Abstract
Walking is a fundamental aspect of human movement, and understanding how irregular surfaces impact gait is crucial. Existing gait research often relies on laboratory settings with ideal surfaces, limiting the applicability of findings to real-world scenarios. While some irregular surface datasets exist, they are often small or lack biomechanical gait data. In this paper, we introduce a new irregular surface dataset with 134 participants walking on surfaces of varying irregularity, equipped with inertial measurement unit (IMU) sensors on the trunk and lower right limb (foot, shank, and thigh). Collected during the North American Congress on Biomechanics conference in 2022, the dataset aims to provide a valuable resource for studying biomechanical adaptations to irregular surfaces. We provide the detailed experimental protocol, as well as a technical validation in which we developed a machine learning model to predict the walking surface. The resulting model achieved an accuracy score of 95.8%, demonstrating the discriminating biomechanical characteristics of the dataset’s irregular surface gait data.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background & Summary
Walking is a fundamental yet challenging feature of human movement, allowing individuals to freely explore their environment. This challenge may be increased where irregular surface topographies are encountered. Unfortunately, gait research is almost exclusively conducted in laboratories with ideal surfaces, making the extrapolation to how irregular surfaces affect movement difficult. Past projects1,2,3,4, including work from our group5,6, have implemented irregular surfaces in laboratories, providing insights into biomechanical adaptations and clinical applications; however, the data associated with these studies are not publically available. Few irregular surface gait datasets are freely available in public repositories. Most recently, Laschowski et al.7 published a dataset of wearable camera images during human movement in different environments. While these data are relevant for image classification and other engineering applications8, they do not provide any biomechanical gait data related to the participants. In 2020, Luo et al.9 released a dataset comprised of 30 young healthy participants walking over several outdoor surfaces while fit with inertial measurement unit (IMU) sensors on their lower limbs. Our group has used that dataset to develop IMU-based machine learning classification algorithms10,11 and to assess biomechanical adaptations to irregular surfaces12. It has also been widely cited and used by others (for example, see13,14). Nonetheless, the dataset is rather small, limiting applications, especially for deep learning projects.
Here, we present a new irregular surface dataset comprising 134 participants walking over surfaces with 4 levels of irregularity while fit with IMU sensors on the trunk and right lower limb (foot, shank, and thigh). The data were collected over three days in the congress hall of the North American Congress on Biomechanics (NACOB) conference in 2022. This data descriptor manuscript describes the methods used to collect and process this publically available dataset. Moreover, we conduct a technical validation, showing that the data contain sufficient information for machine learning classifiers or biomechanical kinematic analyses and share associated usage notes and code, allowing others to reproduce our work.
Methods
Participants
162 participants were recruited from the NACOB conference attendees from August 22–25th, 2022, in Ottawa, Canada. Inclusion criteria were purposely broad; only participants who affirmed being unable to safely walk over irregular surfaces were excluded. Any gait pathologies or relevant impairments were noted and retained in the metadata of this data descriptor. Participants signed a consent form prior to participation, allowing the inclusion of their anonymized data in a public repository. The form was available at our conference kiosk, providing participants with ample time to review its content prior to participation. The University of Montreal ethics committee (Comité d’éthique de la recherche clinique of the Université de Montréal) approved the study under the project number 2022-1557. The data underwent rigorous inspection to identify inconsistencies, errors, and missing information. Notably, some trials exhibited malfunctions in all 4 sensors (4 participants), while a few encountered issues, such as non-recording of the video (3 participants) or absence of synchronization video (21 participants). Those trials were subsequently discarded. It is worth noting that trials where 1, 2 or 3 sensor malfunctions occurred were kept to provide as much raw data as possible. The final dataset comprises 134 participants. Anthropometric data for these remaining participants are presented in Table 1.
Experimental protocol
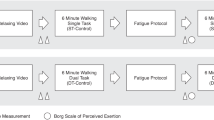
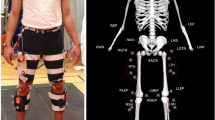
First, anthropometric data (age, height, weight, limb length) were measured (Table 1). Second, participants were fit with wireless IMUs (Dot, Xsens Tech. B.V., Enschede, Netherlands) at the lower back and right thigh, shank, and foot. Sensor data were collected at 120 Hz. These specific sensors were selected given Xsens’ reputation for highly accurate and reliable gait outputs15. Moreover, the Dot system includes an open-software development kit (SDK) allowing for the use of bespoke smartphone data collection applications. Finally, participants were instructed to walk back and forth at their own pace on a circuit of five different surface types: 1 = irregular (low), 2 = artificial grass, 3 = artificial paving stone, 4 = irregular (high), 5 = flat industrial carpet (Figs. 1, 2) for a total duration of 2 minutes. Irregular surfaces (low and high) were manufactured by Terrasensa (Otto Bock HealthCare GmbH, Duderstadt, Germany) and are primarily used in physiotherapy clinics for gait rehabilitation purposes. Both surfaces are shock-absorbing with an average height of 5 centimeters for the high irregular surface and 2 centimeters for the low irregular surface. The remaining surfaces were acquired from local hardware stores. The average height of the artificial grass blades was 2 centimeters. The paving stone surface had only minor height variations (approximately 0.2 centimeters). Surfaces were selected since they were relatively easy to transport to and implement at the conference venue and, based on previous research, should drive subtle, but important biomechanical gait adaptations, compared to the standard surface6. Video recordings (GoPro, Inc. San Mateo, USA) were collected at 120 Hz for each trial. The video recordings allowed for post-hoc partitioning of the trials by surface and thus permitted for quicker data collection (a single continuous recording vs multiple starts and stops). Since the participants walked at their own pace, it was expected that the duration on each surface would not be the same across all participants. On average, 16.6% of the time was spent on surface 1, 25.0% on surface 2, 24.1% on surface 3, 22.8% on surface 4, and, 11.5% on surface 5.
Video annotation
To annotate the recorded videos for each trial, we developed an open-source graphical tool (https://github.com/oussema-dev/video_annotation). The annotation process involved marking the start and end of surface contact. The initiation of contact was identified when the heel of the right foot made first contact with the surface, and the termination was marked when the toe concluded its last contact. This implies that 180° turning manoeuvers required for participants to travel from one surface to the next have been excluded from the dataset. Two annotators performed this task in parallel for the same videos, and a third annotator (O.J.) cross-validated their annotations following these rules: (1) trials with annotation discrepancies exceeding 120 frames between annotators (equivalent to one second) were re-annotated. Remaining annotations for the rest of the trial were aligned with the annotator closest to the corrected segment, (2) trials with annotation discrepancies less than 120 frames, are tagged following annotations from either annotator 1 or 2 (at random).
Data synchronization
The next step involved determining the difference in start times between the IMU data collection and the video recordings to adjust the offset of video annotations. To do so, we took note of the timestamp at which both the video and the IMU sensors started recording. The IMU sensors were triggered using an iPhone, and the GoPro was triggered using a laptop. Both the laptop and the iPhone clocks used the same server for system clock synchronization. Typically, the sensors started recording before the GoPro, resulting in the sensor’s timestamp being less than the video’s timestamp. The synchronization was achieved by subtracting these timestamps to determine the offset, which allowed us to align the video and sensor data accurately. On average, the offset was found to be 719 frames (corresponding to nearly 6 seconds). Subsequently, the sensor data of each trial were trimmed according to the determined offset, ensuring precise synchronization between the video and sensor data.
Data formatting
The formatting phase encompassed the concatenation of sensor data with their respective annotations and anthropometric data for each participant. It is noteworthy that missing anthropometric data from 5 (3.7%) participants who did not provide this information during the data collection were imputed with NaN values. An additional step, for some trials, involved aligning the data by adding missing sensor data columns (imputed with NaN values) to ensure column dimension uniformity across all trials. Specifically, 5 (3.7%) participants lacked foot sensor data, 7 (5.2%) lacked shank sensor data, 10 (7.5%) lacked thigh sensor data, and 7 (5.2%) lacked trunk sensor data. Figure 3 illustrates these steps diagrammatically.
Steps performed for the processing of the raw data files. Missing anthropometric data were imputed with NaN values and merged with surface annotations and raw data files. The align data step added missing sensor data columns if necessary (imputed with NaN values), ensuring uniform column dimensions across all trials.
Data Records
All published raw data are fully anonymized and are available in the raw_data folder on figshare16. Each participant’s trial data file is named P_x.csv, where x denotes the participant number. The first 24 columns are IMU data, column 25 is the participant ID, columns 26 to 32 are the anthropometric data, and the last column “class” is the surface being walked on. Table 2 describes the different data columns in each file. We also provide the concatenated_data.csv file under the data folder. This file combines all participant data making it easier to run machine learning tasks directly rather than processing each participant file individually. The same data structure as the raw data is kept. To summarize the data according to each surface in a visual manner, we plotted radar charts of the mean, max, and min resultant (magnitude) vectors of the acceleration and gyroscope for the different sensor placements (foot, shank, thigh, and trunk) (Fig. 4).
Technical Validation
For technical validation, we used the concatenated_data.csv file (generated using the provided concatenation script, see the usage notes) to extract 8 statistical features (e.g. mean, minimum, maximum, standard deviation, interquartile range, median absolute deviation, area under the curve, and signed area under the curve) for each IMU data column using python functions implemented within the pandas17, scipy18, and numpy19 packages. This process resulted in 192 features (24 × 8). For the purposes of this demonstration, we developed binary classification models: irregular (surface 1 and 4) vs flat (surface 5). Subsequently, the dataset was utilized to train an XGBoost model. This architecture was chosen for its ability to model complex relationships, handle missing data, and its robust performance20. The model was coupled with an inter-subject (subject-wise) splitting approach to partition the dataset into training and testing sets, ensuring that all trials corresponding to the same participant resided within a singular set21. Prior to implementing the XGBoost model, we used filter methods for feature reduction. During this step, we discarded highly correlated features, features with invariant values across the dataset, and features with a low variance (0.1). The resulting model yielded an accuracy score of 95.8% on the test set.
Usage Notes
The raw data is stored within the raw_data folder and can be imported using the read_data.py Python script. To streamline dataset manipulation, we provide three additional Python scripts: A concatenation script (concatenate.py) combines the raw data files vertically and generates a CSV file named concatenated_data.csv. A class management Script (fuse_classes.py) allows users to exclude specific surfaces from the dataset and/or fuse surfaces into the same class. The script modifies the concatenated_data.csv file. Finally, the calculate_statistical_features.py script extracts statistical features per signal and surface segment from the concatenated data. The output is saved as a CSV file named statistical_features.csv. The resulting data files are stored within the data folder. The code related to training the feature-based model (xgb_statistical_features.py) is provided in the repository.
Code availability
All Python code scripts are publicly accessible on the project’s github repository at https://github.com/oussema-dev/Nacob_walking_surface_classification.
References
Gates, D. H., Wilken, J. M., Scott, S. J., Sinitski, E. H. & Dingwell, J. B. Kinematic strategies for walking across a destabilizing rock surface. Gait & Posture 35, 36–42 (2012).
Marigold, D. S. & Patla, A. E. Age-related changes in gait for multi-surface terrain. Gait & Posture 27, 689–696, https://doi.org/10.1016/j.gaitpost.2007.09.005. Number: 4 (2008).
Menz, H. B., Lord, S. R. & Fitzpatrick, R. C. Acceleration patterns of the head and pelvis when walking on level and irregular surfaces. Gait Posture 18, 35–46 (2003).
Thies, S. B., Richardson, J. K. & Ashton-Miller, J. A. Effects of surface irregularity and lighting on step variability during gait: A study in healthy young and older women. Gait & Posture 22, 26–31, https://doi.org/10.1016/j.gaitpost.2004.06.004. Number: 1 (2005).
Dixon, P. et al. Gait adaptations of older adults on an uneven brick surface can be predicted by age-related physiological changes in strength. Gait & Posture 61, 257–262, https://doi.org/10.1016/j.gaitpost.2018.01.027 (2018).
Dussault-Picard, C., Cherni, Y., Ferron, A., Robert, M. T. & Dixon, P. C. The effect of uneven surfaces on inter-joint coordination during walking in children with cerebral palsy. Sci. Reports 13, 21779, https://doi.org/10.1038/s41598-023-49196-w (2023).
Laschowski, B., McNally, W., Wong, A. & McPhee, J. ExoNet Database: Wearable Camera Images of Human Locomotion Environments. Front. Robotics AI 7, 562061, https://doi.org/10.3389/frobt.2020.562061 (2020).
Laschowski, B., McNally, W., Wong, A. & McPhee, J. Preliminary Design of an Environment Recognition System for Controlling Robotic Lower-Limb Prostheses and Exoskeletons. In 2019 IEEE 16th International Conference on Rehabilitation Robotics (ICORR), 868–873, https://doi.org/10.1109/ICORR.2019.8779540 (IEEE, Toronto, ON, Canada, 2019).
Luo, Y. et al. A database of human gait performance on irregular and uneven surfaces collected by wearable sensors. Sci. Data 7, 1–9 (2020).
Lam, G., Rish, I. & Dixon, P. C. Estimating individual minimum calibration for deep-learning with predictive performance recovery: An example case of gait surface classification from wearable sensor gait data. J. Biomech. 111606, https://doi.org/10.1016/j.jbiomech.2023.111606 (2023).
Shah, V., Flood, M. W., Grimm, B. & Dixon, P. C. Generalizability of deep learning models for predicting outdoor irregular walking surfaces. J. Biomech. 139, 111159, https://doi.org/10.1016/j.jbiomech.2022.111159 (2022).
Ippersiel, P., Shah, V. & Dixon, P. The impact of outdoor walking surfaces on lower-limb coordination and variability during gait in healthy adults. Gait & Posture S0966636221004859, https://doi.org/10.1016/j.gaitpost.2021.09.176 (2021).
Hu, B., Li, S., Chen, Y., Kavi, R. & Coppola, S. Applying deep neural networks and inertial measurement unit in recognizing irregular walking differences in the real world. Appl. Ergonomics 96, 103414, https://doi.org/10.1016/j.apergo.2021.103414 (2021).
Sikandar, T. et al. Evaluating the difference in walk patterns among normal-weight and overweight/obese individuals in real-world surfaces using statistical analysis and deep learning methods with inertial measurement unit data. Phys. Eng. Sci. Medicine 45, 1289–1300, https://doi.org/10.1007/s13246-022-01195-3 (2022).
Cudejko, T., Button, K. & Al-Amri, M. Validity and reliability of accelerations and orientations measured using wearable sensors during functional activities. Sci. reports 12, 14619. Place: England (2022).
Jlassi, O. data records. Figshare https://doi.org/10.6084/m9.figshare.25100018 (2024).
pandas development team, T. pandas-dev/pandas: Pandas. Zenodo https://doi.org/10.5281/zenodo.3509134 (2020).
Virtanen, P. et al. SciPy 1.0: Fundamental Algorithms for Scientific Computing in Python. Nat. Methods 17, 261–272, https://doi.org/10.1038/s41592-019-0686-2 (2020).
Harris et al. Array programming with NumPy. Nature 585, 357–362, https://doi.org/10.1038/s41586-020-2649-2 (2020).
Chen, T. & Guestrin, C. Xg boost: A scalable tree boosting system. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining (ACM, New York, NY, USA, 2016).
Saeb, S., Lonini, L., Jayaraman, A., Mohr, D. & Kording, K. The need to approximate the use-case in clinical machine learning. GigaScience 6 (2017).
Acknowledgements
All authors acknowledge support from the North-American Conference on Biomechanics (NACOB) committee for providing us space to perform data collection. We also acknowledge Guillaume Lam, Sahar Mohammadyari Gharehbolagh, and Gabrielle Thibault for help with data collection efforts as well as Elvige Fegni-Ndam and Gabriel Lalonde for supplying data annotation. P.C.D. acknowledges support from the Natural Sciences and Engineering Research Council of Canada (NSERC) Discovery grant (RRGPIN-2022-04217) and the Fonds de Recherche Québec Santé (FRQS) research scholar award (Junior 1). V.S. is funded by TransMedTech and its primary funding partner, the Canada First Research Excellence Fund.
Author information
Authors and Affiliations
Contributions
O.J. conducted the data verification, annotation, preprocessing, and technical validation. V.S. developed the experimental protocol and collected the data. P.C.D. supervised the study. O.J. and P.C.D. drafted the manuscript. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jlassi, O., Shah, V. & Dixon, P.C. The NACOB multi-surface walking dataset. Sci Data 11, 880 (2024). https://doi.org/10.1038/s41597-024-03683-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41597-024-03683-5
- Springer Nature Limited