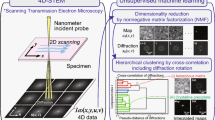
Abstract
Four-dimensional scanning transmission electron microscopy, coupled with a wide array of data analytics, has unveiled new insights into complex materials. Here, we introduce a straightforward unsupervised machine learning approach that entails dimensionality reduction and clustering with minimal hyperparameter tuning to semi-automatically identify unique coexisting structures in metallic alloys. Applying cepstral transformation to the original diffraction dataset improves this process by effectively isolating phase information from potential signal ambiguity caused by sample tilt and thickness variations, commonly observed in electron diffraction patterns. In a case study of a NiTiHfAl shape memory alloy, conventional scanning transmission electron microscopy imaging struggles to accurately identify a low-contrast precipitate at lower magnifications, posing challenges for microscale analyses. We find that our method efficiently separates multiple coherent structures while using objective means of determining hyperparameters. Furthermore, we demonstrate how the clustering result facilitates more robust strain mapping to provide immediate and quantitative structural insights.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
In the recent decade, four-dimensional scanning transmission electron microscopy (4D-STEM) has become an increasingly popular technique for efficient data acquisition on complex materials at a wide range of length scales with high spatial resolution1,2,3,4,5,6,7. A nanosized electron probe is rastered across a two-dimensional (2D) region within a sample, and a 2D electron diffraction pattern is recorded for each scan position. The emergence of 4D-STEM is largely owed to the advent of ultrafast direct electron detectors, which feature a high readout speed and dynamic range8,9,10. Users are able to collect several gigabytes (GBs) of data in minutes, providing access to the entire angular range of electron scattering signals and, thus, a plethora of structural and functional property information. Key information extracted by 4D-STEM experiments include but is not limited to charge density mapping, three-dimensional (3D) structural information, polar skyrmion visualization, medium-range ordering in metallic glasses, and in-situ nanoparticle phase evolution3,4,5,11,12,13,14,15,16,17,18. Evidently, 4D-STEM stands at the frontier of materials characterization, offering a vast array of limitless opportunities. Swiftly collecting extensive, high-quality 4D datasets has now become standard practice with the availability of several open-source data analysis packages, thus underscoring the growing importance of efficient big data analysis19,20,21,22,23,24.
Unsupervised machine learning (USML) can alleviate the challenge of automating data analysis when dealing with new and complex material systems. There is no need for labeling or training, which alleviates much of the human effort needed to support supervised learning models25,26,27,28,29,30,31,32,33. The umbrella of USML methods encompasses dimensionality reduction, clustering, and even unsupervised neural networks like generative adversarial networks34,35,36,37,38,39,40,41. Current literature on utilizing USML for STEM data has shown promising results in distinguishing the stacking order of nanosized islands, visualizing defects in 2D materials, and examining fine edge structures35,38,39,42,43,44,45,46,47,48,49. Typically, some preprocessing is performed to first extract features of interest, such as specific Bragg diffracted discs or a targeted energy loss regime, to then use for clustering39,42,46. However, as we continue to emphasize, this is only applicable when the user already knows which features to look for in the first place.
In this paper, we present a novel methodology to enhance USML performance in characterizing complex microstructures with 4D-STEM using cepstral analysis. It is applicable across different datasets with minimal hyperparameter tuning and prior knowledge required. The algorithm is employed for a NiTiHf-based shape memory alloy (SMA), serving as a representative case study. SMAs are characterized by their ability to recover their original structure upon heating after significant applied strain50. NiTiHf-based SMAs, in particular, exhibit precipitation strengthening with the formation of coherent precipitates and are suitable for high-temperature applications due to their high martensitic transformation temperatures, which has opened up much interest from the aerospace and automotive sectors51,52. Specifically, the precipitation strengthening originates from the Ni(Ti0.6Hf0.4) Han phase precipitates embedded in the NiTi B2 matrix phase that have been identified for NiTiHf systems in previous studies53,54,55,56,57. In pursuit of augmenting their mechanical strength, the incorporation of Al into NiTiHf SMAs has been explored, which leads to the formation of a secondary precipitate, the Ni2TiAl Heusler phase58,59. In a recent investigation by Hsu et al. on a Ni50Ti25-xHf20Alx system, alloying Al resulted in the coexistence of Han and Heusler precipitates while increasing mechanical strength60. However, increasing Al content was also directly correlated to lowering martensitic transformation temperature, which is counterproductive for high-temperature applications. The complexity of the problem deepens with the lack of knowledge regarding other potentially coexisting phases in this quaternary alloy. Therefore, the crux of the materials design problem lies in optimizing the competing desirable properties and correlating them to their structural origins, including precipitate formation and strain. This task requires testing a range of composition and processing combinations, each of which will produce several 4D datasets. The magnitude of this investigation mandates an automated and universally applicable data analytics methodology capable of consistently distinguishing between various phases across multiple datasets.
Results and discussion
Initial investigation of Ni50Ti21Hf20Al4 with STEM
A low-magnification HAADF-STEM image in Fig. 1a shows an array of vertically aligned, needle-shaped, Han phase precipitates embedded in the B2 matrix. Due to the relatively high content of heavy Hf atoms, the Han phase precipitates are denoted by higher image intensities as compared to the B2 matrix. Initial investigation reveals three distinct atomically resolved structures at the edge of one precipitate shown in Fig. 1b labeled as “1”, “2”, and “3”. In order to properly identify these structures, we use high-magnification HAADF-STEM images, which are displayed in the top row of Fig. 1c. These three phases are determined to be the B2 matrix, Han precipitate, and Heusler precipitate, and their corresponding nanobeam electron diffraction (NBED) patterns are given in the bottom row of Fig. 1c. Both image and diffraction data unambiguously isolated three unique phases and determined their orientation relationship given by the coordinate axes in Fig. 1c. Notably, HAADF-STEM imaging revealed that both precipitates, Han and Heusler, maintain structural coherency with the B2 matrix as well as each other. Additionally, energy dispersive x-ray spectroscopy (EDS) analysis provided insight into the chemical variations across the different phases. An EDS line scan in Fig. S1 is drawn across the edge of a Han precipitate, from which the atomic structure of the Heusler phase is visible and correlates to an increased Al concentration and decreased Ti concentration.
a Low-magnification HAADF-STEM image showing an array of needle-shaped precipitates embedded in the matrix of the SMA sample. b High-magnification HAADF-STEM image around the red box in (a) where three unique structures are atomically resolved labeled as “1”, “2”, and “3”. c Structures “1”, “2”, and “3” are displayed from left to right as Matrix, Han, and Heusler. A high-magnification HAADF-STEM image, overlaying Vesta model, and corresponding NBED pattern are shown. The scale bars for the HAADF-STEM images and NBED patterns are 5 Å and 10 nm−1, respectively.
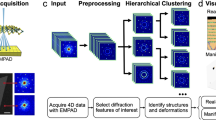
Characterizing the Heusler phase formation is key to understanding this SMA system as it is directly linked to the addition of Al, and thus linked with improved strengthening and lowered martensitic transformation temperatures60. However, the Heusler phase exhibits significantly diminished contrast relative to the B2 matrix due to similar atomic arrangements and constituent elements with only minute addition of a light element, Al, to the Heusler phase. As a result, identifying nanoscale coherent Heusler phase precipitates at larger length scales, and thus lower magnifications, is challenging via imaging alone, albeit achieving atomic resolution. Note that the Heusler phase was not distinguishable in the low-magnification HAADF-STEM image in Fig. 1a and only more discernable upon increasing magnification in Fig. 1b, c by examining the subtle difference in atomic arrangements. Consequently, applying image segmentation methods to the low-magnification HAADF-STEM images in order to quantify the presence of the Heusler phase did not seem properly viable as relatively high contrast features have been used for these types of identification tasks in other studies29,30,61,62,63. On the other hand, the NBED patterns in the bottom row of Fig. 1c exhibit better contrast between the three phases due to their distinction from one another, which led us to employ 4D-STEM to further this study. The 4D-STEM workflow, schematized in Fig. 2, produced 4D diffraction datasets that were subject to either principal component analysis (PCA) or non-negative matrix factorization (NMF) for dimensionality reduction, which was followed by K-means clustering to gauge the performance on phase segregation. Afterward, a cepstral transformation was applied to these diffraction datasets, and this 4D cepstrum stack was then subject to the same USML algorithm to compare their results.
At each electron beam scan position, a nanobeam electron diffraction (NBED) pattern is acquired by the EMPAD and stored in a 4D dataset. PCA or NMF with K-means clustering is applied to the NBED dataset to evaluate phase segregation capability. A cepstral transform is then applied to the NBED dataset and run through the same clustering algorithm to compare the performance of all four combinations.
Trial 1: PCA and NMF + K-means clustering on original NBED dataset
The process starts by normalizing all diffraction patterns acquired from the 4D-STEM scan region visualized in the virtual annular dark-field (vADF) image in Fig. 3a, where the Han precipitate stands out with its darker contrast. We then apply PCA and NMF to decompose the dataset into five components. We established the choice of number of components by first decomposing the dataset into n components, with n ranging from two to ten. We then calculate the root-mean-squared error (RMSE) between the original and decomposed datasets for an average of ten runs per value of n. Figure S2 shows an example of one of these RMSE vs n plots. Note that as n > 5, there is an onset of a plateau that indicates a worsened tradeoff between gaining more accuracy and longer processing times.
a Virtual annular dark-field image acquired directly from the EMPAD’s software showing the field-of-view of a single Han precipitate. Note that this precipitate is the same one at the center of Fig. 1a. b, d The NBED + PCA and NBED + NMF clustering results, respectively, and c, e the corresponding cluster-averaged NBED patterns. Scale bars are 10 nm−1.
The unfavorable clustering results from this initial trial are shown in Fig. 3b and 3d, respectively, where the automatically determined structural patterns do not have any inherent correlation to the material itself. The respective K values were determined by the elbow point method, which is described in Methods and displayed in Fig. S3a. There is a likeness to the needle-shaped Han precipitate (Fig. 3a), but the dark blue that represents the Han precipitate cluster also appears in the outer regions where the matrix and Heusler should reside based on our initial STEM imaging study. As a means to quantitatively gauge the clustering performance, cluster-averaged NBED patterns are displayed next to the NBED + PCA and NBED + NMF cluster maps in Fig. 3c and 3e, respectively. It is clear that while the distinction of the Han phase NBED stands out in Cluster 1 and Cluster 4 for the PCA and NMF trials, respectively, the algorithm matches up seemingly random parts of the non-Han precipitate regions as being part of the same clusters. Aside from slight differences in intensities, Clusters 0 and 2 in Fig. 3c, Clusters 0 and 3 in Fig. 3e, and Clusters 1 and 2 in Fig. 3e are very similar to each other. Unfortunately, this makes the actual post-clustering phase identification a nontrivial task.
In order to better understand these results, Fig. 4 shows the NBED + NMF clustering result with two individual NBED patterns from opposite sides of the Han precipitate that were partitioned into separate clusters. The NBED patterns revealed that both areas correspond to the B2 Matrix phase, yet they were still separated by the algorithm. This is likely due to the uneven intensity distributions present in these NBED patterns, which are typically observed in electron diffract data and arise from local sample tilting or bending. The red arrows in the NBED patterns point to the pattern’s center of mass and indicate that the two patterns have opposite intensity distributions. This indeed implies that a key feature of the data is the local sample bending around the Han precipitate. This was nonideal for the purpose of this case study, as we would like the algorithm to identify features that would normally be used to distinguish phases, such as the position and symmetry of peaks in the diffraction patterns rather than the intensity variations. This was a significant challenge to encounter in that one cannot simply force the dimensionality reduction algorithms to prioritize certain parts or features in the dataset.
Prior NMF + NBED clustering result from Fig. 3d with an individual diffraction pattern from the top left (light blue cluster) and top right (orange cluster) region near the Han precipitate. The red arrows point toward the pattern intensity’s center-of-mass, indicating local sample bending around the Han precipitate.
Trial 2: Comparing USML methods on cepstral transformed 4D dataset
The cepstral transform of electron diffraction data was first proposed by Padgett et al. as a means to precisely quantify strain maps from diffraction data that may be subject to unfavorable, complex multiple scattering effects64. This study found that the cepstral transformation effectively decouples lattice information from intensity variations caused by tilt and thickness. The cepstrum pattern, CP, can be calculated from an electron diffraction pattern, \(I(\mathop{k}\limits^{ \rightharpoonup })\), using Eq. 1 where \({\mathscr{F}}\) stands for a Fourier transform.
Since the diffraction patterns represent a crystal’s signal in reciprocal space, the peaks in the cepstrum pattern represent interatomic spacings within the real space structure. This technique has been used across a wide variety of studies in this field, from strain mapping to imaging local short-range ordering in high entropy alloys64,65,66,67,68,69,70. The significant motivation provided by these early works encourages the application of this transformation method to the SMA dataset to see if the isolation of phase information from those of sample bending would be reflected in the subsequent clustering results.
The corresponding Cepstrum + PCA and Cepstrum + NMF cluster maps in Fig. 5a and 5c depict a clear, sharp boundary around the Han precipitate with no mixing into other regions like in Fig. 3b and 3d. For the Cepstrum + NMF cluster map in particular, Cluster 2 appears to visually match well with the expectation from initial HAADF-STEM imaging that the Heusler phase tends to be located in pockets at the side edges of the Han precipitate. Quantitatively, the values in the inertia plots for Cepstrum + PCA and Cepstrum + NMF in Fig. S3b are about two orders of magnitude lower than those for the original diffraction dataset, indicating a much tighter K-means clustering output for both the PCA and NMF case, but more so for the latter. The clustering results were once again validated by taking both averaged diffraction and cepstrum patterns for each cluster map in Fig. 5b and 5d. While the averaged NBED patterns in Fig. 5b are now more distinct from each other, the averaged cepstrum patterns for Clusters 0 and 2 are very similar. On the other hand, the results in Fig. 5d show no significant signs of overlapping or mixed signals between phases from either the NBED or Cepstrum patterns, indicating a highly precise clustering result for the Cepstrum + NMF combination. This is determined by comparing these averaged patterns with the individual representative patterns collected from each phase, as seen in Fig. S4, and noting that the averaged patterns do not exhibit irregular or unexpected ordering. Furthermore, a cepstrum vDF image in Fig. S5a, using spots unique to the Heusler cepstrum pattern in Fig. S5b, shows the expected locations of the Heusler phase that once again match well with the layout of Cluster 2 in the Cepstrum + NMF cluster map. The phase mixing in the Cepstrum + PCA cluster map may be attributed to the orthogonality constraint, as noted in a prior study comparing PCA vs NMF for grain mapping in Au-Pd nanoparticles35. This orthogonality constraint allows negative values in the PCA components, which are nonphysical with respect to electron diffraction patterns. Therefore, the components highlighted by the PCA method cannot be linked to any physical crystal structure. To this extent, NMF is more well-suited for applications in electron diffraction data analysis. A similar conclusion was reached in another study that applied unsupervised machine learning to scanning precession electron diffraction data for semi-automatically separating twinned structures45.
Moreover, the improved quality of the clustering result for the Cepstrum + NMF combination compared to that of the NBED + NMF combination can be mainly attributed to the cepstral analysis of the original dataset. This is seen in Fig. 6, where the two cepstrum patterns corresponding to the two NBED patterns in Fig. 4 are displayed with the Cepstrum + NMF cluster map. Clearly, while both diffraction patterns in Fig. 4 exhibit a level of uneven intensity distribution due to sample bending, the cepstrum patterns are centrosymmetric. This result shows how the cepstral transform successfully decouples the lattice structure from the effects of bending. This augmented clustering result can now be used to obtain a more objective map calculation since we have precise access to the locations of particular phases we may want to prioritize as reference structures.
An objective selection of a reference for strain mapping aided by clustering
A fundamental step of strain mapping is the selection of a reference structure, which is normally a user-subjective step. While this step may be relatively straightforward in an epitaxial thin film scenario where the substrate is typically the reference71,72, it is complicated in this SMA system due to the existence of complex microstructures. Nonetheless, through the application of the aforementioned USML workflow, users gain precise access to all data points associated with any given cluster. Therefore, one can readily opt for any cluster as a reference and subsequently average the measured lattice parameters to yield a statistically robust reference lattice. This reference lattice will be self-consistent with every iteration of the workflow. Additionally, it is strongly suggested to omit boundary points to avoid extracting reference lattice information from mixed signals. For the current study, this is accomplished by the application of a 2D convolution over the cluster map using a kernel size of 11 × 11, which corresponds to a roughly 16.5 × 16.5 nm2 region in this scan region.
The resulting strain maps for this scan region are shown in Fig. 7 using the B2 matrix points in Cluster 0 of Fig. 5c to build the reference lattice using the peak pairs method discussed in Methods. Based on the known structure models, the X-direction lattice parameters of the Han and Heusler phase are expected to be larger and smaller than that of the matrix, respectively, which is reflected in Fig. 773,74. Interestingly, the εxx strain map contains similar patterns to those observed in the Cepstrum +NMF cluster map with a clear demarcation of the Han precipitate and Heusler phase locations, which adds to the validation of our clustering algorithm’s performance. In order to quantify the precision of the strain measurement, a subset of matrix lattice parameters measurements from an assumed homogenous region is taken to calculate the standard deviation as a representation of error. The standard deviations of strain in the X-direction and Y-direction are 0.0020 and 0.0017 (or 0.2% and 0.17%), respectively. Based on work done by Yu and colleagues75, the strain fields are likely linked to the formation mechanisms of the precipitates, namely their equilibrium shape and size, which subsequently affects the bulk properties. Hence, possessing this dependable strain calculation method that seamlessly adapts from the clustering results is of utmost value.
Application of USML to a larger field-of-view
We now apply the proposed USML method to a dataset acquired from a larger field of view without prior investigation of the possible existing structures in this region. Enhancing the ability to apply microscale statistical analysis of phase volume fraction, capture complex microstructures, and strain modulation, rather than nano or atomic scale, holds profound significance for metallurgical research. Being able to precisely extract the locations of the secondary precipitate, despite their relatively small volume, with lower spatial resolution scans would prove invaluable.
A vADF image of the larger scan region, using NBED pattern intensities, is displayed in Fig. 8a along with its Cepstrum + NMF clustering result in Fig. 8b for K = 4 (determined by elbow point method, Fig. S6). There are clear boundaries around the needle-shaped precipitates, and the average NBED and cepstrum patterns in Fig. 8c show little sign of intermixing between phases. Clusters 0, 1, and 2 were quickly identified as the B2, Han, and Heusler phases initially described in Fig. 1. Interestingly, a fourth cluster is automatically segregated from the rest of the previously known phases such that two different types of needle-shaped precipitates appear to coexist as Clusters 1 and 3. Cluster 3, upon further investigation, was identified as the [001] Han phase, a 90° rotated version of the [010] Han phase structure initially observed (Fig. S7). Therefore, a noteworthy finding from this study is the coexistence of both Han precipitate orientation relationships within this SMA system. This emphasizes the potential of this data analysis technique, as it automatically identified the new orientation relationship between the B2 matrix and Han precipitates without extensive surveying and imaging of the sample in the TEM. Moreover, the new finding holds great significance in understanding this material system, since varying orientation relationships affect interfacial structures and are likely to have an impact on the macroscopic properties of interest (e.g., martensitic transformation pathways and shape recovery performance)56,57,76,77,78. The strain maps for this dataset, which show consistent features with the cluster map and the strain maps in Fig. 7, are provided in Supplementary Fig. S8.
a vADF image of the large FoV scan region showing several needle-shaped precipitates embedded in the matrix. b Cepstrum + NMF clustering result for K = 4, which was automatically determined again by the elbow point method (Fig. S6). c Averaged NBED and cepstrum patterns from (b). Scale bars are 10 nm−1 for the NBED patterns and 5 Å for the cepstrum patterns.
In summary, we provide a simple and effective USML workflow that demonstrates how the combination of cepstral analysis, NMF, and K-means clustering is an ideal combination for visualizing the arrangement of coherent, heterogeneous structures at small to large length scales in the TEM. The performance was judged based on the visual quality of the clustering maps, good agreement with the cepstrum vDF image, and the lack of phase intermixing in the cluster-averaged diffraction and cepstrum patterns. This analysis can be performed semi-automatically with objective means of determining just two input parameters, the number of components and number of clusters, and minimal prior knowledge of the material system. Additionally, we establish the usage of the clustering results to declare an objective reference lattice from which more robust strain maps can be collected. The εxx strain map, in particular, featured structural patterns that matched very well with those in the Cepstrum + NMF cluster map, thus furthering our confidence in the effectiveness of this USML method. We expect that this data analysis method will excel at accelerating systematic studies on novel multiphase systems and improving our understanding of fundamental processing-structure-property relationships. As such, we strongly encourage the use of the aforementioned USML workflow in other complex materials to gain greater insight into the correlations between structural formation mechanisms and macroscopic properties of interest to inform better materials design considerations.
Moving forward, we note that this particular workflow has performed well for the phase segregation task thanks to the innate properties of the cepstral analysis, but not when precise orientation determination is needed. In that case, using the original NBED 4D dataset would be better suited and may be augmented by applying energy filtering79,80. The SMA study itself would also benefit from expanding on the cepstral analysis through the application of different cepstrums. This method can offer improved visualization and insight into local ordering within disordered material systems, especially in alloys with nonuniform chemical distritbuion65,66,67,69,81.
Methods
Ni50Ti26Hf20Al4 shape memory alloy fabrication
The Ni50Ti26Hf20Al4 alloy was fabricated by arc-melting under an ultra-high purity argon atmosphere. The raw metals from Alfa Aesar, Ni wires (99.5% purity), Ti wires (99.7% purity), Hf wires (99.7% purity), and Al wires (99.999% purity) were weighed and melted into buttons. The buttons were flipped six times to ensure homogeneity. The as-cast material was vacuum-sealed in a quartz tube, solution-treated at 950 °C for 100 h and quenched in oil. The same encapsulation procedure was followed to age the alloy at 600°C for 100 h and oil-quenched once again. Differential scanning calorimetry (DSC) was used to determine the martensitic transformation temperature for this particular composition, which was found to be below the minimum scan temperature of −60 ˚C. This sample is used as a reference point to which future studies on other NiTiHfAl SMAs can be compared.
STEM experiments and sample preparation
The lamella for STEM and 4D-STEM analysis was prepared using standard focused ion beam (FIB) procedures with 30 kV coarse thinning followed by cleaning at 5 kV and 2 kV. Prior to FIB preparation, the alloy specimens were successively polished using 1200-grit SiC papers and diamond slurry of 3, 1, and 0.3 μm. High-angle annular dark-field (HAADF) imaging in STEM was performed on an aberration-corrected Themis Z STEM (Fisher Scientific) at 200 kV with a probe semiconvergence angle of 22 mrad, probe current of 25 pA, camera length of 115 mm, and detector inner cutoff collection angle of 58 mrad. 4D-STEM experiments were also performed on the Themis Z, which is equipped with an electron microscope pixel array detector (EMPAD). The accelerating voltage was 200 kV with an electron probe size (full width at half maximum) of ~1.5 nm using a probe semiconvergence angle of 0.94 mrad, camera length of 360 mm, and C2 aperture size of 50 μm. Two 4D datasets were acquired using scan step sizes of 0.40 nm and 0.68 nm to cover 163.8 × 63.7 nm2 and 213 × 213 nm2 regions, respectively. The exposure time was set to 10 ms, and the screen current was set such that the EMPAD was subject to a current density of less than 2 pA/pixel.
Principal component analysis
Principal component analysis (PCA) operates by finding a set number of orthogonal components that are linear combinations of the original data while retaining maximum variance36. The orthogonality constraint makes PCA more visually interpretable in 2D and 3D representations, but it also allows for negative values that are nonphysical in the context of electron diffraction patterns. The PCA algorithm seeks to factorize the matrix X into a product of three other matrices, C, D, and V. First, X is centered at its mean, which allows PCA to be performed using singular value decomposition as in Eq. 2 where X is decomposed to a number of components r.
D is a diagonal matrix where the elements along the diagonal are arranged in descending order. The square of these values gives the variances explained by the corresponding principal component. Essentially, the product of C and D, we denote this as P, gives the principal components in the columns while the values in V are the weights, or coefficients, that dictate each component’s contribution to a corresponding original data point in X.
Nonnegative matrix factorization
Nonnegative matrix factorization (NMF) learns the parts of an image without an orthogonality constraint while keeping all elements positive37. This results in components that are physically interpretable as electron diffraction patterns since negative electron counts are not possible. In the original work by Lee and Seung37, it was shown that NMF results in components that are mostly sparse (empty) while holding only a specific feature, such as an eyebrow or chin in the case of facial image analysis. The matrix factorization with NMF is similar to that of PCA:
Mathematically, NMF is defined by Eq. 3 above, where the two matrices W and H are analogous to P and V in PCA, respectively. NMF learns the best representation of the dataset when reduced to a lower rank r by minimizing the difference between the original data X and the product of the basis vectors W and corresponding weights H.
K-means clustering
K-means clustering minimizes the Euclidian distance between data points and their respective cluster centers82. After the entire dataset is reduced down to its optimal number of components from either PCA or NMF, each set of components is clustered using the K-means algorithm. The elbow method is used to pick the optimal value of K due to our lack of knowledge of all possible coexisting phases. First, the within-sum-of-squares (WSS), or inertia, is plotted against a range of K values to determine the optimal choice for number of clusters. The elbow point is the K-value where incrementing K to K + 1 does not provide as much optimization as incrementing K – 1 to K. In other words, the elbow point of the inertia curve is where the second derivative initially becomes negative, or the slope decreases. These plots are made such that each point representing an inertia value for a value of K is an average of 10 individual runs with random initializations. In cases where the elbow point is unclear, we suggest erring toward a higher K-value since two or more clusters that are found to be of the same phase can simply be conjoined after the fact. On the contrary, underestimating the K-value can lead to missing important unique structures that may otherwise only be distinguishable by subtle features.
Strain mapping—peak pairs method
The peak pairs analysis method is utilized to create strain maps, which was first introduced by Galindo et al.71. Lattice parameters are calculated for every pattern in sub-pixel units, and a portion of those are used to define a reference lattice with two basis vectors. The selection of reference lattice points is often subjective, but it is later described how the clustering results can be used to consistently form ideal references. From here, the longitudinal and shear strain can be calculated using the equations below.
Where exx, eyy, and exy/eyx are matrices containing the longitudinal x-strain, longitudinal y-strain, and shear strains, respectively, while u and v contain the actual peak positions. The vectors a and b denote the reference lattice and do not change after they are defined.
Data availability
The 4D-STEM datasets used to support the findings in this work will be made available in a repository at https://zenodo.org/communities/kimgroup-uf/.
Code availability
The custom Python scripts used for this work will be available upon request to the corresponding author.
References
Ophus, C. Four-dimensional scanning transmission electron microscopy (4D-STEM): from scanning nanodiffraction to ptychography and beyond. Microsc. Microanal. 25, 563–582 (2019).
Rauch, E. F. & Véron, M. Methods for orientation and phase identification of nano-sized embedded secondary phase particles by 4D scanning precession electron diffraction. Acta Crystallogr. B Struct. Sci. Cryst. Eng. Mater. 75, 505–511 (2019).
Shukla, A. K. et al. Effect of composition on the structure of lithium- and manganese-rich transition metal oxides. Energy Environ. Sci. 11, 830–840 (2018).
Chen, W. et al. Formation and impact of nanoscopic oriented phase domains in electrochemical crystalline electrodes. Nat. Mater. 22, 92–99 (2023).
Han, Y. et al. Strain mapping of two-dimensional heterostructures with subpicometer precision. Nano Lett. 18, 3746–3751 (2018).
Thronsen, E. et al. Studying GPI zones in Al-Zn-Mg alloys by 4D-STEM. Mater. Charact. 185, 111675 (2022).
Bustillo, K. C. et al. 4D-STEM of beam-sensitive materials. Acc. Chem. Res. 54, 2543–2551 (2021).
Nord, M. et al. Fast pixelated detectors in scanning transmission electron microscopy. Part I: Data acquisition, live processing, and storage. Microsc. Microanal. 26, 653–666 (2020).
Paterson, G. W. et al. Fast pixelated detectors in scanning transmission electron microscopy. Part II: Post-acquisition data processing, visualization, and structural characterization. Microsc. Microanal. 26, 944–963 (2020).
Tate, M. W. et al. High dynamic range pixel array detector for scanning transmission electron microscopy. Microsc. Microanal. 22, 237–249 (2016).
Gao, W. et al. Real-space charge-density imaging with sub-ångström resolution by four-dimensional electron microscopy. Nature 575, 480–484 (2019).
Chen, Z. et al. Electron ptychography achieves atomic-resolution limits set by lattice vibrations. Science 372, 826–831 (2021).
Das, S. et al. Observation of room-temperature polar skyrmions. Nature 568, 368–372 (2019).
Wang, S., Eldred, T. B., Smith, J. G. & Gao, W. AutoDisk: automated diffraction processing and strain mapping in 4D-STEM. Ultramicroscopy 236, 113513 (2022).
Mukherjee, D., Gamler, J. T. L., Skrabalak, S. E. & Unocic, R. R. Lattice strain measurement of core@shell electrocatalysts with 4D scanning transmission electron microscopy nanobeam electron diffraction. ACS Catal. 10, 5529–5541 (2020).
Ophus, C. et al. Automated crystal orientation mapping in py4DSTEM using sparse correlation matching. Microsc. Microanal. 28, 390–403 (2022).
Im, S. et al. Direct determination of structural heterogeneity in metallic glasses using four-dimensional scanning transmission electron microscopy. Ultramicroscopy 195, 189–193 (2018).
Im, S. et al. Structural heterogeneity, ductility, and glass forming ability of Zr-based metallic glasses. SSRN J. https://doi.org/10.2139/ssrn.3683539 (2020).
Cautaerts, N. et al. Free, flexible and fast: orientation mapping using the multi-core and GPU-accelerated template matching capabilities in the Python-based open source 4D-STEM analysis toolbox Pyxem. Ultramicroscopy 237, 113517 (2022).
Savitzky, B. H. et al. py4DSTEM: a software package for multimodal analysis of four-dimensional scanning transmission electron microscopy datasets. Microsc. Microanal. 27, 712–743 (2021).
Savitzky, B. H. et al. py4DSTEM: open source software for 4D-STEM data analysis. Microsc. Microanal. 25, 124–125 (2019).
Clausen, A. et al. LiberTEM: software platform for scalable multidimensional data processing in transmission electron microscopy. JOSS 5, 2006 (2020).
Ziatdinov, M., Ghosh, A., Wong, C. Y. & Kalinin, S. V. AtomAI framework for deep learning analysis of image and spectroscopy data in electron and scanning probe microscopy. Nat. Mach. Intell. 4, 1101–1112 (2022).
De La Pena, F. et al. Electron microscopy (big and small) data analysis with the open source software package HyperSpy. Microsc. Microanal. 23, 214–215 (2017).
O’Shea, K. & Nash, R. An introduction to convolutional neural networks. Preprint at http://arxiv.org/abs/1511.08458 (2015).
Smith, L. N. A disciplined approach to neural network hyper-parameters: Part 1—learning rate, batch size, momentum, and weight decay. Preprint at http://arxiv.org/abs/1803.09820 (2018).
Yuan, R., Zhang, J., He, L. & Zuo, J.-M. Training artificial neural networks for precision orientation and strain mapping using 4D electron diffraction datasets. Ultramicroscopy 231, 113256 (2021).
Zhang, C., Feng, J., DaCosta, L. R. & Voyles, P. M. Atomic resolution convergent beam electron diffraction analysis using convolutional neural networks. Ultramicroscopy 210, 112921 (2020).
Roberts, G. et al. Deep learning for semantic segmentation of defects in advanced STEM images of steels. Sci. Rep. 9, 12744 (2019).
Lin, R., Zhang, R., Wang, C., Yang, X.-Q. & Xin, H. L. TEMImageNet training library and AtomSegNet deep-learning models for high-precision atom segmentation, localization, denoising, and deblurring of atomic-resolution images. Sci. Rep. 11, 5386 (2021).
Goodge, B. H. et al. Disentangling coexisting structural order through phase lock-in analysis of atomic-resolution STEM data. Microsc. Microanal. 28, 404–411 (2022).
Lee, C.-H. et al. Deep learning enabled strain mapping of single-atom defects in two-dimensional transition metal dichalcogenides with sub-picometer precision. Nano Lett. 20, 3369–3377 (2020).
Aguiar, J. A., Gong, M. L., Unocic, R. R., Tasdizen, T. & Miller, B. D. Decoding crystallography from high-resolution electron imaging and diffraction datasets with deep learning. Sci. Adv. 5, eaaw1949 (2019).
Khan, A., Lee, C.-H., Huang, P. Y. & Clark, B. K. Leveraging generative adversarial networks to create realistic scanning transmission electron microscopy images. NPJ Comput. Mater. 9, 85 (2023).
Allen, F. I. et al. Fast grain mapping with sub-nanometer resolution using 4D-STEM with grain classification by principal component analysis and non-negative matrix factorization. Microsc. Microanal. 27, 794–803 (2021).
Jolliffe, I. T. & Cadima, J. Principal component analysis: a review and recent developments. Philos. Trans. R. Soc. A. 374, 20150202 (2016).
Lee, D. D. & Seung, H. S. Learning the parts of objects by non-negative matrix factorization. Nature 401, 788–791 (1999).
Kalinin, S. V. et al. Unsupervised machine learning discovery of structural units and transformation pathways from imaging data. APL Mach. Learn. 1, 026117 (2023).
Ryu, J. et al. Dimensionality reduction and unsupervised clustering for EELS-SI. Ultramicroscopy 231, 113314 (2021).
Wang, N., Freysoldt, C., Zhang, S., Liebscher, C. H. & Neugebauer, J. Segmentation of static and dynamic atomic-resolution microscopy data sets with unsupervised machine learning using local symmetry descriptors. Microsc. Microanal. 27, 1454–1464 (2021).
Uesugi, F. et al. Non-negative matrix factorization for mining big data obtained using four-dimensional scanning transmission electron microscopy. Ultramicroscopy 221, 113168 (2021).
Bruefach, A., Ophus, C. & Scott, M. C. Robust design of semi-automated clustering models for 4D-STEM datasets. APL Mach. Learn. 1, 016106 (2023).
Mu, X., Chen, L., Mikut, R., Hahn, H. & Kübel, C. Unveiling local atomic bonding and packing of amorphous nanophases via independent component analysis facilitated pair distribution function. Acta Mater. 212, 116932 (2021).
Nalin Mehta, A. et al. Unravelling stacking order in epitaxial bilayer MX2 using 4D-STEM with unsupervised learning. Nanotechnology 31, 445702 (2020).
Martineau, B. H., Johnstone, D. N., Van Helvoort, A. T. J., Midgley, P. A. & Eggeman, A. S. Unsupervised machine learning applied to scanning precession electron diffraction data. Adv. Struct. Chem. Imaging 5, 3 (2019).
Shi, C. et al. Uncovering material deformations via machine learning combined with four-dimensional scanning transmission electron microscopy. NPJ Comput. Mater. 8, 114 (2022).
Bruefach, A., Ophus, C. & Scott, M. C. Analysis of interpretable data representations for 4D-STEM using unsupervised learning. Microsc. Microanal. 28, 1998–2008 (2022).
Kimoto, K. et al. Unsupervised machine learning combined with 4D scanning transmission electron microscopy for bimodal nanostructural analysis. Sci. Rep. 14, 2901 (2024).
Wen, H., Luna-Romera, J. M., Riquelme, J. C., Dwyer, C. & Chang, S. L. Y. Statistically representative metrology of nanoparticles via unsupervised machine learning of TEM images. Nanomaterials 11, 2706 (2021).
Otsuka, K. & Kakeshita, T. Science and technology of shape-memory alloys: new developments. MRS Bull. 27, 91–100 (2002).
Karaca, H. E., Acar, E., Tobe, H. & Saghaian, S. M. NiTiHf-based shape memory alloys. Mater. Sci. Technol. 30, 1530–1544 (2014).
Hartl, D. J. & Lagoudas, D. C. Aerospace applications of shape memory alloys. Proc. Inst. Mech. Eng. G J. Aerosp. Eng. 221, 535–552 (2007).
Han, X. D., Wang, R., Zhang, Z. & Yang, D. Z. A new precipitate phase in a TiNiHf high temperature shape memory alloy. Acta Mater. 46, 273–281 (1998).
Yang, F. et al. Structure analysis of a precipitate phase in an Ni-rich high-temperature NiTiHf shape memory alloy. Acta Mater. 61, 3335–3346 (2013).
Coughlin, D. R. et al. Characterization of the microstructure and mechanical properties of a 50.3Ni–29.7Ti–20Hf shape memory alloy. Scr. Mater. 67, 112–115 (2012).
Coughlin, D. R., Casalena, L., Yang, F., Noebe, R. D. & Mills, M. J. Microstructure–property relationships in a high-strength 51Ni–29Ti–20Hf shape memory alloy. J. Mater. Sci. 51, 766–778 (2016).
Stebner, A. P. et al. Transformation strains and temperatures of a nickel–titanium–hafnium high temperature shape memory alloy. Acta Mater. 76, 40–53 (2014).
Jung, J., Ghosh, G. & Olson, G. B. A comparative study of precipitation behavior of Heusler phase (Ni2TiAl) from B2-TiNi in Ni–Ti–Al and Ni–Ti–Al–X (X = Hf, Pd, Pt, Zr) alloys. Acta Mater. 51, 6341–6357 (2003).
Jung, J., Ghosh, G., Isheim, D. & Olson, G. B. Precipitation of Heusler phase (Ni2TiAl) from B2-TiNi in Ni-Ti-Al and Ni-Ti-Al-X (X = Hf, Zr) alloys. Met. Mater. Trans. A 34, 1221–1235 (2003).
Hsu, D. H. D. et al. The effect of aluminum additions on the thermal, microstructural, and mechanical behavior of NiTiHf shape memory alloys. J. Alloy. Compd. 638, 67–76 (2015).
Horwath, J. P., Zakharov, D. N., Mégret, R. & Stach, E. A. Understanding important features of deep learning models for segmentation of high-resolution transmission electron microscopy images. NPJ Comput. Mater. 6, 108 (2020).
Akers, S. et al. Rapid and flexible segmentation of electron microscopy data using few-shot machine learning. NPJ Comput. Mater. 7, 187 (2021).
Calvino, J. J., López-Haro, M., Muñoz-Ocaña, J. M., Puerto, J. & Rodríguez-Chía, A. M. Segmentation of scanning-transmission electron microscopy images using the ordered median problem. Eur. J. Oper. Res. 302, 671–687 (2022).
Padgett, E. et al. The exit-wave power-cepstrum transform for scanning nanobeam electron diffraction: robust strain mapping at subnanometer resolution and subpicometer precision. Ultramicroscopy 214, 112994 (2020).
Hsiao, H.-W. et al. Data-driven electron-diffraction approach reveals local short-range ordering in CrCoNi with ordering effects. Nat. Commun. 13, 6651 (2022).
Shao, Y.-T. et al. Cepstral scanning transmission electron microscopy imaging of severe lattice distortions. Ultramicroscopy 231, 113252 (2021).
Pidaparthy, S., Ni, H., Hou, H., Abraham, D. P. & Zuo, J.-M. Fluctuation cepstral scanning transmission electron microscopy of mixed-phase amorphous materials. Ultramicroscopy 248, 113718 (2023).
Hou, H., Pidaparthy, S., Ni, H. & Zuo, J.-M. Fluctuation component analysis-based K-means clustering in 4D-STEM of heterogeneous materials. Microsc. Microanal. 29, 687–688 (2023).
Yin, K., Hsiao, H.-W., Feng, R., Liaw, P. K. & Zuo, J.-M. Deformation defects characterization in short-range ordered CrCoNi using fast electron detectors and 4D-STEM. Microsc. Microanal. 29, 251–253 (2023).
Bolhuis, M., van Heijst, S. E., Sangers, J. J. M. & Conesa-Boj, S. 4D‐STEM nanoscale strain analysis in van der Waals materials. Small Sci. 4, 2300249 (2024).
Galindo, P. L. et al. The Peak Pairs algorithm for strain mapping from HRTEM images. Ultramicroscopy 107, 1186–1193 (2007).
Zuo, J.-M. et al. Lattice and strain analysis of atomic resolution Z-contrast images based on template matching. Ultramicroscopy 136, 50–60 (2014).
Santamarta, R. et al. TEM study of structural and microstructural characteristics of a precipitate phase in Ni-rich Ni–Ti–Hf and Ni–Ti–Zr shape memory alloys. Acta Mater. 61, 6191–6206 (2013).
Oh-ishi, K., Horita, Z. & Nemoto, M. Phase separation and lattice misfit in NiAl-Ni2TiAl-NiTi system. Mater. Trans. JIM 38, 99–106 (1997).
Yu, T. et al. H-phase precipitation and its effects on martensitic transformation in NiTi-Hf high-temperature shape memory alloys. Acta Mater. 208, 116651 (2021).
Timofeeva, E. E. et al. Effect of one family of Ti3Ni4 precipitates on shape memory effect, superelasticity and strength properties of the B2 phase in high-nickel [001]-oriented Ti-51.5 at.%Ni single crystals. Mater. Sci. Eng. A 832, 142420 (2022).
Kim, H. et al. Elucidating the role of a unique step-like interfacial structure of η4 precipitates in Al-Zn-Mg alloy. Sci. Adv. 9, eadf7426 (2023).
Xia, C. et al. Orientation relationships between precipitates and matrix and their crystallographic transformation in a Cu–Cr–Zr alloy. Mater. Sci. Eng. A 850, 143576 (2022).
Miller, B. K., Schaffer, B. & Pakzad, A. Continuous 4D STEM recording and visualization for in-situ experiments. Microsc. Microanal. 29, 271–271 (2023).
Kazmierczak, N. P. et al. Strain fields in twisted bilayer graphene. Nat. Mater. 20, 956–963 (2021).
Zuo, J.-M. et al. Electron microscopy of electrochemical degradation in energy materials across multiple length scales: challenges and opportunities. Microsc. Microanal. 29, 1272–1273 (2023).
Macqueen, J. Some methods for classification and analysis of multivariate observations. In Proc. Fifth Berkeley Symposium on Mathematical Statistics and Probability, 1, 281–297 (1967).
Acknowledgements
This work was supported by the U.S. National Science Foundation (Grant No. DMR 2226478). The authors acknowledge the Research Service Centers at the Herbert Wertheim College of Engineering at the University of Florida. T.Y. acknowledges the University of Florida Graduate School Fellowship.
Author information
Authors and Affiliations
Contributions
Experiments were performed by T.Y. and E.H. under the supervision of H.K.; T.Y. developed the unsupervised machine learning algorithm under the supervision of H.K.; Y.Y., F.C.G., and M.V.M. synthesized and provided the shape memory alloy sample.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
41524_2024_1414_MOESM1_ESM.docx
Unsupervised Machine Learning and Cepstral Analysis with 4D-STEM for Characterizing Complex Microstructures of Metallic Alloys
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yoo, T., Hershkovitz, E., Yang, Y. et al. Unsupervised machine learning and cepstral analysis with 4D-STEM for characterizing complex microstructures of metallic alloys. npj Comput Mater 10, 223 (2024). https://doi.org/10.1038/s41524-024-01414-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41524-024-01414-3
- Springer Nature Limited