DB2)52. In addition to these secondary structures, SL and DB have been shown to form pseudoknots (PKs), thus conferring an even more complex 3D structure56,57,58. Domain 3 is the shortest and most conserved, and consists of a small hairpin (sHP) followed by a terminal 3′SL structure which is particularly stable. This domain is crucial for viral replication as it allows the circularization of the genome52,55. Collectively, the different domains within the 3′–UTR play a vital role in the life cycle of flaviviruses in both mosquitoes and mammals.
A Canonical secondary structures of the flavivirus 3′–UTR, based on WNVKunj54. B Configurations of the USUV 3′–UTR domains inserted in place of the corresponding parental domains within the 3′–UTR of the WNV replicon. C HEK 293 T cells stably expressing IFI6, SHFL or ISG20 were infected for 48 h with WNV or chimeric RVPs described in panel B (MOI of 1). Infection was assessed by flow cytometry. D RT-qPCR analysis of rhISG20WT exonuclease assay of total RNA extracted from HEK 293 T cells infected for 24 h with WNV or chimeric RVPs described in panel B (MOI of 0.1) and treated with rhISG20WT for 1 h. The relative amount of RNA was normalized to the geometric mean of three housekeeping genes (RPL13A, ACTB and B2M), following the 2–ΔΔCt method. Data are presented as a percentage of the relative amount of RNA compared to untreated samples. E Secondary structure diagrams of WNV and USUV DB2 fragments, obtained by hSHAPE on the full-length 3′–UTR of WNV and USUV (presented in Supplementary Fig. 4) with overlay of hSHAPE reactivities. Position numbering starts at +1 after stop codon. Arrowheads indicate the region further analyzed in the exonuclease assay shown in panel I. The black lines indicate the nucleotides involved in pseudoknot formation (PK), while the green shaded lines show the complementary sequence (CS) present in the regions flanking USUV DB2. F Genomic organization of USUV AF2 DB2 and USUV EU5 DB2 regions inserted in place of parental DB2 within the 3′–UTR of the WNV replicon. G HEK 293 T cells stably expressing ISG20 were infected for 48 h with WNV or chimeric RVPs described in panel F (MOI of 1). Infection was assessed by flow cytometry. H RT-qPCR analysis of rhISG20WT exonuclease assay of total RNA extracted from HEK 293 T cells infected for 24 h with WNV or chimeric RVPs described in panel F (MOI of 0.1) and treated with rhISG20WT for 1 h. The relative amount of RNA was normalized to the geometric mean of the three housekeeping (RPL13A, ACTB and B2M), following the 2–ΔΔCt method. Data are presented as a percentage of the relative amount of RNA compared to untreated samples. I Experimental procedure for exonuclease assay on 5′ end 32P-radiolabelled WNV L2 and USUV AF2 DB2 RNA fragments incubated with rhISG20 and electrophoretic analysis. The RNA sequences used are indicated between the arrowheads in panel E and are also described in the Materials and methods section. Template sizes (nucleotides) are indicated. J Quantification of the specific WNV L2 DB2 and USUV AF2 DB2 RNA substrate 3′ end excision by rhISG20. K Proposed model for USUV escape to ISG20-mediated degradation. Data are presented as individual biological replicates and mean ± SD. ***, P ≤ 0.001; **, P ≤ 0.01; *, P ≤ 0.05; ns, P > 0.05 (two-way ANOVA followed by Tukey’s HSD post hoc). Exact P-values are provided in Supplementary Table 7. Abbreviations: SL, stem-loop; DB, dumbbell; PK, pseudoknot; CS, complementary sequence; sHP, short hairpin; rhISG20, recombinant human ISG20; ND, no data. Source data are provided as a Source Data file.
In addition to USUV’s unique resistance to ISG20, it is important to note that the 3′–UTR of flaviviruses is notoriously resistant to the 5′−3′ exonuclease XRN1. Indeed, the incomplete degradation of viral RNA by XRN1, which stalls on the folded RNA structures present in the 3′–UTRs, generates small 3′–UTR-derived non-coding RNAs, which are known as subgenomic flaviviral RNAs (sfRNAs)59,60,61,62. Numerous studies have focused on the structure and functions of sfRNAs, which play multiple roles in facilitating viral pathogenicity63, including by inhibiting the IFN response in target cells64.
Since we determined that USUV resistance to ISG20 was conferred by its 3′–UTR, we sought to determine whether one of its 3 domains was specifically responsible for this phenotype. To do this, we engineered chimeric WNV L2 replicons containing progressively shorter segments of the 3′–UTR of USUV. The chimeras generated included either all three domains of USUV 3′–UTR (USUV D1/D2/D3, i.e. the previously described 3′–USUV), the last two domains (USUV D2/D3), or only the last domain (USUV D3). The resulting chimeric RVPs were then compared to the wild-type WNV RVPs (Fig. 5B).
As expected, overexpression of IFI6 or SHFL effectively blocked the infection of all RVPs (Fig. 5C), and overexpression of ISG20 inhibited WNV RVPs but not the chimeric USUV D1/D2/D3 RVPs (Fig. 5C), consistent with our previous findings (Fig. 4I). The chimeric USUV D2/D3 RVPs retained this resistance, whereas the USUV D3 RVPs were inhibited by ISG20 overexpression (Fig. 5C). This finding was once again confirmed by assessing the sensitivity of replicon RNAs extracted from RVP-infected HEK 293 T to rhISG20-mediated degradation (Fig. 5D). Taken together, these results therefore indicate that resistance to ISG20 is primarily mediated by the domain 2 of the USUV 3′–UTR, in agreement with the block located around 100 nucleotides upstream of the 3′ end of the USUV 3′–UTR suggested by our previous observations (Fig. 4E).
We then used benzoyl cyanide (BzCN) to analyze by high-throughput selective 2′–hydroxyl acylation analyzed by primer extension (hSHAPE) the detailed secondary structure of USUV and WNV 3′–UTR RNAs. We used our previously generated 3′–UTR chimeric replicons (Fig. 4F) as templates for consistency with our functional investigations. SHAPE experiments interrogate RNA structural dynamics using electrophilic reagents that preferentially acylate the 2′–hydroxyl (2′OH) ribose group of flexible nucleotides that most often correspond to single-stranded RNA65,66. 2′OH acylation leads to a block to reverse transcription that can be quantified using fluorescent oligonucleotides on a capillary electrophoresis device67. First, we analyzed the SHAPE reactivities of WNV and USUV 3′–UTR RNAs and showed that the deduced secondary structure models (Supplementary Fig. 4) are rather consistent with previous flavivirus 3′–UTR models (Fig. 5A) and consists of three autonomously folded domains51,52,53,54,55,59.
Based on their homology to sequences in other mosquito-borne flaviviruses, several nucleotides in the large bulge of SL1 (WNV 89 – 95) and in the lower stem of SL1 (WNV 113 – 119) are predicted to participate in the formation of an RNA pseudoknot (PK). In our conditions, these positions react with BzCN (Supplementary Fig. 4) arguing against the formation of a stable PK60,68. However, such a structure may be transient or unstable under the probing conditions. Domain 2 is a conserved region and our chemical data analyses suggest the presence of the DB structures (DB1 and DB2) and the formation of the two PK motifs: for WNV, PKs involve nucleotides 335 – 339 (DB1) and 411 – 415 (DB2) which base pair with nucleotides 461 – 435 and 473 – 477, respectively; and for USUV, PKs involved nucleotides 410 – 414 (DB1) and 490 – 494 (DB2) which base pair with nucleotides 542 – 546 and 549 – 553, respectively (Fig. 5E). Finally, our chemical probing data concerning domain 3 are consistent with the formation of a previously identified stem-loop structure and small upstream hairpin at the 3′ terminus of the WNV and USUV RNAs.
These hSHAPE data did not identify any notable differences in the RNA conformation of domain 2 between USUV and WNV besides a complementary sequence (CS) consisting of four nucleotides on either side of USUV DB2 that is absent in the WNV 3′–UTR (Fig. 5E). This suggests that USUV DB1 and/or DB2 may adopt a distinctive 3D structure that is resistant to ISG20-mediated degradation. Given that DB2 is the first that ISG20 will encounter and has the additional CS, we investigated whether this structure would be sufficient to confer WNV resistance to ISG20. We therefore replaced the DB2 of the WNV L2 replicon with that of USUV AF2 (Supplementary Fig. 5 and Fig. 5F). The phenotype of this chimeric replicon was assessed in HEK 293 T overexpressing ISG20 (Fig. 5G), and with rhISG20 treatment of total RNA extracted from infected cells (Fig. 5H). As a control, we also assessed the ISG20 sensitivity of a WNV L2 replicon containing the ISG20-sensitive USUV EU5 DB2 (Supplementary Fig. 5 and Fig. 5F). Systematically, the insertion of USUV AF2 DB2 resulted in significant resistance to ISG20, whereas that of USUV EU5 DB2 did not (Fig. 5G, H), indicating that USUV AF2 DB2 is sufficient to confer resistance to ISG20.
To confirm these observations and visualize this difference in susceptibility to ISG20-mediated degradation between WNV L2 and USUV AF2 DB2s, radiolabeled synthetic RNAs corresponding to the DB2 regions of WNV L2 (i.e. nucleotides 386 – 458, in-between arrowheads on Fig. 5E) and USUV AF2 (i.e. nucleotides 461 – 542, in-between arrowheads on Fig. 5E), were incubated with rhISG20 and their decay was analyzed on urea-PAGE in a time course experiment (Fig. 5I). As a control, we aimed to treat the sHP/3′SL sequences of both viruses with rhISG20, but this domain was barely degraded in our experimental conditions (Supplementary Fig. 6). The level of both structured RNA substrates decreased over time, as expected (Fig. 5I, J). However, while WNV L2 DB2 substrate decayed linearly, USUV AF2 DB2 exhibited a distinct pattern with intermittent halts. For instance, after 5 minutes, the USUV AF2 DB2 substrate lightened by a few nucleotides, as shown with a black arrowhead (Fig. 5I), suggesting that ISG20 initiated degradation but paused after a short sequence. After 15 minutes and beyond, the differences became more pronounced: USUV AF2 RNA 3′ end excision consistently lagged behind that of WNV DB2 (Fig. 5J). These findings further confirm that USUV AF2 DB2 is intrinsically more resistant to ISG20-mediated degradation than its WNV L2 counterpart.
Altogether, our results demonstrate that the main determinant of USUV resistance to ISG20 is the DB2 structure located in the 3′–UTR of its genome.
Discussion
Although USUV has been described to be sensitive to IFN13,14,15, no studies have yet identified which ISGs inhibit its replication. To investigate this, we tested the main ISGs that are known to inhibit WNV replication, given the close phylogenetic relationship between the two viruses. As expected, most of these ISGs were effective in inhibiting USUV replication, including IFI6, IFIT1, and SHFL. Surprisingly, neither RSAD2 (Viperin) nor IFITMs were found to block WNV or USUV infection in our mini-screen, despite their well-documented anti-flaviviral activity19,20,39,40. It is possible that the use of wild-type viruses instead of reporter virus-like particles, as in most previous studies19,20,39,40, could explain these contradictory observations. But the most striking result that emerged from our comparative mini-screen was that USUV and WNV have entirely different sensitivity to ISG20.
Initially discovered in 1997 as a novel ISG69, ISG20 was later identified as a 3′–5′ exonuclease able preferentially to degrade single-stranded RNA in vitro30. By 2003, it was established that ISG20 is an IFN–I effector that effectively inhibits replication of unrelated RNA viruses such as vesicular stomatitis virus (Rhabdoviridae)33,35, influenza virus (Orthomyxoviridae)33,34, and encephalomyocarditis virus (Picornaviridae)33,36. Subsequently, several other viruses, including flaviviruses, were identified as sensitive to ISG2019,36,37. However, our work revealed USUV as an exception among flaviviruses, as it is the sole tested virus within this genus to exhibit resistance to the antiviral effects of human ISG20. This resistance phenotype is highly conserved among the different USUV strains, since we have found that all USUV isolates were resistant to ISG20, including EU1, EU2, EU3, AF2 and AF3. The only exception was EU5, which proved to be sensitive to ISG20. The EU5 lineage comprises a few avian viruses isolated in Germany in 2016, and it has been proposed that this minor viral cluster be classified as the 5th European lineage, which included only 4 at the time8,9,10. However, none of these viruses has ever been associated with infections in humans or even other mammals, so it is unlikely that they have undergone any selection pressure towards human ISGs.
We characterized USUV resistance to ISG20 and discovered that this resistance is not due to the recruitment of viral or cellular proteins to viral RNA, nor to the presence of post-transcriptional modifications. Instead, it is an inherent characteristic of the USUV genome, which is naturally resistant to ISG20-mediated degradation. By constructing chimeric WNV replicons containing different fragments of the USUV genome, we were able to map the determinant of this resistance within the domain 2 of the USUV 3′–UTR. Furthermore, we successfully identified the smallest domain capable of conferring WNV resistance as USUV DB2. Since previous studies have proposed that ISG20 struggles to break down thermodynamically stable RNA structures, such as stem-loops30,46,47, we first hypothesized that USUV DB2 would have a particular secondary structure, rich in double-strand pairing, which would explain why it was not degraded by ISG20. However, hSHAPE data revealed that the secondary structure of USUV DB2 was in fact broadly equivalent to that found in the WNV genome, therefore suggesting that this domain may adopt a particular 3D structure that prevents it from being degraded by ISG20. Another possible explanation could be that it is during the degradation of USUV 3′–UTR that an RNA structure resistant to ISG20 is generated (Fig. 5K). Indeed, if we compare the sequences located at the base of the DB2, we noticed that USUV has a complementary sequence (CS) comprised of four nucleotides on either side of the DB2, whereas this is not the case for WNV (Fig. 5E). It is therefore possible that, during the degradation of USUV RNA, ISG20 breaks PK3 and PK4, which connect DB1 and DB2 to the downstream sequence, respectively, allowing the complementary sequences to pair, generating a longer double-stranded RNA sequence that ISG20 would have difficulty degrading (Fig. 5K). Neither WNV nor USUV EU5 would be able to form such structures in the absence of complementary sequences at the base of DB2 (Supplementary Fig. 5). Although speculative, this model may explain how, despite an apparently identical secondary structure to WNV, the unique context provided by the flanking sequences of the USUV DB2 would confer resistance to digestion by ISG20.
The fact that we were able to confer resistance to ISG20 by replacing only a short 86 nucleotide sequence in its 3′–UTR raises the question of whether another flavivirus could acquire this resistance spontaneously. This may happen through mutations, or, more likely, through RNA recombination events70,71,72,73. In this respect, WNV and USUV co-circulate in many European countries74,75 and cases of co-infection of birds have been reported76, suggesting that recombination events between these two viruses are theoretically possible.
Yet, a crucial question remains: if ISG20 possesses the capacity to hinder the replication of all flaviviruses, why has only USUV evolved an evasion strategy while other flaviviruses have not? One plausible explanation is that USUV might lack the robust mechanisms exhibited by other flaviviruses to inhibit viral sensing, IFN synthesis, and IFN signaling77,78,79,80. Studies have shown that USUV induces significantly higher levels of IFN in human cells and is more sensitive to the antiviral effects of IFN compared to WNV14,15,81. Therefore, if USUV fails to effectively counteract the IFN response, its survival could be dependent on developing resistance to key antiviral ISGs. To test this hypothesis, it would be necessary to evaluate the ability of USUV proteins to antagonize the IFN response, mirroring previous investigations conducted on major pathogenic flaviviruses14,15,81. However, it is also possible that the mutations identified in the 3′–UTR of USUV are the result of an adaptation to its two main hosts, namely mosquitoes and wild birds, and that the resistance they confer to human ISG20 is fortuitous. One way of assessing whether ISG20 resistance confers a selective advantage on USUV or is incidental, would be to evaluate the sensitivity of USUV to avian ISG20.
While previous studies have proposed that ISG20 struggles to break down thermodynamically stable secondary structures such as stem-loops30,46,47, our work is the first to identify this as an actual strategy employed by a virus to escape ISG20. This original strategy developed by USUV contrasts with what has been described for HBV, for which it is precisely a stem-loop that enables ISG20 degradation. Indeed, ISG20 is recruited to HBV RNA via a direct interaction with a stem-loop called Epsilon, prior to viral RNA degradation46.
USUV is not the only virus to overcome ISG20, since we recently reported that HIV-1 can also resist ISG20-mediated degradation, but through a distinct mechanism. Specifically, our findings revealed that HIV-1 utilizes the FTSJ3 methyltransferase to introduce 2′-O-methylations into its genome82. This modification hinders the access of viral RNA to the catalytic site of ISG20 through steric hindrance32,83. The development of distinct strategies by unrelated viruses to evade ISG20-mediated restriction highlights the great pressure imposed by this cellular exonuclease on RNA viruses.
In the complex environment of an infected cell, the virus engages in a race against antiviral factors. Our findings reveal that, when compared to WNV, USUV DB2 inherently displays greater, but not absolute, resistance to ISG20. On the other hand, the sHP/3′SL sequences of both viruses completely resist ISG20 in vitro. However, within the dynamic framework of an entire genome, these sequences prove ineffective in safeguarding the remainder of the viral genome. The temporal delay that USUV DB2 introduces to the ISG20 degradation may prove sufficient for replication to regain momentum within the context of an infection.
While it was commonly accepted that ISG20 restriction depended solely on its exonuclease activity, recent studies proposed that ISG20 binds to viral RNA, blocking its translation without causing degradation35,38. Our current study, along with our prior research on HIV-132 demonstrates that ISG20 primarily exerts its antiviral activity through the direct degradation of viral RNA. This degradation was not limited to in vitro conditions but was also observed in infected cells. However, we cannot formally rule out the possibility that ISG20 may also inhibit translation in infected cells, independently of its ability to degrade RNA.
The exact mechanism by which ISG20 degrades viral RNAs while sparing cellular RNAs remains an open question. Although certain viruses that are sensitive to ISG20 have polyadenylated genomes (e.g., picornaviruses or retroviruses), the presence of a polyA tail seems necessary but not sufficient to escape ISG20-mediated degradation47. For viruses with a highly structured 3′ non-coding region instead of a polyA tail, such as flaviviruses, the nature of RNA structures influences their sensitivity to ISG20. Despite ISG20’s ability to degrade both viral and cellular RNAs in vitro30, we observed a more severe degradation of the genome of an ISG20-sensitive virus than that of a resistant virus, or of cellular mRNAs. This implies that ISG20 alone does not specifically discriminate between self and non-self RNA but rather succeeds or fails in degrading different RNAs. Overall, these findings indicate that the susceptibility of an RNA viral genome to ISG20-mediated degradation is likely influenced by structural, genetic, and epigenetic determinants, and further research will be necessary to fully elucidate these mechanisms.
Methods
Cell lines
HEK 293 T (CRL-11268), Vero E6 (CRL-1586), Huh7.584, HeLa (CCL-2), A549 (CCL-185) HT1080 (CCL-121), and C6/36 cells (CRL-1660) were purchased from the American Type Culture Collection (ATCC). HEK 293FT (R70007) cells were purchased from Thermo Fisher Scientific. All cells, except C6/36, were cultured in complete high-glucose Dulbecco′s modified Eagle′s medium (DMEM, Gibco) supplemented with 10% fetal bovine serum (FBS, Serana), and 1% Penicillin/Streptomycin (Gibco), cells were kept at 37 °C with 5% CO2.
C6/36 cells were cultured in Leibovitz′s L15 medium (Gibco) supplemented with 10% FBS, 1% Penicillin/Streptomycin, 1% tryptose phosphate broth (Gibco), and 1% non-essential amino acids (Gibco), cells were kept at 28 °C with no CO2.
Viruses
Usutu virus (USUV) Europe 2 (TE20421/Italy/2017) was kindly provided by Giovanni Savini (Istituto Zooprofilattico Sperimentale dell′Abruzzo e del Molise, Teramo, Italy). USUV Europe 1 (Vienna 2001-blackbird) was provided by INIA Madrid (Spain). USUV Europe 5 (BNI507/2016/Germany) was provided by the Bernhard Nocht Institute for Tropical Medicine, Hamburg (Germany). USUV Europe 3 (USUV-HautRhin7315/France/2015), USUV Africa 2 (Rhône2705/France/2015), USUV Africa 3 (USUV-HauteVienne4997/France/2018) and West Nile virus (WNV) lineage 2 (WNV-6125/France/2018) were provided by ANSES (National Agency for Food, Environmental and Occupational Health Safety, France). WNV lineage 1 (WN-Tunisia-1997 PaH001) was provided by Isabelle Leparc-Goffart (French National Reference Center on Arboviruses, Marseille, France).
Dengue virus 1 (UVE/DENV-1/2012/VC/16692), Dengue virus 2 (UVE/DENV-2/2014/FR/CNR_26104), Dengue virus 3 (UVE/DENV-3/2015/TH/7716), Dengue virus 4 (UVE/DENV-4/2014/HT/6169) and Zika virus (strains H/PF/2013 and MRS/OPY/Martinique/PaRi/2015) were kindly provided by Xavier de Lamballerie (Unité des Virus Emergents, Aix Marseille Université, Marseille, France), through the European Virus Archive GLOBAL (EVA-GLOBAL) project.
All viruses were amplified on C6/36 cells. Viral stocks were prepared by infecting 80% confluent C6/36 cells for 2 h, the inoculum was then removed, fresh medium was added on the cells, and cellular supernatants were collected 5 (USUV and WNV), 7 (DENV), or 10 (ZIKV) days after infection when cytopathic effect was observed. Viral titers were determined by TCID50 using the Spearman–Kärber method and flow cytometry85. Information on strain accession numbers, number of passages, and titers of the viral pools are detailed in Supplementary Table 1.
Infections were carried out by inoculating cells with a low volume of viral dilution, at an appropriate multiplicity of infection (MOI) as detailed in figure legends, in serum-free DMEM for 2 h. The inoculum was then removed, cells were washed with phosphate-buffered saline (PBS, Gibco), and cultured in fresh DMEM, 2% FBS, 1% Penicillin/Streptomycin.
ISG overexpression
ISG overexpression was performed using lentiviral constructions from the SCRPSY library (GenBank accession no. KT368137) that express a red fluorescent protein (TagRFP) and puromycin resistance, as described previously41. Constructions carrying IFI6, SHFL or ISG20 were co-transfected with VSV-G (pMD2.G, Addgene accession no. 12259), and HIV-1 gag-pol (pCMVR8.74, Addgene accession no. 22036) in HEK 293 T cells using polyethylenimine (PEI, Polysciences). Culture medium was replaced 8 h post-transfection and supernatants were collected 48 h post-transfection. Lentivector titers were determined by flow cytometry.
For the transient ISG expression mini-screen, HEK 293 T cells were transduced with lentivectors for 72 h, at a multiplicity leading to TagRFP protein (and hence ISG) expression in around 50% of cells. Subsequently, cells were infected with 1 TCID50 of either WNV L2 or USUV AF2 for 48 h, resulting in around 50% of infected cells. Four distinct populations can therefore be observed by flow cytometry: double-negative, TagRFP-negative/infected, TagRFP-positive/uninfected, and double-positive. The proportion of double-positive cells (ISG-expressing cells that are infected) thus reveals the antiviral effect of the ISG.
For stable expression experiments, cells were transduced with lentivectors for 48 h and submitted to 1 µg/mL puromycin selection for at least 14 days, with additional cell sorting on a FACSAria II (Becton Dickinson) to ensure clean populations.
Stimulation of ISG expression by interferon
Confluent cell monolayers were cultured in complete DMEM with 10 or 100 U/mL of universal type I interferon (PBL Assay Science) for 8 h to induce the expression of ISGs. Cells were then washed with PBS to remove interferon and used for further analysis.
Flow cytometry
For TagRFP or eGFP expression quantification, cells were detached with 5 min trypsin treatment (Gibco) and pelleted for 5 min at 500 × g. Cells were washed with cold PBS, pelleted, and fixed with 4% formaldehyde in PBS for 15 min. Cells were pelleted and washed with PBS to remove excess of formaldehyde.
For flavivirus infection quantification, infected cells were detached with trypsin, pelleted and fixed like previously described. Cells were then incubated with 0.5 µg/mL of the pan-flavivirus anti-Env 4G2 primary antibody (Novus Biologicals) in PBS, 1% bovine serum albumin (BSA, Thermo Fisher Scientific) and 0.05% saponin for 2 h. Cells were then washed twice with PBS and incubated with 1 µg/mL of anti-mouse secondary antibody coupled with Alexa Fluor 488 (Thermo Fisher Scientific) in PBS, 1% BSA and 0.05% saponin. Cells were then washed again with PBS.
Flow cytometry was performed with a Novocyte flow cytometer (Agilent), analysis was performed with the Novoexpress software (Agilent), visualization of the data was performed with FlowJo v10 for Windows (BD Biosciences).
Western blot
Confluent cell monolayers were washed with PBS, lysed in ice-cold RIPA buffer (25 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.5% sodium deoxycholate, 1% NP-40, 1 mM EDTA, 2.5 mM sodium pyrophosphate, 1 mM sodium orthovanadate, and 1 mM β-Glycerophosphate) for 15 min with periodic scrapping, and centrifuged at 15,000 × g for 10 min to remove insoluble debris. Samples were heat-denatured denatured at 95 °C for 10 min in 4X Laemmli buffer (250 mM Tris-HCl [pH 7], 8% sodium dodecyl sulfate [SDS], 40% glycerol, 10% β-mercaptoethanol, and 0.005% bromophenol blue) underwent SDS polyacrylamide gel electrophoresis (SureCast Gel Handcast System, Thermo Fisher Scientific), followed by transfer onto a 0.45 µm nitrocellulose membrane (Amersham). Membranes were saturated in PBS, 0.05% Tween 20 (PBST) with 10% fat-free milk for 30 min. Primary antibodies and secondary antibodies (Supplementary Table 2) were diluted in PBST, 1% BSA. Protein visualization was achieved by HRP activity (Immobilon Forte Western HRP substrate, Merck) on a ChemiDoc imaging system (Bio-Rad). Images were analyzed with ImageJ.
Real-time quantitative RT-PCR (RT-qPCR)
Cellular RNAs were extracted using the RNeasy Mini kit (Qiagen) following the manufacturer′s instructions. RNA concentration and purity were evaluated by spectrophotometry (NanoDrop 2000c, Thermo Fisher Scientific). A maximum of 500 ng of RNA were reverse transcribed with both oligo dT and random primers using a PrimeScript RT Reagent Kit (Perfect Real Time, Takara Bio Inc.) in a 10 µL reaction. Real-time PCR reactions were performed in duplicate using Takyon ROX SYBR MasterMix blue dTTP (Eurogentec) on an Applied Biosystems QuantStudio 5 (Thermo Fisher Scientific).
Cellular transcripts were quantified with primers which hybridize the cDNA sequences (Supplementary Table 3). Viral genomes were quantified with primers within the 3′–UTR regions (Supplementary Table 3). Plasmids into which USUV and WNV amplicons have been cloned were used for absolute quantification of viral cDNA. Wild-type and chimeric WNV replicon genomes were quantified with primers which hybridize within the NS5 of WNV (Supplementary Table 3). Quantifications were performed with the following program: 3 min at 95 °C, 40 cycles of 15 s at 95 °C, 20 s at 60 °C and 20 s at 72 °C. Melting curves were also assessed.
Selective ISG knockdown
siRNAs targeting the transcripts encoding PPIB, IFI6, SHFL and ISG20 were ordered from Horizon Discovery as pools of four individual siRNAs (Dharmacon ON-TARGETplus SMARTpools) and transfected into A549 cells using HiPerFect Transfection Reagent (Qiagen), according to the manufacturer’s instructions. Knockdown efficiency was evaluated by RT-qPCR 40 h post-transfection.
CRISPR/Cas9 ISG20 knockout
Synthetic guide RNA sequences targeting ISG20 were ordered from Thermo Fisher Scientific, (TrueGuide Synthetic sgRNA, CRISPR679065_SGM: TAATCGGTGATCTCTCCCTC AGG and CRISPR679068_SGM: TCTACGACACGTCCACTGAC AGG). Non-targeting guide RNA was ordered from Thermo Fisher Scientific (TrueGuide sgRNA Negative Control, non-targeting 1). Guide RNA sequences were co-transfected in A549 cells using Cas9 Lipofectamine CRISPRMAX system with v2 TrueCut Cas9 enzyme. 48 h post-transfection, the cells were harvested and used to derivate clones using limiting dilution cloning.
Gene editing of one A549 IS20KO clone displaying normal growth kinetic, and absence of detectable ISG20 protein expression in Western blot was confirmed by amplifying ISG20 transcript from cell cDNA using ISG20 mRNA primers (Supplementary Table 4). Amplicons were cloned in PCR Blunt II-TOPO (Thermo Fisher Scientific), and ± 10 colonies were picked and sent for Sanger sequencing using M13 primers (Eurofins Genomics).
Complementation of A549 IS20KO cells with wild-type or mutant ISG20
Plasmids designed to express 3X-FLAG tagged, codon optimized, wild-type, catalytic mutant (D94A) and 2′-O-methylation insensitive double mutant (R53A/D90A) of ISG20 were previously described86. ISG20 coding sequences were amplified using ISG20 addback primers (Supplementary Table 4) designed to add NheI/EcoRV restriction sites, and amplicons were cloned into PLX_307 lentiviral vector (Addgene accession no. 184492) digested with NheI/EcorV to generate the PLX_307-ISG20-3X-FLAG plasmids. Lentiviral particles were produced in HEK 293FT cells co-transfected with VSV-G (pCMV-VSV-G opt, obtained from Pierre Charneau, Institut Pasteur, Paris, France), and HIV-1 gag-pol (psPAX2, Addgene accession no. 12260) expressing plasmids along with PLX_307-ISG20-3X-FLAG vectors using Trans IT-293 (Mirus) following the manufacturer’s instructions. PLX_307-GFP vector was used as control. Culture medium was replaced 24 h post-transfection and supernatants were collected 48 h post-transfection. A549 IS20KO were transduced with lentivectors for 48 h and submitted to 1 µg/mL puromycin selection for 10 days.
Transient expression of wild-type or mutant ISG20
Plasmids expressing 1X-FLAG tagged wild-type and catalytic mutant (D94A) of ISG20 (kind gift of Andrea Cimarelli, Center International de Recherche en Infectiologie [CIRI], France) were previously described35. The plasmid designed to express 3X-FLAG tagged, codon optimized, 2′-O-methylation insensitive double mutant (R53A/D90A) was previously described86. The plasmids were transfected in HEK 293 T using PEI. pcDNA3.1-eGFP was used as a control. Viral infection and transgenic expression of the constructs by Western blot were performed at 48 h post-transfection.
Production of wild-type and chimeric WNV reporter virus particles (RVPs)
The constructs (WNVII Rep-G/Z and WNV NY99 CprME) used to generate the eGFP-encoding WNV RVPs were kindly provided by Theodore Pierson (Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, Maryland, USA). Chimeras of WNVII Rep-G/Z with fragments of the USUV 3′–UTR were generated by PCR using Q5 high fidelity DNA polymerase (New England Biolabs) using insert/backbone primers listed in Supplementary Table 4 and assembled using GeneArt Gibson Assembly HiFi Master Mix (Thermo Fisher Scientific) following the manufacturer’s recommendations.
First, the full-length USUV AF2 3′–UTR was inserted into WNVII Rep-G/Z. USUV AF2 3′–UTR insert was amplified by touch-up PCR on cDNA from USUV AF2-infected HEK 293 T cells with the following program: 30 s at 98 °C followed by 35 cycles of 10 s at 98 °C, 20 s at 57 °C ( + 1 °C every cycle until reaching 68 °C) and 20 s at 72 °C. Final extension was allowed for an additional 2 min. 3′–UTR-lacking WNVII Rep-G/Z backbone was generated by PCR on WNVII Rep-G/Z with the following program: 30 s at 98 °C followed by 35 cycles of 10 s at 98 °C and 6 min at 72 °C, with a final extension of 2 min.
Next, functional domains within the 3′–UTR of WNVII Rep-G/Z were replaced with those from USUV. The USUV AF2 D2D3, D3, and DB2 inserts were generated by PCR on the USUV 3′–UTR-containing WNVII Rep-G/Z chimera with the following program: 30 s at 98 °C, 35 cycles of 10 s at 98 °C, 20 s at 72 °C (D2D3), 58 °C (D3), or 65 °C (DB2), and 20 s at 72 °C, with a final extension of 2 min. USUV EU5 DB2 was amplified by PCR on cDNA from USUV EU5-infected HEK 293 T cells with the following program: 30 s at 98 °C, 35 cycles of 10 s at 98 °C, 20 s at 70 °C, and 20 s at 72 °C, with a final extension of 2 min. The corresponding WNVII Rep-G/Z backbones lacking these domains were generated by PCR on WNVII Rep-G/Z with the following program: 30 s at 98 °C, 35 cycles of 10 s at 98 °C, and 6 min at 72 °C, with a final extension of 2 min.
All plasmids were amplified in Stbl2 Max Efficiency bacteria (Thermo Fisher Scientific) at 30 °C and analyzed by sequencing (Plasmidsaurus and Eurofins Genomics). Production of RVPs was performed as described previously87. Briefly, HEK 293 T cells were transfected with the plasmids encoding the replicons and the WNV structural proteins (WNV NY99 CprME) using PEI in a 3:1 (structural:replicon) ratio. RVP-containing supernatants were harvested 48 h post-transfection, filtered, concentrated by ultracentrifugation at 60,000 × g on an Optima XE-90 centrifuge with a SW 32 Ti rotor (Beckam Coulter), and stored at −80 °C. Viral titers were determined by flow cytometry.
Fluorescent microscopy
HEK 293 T cells plated on poly-D-lysine-treated (Thermo Fisher Scientific) glass coverslips were infected with wild-type or chimeric RVPs for 48 h. Cells were fixed with 4% formaldehyde in PBS for 15 min, followed by nuclear staining using 1 μg/ml Hoechst 33342 (Thermo Fisher Scientific) in PBS for 10 min. Coverslips were mounted onto glass slides using ProLong Diamond Antifade mounting medium (Thermo Fisher Scientific). Fluorescence was assessed for eGFP and Hoechst using a Thunder imaging system (Leica Microsystems). Images were analyzed with ImageJ.
In vitro transcription of full-length USUV and WNV 3′–UTRs
A T7 initiation site-containing linear DNA template of the 3′–UTR of WNV L2 or USUV AF2 was generated by elongation PCR using Q5 high fidelity DNA polymerase on the previously described replicons (wild-type or USUV 3′–UTR-containing WNVII Rep-G/Z) using appropriate primers (Supplementary Table 5) with the following program: 30 s at 98 °C followed by 40 cycles of 10 s at 98 °C, 20 s at 68.5 °C and 30 s at 72 °C. Final extension was allowed for an additional 2 min. The templates were run on a 1% agarose DNA gel (40 mM Tris-Acetate [pH 8.3], 1 mM EDTA) prior to purification (NucleoSpin Gel and PCR Clean‑up, Macherey-Nagel), concentration and purity were evaluated by spectrophotometry.
The in vitro transcription was performed on 1 µg of DNA template with T7 RNA polymerase (New England Biolabs) in presence of murine RNase inhibitor (New England Biolabs) according to the manufacturer’s instructions. The reaction was allowed for 16 h at 37 °C. The DNA template was removed with RNase-free DNase I treatment (New England Biolabs) for 15 min at 37 °C. RNA was then purified (Monarch RNA cleanup, New England Biolabs), concentration and purity were evaluated by spectrophotometry, and the samples were stored at −80 °C until analysis.
hSHAPE analysis of USUV and WNV 3′–UTRs
For hSHAPE experiments, in vitro transcribed WNV and USUV full-length 3′–UTR RNA (see above) were purified by exclusion chromatography on a TSKgel G4000SW column as previously described88. RNAs integrity and purity were confirmed by electrophoresis on an 8% polyacrylamide denaturing gel. For each RNA, benzoyl cyanide (BzCN) was used to acylate the 2’-hydroxyl group of the unconstrained nucleotides in the RNA structure, followed by interrogation of each nucleotide using two sets of labeled primers (Supplementary Table 6). One primer within each set was labeled with either VIC (AS 1 & AS 3) or NED (AS 2 & AS 4) for USUV 3′–UTR RNA and VIC (AS 5 & AS 7) or NED (AS 6 & AS 8) for WNV 3′–UTR. The NED-labeled primers from each set were used to prepare a ddG sequencing ladder from the untreated RNA samples. The VIC-labeled primers were used for reverse transcription of the modified or DMSO-treated control RNAs.
3′–UTR RNA modification with BzCN. One pmol of in vitro transcribed 3′–UTR RNA was denatured at 90 °C for 2 min and then cooled on ice for 2 min, followed by the addition of excess yeast tRNA (2 μg) and RNasin (5 U) in 10 μL HEPES Buffer (30 mM HEPES pH 8, 300 mM KCl, 5 mM MgCl2). The RNA was then folded at 37 °C for 20 min and modified by 3 μL of 300 mM BzCN in DMSO for one min at room temperature. After adding 82 μL of water, the chemically modified RNA was ethanol precipitated and resuspended in 7 μL of water. Similarly, for the control (unmodified RNA sample), 3 μL of anhydrous DMSO was added to the folded RNA instead of BzCN and treated similarly.
cDNA synthesis and analysis. For elongation of both the modified and control samples, 1 μL of AS 1, AS 3, AS 5 or AS 7 (1 μM) were added to the resuspended RNA and incubated at 90 °C for 2 min, then cooled on ice for 2 min. 2 μL of commercial 5X RT buffer was added to each of the samples and incubated at room temperature for 10 min. Reverse transcription was initiated by addition of 10 μL of the elongation mix (2 μL of 5X RT Buffer, 0.6 μL of 25 mM dNTP, 7.2 μL H2O and 2 U of AMV RT (Life Science)) and incubation at 42 °C for 50 min and 60 °C for 10 min. For the ddG sequencing ladder, 2 pmol of untreated RNA and 1 μL of the AS 2, AS 4, AS 6 or AS 8 (2 μM) were used and treated as above except for the composition of the sequencing mix (2 μL of 5X RT Buffer, 1 μL of 100 μM ddGTP, 6 μL of G10 (0.25 mM dGTP, 1 mM dATP, 1 mM dCTP, 1 mM dTTP), 0.6 μL H2O and 4 U of AMV RT (Life Science)). For each experiment, 80 μL of water were added and cDNA were extracted using Roti Aqua-Phenol/Chloroform/Isoamyl alcohol (Carl Roth). The aqueous phase of modified or unmodified samples were pooled with the aqueous phase of the ddG sequencing ladder. The samples were then ethanol precipitated and resuspended in 10 μL of HiDi Formamide (ABI). The samples were then denaturated 5 min at 90 °C, cooled on ice for 5 min, centrifuged, and loaded onto a 96-well plate for sequencing (Applied Biosystems 3130xl genetic analyzer).
The electropherograms obtained were analyzed with QuShape algorithm89 to extract reactivity data for each nucleotide. The mean reactivity data from three independent experiments were applied as constraints to the RNA sequence in RNAstructure90 (version 6.1) as well as a maximum pairing distance of 250 nts in order to avoid artifactual pairing of 5′ and 3′ ends of the RNAs.
Synthesis of radiolabeled 3′–UTR fragments
Synthetic RNAs corresponding to WNV DB2 element (5′–AAGUGCACGGCCCAACUUGGCUGAAGCUGUAAGCCAAGGGAAGGACUAGAGGUUAGAGGAGACCCCGUGCCAA, 73 nucleotides), or USUV DB2 element (5′–AACUUAGGUGCGGCCCAAGCCGUUUCCGAAGCUGUAGGAACGGUGGAAGGACUAGAGGUUAGAGGAGACCCCGCAUCAUAAGCA, 84 nucleotides) were purchased from Biomers. The RNAs were radiolabeled at their 5′ end using T4 polynucleotide kinase (New England Biolabs) and [γ32P]-ATP (Perkin Elmer) according to the manufacturer’s instructions. Radiolabeled RNAs corresponding to the 3′–SL of WNV (5′–GACACCUGGGAUAGACUAGGGGAUCUUCUGCUCUGCACAACCAGCCACACGGCACAGUGCGCCGACAUAGGUGGCUGGUGGUGCUAGAACACAGGAUCU, 99 nucleotides), or USUV (5′–AACUUAGGUGCGGCCCAAGCCGUUUCCGAAGCUGUAGGAACGGUGGAAGGACUAGAGGUUAGAGGAGACCCCGCAUCAUAAGCA, 84 nucleotides) were acquired as a control.
ISG20 exonuclease assays
Expression and purification of recombinant human wild-type or R53A/D90A mutant ISG20 (rhISG20) were previously described86.
Quantitative ISG20 RNA degradation assays were conducted by incubating 100 ng of total RNA from HEK 293 T cells infected with WNV, USUV or RVPs for 24 h, or 25 ng of in vitro transcribed full-length 3′–UTR RNAs, with 31.25–1000 nM wild-type or R53A/D90A mutant rhISG20 in an optimized buffer (50 mM Tris-HCl [pH 7], 2.5 mM MnCl2, 1 mM β-mercaptoethanol, 0.1 mM DTT and 0.1% triton) for 1 h at 37 °C. The reaction was halted by inactivating ISG20 for 5 min at 70 °C, and the digestion products were stored at −80 °C until analysis by RT-qPCR.
Qualitative assessment of ISG20 degradation of in vitro transcribed flavivirus 3′–UTR RNA was conducted by incubating 1 µg of full-length 3′–UTR RNAs with 500, 2500 or 3000 nM wild-type or R53A/D90A mutant rhISG20 (indicated in figure legends) in the optimized buffer for 1 h at 37 °C. Incubation with 1 µg/mL RNase A (Qiagen) for 5 min at 25 °C served as a control. The reaction was halted by inactivating ISG20 for 5 min at 70 °C or addition of 40 U of murine RNase inhibitor (New England Biolabs). Digestion products were denatured in gel loading buffer II with SYBR Gold (Thermo Fisher Scientific) by heating for 15 min at 55 °C and then analyzed on a 1% agarose RNA gel (20 mM MOPS [pH 7], 5 mM sodium acetate, 1 mM EDTA, and 4% formaldehyde). Alternatively, digestion products were analyzed on 2% agarose RNA gel and stained with GelRed nucleic acid gel stain (Merck). Images were acquired using UV transillumination (E-Box, Vilber) and analyzed with ImageJ.
Qualitative assessment of ISG20 degradation of radiolabeled 3′–UTR fragments was conducted by incubating 500 nM of substrate with 5 nM rhISG20 in the optimized buffer for 0, 1, 5, 15, 30, and 60 min at 37 °C. 5 µL of suspension was taken and blocked in 15 µL of loading buffer containing 96% formamide and 10 mM EDTA. The digested products were loaded on 7 M urea-containing 14% polyacrylamide gels (acrylamide/bisacrylamide, 19:1 ratio) buffered with Tris-Taurine-taurine-EDTA and run at 65 W. Results were visualized by phosphor imaging using a Typhoon-9410 variable-mode scanner (GE Healthcare). RNA degradation was quantified using the FujiImager and Image Gauge analysis software.
In silico analysis of the 3′–UTR features
The 3′–UTR sequences of the previously described replicons (wild-type or USUV 3′–UTR-containing WNVII Rep-G/Z, see above) were used for the prediction and comparison of post-transcriptional methylations and secondary structure folding.
Potential m6A sites were predicted using the online tool SRAMP91 in mature RNA mode and using the HEK 293 cell model. Only high confidence sites were considered. The potential 2′-O methylation sites were predicted using the online tool NmSEER V2.092 using the HEK 293 cell model. Only sites with a score >0.25 were considered. Secondary structure of RNA was predicted using the ViennaRNA Package 2.0 for Linux93 and confirmed with the Mfold web server94 in a literature-oriented approach. RNA 2D Structures were visualized with either Structure Editor graphical tool, a module of the RNAstructure software, or RNAcanvas95.
Quantification and statistical analyses
Graphical representations and statistical analyses were performed using GraphPad Prism version 10 for Windows (GraphPad Software). To analyze two conditions, two-tailed Mann-Whitney tests were performed. To analyze multiple conditions, a normality test was first performed, data were then analyzed with one-way or two-way analyses of variance (ANOVA) followed by the appropriate multiple comparison analysis (indicated in figure legends).
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
All data supporting the findings of this study are provided in Supplementary Information or Source Data file. Source data are provided with this paper.
References
Pierson, T. C. & Diamond, M. S. The continued threat of emerging flaviviruses. Nat. Microbiol. 5, 796–812 (2020).
Ashraf, U. et al. Usutu virus: an emerging flavivirus in Europe. Viruses 7, 219–238 (2015).
Clé, M. et al. Usutu virus: a new threat? Epidemiol. Infect. 147, e232 (2019).
Roesch, F., Fajardo, A., Moratorio, G., & Vignuzzi, M. Usutu virus: an arbovirus on the rise. Viruses 11. https://doi.org/10.3390/v11070640 (2019).
Woodall, J. The viruses isolated from arthropods at the East African Virus Research Institute in the 26 years ending December 1963. Proc. E Afr. Acad. 2, 141–146 (1964).
Weissenböck, H. et al. Emergence of Usutu virus, an African mosquito-borne flavivirus of the Japanese encephalitis virus group, central Europe. Emerg. Infect. Dis. 8, 652–656 (2002).
Gill, C. M. et al. Usutu virus disease: a potential problem for North America? J. Neurovirol. 26, 149–154 (2020).
Vilibic-Cavlek, T. et al. Epidemiology of Usutu virus: the European scenario. Pathogens 9, 699 (2020).
Engel, D. et al. Reconstruction of the evolutionary history and dispersal of Usutu virus, a neglected emerging arbovirus in Europe and Africa. mBio 7, https://doi.org/10.1128/mbio.01938-15 (2016).
Cadar, D. et al. Widespread activity of multiple lineages of Usutu virus, western Europe, 2016. Eur. Surveill. 22, 30452 (2017).
Khare, B. & Kuhn, R. J. The Japanese encephalitis antigenic complex viruses: from structure to immunity. Viruses 14, 2213 (2022).
Cadar, D. & Simonin, Y. Human Usutu virus infections in Europe: a new risk on horizon? Viruses 15, 77 (2023).
Anderson, J. F. & Rahal, J. J. Efficacy of interferon alpha-2b and ribavirin against West Nile virus in vitro. Emerg. Infect. Dis. 8, 107–108 (2002).
Scagnolari, C. et al. Usutu virus growth in human cell lines: induction of and sensitivity to type I and III interferons. J. Gen. Virol. 94, 789–795 (2013).
Cacciotti, G. et al. Variation in interferon sensitivity and induction between Usutu and West Nile (lineages 1 and 2) viruses. Virology 485, 189–198 (2015).
Katze, M. G., He, Y. & Gale, M. Viruses and interferon: a fight for supremacy. Nat. Rev. Immunol. 2, 675–687 (2002).
Schneider, W. M., Chevillotte, M. D. & Rice, C. M. Interferon-stimulated genes: a complex web of host defenses. Annu. Rev. Immunol. 32, 513–545 (2014).
Schoggins, J. W. Interferon-stimulated genes: what do they all do? Annu. Rev. Virol. 6, 567–584 (2019).
Jiang, D. et al. Identification of five interferon-induced cellular proteins that inhibit West Nile virus and dengue virus infections. J. Virol. 84, 8332–8341 (2010).
Schoggins, J. W. et al. A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472, 481–485 (2011).
Liu, B. et al. Overlapping and distinct molecular determinants dictating the antiviral activities of TRIM56 against flaviviruses and coronavirus. J. Virol. 88, 13821–13835 (2014).
Suzuki, Y. et al. Characterization of RyDEN (C19orf66) as an interferon-stimulated cellular inhibitor against dengue virus replication. PLoS Pathog. 12, e1005357 (2016).
Richardson, R. B. et al. A CRISPR screen identifies IFI6 as an ER-resident interferon effector that blocks flavivirus replication. Nat. Microbiol. 3, 1214–1223 (2018).
Dukhovny, A. et al. A CRISPR activation screen identifies genes that protect against Zika virus infection. J. Virol. 93, e00211–e00219 (2019).
Valdez, F. et al. Schlafen 11 restricts flavivirus replication. J. Virol. 93, e00104-19 (2019).
Lesage, S. et al. Discovery of genes that modulate flavivirus replication in an interferon-dependent manner. J. Mol. Biol. 434, 167277 (2022).
Balinsky, C. A. et al. IRAV (FLJ11286), an interferon-stimulated gene with antiviral activity against dengue virus, interacts with MOV10. J. Virol. 91, e01606–e01616 (2017).
Moser, M. J., Holley, W. R., Chatterjee, A. & Mian, I. S. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 25, 5110–5118 (1997).
Shevelev, I. V. & Hübscher, U. The 3′ 5′ exonucleases. Nat. Rev. Mol. Cell Biol. 3, 364–376 (2002).
Nguyen, L. H., Espert, L., Mechti, N. & Wilson, D. M. The human interferon- and estrogen-regulated ISG20/HEM45 gene product degrades single-stranded RNA and DNA in vitro. Biochemistry 40, 7174–7179 (2001).
Espert, L. et al. Interferon-induced exonuclease ISG20 exhibits an antiviral activity against human immunodeficiency virus type 1. J. Gen. Virol. 86, 2221–2229 (2005).
El Kazzi, P. et al. Internal RNA 2’O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res. 51, 2501–2515 (2023).
Espert, L. et al. ISG20, a new interferon-induced RNase specific for single-stranded RNA, defines an alternative antiviral pathway against RNA genomic viruses. J. Biol. Chem. 278, 16151–16158 (2003).
Qu, H. et al. Influenza A Virus-induced expression of ISG20 inhibits viral replication by interacting with nucleoprotein. Virus Genes 52, 759–767 (2016).
Wu, N. et al. The interferon stimulated gene 20 protein (ISG20) is an innate defense antiviral factor that discriminates self versus non-self translation. PLOS Pathog. 15, e1008093 (2019).
Zhou, Z. et al. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409, 175–188 (2011).
Ding, J. et al. Placenta-derived interferon-stimulated gene 20 controls ZIKA virus infection. EMBO Rep. 22, e52450 (2021).
Weiss, C. M. et al. The interferon-induced exonuclease ISG20 exerts antiviral activity through upregulation of type I interferon response proteins. mSphere 3, e00209–e00218 (2018).
Brass et al. The IFITM proteins mediate cellular resistance to influenza A H1N1 virus, West Nile Virus, and Dengue Virus. Cell 139, 1243–1254 (2009).
Perreira, J. M., Chin, C. R., Feeley, E. M. & Brass, A. L. IFITMs restrict the replication of multiple pathogenic viruses. J. Mol. Biol. 425, 4937–4955 (2013).
Kane, M. et al. Identification of interferon-stimulated genes with antiretroviral activity. Cell Host Microbe 20, 392–405 (2016).
Feng, J. et al. Interferon-stimulated gene (ISG)-expression screening reveals the specific antibunyaviral activity of ISG20. J. Virol. 92, https://doi.org/10.1128/jvi.02140-17 (2018).
Rihn, S.J. et al. TRIM69 inhibits vesicular stomatitis Indiana virus. J. Virol. 93, https://doi.org/10.1128/jvi.00951-19 (2019).
Wickenhagen, A. et al. A prenylated dsRNA sensor protects against severe COVID-19. Science 374, eabj3624 (2021).
Leong, C. R. et al. Interferon-stimulated gene of 20 kDa protein (ISG20) degrades RNA of hepatitis B virus to impede the replication of HBV in vitro and in vivo. Oncotarget 7, 68179–68193 (2016).
Liu, Y. et al. Interferon-inducible ribonuclease ISG20 inhibits hepatitis B virus replication through directly binding to the epsilon stem-loop structure of viral RNA. PLoS Pathog. 13, e1006296 (2017).
Louvat, C. et al. Stable structures or PABP1 loading protects cellular and viral RNAs against ISG20-mediated decay. Life Sci. Alliance 7. https://doi.org/10.26508/lsa.202302233 (2024).
Imam, H., Kim, G.-W., Mir, S. A., Khan, M. & Siddiqui, A. Interferon-stimulated gene 20 (ISG20) selectively degrades N6-methyladenosine modified Hepatitis B Virus transcripts. PLoS Pathog. 16, e1008338 (2020).
Lichinchi, G. et al. Dynamics of human and viral RNA methylation during Zika virus infection. Cell Host Microbe 20, 666–673 (2016).
Dong, H. et al. 2’-O methylation of internal adenosine by flavivirus NS5 methyltransferase. PLoS Pathog. 8, e1002642 (2012).
Bidet, K. & Garcia-Blanco, M. A. Flaviviral RNAs: weapons and targets in the war between virus and host. Biochem J. 462, 215–230 (2014).
Villordo, S. M., Carballeda, J. M., Filomatori, C. V. & Gamarnik, A. V. RNA structure duplications and flavivirus host adaptation. Trends Microbiol. 24, 270–283 (2016).
Ng, W. C., Soto-Acosta, R., Bradrick, S. S., Garcia-Blanco, M. A. & Ooi, E. E. The 5′ and 3′ Untranslated Regions of the Flaviviral Genome. Viruses 9, 137 (2017).
Fernández-Sanlés, A., Ríos-Marco, P., Romero-López, C., & Berzal-Herranz, A. Functional information stored in the conserved structural RNA domains of flavivirus genomes. Front. Microbiol. 8, 546 (2017).
Liu, Y. et al. Structures and functions of the 3′ Untranslated regions of positive-sense single-stranded RNA viruses infecting humans and animals. Front. Cell. Infection Microbiol. 10, 453 (2020).
Shi, P.-Y., Brinton, M. A., Veal, J. M., Zhong, Y. Y. & Wilson, W. D. Evidence for the existence of a Pseudoknot structure at the 3‘ terminus of the flavivirus genomic RNA. Biochemistry 35, 4222–4230 (1996).
Olsthoorn, R. C. & Bol, J. F. Sequence comparison and secondary structure analysis of the 3′ noncoding region of flavivirus genomes reveals multiple pseudoknots. RNA 7, 1370–1377 (2001).
Romero, T. A., Tumban, E., Jun, J., Lott, W. B. & Hanley, K. A. Secondary structure of dengue virus type 4 3′ untranslated region: impact of deletion and substitution mutations. J. Gen. Virol. 87, 3291–3296 (2006).
Akiyama, B. M., Graham, M. E., O′Donoghue, Z., Beckham, J. D. & Kieft, J. S. Three-dimensional structure of a flavivirus dumbbell RNA reveals molecular details of an RNA regulator of replication. Nucleic Acids Res. 49, 7122–7138 (2021).
Chapman, E. G., Moon, S. L., Wilusz, J. & Kieft, J. S. RNA structures that resist degradation by Xrn1 produce a pathogenic Dengue virus RNA. eLife 3, e01892 (2014).
MacFadden, A. et al. Mechanism and structural diversity of exoribonuclease-resistant RNA structures in flaviviral RNAs. Nat. Commun. 9, 119 (2018).
Chapman, E. G. et al. The structural basis of pathogenic subgenomic flavivirus RNA (sfRNA) production. Science 344, 307–310 (2014).
Slonchak, A. & Khromykh, A. A. Subgenomic flaviviral RNAs: What do we know after the first decade of research. Antivir. Res. 159, 13–25 (2018).
Manokaran, G. et al. Dengue subgenomic RNA binds TRIM25 to inhibit interferon expression for epidemiological fitness. Science 350, 217–221 (2015).
Merino, E. J., Wilkinson, K. A., Coughlan, J. L. & Weeks, K. M. RNA structure analysis at single nucleotide resolution by Selective 2‘-Hydroxyl Acylation and Primer Extension (SHAPE). J. Am. Chem. Soc. 127, 4223–4231 (2005).
Badorrek, C. S. & Weeks, K. M. RNA flexibility in the dimerization domain of a gamma retrovirus. Nat. Chem. Biol. 1, 104–111 (2005).
Wilkinson, K. A. et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLOS Biol. 6, e96 (2008).
Sztuba-Solinska, J. et al. Identification of biologically active, HIV TAR RNA-binding small molecules using small molecule microarrays. J. Am. Chem. Soc. 136, 8402–8410 (2014).
Gongora, C. et al. Molecular cloning of a new interferon-induced PML nuclear body-associated protein. J. Biol. Chem. 272, 19457–19463 (1997).
Tolou, H. J. G. et al. Evidence for recombination in natural populations of dengue virus type 1 based on the analysis of complete genome sequences. J. Gen. Virol. 82, 1283–1290 (2001).
Chen, S.-P. et al. Identification of a recombinant dengue virus type 1 with 3 recombination regions in natural populations in Guangdong province, China. Arch. Virol. 153, 1175–1179 (2008).
Chuang, C.-K. & Chen, W.-J. Experimental evidence that RNA recombination occurs in the Japanese encephalitis virus. Virology 394, 286–297 (2009).
Carney, J., Daly, J. M., Nisalak, A. & Solomon, T. Recombination and positive selection identified in complete genome sequences of Japanese encephalitis virus. Arch. Virol. 157, 75–83 (2012).
Nikolay, B. A review of West Nile and Usutu virus co-circulation in Europe: how much do transmission cycles overlap? Trans. R. Soc. Trop. Med. Hyg. 109, 609–618 (2015).
Zannoli, S. & Sambri, V. West Nile Virus and Usutu Virus co-circulation in Europe: epidemiology and implications. Microorganisms 7, 184 (2019).
Santos, P. D. et al. Co-infections: simultaneous detections of West Nile virus and Usutu virus in birds from Germany. Transbound. Emerg. Dis. 69, 776–792 (2022).
Gack, M. U. & Diamond, M. S. Innate immune escape by Dengue and West Nile viruses. Curr. Opin. Virol. 20, 119–128 (2016).
Cumberworth, S. L., Clark, J. J., Kohl, A. & Donald, C. L. Inhibition of type I interferon induction and signalling by mosquito-borne flaviviruses. Cell Microbiol. 19, e12737 (2017).
Miorin, L., Maestre, A. M., Fernandez-Sesma, A. & García-Sastre, A. Antagonism of type I interferon by flaviviruses. Biochem Biophys. Res Commun. 492, 587–596 (2017).
Zoladek, J. & Nisole, S. Mosquito-borne flaviviruses and type I interferon: catch me if you can! Front Microbiol. 14, 1257024 (2023).
Martin, M.-F. et al. Usutu Virus escapes langerin-induced restriction to productively infect human Langerhans cells, unlike West Nile virus. Emerg. Microbes Infect. 11, 761–774 (2022).
Ringeard, M., Marchand, V., Decroly, E., Motorin, Y. & Bennasser, Y. FTSJ3 is an RNA 2′-O-methyltransferase recruited by HIV to avoid innate immune sensing. Nature 565, 500–504 (2019).
Decombe, A., El Kazzi, P. & Decroly, E. Interplay of RNA 2′-O-methylations with viral replication. Curr. Opin. Virol. 59, 101302 (2023).
Blight, K. J., McKeating, J. A. & Rice, C. M. Highly permissive cell lines for subgenomic and genomic hepatitis C Virus RNA replication. J. Virol. 76, 13001–13014 (2002).
Grigorov, B., Rabilloud, J., Lawrence, P. & Gerlier, D. Rapid titration of measles and other viruses: optimization with determination of replication cycle length. PLoS One 6, e24135 (2011).
El Kazzi, P. et al. Internal RNA 2′O-methylation in the HIV-1 genome counteracts ISG20 nuclease-mediated antiviral effect. Nucleic Acids Res. 51, 2501–2515 (2022).
Pierson, T. C. et al. A rapid and quantitative assay for measuring antibody-mediated neutralization of West Nile virus infection. Virology 346, 53–65 (2006).
Gilmer, O. et al. Structural maturation of the HIV-1 RNA 5′ untranslated region by Pr55Gag and its maturation products. RNA Biol. 19, 191–205 (2022).
Karabiber, F., McGinnis, J. L., Favorov, O. V. & Weeks, K. M. QuShape: rapid, accurate, and best-practices quantification of nucleic acid probing information, resolved by capillary electrophoresis. RNA 19, 63–73 (2013).
Reuter, J. S. & Mathews, D. H. RNAstructure: software for RNA secondary structure prediction and analysis. BMC Bioinforma. 11, 129 (2010).
Zhou, Y., Zeng, P., Li, Y.-H., Zhang, Z. & Cui, Q. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res. 44, e91 (2016).
Zhou, Y., Cui, Q. & Zhou, Y. NmSEER V2.0: a prediction tool for 2′-O-methylation sites based on random forest and multi-encoding combination. BMC Bioinforma. 20, 690 (2019).
Lorenz, R. et al. ViennaRNA Package 2.0. Algorithms Mol. Biol. 6, 26 (2011).
Zuker, M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31, 3406–3415 (2003).
Johnson, P.Z. & Simon, A.E. RNAcanvas: interactive drawing and exploration of nucleic acid structures. Nucleic Acids Res. gkad302. https://doi.org/10.1093/nar/gkad302 (2023).
Acknowledgements
This work was supported by the Agence Nationale de la Recherche (ANR-21-CE15-0041, to S.N. and Y.S., ANR-17-CE15-0029 to E.D., and ANR-20-CE11-0024 to E.D. and N.J.), the UK Medical Research Council (MC_UU_12014/10, MR/K024752/1 and MR/P022642/1 to S.J.W.) and the Foundation for Medical Research (FRM-REPLI80C/U160, FDT202204014965 to E.D.). J.Z. was supported by a grant from the Agence Nationale de la Recherche (ANR-21-CE15-0041-03). M.C. and I.B. are recipients of doctoral grants from the French Ministry of Higher Education and Research. We acknowledge the imaging facility of Montpellier (MRI), member of the national infrastructure France-BioImaging supported by the Agence Nationale de la Recherche (ANR-10-INBS-04, “Investissements d′avenir”). This publication was supported by the European Virus Archive GLOBAL (EVA-GLOBAL) project that has received funding from the European Union′s Horizon 2020 research and innovation program under grant agreement No 871029. We thank Karim Majzoub and Pierre Khalfi (IGMM, Montpellier, France) for helpful discussions and Nathalie J. Arhel (IRIM, Montpellier, France) for critical reading of the manuscript.
Author information
Authors and Affiliations
Contributions
Conceptualization: S.N. and J.Z., methodology: S.N., J.Z., Y.S., N.J., E.D., J.-C.P. and S.J.W., investigation: J.Z., P.E.K., V.C., V.V.-B., M.C., S.R., and L.R., resources: V.C., I.B., Y.S., N.J., E.D., and S.J.W., visualization: J.Z., P.E.K., V.C., and V.V.-B. formal analysis: J.Z., funding acquisition: S.N., N.J., E.D., J.-C.P., and S.J.W., supervision: S.N., writing - original draft: J.Z. and S.N., writing - review & editing: J.Z., P.E.K., V.C., V.V.-B., M.C., E.D., S.R., I.B., L.R., Y.S., N.J., E.D., J.-C.P., S.J.W., and S.N. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Nature Communications thanks the anonymous reviewer(s) for their contribution to the peer review of this work. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Source data
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zoladek, J., El Kazzi, P., Caval, V. et al. A specific domain within the 3′ untranslated region of Usutu virus confers resistance to the exonuclease ISG20. Nat Commun 15, 8528 (2024). https://doi.org/10.1038/s41467-024-52870-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41467-024-52870-w
- Springer Nature Limited