Abstract
The interim analysis of the CIRCULATE-Japan GALAXY observational study demonstrated the association of circulating tumor DNA (ctDNA)-based molecular residual disease (MRD) detection with recurrence risk and benefit from adjuvant chemotherapy (ACT) in resectable colorectal cancer (CRC). This updated analysis with a 23-month median follow-up, including 2,240 patients with stage II–III colon cancer or stage IV CRC, reinforces the prognostic value of ctDNA positivity during the MRD window with significantly inferior disease-free survival (DFS; hazard ratio (HR): 11.99, P < 0.0001) and overall survival (OS; HR: 9.68, P < 0.0001). In patients who experienced recurrence, ctDNA positivity correlated with shorter OS (HR: 2.71, P < 0.0001). The significantly shorter DFS in MRD-positive patients was consistent across actionable biomarker subsets. Sustained ctDNA clearance in response to ACT was an indicator of favorable DFS and OS compared to transient clearance (24-month DFS: 89.0% versus 3.3%; 24-month OS: 100.0% versus 82.3%). True spontaneous clearance rate with no clinical recurrence was 1.9% (2/105). Overall, our findings provide evidence for the utility of ctDNA monitoring for post-resection recurrence and mortality risk stratification that could be used for guiding adjuvant therapy.
Similar content being viewed by others
Main
Circulating tumor DNA (ctDNA) has emerged as a minimally invasive and reliable prognostic biomarker for detecting post-surgical molecular residual disease (MRD) and predicting recurrence risk in colorectal cancer (CRC)1,2,3,4. Despite the established association between MRD and recurrence, the prognostic effect of MRD on overall survival (OS) remains unclear due to a lack of prospective evidence. Furthermore, although targeted therapies could be considered for adjuvant treatment intensification in patients with MRD-positive CRC, the prognostic and predictive value of ctDNA-based MRD detection according to actionable biomarkers is yet to be established. Additionally, the potential of MRD clearance as a surrogate endpoint of treatment efficacy and clinical outcomes in CRC, as demonstrated in leukemia and multiple myeloma5,6, warrants further investigation. To address these critical knowledge gaps, a large prospective trial with sufficient follow-up is needed.
We previously published an interim analysis from the GALAXY study, the observational arm of the ongoing, multinational (Japan and Taiwan), prospective, multicenter CIRCULATE-Japan study (UMIN000039205)3. The GALAXY study is designed to monitor ctDNA-based MRD status for patients with clinical stage II–IV or relapsed CRC after curative-intent surgery7. In our initial report involving 1,039 patients, we showed a significant correlation between post-surgical ctDNA positivity and elevated recurrence risk, compared to other high-risk clinicopathological features, and a larger benefit from adjuvant chemotherapy (ACT) in patients with MRD-positivity3. The patients were followed for a median of 16.7 months (range, 0.5–24.8 months). Here we report updated results from the GALAXY study in an expanded cohort of 2,240 patients with stage II–III resectable colon cancer or stage IV CRC with extended follow-up (median, 23 months; range, 2–49 months). We aimed to comprehensively evaluate the prognostic and predictive value of post-surgical ctDNA detection and its association with disease-free survival (DFS) and OS. We further investigated whether ctDNA MRD positivity predicts outcomes based on actionable biomarker stratification and whether ctDNA clearance in response to ACT informs treatment efficacy.
Results
Patient characteristics
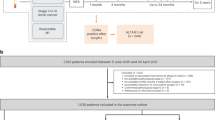
Of 6,061 patients with surgically resectable CRC enrolled between 8 May 2020 and 31 March 2024, 2,240 patients with stage IV rectal or any stage colon cancer were included in this analysis (Fig. 1a) with a total of 13,429 plasma samples. The median patient follow-up was 23 months (range, 2–49 months) as of 3 June 2024. Patient characteristics are summarized in Fig. 1b. Extended Data Table 1a,b describe characteristics for patients with stage I–III and stage IV disease, respectively, and Extended Data Table 1c lists organs involved in metastasis for patients with stage IV disease.
ctDNA status in the MRD and surveillance windows
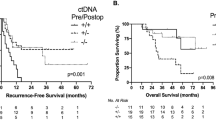
ctDNA status during the MRD window was available for 2,110 patients. With one patient having an OS event before the landmark (date of MRD timepoint), a total of 2,109 patients were included in the outcomes analysis. Of these 2,109 patients, 15.93% (336/2,109) were MRD positive, of whom 78.27% (263/336) experienced recurrence, whereas only 13.14% (233/1,773) of MRD-negative patients experienced recurrence (hazard ratio (HR): 11.99, 95% confidence interval (CI): 10.02–14.35, P < 0.0001; Fig. 2a), demonstrating 24-month DFS (from the landmark timepoint) of 20.57% (95% CI: 16.14–25.37%) versus 85.10% (95% CI: 83.20–86.90%), respectively. Remarkably, 36-month DFS for MRD-positive and MRD-negative patients were 16.7% (95% CI: 12.1–21.9%) versus 83.5% (95% CI: 81.2–85.6%). The association of MRD positivity with a significantly increased risk for recurrence was observed across all pathological stages (Extended Data Fig. 1a,b). Extended Data Fig. 2a,b shows DFS analysis with similar trends for stage I–III (HR: 15.97, 95% CI: 12.51–20.37, P < 0.0001) and stage IV (HR: 5.83, 95% CI: 4.44–7.64, P < 0.0001). Next, MRD positivity was found to be significantly associated with worse OS compared to MRD-negative patients (HR: 9.68, 95% CI: 6.33–14.82, P < 0.0001; Fig. 2b), demonstrating a 24-month OS of 83.65% (95% CI: 77.84–88.06%) versus 98.50% (95% CI: 97.70–99.10%), respectively, and a 36-month OS of 71.80% (95% CI: 63.40–78.60%) versus 96.0% (95% CI: 94.30–97.20%), respectively. Extended Data Fig. 2c,d shows OS analysis with similar trends for stage I–III (HR: 9.64, 95% CI: 5.47–16.99, P < 0.0001) and stage IV (HR: 6.24, 95% CI: 3.21–12.11, P < 0.0001).
a,b, Kaplan–Meier estimates for DFS (a) and OS (b) stratified by ctDNA status during the MRD window (negative versus positive). P = 7.55 × 10–162 (a); P = 1.25 × 10–25 (b). P value for the bar plots showing the association between ctDNA status and DFS or OS events was calculated based on the two-sided chi-square test: P = 4.11 × 10−146 (a); P = 6.97 × 10−29 (b). c,d, Forest plot depicting the multivariate analysis (including ctDNA status during MRD window and other clinicopathological factors) for DFS (c) and OS (d) in patients with CRC. P = 1.10 × 10–96 (c, ctDNA); P = 1.43 × 10−4 (c, MSI); P = 2.55 × 10−15 (d, ctDNA); P = 3.56 × 10−5 (d, BRAF V600E). e,f, Kaplan–Meier estimates for DFS (e) and OS (f) stratified by ctDNA status during the surveillance window (negative versus positive). P = 1.21 × 10−163 (e); P = 1.20 × 10−20 (f). Two-sided chi-square test for bar plots: P = 1.19 × 10−217 (e); P = 1.10 × 10−29 (f). g, Forest plot depicting the multivariate analysis (including ctDNA status during the surveillance window and other clinicopathological factors) for DFS in patients with CRC. P = 6.79 × 10−118 (g, ctDNA); P = 1.88 × 10−4 (g, sex). HRs and 95% CIs were calculated using the Cox proportional hazard model; P values were calculated using the two-sided log-rank test (a,b,e,f). Various prognostic factors and their association with DFS, as indicated by HR, were analyzed across the cohort using the two-sided Wald chi-squared test (c,d,g). The unadjusted HRs (squares) and 95% CIs (horizontal lines) are shown for each prognostic factor; the vertical dotted line represents the null hypothesis (c,d,g). The DFS and OS analyses in the MRD window were landmarked from the date of the MRD timepoint, and the analyses in the surveillance window were landmarked at 10 weeks after surgery. Median DFS/OS and percentage DFS and OS at 24 months, 30 months and 36 months were estimated from the landmark timepoint. AIC, Akaike information criterion; mDFS, median DFS; mOS, median OS; NR, not reached.
To further evaluate the prognostic value of ctDNA when compared to other clinicopathological risk factors, we performed a multivariate analysis in the MRD window. ctDNA positivity was observed to be the single most significant prognostic factor associated with inferior DFS (HR: 12.08, 95% CI: 9.56–15.27, P < 0.001; Fig. 2c) and OS (HR: 9.87, 95% CI: 5.60–17.40, P < 0.001; Fig. 2d). The other clinicopathological factors prognostic of both poor DFS and OS included the presence of BRAF V600E mutation (DFS: HR: 2.03, 95% CI: 1.19–3.45, P = 0.009; OS: HR: 7.44, 95% CI: 2.87–19.27, P < 0.001), lymph node positivity (DFS: HR: 1.56, 95% CI: 1.19–2.03, P = 0.001; OS: HR: 2.19, 95% CI: 1.12–4.30, P = 0.022) and the presence of RAS mutations (DFS: HR: 1.43, 95% CI: 1.14–1.80, P = 0.002; OS: HR: 1.93, 95% CI: 1.07–3.48, P = 0.029; Fig. 2c,d). Higher pathological T stage was significantly associated with inferior DFS (HR: 1.67, 95% CI: 1.05–2.65, P = 0.031; Fig. 2c) but not with inferior OS.
In the surveillance window, 1,791 and 1,794 patients were included in the DFS and OS analysis, respectively (Fig. 1a). Compared to patients who were serially ctDNA negative, patients with ctDNA positivity at any timepoint were approximately 34 times more likely to recur (HR: 33.56, 95% CI: 26.07–43.20, P < 0.0001; 24-month DFS: 8.93% (95% CI: 5.56–13.27%) versus 93.20% (95% CI: 91.50–94.50%); Fig. 2e) and also exhibited a significantly shorter OS (HR: 19.51, 95% CI: 10.44–36.44, P < 0.0001; 24-month OS: 83.20 (95% CI: 76.50–88.10%) versus 99.30% (95% CI: 98.40–99.70%); Fig. 2f). Additionally, in the multivariate analysis, ctDNA positivity during surveillance was the strongest prognostic factor associated with poor DFS (HR: 44.24, 95% CI: 32.07–61.03, P < 0.001; Fig. 2g).
Extended Data Fig. 3a represents a Sankey plot demonstrating the percentage of patients with MRD positivity or negativity, the percentage of patients with subsequent ctDNA status during surveillance (after ACT or observation) and the percentage of each group with confirmed radiological recurrence. Furthermore, we analyzed a subgroup of patients for whom ctDNA status in both MRD and surveillance windows was available (n = 1,664). Notably, 84.33% (253/300) of patients who were ctDNA positive in the surveillance window recurred, whereas only 5.27% (72/1,364) of those who were ctDNA negative in that window recurred (Extended Data Fig. 3b), which is consistent with the recurrence rates observed in the overall surveillance window analysis cohort (84.19% versus 6.01%; Fig. 2e). Longitudinally, ctDNA positivity preceded radiological recurrence by a median of 5.91 months (range, 0–33.15 months).
Together, our updated analysis demonstrates that MRD positivity is the most significant predictor not only of DFS but also of OS, outperforming conventional clinicopathological characteristics, microsatellite instability (MSI) status and BRAF V600E mutations.
Impact of ctDNA status on post-recurrence survival
We found that ctDNA positivity was associated with not only an increased risk of recurrence but also a higher risk of mortality in patients with resected CRC. However, it is crucial to determine whether the association between ctDNA positivity and mortality is solely due to a higher incidence of clinical recurrence or if ctDNA positivity itself has an impact on overall prognosis, independent of recurrence. To address this question, we investigated the association between ctDNA status and survival outcomes of patients who experienced radiological recurrence during the follow-up (n = 500; Fig. 3a). Figure 3b shows the association of percentage of patients with ctDNA positivity and negativity during the MRD window and any time after surgery with the site of recurrence.
a, CONSORT diagram depicting patients included in the OS cohorts of patients with radiological recurrence. b, Bar plot showing the association of percentage of patients with ctDNA positivity and negativity during the MRD window and any time after surgery with the site of recurrence. c, ctDNA status in the MRD window is predictive of OS across all recurrence sites in post-surgical patients with CRC. Note: Given that 226 of the 500 patients with radiological recurrence had metastatic involvement at more than one site, these individual patients were counted in more than one category. d,e, Kaplan–Meier estimates for OS stratified by ctDNA status during the MRD window (d) or surveillance window (e) of patients with CRC who had radiological recurrence during follow-up. P = 9.89 × 10−5 (d). HRs and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. The OS analysis in the MRD window was landmarked from the date of the MRD timepoint, and the analysis in the surveillance window was landmarked at 10 weeks after surgery. Median OS and percentage OS at 24 months were estimated from the landmark timepoint. ctDNA−, ctDNA negative; ctDNA+, ctDNA positive; LN, lymph node.
ctDNA positivity was reported to be associated with the recurrence site8,9, and patients with lung recurrence, which is associated with a better prognosis10, were less likely to have ctDNA positivity. However, the HRs for OS between recurrent patients with ctDNA positivity and negativity in the MRD window were observed to be consistent regardless of the site of recurrence (Fig. 3c). Patients with ctDNA positivity during the MRD and surveillance windows had significantly inferior OS than their ctDNA-negative counterparts (MRD window: HR: 2.71, 95% CI: 1.64–4.47, P < 0.0001, Fig. 3d; surveillance window: HR: 8.40, 95% CI: 2.03–34.78, P = 0.003, Fig. 3e). Similarly, ctDNA positivity during the MRD and surveillance windows was observed to be significantly associated with poor post-recurrence survival (PRS) (MRD window: HR: 1.97, 95% CI: 1.20–3.24, P = 0.008, median follow-up after recurrence: 11.75 months, Extended Data Fig. 4a; surveillance window: HR: 6.57, 95% CI: 1.59–27.21, P = 0.009, median follow-up after radiological recurrence: 11.48 months, Extended Data Fig. 4b).
Long-term survival can be expected for patients with oligometastatic disease (OMD) if metastasectomy is achieved. In line with the observation above where ctDNA-positive patients experienced poor PRS, more recurrent patients with ctDNA-negative status (41.1%) underwent curative-intent resection for OMD when compared to ctDNA-positive patients (29.9%, P = 0.014; Extended Data Fig. 4c).
MRD status is prognostic of DFS across actionable biomarkers
In our cohort of 2,240 patients with CRC, the following actionable biomarkers were identified: wild-type (WT) RAS/BRAF in 1,125 (50.22%) patients, high tumor mutational burden (TMB) in 230 (10.27%) patients, MSI high in 215 (9.55%) patients, BRAF V600E in 178 (7.94%) patients, KRAS G12C in 49 (2.19%) patients, ERBB2 amplification in 36 (1.61%) patients, TP53 Y220C in 24 (1.08%) patients, NTRK1/2/3 fusion in two (0.09%) patients and RET fusion in one (0.05%) patient (Fig. 4a). MSI-high status overlapped with high TMB and the presence of BRAF V600E mutation in 99.5% (214/215) and 52.6% (113/215) of patients, respectively.
a, Prevalence of genomic biomarkers in our cohort. b,c, Kaplan–Meier estimates for DFS stratified by the presence of genomic biomarkers. P = 2.17 × 10−7 (c, TMB high); P = 3.62 × 10−7 (c, MSI high). Two-sided chi-square test for bar plot: P = 3.04 × 10−17 (b). d, ctDNA detection percentage rates in the MRD window by genomic biomarkers. Error bars represent 95% CI. e,f, Kaplan–Meier estimates for DFS stratified by the ctDNA status in the MRD window and genomic biomarkers. The unadjusted HRs (circles) and 95% CIs (horizontal lines) are shown for each biomarker; the vertical dashed line represents the null hypothesis (f). P = 8.75 × 10−12 (e, BRAF V600E); P = 7.55 × 10−162 (f, all); P = 8.75 × 10−12 (f, BRAF V600E); P = 3.35 × 10−5 (f, ERBB2); P = 1.01 × 10−6 (f, KRAS G12C); P = 3.37 × 10−8 (f, MSI high); P = 1.33 × 10−7 (f, TMB high); P = 4.75 × 10−74 (f, RAS/BRAF WT). Two-sided chi-square test for bar plot: P = 1.01 × 10−15 (e). HRs and 95% CIs were calculated using the Cox proportional hazard model; P values were calculated using the two-sided log-rank test (c,e,f). Analyses in d–f were landmarked from the date of the MRD timepoint. Median DFS and percentage DFS at 24 months were estimated from the landmark timepoint. Statistical comparisons of baseline characteristics by the presence of each actionable biomarker were performed using Fisher’s exact test for categorical variables and the Mann–Whitney test for continuous variables. Multivariate logistic regression analysis was used to test the association of post-surgical MRD detection with actionable biomarkers as well as baseline characteristics.
Compared to patients with WT RAS/BRAF, those with TMB-high or MSI-high status had significantly superior DFS (TMB high: HR: 0.19, 95% CI: 0.10–0.35, P < 0.0001; MSI high: HR: 0.16, 95% CI: 0.08–0.32, P < 0.0001), whereas those with KRAS G12C mutation had significantly inferior DFS (HR: 2.18, 95% CI: 1.37–3.48, P = 0.0011) (Fig. 4b,c). Compared to patients with WT RAS/BRAF, those with BRAF V600E mutation showed better DFS, whereas those with ERBB2 amplification or TP53 Y220C mutation tended to have worse DFS (Fig. 4b,c). Similar trends were observed in a subanalysis of patients with stage I–III disease (Extended Data Fig. 5a). Of patients with stage IV disease, KRAS G12C mutation was associated with worse DFS compared to WT RAS/BRAF, whereas the other comparisons were not statistically significant (Extended Data Fig. 5b). Patients with MSI-high and TMB-high disease were also observed to have significantly better DFS when compared to patients with microsatellite stable (MSS) (Extended Data Fig. 5c) and TMB-low (Extended Data Fig. 5d) disease, respectively.
ctDNA positivity during the MRD window varied from 2.8% in patients with MSI-high disease to 27.8% in those with ERBB2 amplification (Fig. 4d). Notably, we observed that MRD positivity was significantly associated with worse DFS compared to MRD negativity consistently across all actionable biomarkers, except for TP53 Y220C, with HR values ranging from 11.00 (KRAS G12C) to 245.19 (BRAF V600E) (Fig. 4e,f). Remarkably, BRAF V600E patients with MRD negativity had a very low recurrence rate when compared to those who were MRD positive (7.89% (12/152) versus 100% (11/11); Fig. 4e). A similar trend was observed for TP53 Y220C; however, it did not reach statistical significance (Fig. 4f).
Upon further analysis of BRAF and MSI relative to ctDNA status, we noted that BRAF V600E/MSI had very low MRD positivity at 0.94%, whereas BRAF V600E/MSS MRD positivity was much higher at 16.94%. Of MRD-negative patients, DFS was more dependent on MSI status, with patients with MSI-high disease having better outcomes than patients with MSS disease and with no significant difference observed based on BRAF status between these two groups (Extended Data Fig. 6). ctDNA detection rate in the MRD window, pathological stage and tumor location distribution in BRAF V600E/MSI and BRAF V600E/MSS subgroups are shown in Supplementary Table 1.
MRD status identifies patients likely to benefit from ACT
To further verify the effect of ACT on MRD-positive and MRD-negative patients with extended follow-up, we implemented an analysis with similar criteria as in our previous publication3. In brief, a landmark at 2 months after surgery was implemented to address the immortal time bias, and HR was adjusted for other confounding factors (age, sex, pathological stage and performance status). Of 1,349 patients with high-risk pathological stage II or stage III colon cancer included in this analysis, 14.23% (192/1,349) were ctDNA positive during the MRD window, 75.52% (145/192) of whom received ACT, whereas 85.77% (1,157/1,349) were ctDNA negative during the MRD window, 49.35% (571/1,157) of whom received ACT. We observed that MRD-positive patients derived significant benefit from ACT (adjusted HR: 0.23, 95% CI: 0.15–0.35, P < 0.0001; clinical recurrence rate: 60.68% (88/145) for the ACT group versus 95.74% (45/47) for the observation group; Extended Data Fig. 7a). Conversely, no statistically significant benefit from ACT was observed for MRD-negative patients (adjusted HR: 0.70, 95% CI: 0.46–1.06, P = 0.091; clinical recurrence rate: 9.63% (55/571) for the ACT group versus 8.53% (50/586) for the observation group; Extended Data Fig. 7b).
Similar findings were observed when patients with high-risk stage II and stage III disease were analyzed separately (Extended Data Fig. 7c,f). In high-risk stage II patients, we further analyzed the benefit of ACT in T3N0 and T4N0 separately, given the concern for de-escalating ACT in T4N0 patients. Although MRD-negative patients with T4N0 disease had inferior survival than T3N0 patients, neither cohort derived any statistically significant benefit from ACT (Extended Data Fig. 8a,b).
Finally, in patients with resected stage IV CRC who were MRD positive, we observed that patients in the observation group had significantly worse DFS than those who received ACT, regardless of whether they received neoadjuvant chemotherapy (NAC). However, the magnitude of ACT benefit was more pronounced in chemotherapy-naive patients who underwent upfront surgery than in patients who received NAC (Extended Data Fig. 9a,b). Conversely, in patients with stage IV CRC who were MRD negative, no benefit from ACT was observed, regardless of whether NAC was given (Extended Data Fig. 9c,d).
Association of ctDNA clearance at 3 months and 6 months with DFS and OS
We assessed whether ctDNA clearance on ACT (from MRD assessment to 3-month and 6-month timepoints, respectively) in MRD-positive patients was predictive of treatment efficacy and outcomes. Of 185 MRD-positive patients who received ACT, ctDNA status at 3-month and 6-month timepoints was available for 171 and 113 patients, respectively. We observed that, compared to patients who remained ctDNA positive despite receiving ACT, those with ctDNA clearance on ACT had superior DFS and OS, respectively (clearance at 3 months: DFS: HR: 5.38, 95% CI: 3.59–8.04, P < 0.0001, Fig. 5a and OS: HR: 3.76, 95% CI: 1.53–9.24, P = 0.004, Fig. 5b; clearance at 6 months: DFS: HR: 11.12, 95% CI: 6.09–20.29, P < 0.0001, Fig. 5c and OS: HR: 6.33, 95% CI: 1.58–25.37, P = 0.009, Fig. 5d).
a,b, Kaplan–Meier estimates for DFS (a) and OS (b) stratified by ctDNA clearance status from MRD window to 3-month timepoint in MRD-positive patients receiving ACT. This analysis was landmarked at the date of 3-month timepoint. P = 2.67 × 10−16 (a). Two-sided chi-square test for bar plot: P = 4.65 × 10−10 (a). c,d, Kaplan–Meier estimates for DFS (c) and OS (d) stratified by ctDNA clearance status from MRD window to 6-month timepoint in MRD-positive patients receiving ACT. P = 4.24 × 10−15 (c). Two-sided chi-square test for bar plot: P = 3.62 × 10−9 (c). HRs and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. This analysis was landmarked at the date of 6-month timepoint. Analyses were performed using R software version 4.4.0. Median DFS and percentage DFS at 24 months were estimated from the landmark timepoint.
ctDNA clearance patterns and molecular recurrence
We next examined the ctDNA clearance patterns in response to ACT in MRD-positive patients. Of the 185 MRD-positive patients who received ACT, longitudinal ctDNA timepoints after MRD were not available for 2.16% (4/185) of patients. For those with available ctDNA after MRD, 37.56% (68/181) of patients had sustained ctDNA clearance and 32.04% (58/181) had transient ctDNA clearance, whereas 30.39% (55/181) of patients did not clear their ctDNA. We then evaluated the association of the ctDNA clearance pattern with DFS. Compared to patients with sustained ctDNA clearance (events: 10.29% (7/68)), patients with transient clearance or no clearance had significantly shortened DFS and OS (transient clearance (DFS: HR: 19.72, 95% CI: 8.61–45.17, P < 0.0001, events: 86.21% (50/58); OS: HR: 25.51, 95% CI: 3.10–3,314.73, P = 0.0007) and no clearance (DFS: HR: 124.76, 95% CI: 51–305.24, P < 0.0001, events: 100% (55/55); OS: HR: 75.62, 95% CI: 10.22–9,650.93, P < 0.0001, respectively)) (Fig. 6a,b).
a,b, Kaplan–Meier estimates for DFS (a) and OS (b) stratified by ctDNA clearance patterns (no clearance versus transient clearance versus sustained clearance) in MRD-positive patients receiving ACT. *Based on Firth’s penalized maximum likelihood; P value from log-rank test. P = 1.78 × 10–12 (a, transient clearance); P = 3.97 × 10–26 (a, no clearance); P = 3.72 × 10–9 (b, no clearance). Two-sided chi-square test for bar plots: P = 6.69 × 10–28 (a); P = 3.11 × 10–6 (b). c, Among patients with transient clearance who recurred (n = 50), the cumulative incidence plot demonstrates a timeline of patients turning back ctDNA positive (time from surgery). d, Among MRD-negative patients who had molecular recurrence before radiological recurrence (n = 165), the cumulative incidence plot demonstrated a timeline of when the patients turned ctDNA positive. e, Kaplan–Meier estimates for OS stratified by ctDNA MRD status and molecular recurrence during the surveillance window (all-time ctDNA negative versus MRD negative with molecular recurrence versus MRD positive). P = 7.09 × 10−11 (MRD positive). Two-sided chi-square test for bar plot: P = 2.63 × 10–31. f, Kaplan–Meier estimates for OS stratified by timing of molecular recurrence after surgery. HRs and 95% CIs were calculated using the Cox proportional hazard model (a,e) or Cox regression with Firth’s penalized likelihood hazard model (b,f). P values were calculated using the two-sided log-rank test. These analyses were landmarked at the MRD timepoint date and were performed using R software version 4.4.0. Median DFS and OS and percentage DFS and OS at 24 months were estimated from the landmark timepoint.
Among patients who had transient clearance (n = 58) in response to ACT, 86.2% (50/58) experienced clinical recurrence. Of these, 14.0% (7/50), 76.0% (38/50) and 96.0% (48/50) of patients converted back to ctDNA positivity by 6 months, 12 months and 18 months after surgery, respectively (Fig. 6c).
We further evaluated the timeline of molecular recurrence to radiological confirmation among patients who were MRD negative. Of the 1,773 MRD-negative patients, 9.3% (165/1,773) turned ctDNA positive before radiological recurrence (128/165) or, in the absence of radiological recurrence, until the last follow-up (37/165). Of these 165 patients with molecular recurrence, 46.7% (77/165), 81.8% (135/165) and 95.2% (157/165) turned ctDNA positive by 6 months, 12 months and 18 months after surgery, respectively (Fig. 6d). Compared to the patients who remained serially ctDNA negative, MRD-negative patients who had molecular recurrence during the surveillance window had significantly worse OS (HR: 11.88, 95% CI: 5.64–25.00, P < 0.0001), similar to MRD-positive patients (P = 0.063; Fig. 6e). When including all 165 ctDNA-negative patients who experienced molecular recurrence after MRD in a Cox regression analysis with serial ctDNA as a time-dependent variable, ctDNA detection after molecular recurrence was associated with a significantly shorter DFS (HR: 3.05; 95% CI: 1.82–5.11, P < 0.0001). Consistently with these results, patients with early molecular recurrence within fewer than 6 months or between 6 months and 12 months after surgery exhibited inferior OS than those with ctDNA conversion beyond 12 months (HR: 10.32, 95% CI: 1.35–1,325.45, P = 0.019 and HR: 7.58, 95% CI: 0.86–995.92, P = 0.073, respectively; Fig. 6f).
We also examined the rate of ‘spontaneous clearance’ among 151 MRD-positive patients who did not receive ACT. Of those 151 patients, 105 had ctDNA results available after the MRD timepoint, six of whom subsequently cleared their ctDNA. Of these, three patients experienced transient clearance with ctDNA turning back positive during surveillance, all of whom experienced clinical recurrence (two nodal and one peritoneal recurrence). The remaining three patients were noted to have sustained clearance—one of whom received neoadjuvant treatment and had a clinical recurrence in the lung, whereas the other two patients remained ctDNA negative and recurrence free, resulting in a true spontaneous clearance rate with no clinical recurrence of 1.9% (2/105).
Discussion
To date, the prognostic impact of MRD on OS in CRC has remained unclear due to a lack of prospective evidence. Our previous interim analysis, which included 1,039 patients with a median follow-up of 16.7 months, established a significant association between post-surgical ctDNA positivity and elevated recurrence risk3. However, the follow-up duration was insufficient to evaluate the impact of MRD status on OS. In this updated analysis with 2,240 patients and 23-month median follow-up, we demonstrate that ctDNA-based MRD detection is a powerful prognostic biomarker for OS as well as DFS in patients with resected CRC.
In the present analysis, ctDNA positivity emerged as the most significant prognostic factor associated with poor OS, outperforming other well-established clinicopathological features. Furthermore, among patients with radiological recurrence, presence of ctDNA positivity significantly correlated with higher mortality compared to ctDNA negativity, regardless of recurrence sites, and with fewer opportunities for curative metastasectomy. These findings underscore the urgent need for novel therapeutic approaches for this population, both in the adjuvant setting and after clinical recurrence. This updated analysis also strongly validates our previous finding that observation alone may be sufficient for favorable outcomes among MRD-negative patients, whereas MRD-positive patients derive significant benefit from ACT. Among MRD-negative patients, T4N0 poses a challenge due to the concern for transcoelomic spread to the peritoneum, which is known to be a lower shedding site of metastasis. Interestingly, although T4N0 demonstrated slightly inferior outcomes compared to the T3N0 counterparts, ACT did not show any survival benefit after adjusting for age, sex and performance status to mitigate residual bias. This finding provides further support for the randomized, phase 3 VEGA trial in the CIRCULATE-Japan platform, which is currently assessing the non-inferiority of observation alone compared to standard ACT in patients with high-risk stage II or low-risk pathological stage III CRC who are confirmed to be ctDNA negative 4 weeks after surgery7.
The role of MRD as a regulatory surrogate endpoint has been established for leukemia and multiple myeloma. Although previous studies in solid tumors suggested that patients with detectable ctDNA after surgery may benefit from ACT corresponding to ctDNA clearance1,11, these studies were limited by small sample sizes, and the optimal timing of assessment for ctDNA clearance has yet to be established. In the present study, we demonstrate that patients with ctDNA clearance even at 3 months after surgery had significantly better DFS and OS than those without ctDNA clearance. The difference in DFS and OS was even more pronounced when assessed at a 6-month timepoint. These findings highlight the potential of ctDNA clearance as an early indicator of treatment efficacy and a regulatory surrogate endpoint for long-term outcomes and strongly warrant further evaluation for adopting ctDNA-based MRD and ctDNA clearance as surrogate endpoints by conducting meta-analyses across trials.
Although any clearance of ctDNA was associated with improved DFS among MRD-positive patients, ctDNA clearance at a single timepoint may not be enough to achieve a favorable outcome. Our analysis of ctDNA clearance patterns demonstrated that sustained ctDNA clearance in response to ACT was a better indicator of treatment efficacy and favorable outcomes compared to transient clearance (10.29% versus 86.21% clinical recurrences or death). Among patients with transient clearance, more than 60% became ctDNA positive within 9 months from surgery, and almost all turned positive by 18 months after surgery, with roughly 50% of molecular recurrences in the 6–9-month post-surgery timeframe, coinciding with the period after conclusion of ACT. These findings support the concept that chemotherapy suppresses ctDNA shedding and highlights the utility of longitudinal ctDNA monitoring in predicting long-term benefit of ACT and the need to repeat testing once active systemic therapy is complete. Lastly, the low spontaneous clearance rate of 1.9% (2/105) observed in MRD-positive patients further reinforces the high PPV of the personalized tumor-informed assay and the rarity of false-positive results using this approach.
Longitudinal ctDNA testing is also valuable for MRD-negative patients, as the detection of molecular recurrence was found to be associated with poor OS. Upon investigating the clinically recurrent cases, ctDNA-positive recurrences significantly correlate with higher mortality compared to ctDNA-negative recurrences, regardless of recurrence sites. Although the exact mechanism is unknown, it is hypothesized that ctDNA-negative recurrences represent a cohort of patients who have tumors with lower shedding rates due to a more indolent biology and/or potentially sequestered sites. The indolent biology hypothesis is further supported by the fact that more patients with ctDNA-negative recurrences had disease amenable to curative-intent resection of OMD than their ctDNA-positive counterparts. Nonetheless, molecular recurrence in the surveillance setting remains valuable as approximately 30% of recurrences detected by ctDNA were OMD and underwent curative-intent resection with a second chance of cure. This represents a 50% improvement in detecting OMD compared to historical data, which estimates OMD at 20% as reported in a recent meta-analysis by Szturz et. al.12.
A recent study reported the perceived utility of ctDNA testing and dimensions of well-being from patients enrolled in the BESPOKE CRC study (NCT04264702), a multicenter, prospective, observational study investigating the clinical utility of ctDNA for optimal use of ACT and early detection of recurrence in patients with surgically resected CRC4. The results indicate that 96% of patients valued the information received from their ctDNA results, with 73% of patients reporting reduced anxiety about cancer recurrence. Notably, ctDNA-negative patients felt less anxious about cancer recurrence compared to ctDNA-positive patients4. ctDNA-directed treatment change/escalation could be beneficial for MRD-positive patients achieving no or only transient ctDNA clearance in response to ACT as well as for MRD-negative patients later having molecular recurrence. Timely change in treatment can alleviate patient anxiety about cancer recurrence as well as potentially improve survival outcomes. Clinical trials using post-surgical ctDNA status or molecular recurrence can help fulfill the unmet need for a new treatment strategy to improve outcomes of patients not responding to current standard-of-care treatment. Currently, the phase 3 ALTAIR trial in the CIRCULATE-Japan platform is investigating the superiority of trifluridine/tipiracil over placebo on DFS among patients with CRC with molecular relapse and no clinical evidence of recurrence after standard-of-care treatment (3 months of adjuvant CAPOX or observation for low-risk patients with stage II disease)7.
Previous studies in breast and lung cancer demonstrated that, even within the same cancer type, the tumor biology differs according to molecularly stratified subtype, potentially affecting ctDNA detection rates and the utility of ctDNA testing13,14,15. Recent advancements in molecular profiling of CRC also revealed distinct biological behaviors by actionable biomarkers, leading to the development of biomarker-guided therapeutic strategies. It is, therefore, crucial to assess the association between MRD status and prognosis in patients with resectable CRC according to these actionable biomarkers, as this could help advance targeted therapy approaches into early disease. In the present study, we found that, across all actionable biomarkers observed in our cohort, patients with ctDNA positivity in the MRD window had significantly worse DFS. Particularly noteworthy was the low recurrence rate observed in patients with BRAF V600E-mutated CRC who tested negative for ctDNA, at only 7.89%, whereas those with positive ctDNA-based MRD were at a much higher risk for recurrence, resulting in a high HR of over 200. Among patients with MSI-high disease, BRAF V600E mutation did not result in a worse prognosis, in line with previously published data16,17. However, in our study, we additionally observed that, among MSS and MRD-negative patients, BRAF V600E patients had a similar prognosis to BRAF WT. These findings suggest that ctDNA-based MRD detection could be implemented in clinical practice for resectable CRC regardless of actionable biomarkers and advocate for the integration of MRD-driven therapeutic strategies, including targeted agents tailored for patients with CRC with specific actionable biomarkers.
Here we report on prospective ctDNA analysis in a large cohort of patients with long-term follow-up, but our study is associated with some limitations. Although, to our knowledge, this is the largest prospective study to date evaluating the role of ctDNA in resected CRC, the median follow-up of 23 months may still be insufficient to fully capture the long-term impact of ctDNA status on OS. Continued follow-up of this cohort will be necessary to validate our findings and provide more robust estimates of the prognostic value of ctDNA. Moreover, the study design was observational, as a randomized trial of ACT versus observation in patients with post-surgical ctDNA positivity was not feasible in Japan when CIRCULATE-Japan was initiated. However, we observed a clear benefit of ACT among MRD-positive patients, which was mirrored in the recently reported interim analysis of BESPOKE CRC, a United States–based observational trial similar to GALAXY in design4. In addition, the ALTAIR and VEGA trials within the CIRCULATE-Japan platform will investigate the benefit of treatment escalation in MRD-positive patients and non-inferiority of observation alone compared to standard ACT in MRD-negative patients, respectively.
Taken together, the findings of this prospective study support the utility of longitudinal ctDNA monitoring in CRC for identifying patients at high risk of recurrence and/or mortality and who are likely to benefit from ACT. The incorporation of ctDNA clearance assessment into clinical trials has the potential to streamline and assist with efficient drug development. Results from ongoing randomized controlled trials evaluating the efficacy of novel therapeutic strategies in ctDNA-positive patients will be crucial to fully harness the potential of ctDNA as a biomarker in the management of resected CRC.
Methods
Ethics statement
All patients provided written informed consent before participation in the study. The clinical protocol was approved by the institutional review board of the National Cancer Center Japan and authorized by the head of each participating institution. The GALAXY study is registered in the Japan Registry of Clinical Trials (UMIN000039205). The study was conducted in accordance with the Declaration of Helsinki.
Study design and participants
Here we present the updated analysis from the GALAXY study, the observational arm of the ongoing, prospective, multicenter CIRCULATE-Japan study. The GALAXY study is a prospectively conducted large-scale multinational registry designed to monitor ctDNA status for patients with clinical stage II–IV CRC undergoing complete surgical resection3. It serves to screen patients for ctDNA-guided MRD status, leading to their assignment to one of the two randomized ctDNA-guided interventional phase 3 trials: ALTAIR (treatment escalation in MRD-positive patients) and VEGA (treatment de-escalation in MRD-negative patients)3,7. The detailed study protocol is published elsewhere7. The key eligibility criteria include the following: (1) histopathologically confirmed colorectal adenocarcinoma; (2) clinical stage II–IV; (3) scheduled for curative-intent resection; (4) patient aged ≥20 years; and (5) Eastern Cooperative Oncology Group (ECOG) performance status of 0 or 1. Patients diagnosed with other malignancies within the past 5 years were excluded. Patients were treated with ACT at the clinician’s discretion based on clinicopathological characteristics according to established guidelines18,19. Blood samples for ctDNA analysis were collected at 4, 12, 24, 36, 48, 72 and 96 weeks after surgery until recurrence. Computed tomography (CT) imaging was performed every 6 months after surgery. Data collection was performed by input into an electronic data capture system (TrialMaster version 5.0 (update 6), Anju Software).
The key inclusion criteria for this analysis included confirmed pathological stage I–V colon cancer or stage IV/recurrent rectal cancer, R0 resection margins, availability of at least one ctDNA timepoint in the MRD or surveillance window, imaging follow-up for more than 6 months or relapsed any time after resection. The MRD window was defined as 2–10 weeks after surgery and before the start of any adjuvant therapy. The potential clinical applications of analysis in this window include prognostication and adjuvant treatment decision-making. The surveillance window was defined as the time from 4 weeks after ACT, or the end of the MRD window if the patient had no ACT, until the last follow-up or clinical recurrence. Here, the relevant clinical application is early detection of recurrence, which can potentially trigger escalation of surveillance imaging and/or treatment (treat on molecular recurrence). Patients with stage I–III rectal cancer were excluded from the analysis, as the clinical management of these patients differs considerably from those with stage I–III colon cancer. Additionally, patients enrolled in associated interventional phase 3 trials (ALTAIR and VEGA) and patients whose sample(s) failed quality control metrics were excluded.
Tumor-informed ctDNA testing
A clinically validated, personalized, tumor-informed 16-plex polymerase chain reaction (PCR) next-generation sequencing (NGS) assay (Signatera, Natera, Inc.) was used for the detection and quantification of ctDNA in blood samples3. In brief, formalin-fixed, paraffin-embedded tumor tissue from surgical resection or biopsy samples and matched normal DNA extracted from a peripheral blood sample were processed for whole-exome sequencing to identify and track up to 16 patient-specific and tumor-specific somatic single-nucleotide variants (SNVs) in the associated patient’s plasma using a multiplex PCR-based NGS approach1. Cell-free DNA was extracted from patient plasma (median, 9.7 ml; range, 1.3–13.2 ml) at a given timepoint and was used to detect ctDNA. Plasma samples with at least two tumor-specific variants detected above a pre-defined threshold were defined as ctDNA positive. The pre-defined threshold is based on Natera’s proprietary variant calling method wherein detecting at least two out of 16 variants ensures the optimal analytical performance of the assay with greater than 95% sensitivity at 0.01% mean variant allele frequency and with 99.7% specificity20. ctDNA concentration was reported as mean tumor molecules per milliliter (MTM/ml) of plasma.
Biomarker analysis
Actionable biomarkers for CRC were defined as biomarkers with OncoKB therapeutic levels of 1–3 for CRC or all solid tumors (Supplementary Table 3). These biomarkers were identified through whole-exome sequencing of tumor tissue performed as part of the tumor-informed ctDNA testing workflow. Due to the low frequency of NTRK and RET fusion, these fusions were not used for the analysis of the association between ctDNA MRD and actionable biomarkers.
Statistical analysis
The primary endpoints were DFS and OS. DFS was defined as the time between the date of landmark and the date of diagnosis with recurrence or death due to any cause or the latest radiological assessment. For DFS, an event was defined as recurrence or death due to any cause. Recurrence was determined based on diagnostic imaging or any other diagnostic procedure if imaging was not confirmative (for example, colonoscopy to diagnose local recurrence). OS was defined as the time between the date of landmark and the date of death due to any cause or latest clinical follow-up. The chi-squared test was used to compare categorical variables. Survival analyses were carried out using R software version 4.4.0 using the survival (version 3.7.0), survminer (version 0.4.9) and coxphf (version 1.13.4) packages. The exact P values from Cox regression hazard models were calculated using the Rmpfr (version 0.9.5) package. The Kaplan–Meier method was used to estimate the survival distribution. Differences between groups were tested using the log-rank test. A multivariable Cox proportional hazard model was used to assess prognostic factors associated with DFS. Clinically relevant cutoffs were applied for demographic variables, wherever appropriate.
MRD analyses were landmarked at the date of MRD timepoint to account for immortal time bias. Surveillance analyses were landmarked at 10 weeks after surgery, because patients with CRC typically receive an adjuvant therapy around 8–10 weeks after surgery per current clinical guidelines. Thus, 10-week landmark ensures that both the ACT-treated and the non-ACT-treated (observation) populations are alive and free of clinical events independent of whether they had ctDNA timepoint and adjuvant treatment.
The secondary endpoint was ctDNA clearance after ACT, and the exploratory endpoint was molecular recurrence analysis (conversion of ctDNA status to positive post-MRD window in ctDNA MRD-negative patients). Cox regression was used to compare the survival benefit between the ACT and observation groups; was landmarked at 2 months after surgery; and was adjusted for age, sex, stage, performance status and MSI status. ctDNA clearance at 3 months and at 6 months was defined as ctDNA clearance from MRD timepoint to the 3-month (70–112 days after surgery) and the 6-month (160–200 days after surgery) timepoints, respectively, in patients treated with ACT. The definition of clearance at 6 months was independent of the clearance status at 3 months. ctDNA clearance analyses were landmarked at 3 months and 6 months, respectively. In patients who were ctDNA positive during the MRD window and received ACT, sustained clearance was defined as ctDNA clearance that was achieved after MRD window and persisted for two or more subsequent ctDNA timepoints, whereas transient clearance was defined as ctDNA clearance after MRD that reverted to ctDNA positive at any subsequent timepoint. The association of ctDNA-based MRD detection with recurrence risk according to each actionable biomarker was evaluated by comparing DFS between patients with a specific actionable biomarker who had either positive or negative ctDNA at the MRD timepoint. P values less than 0.05 were considered statistically significant.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
The authors declare that all relevant, non-proprietary data used to conduct the analyses are available within the article. Supplementary Table 2 includes the de-identified raw data of patient characteristics, outcomes (DFS and OS) and complete ctDNA results (at the MRD window, 3-month and 6-month timepoints and surveillance windows, ctDNA clearance after MRD window and molecular recurrence). To protect the privacy and confidentiality of patients in this study under the Japanese Act on the Protection of Personal Information, identifiable clinical data are not made publicly available in a repository or in the supplementary material of the article but can be requested at any time from the corresponding author (T.Y. (tyoshino@east.ncc.go.jp) or E.O. (oki.eiji.857@m.kyushu-u.ac.jp)). Any requests will be reviewed within a timeframe of 2–3 weeks by the CIRCULATE-Japan study steering committee to verify whether the request is subject to any intellectual property or confidentiality obligations. All data shared will be de-identified and will be provided to researchers with access limited for scientific verification purposes and with strict prohibitions on secondary use.
Code availability
The fully documented code and raw outputs for the R statistical computing environment for analyses related to this manuscript are deposited at the GitHub repository and can be accessed at https://github.com/Natera-TMED/Nakamura-et-al_CIRCULATE-JP-Galaxy-2024.git.
References
Reinert, T. et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol. 5, 1124–1131 (2019).
Loupakis, F. et al. Detection of molecular residual disease using personalized circulating tumor DNA assay in patients with colorectal cancer undergoing resection of metastases. JCO Precis. Oncol. 5, PO.21.00101 (2021).
Kotani, D. et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat. Med. 29, 127–134 (2023).
Kasi, P. M. et al. Circulating tumor DNA (ctDNA) for informing adjuvant chemotherapy (ACT) in stage II/III colorectal cancer (CRC): interim analysis of BESPOKE CRC study. J. Clin. Oncol. 42, https://doi.org/10.1200/JCO.2024.42.3_suppl.9 (2024).
Berry, D. A. et al. Association of minimal residual disease with clinical outcome in pediatric and adult acute lymphoblastic leukemia: a meta-analysis. JAMA Oncol. 3, e170580 (2017).
Munshi, N. C. et al. A large meta-analysis establishes the role of MRD negativity in long-term survival outcomes in patients with multiple myeloma. Blood Adv. 4, 5988–5999 (2020).
Taniguchi, H. et al. CIRCULATE-Japan: circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 112, 2915–2920 (2021).
Henriksen, T. V. et al. Unraveling the potential clinical utility of circulating tumor DNA detection in colorectal cancer-evaluation in a nationwide Danish cohort. Ann. Oncol. 35, 229–239 (2024).
Radomski, S. N. et al. The utilization of circulating tumor DNA to predict the risk and location of relapse after curative-intent local therapy in oligometastatic colorectal cancer. J. Gastrointest. Surg. 28, 534–537 (2024).
Engstrand, J., Nilsson, H., Stromberg, C., Jonas, E. & Freedman, J. Colorectal cancer liver metastases—a population-based study on incidence, management and survival. BMC Cancer 18, 78 (2018).
Tie, J. et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol. 5, 1710–1717 (2019).
Szturz, P., Bonomo, P. & Vermorken, J. B. Estimating the prevalence of a true oligometastatic disease. Cancers (Basel) 15, 214 (2022).
Abbosh, C. et al. Phylogenetic ctDNA analysis depicts early-stage lung cancer evolution. Nature 545, 446–451 (2017).
Magbanua, M. J. M. et al. Circulating tumor DNA in neoadjuvant-treated breast cancer reflects response and survival. Ann. Oncol. 32, 229–239 (2021).
Magbanua, M. J. M. et al. Clinical significance and biology of circulating tumor DNA in high-risk early-stage HER2-negative breast cancer receiving neoadjuvant chemotherapy. Cancer Cell 41, 1091–1102 (2023).
Ciombor, K. K., Strickler, J. H., Bekaii-Saab, T. S. & Yaeger, R. BRAF-mutated advanced colorectal cancer: a rapidly changing therapeutic landscape. J. Clin. Oncol. 40, 2706–2715 (2022).
André, T. et al. Pembrolizumab in microsatellite-instability-high advanced colorectal cancer. N. Engl. J. Med. 383, 2207–2218 (2020).
Hashiguchi, Y. et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int. J. Clin. Oncol. 25, 1–42 (2020).
Yoshino, T. et al. Pan-Asian adapted ESMO Clinical Practice Guidelines for the diagnosis, treatment and follow-up of patients with localised colon cancer. Ann. Oncol. 32, 1496–1510 (2021).
Coombes, R. C. et al. Personalized detection of circulating tumor DNA antedates breast cancer metastatic recurrence. Clin. Cancer Res. 25, 4255–4263 (2019).
Acknowledgements
We thank all of the patients and their families for their participation as well as all study investigators and study teams for their contributions. CIRCULATE-Japan receives financial support from the Japan Agency for Medical Research and Development (grants 19ck0106447h0002, 21ck0106711h0001, 21ck0106710h0001, 22lk0201164h0001 and 22lk0201148h0002) and the National Cancer Center Research and Development Fund (no. 2021-A-6). The funders had no role in the study design; in data collection, analysis or interpretation; or in the writing of the report. We would like to acknowledge the critical review of statistical methods provided by J. Ensor and manuscript writing assistance provided by C. C. Palsuledesai and M. Malhotra from Natera, Inc.
Author information
Authors and Affiliations
Contributions
Y.N., A.A., M.R., D.K., T.Y. and E.O. conceived and designed the study. Y.N. and H.T. developed the methodology. Y.N., J.W., N.A., K.H., K. Kataoka, M.Y., K. Kato, M.K., Y.K., K.H.Y., S.M., H.Y., K.A., M. Miyo, K.Y., H.E., K.O., A.H., H.B., H.T., I.T., T.K., D.K., M. Mori, T.Y. and E.O. recruited patients and collected the data. Y.N., H.S., H.B., M.C.L., A.A. and T.Y. provided administrative, technical or material support. S.K. managed the data. Y.N., G.L., V.N.A., S.S. and A.J. analyzed and interpreted the data. T.M. contributed to the development of the statistical analysis plan and conducted the statistical analysis. All authors drafted and revised the manuscript. All authors critically revised the manuscript for intellectual content. All authors approved the final version of the submitted report and agree to be accountable for all aspects. All authors verify that this study was done per protocol and vouch for data accuracy and completeness.
Corresponding authors
Ethics declarations
Competing interests
Y.N. reports advisory roles with Guardant Health Pte Ltd., Natera, Inc., Roche Ltd., Seagen, Inc., Premo Partners, Inc., Daiichi Sankyo Co., Ltd., Takeda Pharmaceutical Co., Ltd., Exact Sciences Corporation and Gilead Sciences, Inc.; speakers’ bureau from Guardant Health Pte Ltd., MSD K.K., Eisai Co., Ltd., Zeria Pharmaceutical Co., Ltd., Miyarisan Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., CareNet, Inc., Hisamitsu Pharmaceutical Co., Inc., Taiho Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Chugai Pharmaceutical Co., Ltd., Becton, Dickinson and Company and Guardant Health Japan Corp.; research funding from Seagen, Inc., Genomedia, Inc., Guardant Health AMEA, Inc., Guardant Health, Inc., Tempus Labs, Inc., Roche Diagnostics K.K., Daiichi Sankyo Co., Ltd. and Chugai Pharmaceutical Co., Ltd. J.W. reports receiving honoraria for lectures from Johnson & Johnson, Medtronic, Eli Lilly and Takeda Pharmaceuticals and receiving research funding from Medtronic, AMCO, TERUMO and Stryker Japan, all outside the submitted work. N.A. reports nothing to declare. K.H. reports nothing to declare. K. Kataoka reports speakers’ bureau from Merck Biopharma Co., Ltd., Takeda Pharmaceutical Co., Ltd., Eli Lilly Japan Co., Ltd. and Guardant Health Japan Corp. and research funding from Sysmex. M.Y. reports receipt of personal fees and honoraria for lectures from Medtronic, Johnson & Johnson, INTUITIVE, Eli Lilly, Taiho, Merck KGaA, Takeda and Daiichi Sankyo. K. Kato reports nothing to declare. M.K. reports speakers’ bureau from Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co., Ltd., Eli Lilly and Yakult Honsha. Y.K. reports speakers’ bureau from Guardant Health Pte Ltd., MSD K.K., Merck Biopharma Co., Ltd., Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd., Takeda Pharmaceutical Co. and Ono Pharmaceutical Co., Ltd. K.H.Y. reports payment or honoraria for lectures, presentations and speakers’ bureaus from Takeda Pharmaceutical Co., Ltd., Ono Pharmaceutical Co., Ltd, TTY Biopharm Co. Ltd., OEP Group Co., Ltd., Bristol Myers Squibb, Amgen, Inc., Bayer Co., Ltd., Daiichi Sankyo Co., Ltd., Merck Biopharma Co., Ltd., Novartis Co., Ltd. and CStone Pharmaceuticals Co., Ltd. and advisory roles with Merck & Co., Inc, Pfizer Inc., Pierre Fabre, Ltd. and AstraZeneca. S.M. reports honoraria from Taiho Pharmaceutical Co., Ltd., Chugai Pharmaceutical Co., Ltd. and Eli Lilly. H.Y. reports nothing to declare. K.A. reports nothing to declare. M. Miyo reports nothing to declare. T.M. reports advisory role with Anaut, Inc. and Fujifilm Corporation and speakers’ bureau from AstraZeneca K.K., Chugai Pharmaceutical Co., Ltd. and Miyarisan Pharmaceutical Co., Ltd. K.Y. reports speakers’ bureau from Chugai Pharmaceutical Co., Ltd., Daiichi Sankyo Co., Ltd., Yakult Honsha Co., Ltd., Takeda Pharmaceutical Co., Ltd., Merck Biopharma Co., Ltd., Taiho Pharmaceutical Co., Ltd., Eli Lilly Japan K.K., Ono Pharmaceutical Co., Ltd., MSD Co., Ltd. and Bristol Myers Squibb and research funding from Taiho Pharmaceutical Co., Ltd. H.E. reports honoraria from Amgen, Chugai Pharmaceutical Co., Ltd., Guardant Health Japan Corp., Incyte Japan Co., Ltd., Merck Serono Co., Ltd., Ono Pharmaceutical Co., Ltd. and Konica Minolta Co, Ltd. and research funding from Astellas Pharmaceutical Co., Ltd. K.O. reports speakers’ bureau from Taiho Pharmaceutical Co., Ltd. A.H. reports receiving honoraria for lectures from Medtronic, Johnson & Johnson, Merck Biopharma Co., Ltd., Eli Lilly and Fujifilm Corporation. H.S. reports nothing to declare. S.K. is a full-time employee of EPS Corporation. G.L., V.N.A., S.S. and A.J. are employees of Natera, Inc., with stock or options to own stock. M.C.L. is an employee of Natera, Inc., with stock or options to own stock, and further reports grants/contracts (funding to institution: Mayo Clinic) from Eisai, Exact Sciences Corporation, Genentech, Genomic Health, GRAIL, Menarini Silicon Biosystems, Merck, Novartis, Seattle Genetics and Tesaro; travel support reimbursement from AstraZeneca, Genomic Health and Ionis; and ad hoc advisory board meetings (all funds to Mayo Clinic; no personal compensation) from AstraZeneca, Celgene, Roche/Genentech, Genomic Health, GRAIL, Ionis, Merck, Pfizer, Seattle Genetics and Syndax. A.A. is an employee of Natera, Inc., with a leadership position and stock or options to own stock. M.R. reports employment, leadership position, stock and other ownership interests and consulting and advisory role at Natera, Inc., MyOme and Marble Therapeutics and research funding, patents/royalties/other intellectual property and travel/accommodations/expenses from Natera, Inc. and MyOme. H.B. reports research funding from Ono Pharmaceutical and honoraria from Ono Pharmaceutical, Eli Lilly Japan and Taiho Pharmaceutical. H.T. reports speakers’ bureau from MSD K.K, Merck Biopharma Co., Ltd., Takeda, Taiho, Eli Lilly Japan, Bristol Myers Squibb Japan, Chugai Pharmaceutical, Ono Yakuhin and Amgen and research funding from Takeda and Daiichi Sankyo. I.T. reports speakers’ bureau from Medtronic, Johnson & Johnson, Intuitive, Medicaroid and Eli Lilly and research funding from Medtronic and Sysmex. T.K. reports nothing to declare. D.K. reports honoraria from Takeda, Chugai, Eli Lilly, MSD, Ono Pharmaceutical, Seagen, Guardant Health, Eisai, Taiho, Bristol Myers Squibb, Daiichi Sankyo, Pfizer, Merck Biopharma Co., Ltd. and Sysmex and research funding from Ono Pharmaceutical, MSD, Novartis, Servier, Janssen, IQVIA, Syneoshealth, CIMIC and Cimicshiftzero. M. Mori reports nothing to declare. T.Y. reports honoraria from Chugai Pharmaceutical, Takeda Pharmaceutical, Merck Biopharma Co., Ltd., Bayer Yakuhin, Ono Pharmaceutical and MSD K.K; consulting fee from Sumitomo Corp.; and research funding from Amgen, Bristol Myers Squibb, Chugai Pharmaceutical, Daiichi Sankyo, Eisai, FALCO Biosystems, Genomedia, Medical & Biological Laboratories, Merus N.V., Molecular Health GmbH, MSD, Nippon Boehringer Ingelheim, Ono Pharmaceutical, Pfizer Japan, Roche Diagnostics, Sanofi, Sysmex, Taiho Pharmaceutical and Takeda Pharmaceutical. E.O. reports speakers’ bureau from Chugai Pharmaceutical Co., Ltd., Bristol Meyers Squibb, Ono Pharmaceutical Co., Ltd., Eli Lilly and Takeda Pharmaceutical Co., Ltd.; research funding from Guardant Health, Inc.; and an advisory role with GlaxoSmithKline.
Peer review
Peer review information
Nature Medicine thanks Elena Elez, Yu Shen and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Anna Maria Ranzoni and Ulrike Harjes, in collaboration with the Nature Medicine team.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Extended data
Extended Data Fig. 1 ctDNA status in the MRD window is predictive of DFS across all stages in postsurgical patients with CRC.
a-f. Kaplan-Meier estimates for DFS stratified by ctDNA status during the MRD window (negative versus positive) in patients with pathologic stage I (a), stage II (b), stage III (c), stage IV (d), high-risk stage II (e), and high-risk stage III (f) CRC. P = 1.66 × 10−5 (a); P = 1.05 × 10−33 (b); P = 9.55 × 10−59 (c); P = 3.82 × 10−37 (d); P = 8.30 × 10−30 (e); P = 9.66 × 10−36 (f). Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. This analysis was landmarked from the date of the MRD time point. Median DFS and percent DFS at 24 months were estimated from the landmark time point. Abbreviations: MRD, molecular residual disease. DFS, disease-free survival.
Extended Data Fig. 2 ctDNA status in the MRD window is predictive of DFS and OS among patients with stage I-III colon cancer and stage IV CRC.
a-b. Kaplan-Meier estimates for DFS stratified by ctDNA status in the MRD window among patients with pathological stage I-III colon cancer (a) and pathological stage IV colorectal cancer (b). c-d. Kaplan-Meier estimates for OS stratified by ctDNA status in the MRD window among patients with stage I-III colon cancer (c) and stage IV colorectal cancer (d). Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. This analysis was landmarked from the date of the MRD time point. Median DFS and percent DFS at 24 months were estimated from the landmark time point. Abbreviations: MRD, molecular residual disease. DFS, disease-free survival; OS, overall survival.
Extended Data Fig. 3 ctDNA status in the MRD and surveillance windows and subsequence radiological recurrence status.
a. Sankey plot depicting the percentage of all patients in the cohort (N = 2,240) with MRD-positivity or -negativity, the percentage of patients with subsequent ctDNA status during surveillance (after ACT or observation), and the percentage of each group with confirmed radiological recurrence. b. Among patients with ctDNA status available in both MRD and surveillance windows (N = 1,664), Sankey plot shows the percentage of patients with MRD-positivity or -negativity, the percentage of patients with subsequent ctDNA status during surveillance (after ACT or observation), and the percentage of each group with confirmed radiological recurrence. Abbreviations: MRD, molecular residual disease; NA, not available.
Extended Data Fig. 4 ctDNA status in the MRD and surveillance windows is predictive of post-recurrence survival among patients who recurred.
a-b. Kaplan-Meier estimates for PRS stratified by ctDNA status during the MRD window (a) or surveillance window (b) among patients with CRC who had radiological recurrence during follow-up. Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. All analyses were landmarked at the date of radiological recurrence. Median PRS and percent PRS at 24 months were estimated from the landmark time point. c. Bar plot demonstrating the proportion of ctDNA-negative (N = 90) and ctDNA-positive (N = 79) patients undergoing curative resection for recurrence lesions. Error bars represent 95% confidence intervals. P value was generated based on the two-sided Chi-square test. Abbreviations: MRD, molecular residual disease; PRS, post-recurrence survival.
Extended Data Fig. 5 Association of actionable biomarker status with DFS.
a-b. Kaplan-Meier estimates for DFS stratified by the presence of genomic biomarkers among patients with pathological stage I-III colon cancer (a) and pathological stage IV colorectal cancer (b). *MSI high and TMB high lines overlap with each other. Two-sided Chi-square test for bar plots: P = 5.08 × 10−7 (a) c-d. Kaplan-Meier estimates for DFS stratified by MSI status (MSI High versus MSS; c) and TMB status (TMB high versus TMB low; d) for all patients in this cohort (N = 2,040). P = 9.47 × 10−9 (c); P = 2.96 × 10−9 (d). Two-sided Chi-square test for bar plots: P = 3.27 × 10−12 (c); P = 2.59 × 10−12 (d). Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. This analysis was landmarked from the date of the MRD time point.
Extended Data Fig. 6 Combination of MRD status, BRAF mutation status, and MSI status is predictive of DFS.
a-b. Kaplan-Meier estimates for DFS stratified by BRAF mutation status (wild-type versus V600E) and MSI status (MSI high versus MSS) among patients who were MRD-positive (a) or MRD-negative (b). P = 4.95 × 10−7 (b, BRAF WT & MSI high). Two-sided Chi-square test for bar plots: P = 6.50 × 10−6 (b). Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. This analysis was landmarked from the date of the MRD time point. Median DFS and percent DFS at 24 months are estimated from the landmark time point. Abbreviations: WT, wild-type; MRD, molecular residual disease.
Extended Data Fig. 7 ctDNA-based MRD testing is predictive of the benefit of ACT in postsurgical patients with colon cancer.
a-f. Kaplan-Meier estimates for DFS stratified by adjuvant treatment (observation versus ACT) in MRD-positive and -negative patients with: (a-b) pathological high-risk stage II or stage III, (c-d) high-risk stage II and (e-f) stage III disease. P = 1.43 × 10−12 (a); P = 5.38 × 10−5 (c); P = 8.79 × 10−10 (e). Two-sided Chi-square test for bar plots: P = 1.39 × 10−5 (a). *HR was adjusted by age, sex, stage (A, B), and ECOG performance status. Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. The analysis was landmarked at 2 months post-surgery. Median DFS and percent DFS at 24 months were estimated from the landmark time point. Abbreviations: ACT, adjuvant chemotherapy; MRD, molecular residual disease.
Extended Data Fig. 8 MRD-negative patients with T3N0 or T4N0 disease do not derive benefit from ACT.
a-b. Kaplan-Meier estimates for DFS stratified by adjuvant treatment (observation versus ACT) in MRD-negative patients with T30N0 (a) or T4N0 (b) colon cancer. *HR was adjusted by age, sex and ECOG performance status. Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. The analysis was landmarked at 2 months post-surgery. Median DFS and percent DFS at 24 months were estimated from the landmark time point. Abbreviations: ACT, adjuvant chemotherapy.
Extended Data Fig. 9 MRD-positive patients but not MRD-negative patients with stage IV CRC derive benefit from ACT regardless of whether they received neoadjuvant chemotherapy or not.
a-b. Kaplan-Meier estimates for DFS stratified by adjuvant treatment (observation versus ACT) in MRD-positive patients with stage IV CRC who received NAC (a) or did not receive NAC (b). P = 8.34 × 10−7 (b). c-d. Kaplan-Meier estimates for DFS stratified by adjuvant treatment (observation versus ACT) in MRD-negative patients with stage IV CRC who received NAC (c) or did not receive NAC (d). HR was adjusted by age, sex and ECOG performance status. Hazard ratios (HRs) and 95% CIs were calculated using the Cox proportional hazard model. P values were calculated using the two-sided log-rank test. The analysis was landmarked at 2 months post-surgery. Median DFS and percent DFS at 24 months were estimated from the landmark time point. Abbreviations: NAC, neoadjuvant chemotherapy; ACT, adjuvant chemotherapy.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Nakamura, Y., Watanabe, J., Akazawa, N. et al. ctDNA-based molecular residual disease and survival in resectable colorectal cancer. Nat Med (2024). https://doi.org/10.1038/s41591-024-03254-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41591-024-03254-6
- Springer Nature America, Inc.