Abstract
Chinese rice wine (CRW) is the oldest kind of wine in China and is mainly fermented by wheat Qu and yeast with rice, millet, etc. This gives CRW a unique quality, but the flavor components are complex. Its formation is related to microorganisms, but the link between CRW and microorganisms is poorly understood. Here, we used two kinds of sorghum (JZ22 and JB3, of which JZ22 has a higher tannin content) as the raw materials to brew and determined the structural and functional dynamics of the microbiota by metagenomics and flavor analyses. We detected 106 (JZ22) and 109 (JB3) volatile flavor compounds and 8 organic acids. By correlation analysis, we established 687 (JZ22) and 496 (JB3) correlations between the major flavor compounds and microbes. In JZ22, Blautia, Collinsella, Bifidobacterium, Faecalibacterium and Prevotella had the most correlations with flavor production. In JB3, the top 5 genera were Stenotrophomonas, Bdellovibrio, Solibacillus, Sulfuritalea and Achromobacter. In addition, more esters were detected, and more microorganisms correlated with ester generation in JZ22. This study provides a new idea for the micro ecological diversity of CRW fermented with sorghum. This is of significance for improving the quality and broadening the CRW varieties.
Similar content being viewed by others
Introduction
Chinese rice wine (CRW), a traditional alcohol drink, is fermented from rice with wheat Qu and yeast. It is one of the oldest drinks in the world, along with beer and wine1. CRW is also called “liquid cake” because of its abundant nutrition and pleasant aroma2. Sorghum is a kind of natural and high-nutrition functional food that is rich in dietary fiber, protein, fat, folic acid, ferrum and other microelements. Sorghum is the main raw material for producing spirits. Chinese liquor brewed with sorghum, such as Maotai-flavored liquor, has long enjoyed a good reputation. However, with the changes in China’s policies on the liquor industry and the increase in people’s living standards, high-nutrition, low-alcohol CRWis becoming increasingly popular with consumers. Brewing CRW with sorghum not only increases the value of sorghum but also combines these two nutritious foods together.
Flavor differences can be caused by many factors, such as the raw ingredients used in the fermentation, fermentation conditions, distillation practices, and aging processes3. Xiong used 36 varieties of glutinous rice as brewing materials to study the influence of the nutrients in the raw materials on the wine yield of yellow rice wine4. Different raw materials also greatly affected the production of phenylethanol. Compared with rice as a raw material, more phenylethanol was produced when sorghum and maize were used. The L-phenylalanine content in the raw materials had a positive effect on B-phenylethanol yield5. The flavor compounds were mainly produced by biochemical reactions of microorganisms. Cui found through gene sequencing a kind of microorganism in liquor that produces pyrazine compounds6. Zhang identified the relationship between the flavor substances and aroma-producing microorganisms and performed fermentation by separating and screening Daqu microbes7. Fujii and others obtained mutant strains of yeast by molecular biological techniques and improved the isoamyl acetate content in sake8. Beaumont found that esterase is one of the most important enzymes in the synthesis and hydrolysis of volatile esters. This enzyme is produced by a series of microorganisms, such as Aspergillus, Rhizopus, Bacillus, Lactobacillus, Wickerhamomyces and Saccharomycopsis9. Aspergillus and Bacillus have been reported to be involved in amylase and acid protease production and can increase the amylase and acid protease activities in inoculated Daqu10. The research of De and others showed that some lactic acid bacteria (LAB) are considered to be main functional contributors in proteolysis, lipolysis, and amino acid/lipid catabolism11. In a Chinese rice wine research, a total of 64 volatile compounds were identified and quantified in Shaoxing rice wines12. The bacterial community structures and diversity of the Chinese rice wine varied significantly during different fermentation stages. Lactobacillus (the predominant genus) and Bacillus might take an active part in flavor development in CRW13. Lv and others evaluated the differences in the microbial communities between the “Qu” samples obtained from the two locations in China (Suzhou and Yichang). The microorganism community structures were significantly different14. That research was conducted solely on the analysis of the CRW’s flavor substances and microorganisms. However, the relationship between aroma compounds and bacterial community in CRW has not been fully reported. Therefore, we aim to study the effects of brewing raw materials and microorganisms on flavor compounds in Chinese rice wine.
To achieve this goal, this study monitored bacterial succession by high-throughput sequencing and the volatile compound dynamics by HS-SPME/GC–MS during the brewing process. The relationship between the bacterial community and the volatile compounds was analyzed. In this work, two kinds of sorghum were used. One of them is Jinza No. 22 (JZ22), whose tannin content is 1.57%, and the another is Jinbai No. 3 (JB3), with a tannin content of 0.02%. We would like to research the effect of tannin content on the microbial communities and flavor compounds in the brewing of yellow rice wine. The result will help in the development of CRW.
Results
Changes in acid, reducing sugar and alcohol
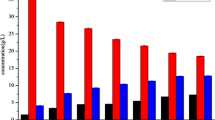
The data in Fig. 1 show that the concentration of acid in both JZ22 and JB3 continued to increase gradually, especially from the 4th to 6th days of fermentation, where there was a increase in JB3. It increased from the 6th to 8th days in JZ22. The alcohol continued to increase gradually and had rapid growth from the 2nd to 4th days in JZ22 and from the 4th to 6th days in JB3. From the 8th to 12th days, it increased gently. The concentration of the reducing sugar reached a peak value on the 2nd day and decreased gradually in JZ22. However, it continued to fall in JB3. Detailed data are attached in Supplementary Table S1.
Changes in organic acid during CRW fermentation
The organic acids identified in Chinese rice wine during fermentation were oxalic acid, tartaric acid, pyruvic acid, malic acid, α-ketoglutaric acid, lactic acid, citric acid and succinic acid (Table S2). In both JZ22 and JB3 samples, oxalic acid, tartaric acid, malic acid, lactic acid, citric acid and succinic acid have a continuing upward trend. Pyruvic acid reached the highest level at the 4th day and then began to decline. However, the content of the pyruvic acid in JZ22 was much higher than JB3. α-Ketoglutaric acid fluctuated greatly, and the changes in the two samples were significantly different. It has been reported that tartaric acid, lactic acid, citric acid, and succinic acid were the primary acids, and the results of this experiment are in agreement with the previous report15.
Analysis of volatile compounds in Chinese rice wine during
Fermentation
A total of 106 volatile compounds were detected in the 7 days’ samples of JZ22, including 16 alkanes, 24 alcohols, 32 esters, 12 ketones, 5 aldehydes, 5 acids, 4 phenols, and 8 other compounds (Table S3a). Among the compounds identified, heneicosane was the major alkane with a high concentration. Among the alcohols detected, 2-methyl-1-propanol, 3-methyl-1-butanol, glycerin, 2,3-butanediol and phenylethyl alcohol had the highest concentrations. Among the esters, hexanoic acid ethyl ester, octanoic acid ethyl ester, decanoic acid ethyl ester, dodecanoic acid ethyl ester, hexadecanoic acid ethyl ester, (E)-9-octadecenoic acid ethyl ester, 9,12-octadecadienoic acid ethyl ester, amyl nitrite and benzoic acid ethyl ester were the major esters with the highest concentrations. Among the ketones, 2-octanone and dihydro-5-propyl-2(3 H)-furanone were the predominant constituents. Among the phenols, 2-methoxy-4-vinylphenol and 2-methoxy-4-vinylphenol had a high relative concentration. On the last day, 47 compounds were detected, including 4 alkanes, 13 alcohols, 21 esters, 1 ketones, 2 aldehydes, 3 phenols, and 3 other compounds. In the 7 days’ samples of JB3, we identified 109 volatile compounds, including 16 alkanes, 26 alcohols, 34 esters, 9 ketones, 6 aldehydes, 4 acids, 4 phenols, and 10 other compounds (Table S3b). 2-Methyl-1-propanol, 3-methyl-1-butanol, glycerin, 2,3-butanediol, phenylethyl alcohol, hexadecanoic acid ethyl ester, (E)-9-octadecenoic acid ethyl ester, 9,12-octadecadienoic acid ethyl ester, amyl nitrite, 2-octanone, 4-ethyl-2-methoxy-phenol and 2-methoxy-4-vinylphenol were the major flavor compounds. On the last day, 47 compounds were detected, including 5 alkanes, 12 alcohols, 14 esters, 2 ketones, 1 aldehyde, 2 phenols, and 3 other compounds.
In comparing the volatile compounds of the two kinds of sorghum yellow rice wine, it can be found that there is not much difference in their quantity and species. However, in some flavor substances, JZ22 is much higher than JB3, such as 2-methyl-1-propanol, 3-methyl-1-butanol, glycerin, 2,3-butanediol, phenylethyl alcohol, octanoic acid ethyl ester, decanoic acid ethyl ester, 2-hydroxy-propanoic acid ethyl ester, tetradecanoic acid ethyl ester, hexadecanoic acid ethyl ester, 9,12-octadecadienoic acid ethyl ester and benzoic acid ethyl ester. They have a unique flavor and are often used in the preparation of flavors and fragrances. However, the 4-ethyl-2-methoxy-phenol and 2-methoxy-4-vinylphenol contents of JB3 were much higher. 4-Ethyl-2-methoxy-phenol has a pungent odor and is often used in perfumes, such as vanilla and whiskey16. Amyl nitrite was detected in both kinds of sorghum yellow wine and is not found in the CRW made from glutinous rice. Amyl nitrite lowers blood pressure and dilates blood vessels17.
Bacterial diversity during different fermentation stages
To explore the intra- and inter-variabilities in the bacterial community among the two kinds of fermentation materials, we estimated a series of alpha diversity indices using the t-test (Table S4). The same data shows that the difference between the early days and the last few days during the fermentation is significant. In comparing JZ22 with JB3, there was a remarkable difference from the 4th to 8th days, especially on the 8th day.
The non-metric multidimensional scaling (NMDS) distribution based on the weighted UniFrac distance described the structures of the bacterial communities for all fermentation samples (Fig. S1). Although there was some overlap between JB3_D6 and JB3_D12, JZ22_D10 and JZ22_D0, there was a distinct separation between the samples of different time points.
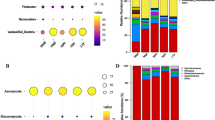
Of all the total reads obtained from the 21 samples of JZ22, 1 059 218 reads passed the quality control filters. A total of 13 bacterial phyla, 23 classes, 50 orders, 74 families, 115 genera and 155 species were identified by matching the sequences of known bacteria. The results showed that, at the phylum level (Fig. 2a), Firmicutes was predominant in all samples, followed by Cyanobacteria and Actinobacteria. At the genus level (Fig. 2b), Bacillus was the most dominant genera, ranging from 83.17% to 94.86% of the total 16 S rRNA gene sequences. Weissella was second only to Bacillus, peaked on the 4th day, and then began to decrease. In the early days of fermentation, Paenibacillus decreased up until the 10th day.
In the JB3 samples, 1 050 616 high-quality sequences were obtained with Illumina MiSeq analysis. The 16S rRNA gene sequences indicated that 153 species of bacteria participated in the fermentation process and were distributed among the 13 phyla, 24 classes, 47 orders, 73 families and 119 genera. At the phylum level (Fig. 2a), same as JZ22, Firmicutes, Proteobacteria and Actinobacteria were the top three most abundant. Bacillus occupied the main position at the genus level (Fig. 2b). Brevibacillus was the second most dominant genus, and it increased and appeared to peak on the 8th day before slightly decreasing at the end of fermentation. Weissella followed Brevibacillus and peaked on the 4th day.
The bacterial community composition of JZ22 and JB3 was not distinguished at the phylum and family levels, but their changing trends were completely different. At the genus level, Bacillus in JZ22 showed a lower abundance on the 4th day. However, in JB3, Bacillus fell to the lowest level on the 8th day. Genus Brevibacillus played an important role in JB3; however, the proportion of Brevibacillus is very small in JZ22. Ethyl alcohol was the precursor of ethyl ester, and the content of ethyl ester in JZ22 was obviously higher than that in JB3, which may be related to the amount of Weissella13. Detailed information on the composition of bacteria is shown in the attached Table S5.
KEGG annotation exposes metabolic differences between JZ22 and JB3 fermentation
Based on the 16 S rRNA gene sequencing data, PICRUSt was used to predict the function of the microorganisms in the fermented mash, which showed similar gene functions among all 42 samples that were categorized into the 39 level-2 KEGG pathways (Fig. 3). Membrane Transport, Carbohydrate Metabolism,Amino Acid Metabolism, Replication and Repair, Cellular Processes and Signaling, Energy Metabolism, Poorly Characterized, Translation, Metabolism of Cofactors and Vitamins were the dominant KEGG pathways. We found that on day 0, JZ22 was similar to JB3, and there were differences in the metabolism during fermentation, especially in the later stage of fermentation. During fermentation, the ABC transporters, glycolysis/gluconeogenesis and pyruvate metabolism pathways had the highest abundance. Detailed information of the predicted gene functions related to KEGG pathways at levels 3 is shown in the attached Table S6.
Fungal diversity during different fermentation stages
Regarding the fungal communities’ sequences in JZ22, 1 073 598 reads passed the quality control filters. Ten phyla, 17 classes, 18 orders, 21 families, 21 genera and 23 species were identified. At the phylum level (Fig. 4a), approximately 99% of the ITS region sequences could be assigned to the phylum Ascomycota. At the genus level (Fig. 4b), Unclassified Saccharomycetales was the most abundant through the whole fermentation process. Pichia decreased until 8th day and then increased in JZ22. The deep sequencing of the fungal communities obtained a total of 1 085 155 sequences in JB3, including 6 phyla, 12 classes, 13 orders, 17 families, 17 genera and 17 species. At the phylum level (Fig. 4a), Ascomycota was the most dominant phylum, accounting for more than 97.44% (94.94–99.93%). At the genus level (Fig. 4b), Unclassified Saccharomycetales was the most abundant, and Pichia had a sharp drop on the second day and then remained stable. The main fungi in the two CRW were almost the same, but at the genus level, Cladosporium and Trichosporon were only detected in JB3. Detailed information on the composition of fungal is shown in the attached Table S7.
Correlation between microorganisms and flavor compounds
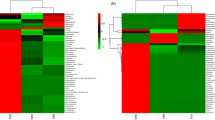
The 28 main flavor substances in the fermentation process were correlated with the change in the microorganisms. In JB3, 44 microorganisms were involved in the production of the major flavor compounds (P < 0.05). Among them, Stenotrophomonas, Bdellovibrio and Solibacillus were related to more than 10 flavor compounds. In JZ22, there were 68 kinds of microorganisms associated with the formation of the major flavor substances (P < 0.05). Blautia, Collinsella, Bifidobacterium, Faecalibacterium, Eubacterium rectale group, Ruminococcus torques group, Anaerococcus, Prevotella_9, Anaerostipes, and Escherichia-Shigella were related to more than 15 flavor compounds. Achromobacter, Eubacterium_ruminantium_group, Lachnospiraceae_NK4A136_group, Subdoligranulum, Bacteroides, Chryseobacterium, Roseburia and Sphingomonas were involved in the formation of 10 to 15 flavor substances. Through association analysis, we can see that there were obviously more main flavor substances formed in the JZ22 brewing process than JB3.
The P value of the correlation analysis of all the flavor compounds and microorganisms was adjusted. We selected a Padjust <0.05 for the data (see Fig. 5). There were 687 correlations in JZ22. Among them, there were 20 alcohols, 24 esters and 3 phenols with a Padjust <0.05, including glycerin with Bifidobacterium; octadecanoic acid ethyl ester with Lactococcus, Saccharomycopsis and Pichia; acetic acid phenylethyl ester with Blautia; amyl nitrite with a complex series of microorganisms; and 2-methoxy-4-vinylphenol and 4-ethyl-2-methoxy-phenol with Bifidobacterium. In JB3, we established 496 correlations. These included 19 alcohols, 24 esters and 3 phenols. For example, octadecanoic acid ethyl ester had a closer correlation with Cryptosporidium, Trichosporon, Galactomyces, Symbiodinium, Cladosporium, Pichia, Cochliobolus, Saccharomycopsis, Lactococcus and Rhizobium. Tetradecanoic acid ethyl ester’s correlation with Catenibacterium and Deinococcus met the conditions. The correlation between Lactococcus and 3-hydroxy-2-butanone satisfied a Padjust <0.05. These correlations have not been reported in other reports and require further experimental verification.
Discussion
In the fermentation process of Chinese rice wine, microorganisms play an important role in the formation of endogenous factors. Comparing the changes in the reducing sugar, total acid and alcohol content with the microbial flora, we found that the variation trends of total acid were similar to Bacillus, which has strong protease, amylase and lipase activity and can produce rich metabolic products and synthesize many organic acids.
Flavor differences may be caused by a number of factors, such as fermentation materials, fermentation conditions and aging processes. Southern rice wine is mostly fermented with glutinous rice, but this experiment used sorghum, which is usually used as raw material in the production of liquor. There are some reports about volatile compounds of Chinese rice wine, and most of them have used headspace solid-phase microextraction followed by gas chromatography–mass spectrometry18. A total of 64 volatile compounds were identified and quantified in Shaoxing rice wines. Xu and others determined the formation of volatiles in the enzymatic extrusion process of rice wine and confirmed its characteristic flavor compounds1. In Shaoxing rice wine, 10 bacterial genera, including Bacillus and Lactobacillus, were found to be predominant12. Similarly, five Bacillus species and three lactic acid bacteria were found to be the dominant bacteria in Hong Qu rice wine, a popular Chinese rice wine19. These microorganisms play important roles in fermentation and the production of special flavors20. Liu’s investigation showed that Bacillus, Leuconostoc, Lactococcus, Weissella, Thermoactinomyces, Pseudomonas, Saccharopolyspora, Staphylococcus, Enterobacter and Lactobacillus were the dominate genera during the Shaoxing rice wine fermentation process12. Although the main flora in this experiment are similar to those in Shaoxing rice wine, the flavor compounds are different. One hundred and six kinds of flavor compounds were detected in JZ22, and 109 were detected in JB3, which are more than in Shaoxing rice wine. This indicated that sorghum could produce more kinds of flavor compounds as raw materials. The two kinds of sorghum used in this experiment were fermented using the same conditions (high tannin content in the JZ22), and the Wheat Qu used the same conditions as well, but the content of the flavor compounds was different. This shows that the difference in the nutrients of the raw materials will affect the flavor of the final wine, which is largely caused by micro ecological changes and microbial metabolism during the fermentation process. In this experiment, JZ22 and JB3 had little difference in the main kinds of esters, but the content of the esters in JZ22 was higher than that in JB3. Through association analysis, we can see that the number of microbial genera involved in the formation of various esters in JZ22 was 2–3 times that of JB3, and the tannin content in JZ22 was also higher than JB3. We can estimate that tannins have important effects on the brewing process, microbial growth and the production of incense. According to previous reports, tannase (tannin acyl hydrolase, EC 3.1.1.20) catalyzes the hydrolysis of ester linkages in hydrolysable tannins and produces glucose and gallic acid21. Although gallic acid is widely distributed in nature, it is difficult for bacteria to use it as a carbon source for growth at neutral or alkaline pH. Only bacteria of the genera Pseudomonas21, Lactobacillus22 and Sphingomonas23 have been reported to be able to utilize free gallic acid as the sole carbon and energy source under aerobic conditions. Through correlation analysis, we found that Sphingomonas was related to the formation of 11 flavor substances in the JZ22 sample and was related to 6 flavor substances in JB3. These results indicate that Sphingomonas may play an important role in the degradation of tannins during the sorghum fermentation process, and the degradation products can produce a variety of aromatic compounds. The aerobic metabolism of gallic acid usually starts with a direct ring-cleavage reaction and the formation of the central intermediate, 4-oxalomesaconic acid, which then undergoes hydration to 4-carboxy-4-hydroxy-2-oxoadipic acid and aldol cleavage to oxaloacetic and pyruvic acids24. As determined by HPLC, the final pyruvate content in JZ22 was 14.09 mg/l, which was much higher than the 1.17 mg/l in JB3. Oxaloacetic and pyruvic acids are an important part of the TCA cycle and microbial carbon metabolism and may stimulate more microorganism involvement in the formation of the aroma. This also explains why the association analysis showed that JZ22 had more microorganisms involved in the formation of the flavor prediction. As seen in Fig. 4, in addition to Bacillus, Weissella occupied the most dominant position at the genus level in JZ22, while Brevibacillus occupied the dominate position in JB3. This may reveal that lactic acid bacteria (LAB) play a role in the metabolism of tannins. Therefore, in the process of brewing, tannins can be added for fermentation in order to increase the flavor of rice wine.
In both JZ22 and JB3, the characteristic flavor substances were 2-methyl-1-propanol, 3-methyl-1-butanol, phenethyl alcohol, decanoic acid ethyl ester, octanoic acid ethyl ester, 9,12-octadecadienoic acid ethyl ester and hexadecanoic acid ethyl ester, which is similar to the conclusion of the research on the volatile flavor substances of yellow rice wine25.
2-Methyl-1-propanol can be used in the preparation of flavors and fragrances and can also be used as a food flavoring agent. In the association of 2-methyl-1-propanol with microorganisms, it was found that the production of 2-methyl-1-propanol in JZ22 was related to Ralstonia, but no association was found in JB3. Studies have shown that Ralstonia can utilize H2O, CO2 and formic acid to generate 2-methyl-1-propanol26. At the end of fermentation, the content of 2-methyl-1-propanol in JZ22 was about two times that of JB3, which may be related to Ralstonia in the JZ22 fermentation system. The content of isobutyl alcohol may be controlled by controlling Ralstonia. Organic acids and other substances constitute the unique aroma of CRW; A proper amount of organic acids can harmonize the taste of yellow rice wine and stabilize the aroma, which will help to enrich and improve the aroma and quality of Chinese rice wine27. In addition, organic acids play an important role in improving intestinal function, preventing fatigue and other health functions. For example, malic acid functions in preventing fatigue, protecting the liver and strengthening heart function28, while citric acid functions in delaying senility, eliminating fatigue and lowering blood pressure29. Through association analysis, we found that Lactococcus, Saccharomycopsis, Cladosporium, Fusicatenibacter, Cochliobolus, and Pichia were related to the production of a variety of organic acids. Among them, Lactococcus mainly is involved in citric acid and lactic acid formation. Saccharomycopsis can ferment galactose, glucose, maltose and other sugars to produce acid. Cladosporium can lead to the decay of plants, such as wheat and grapes, and affects the flavor of wine30. Fusicatenibacter ferments glucose to produce lactic acid, formic acid, acetic acid, and succinic acid31. By controlling the related microorganisms, the organic acid content in the fermented mash can be adjusted properly, and the final aim of improving the quality of rice wine can be achieved.
The volatile flavor compounds in CRW are mainly derived from esters, and the production of esters comes from the following aspects: raw materials, esters and the combination of the fatty acids and alcohols in wheat Qu; the organic acids and amino acids combined with alcohols; and the decomposition, binding and oxidation of rice wine during aging. The aging process of yellow rice wine under natural conditions is quite slow, so the flavor compounds are mainly produced by biochemical reactions during fermentation. In the process of fermentation, the main esters are ethyl esters. The whole aroma of Chinese rice wine mainly depends on the aroma of esters. Ester material mainly comes from the esterification of acids and alcohols, as well as the activities of yeasts and other microorganisms in the brewing process. In addition to ethyl ester, this experiment also detected the amyl nitrite, which was not detected in southern rice wine. Amyl nitrite is a nitrous acid that can relax the vascular smooth muscle, especially the coronary vessels and cerebrovascular vessels, which make the blood vessels dilate and the blood pressure drop32. In JZ22, many more microorganisms were associated with amyl nitrite than in JB3, but both Stenotrophomonas and Roseburia were related to the formation of this compound in both CRWs. Stenotrophomonas can use succinate, malate, citrate, acetate, lactate and propionate as a growth factor33. Roseburia ferments carbohydrates such as glucose, maltose, sucrose, and starch and mainly produces butyric acid. Organic acids can be combined with alcohols to produce esters, and the relationship between these two microorganisms and amyl nitrite requires further study. Correlation is not always causation. As such, the relationship between microorganisms and the flavor substances in Chinese rice wine fermentation is very complex. The generation of a flavor substance may be the result of the synergetic effects of microorganisms.
The OTU abundance table was standardized by PICRUSt according to the KEGG database. In addition, for the pathway, 3 levels of information on metabolic pathways were obtained using PICRUSt, and the abundance tables at each level were obtained. In the high-expression metabolic pathways, ABC transporters are members of a transport system superfamily that is one of the largest and possibly one of the oldest families, with representatives in all extant phyla from prokaryotes to humans34. ABC transporters take up a large variety of nutrients, biosynthetic precursors, trace metals and vitamins, while exporters transport lipids, sterols, drugs, and a large variety of primary and secondary metabolites35. Glycolysis/gluconeogenesis is a series of reactions that degrade glucose and glycogen to pyruvate and ATP. It is a ubiquitous pathway of glucose degradation in all living organisms. Most monosaccharides, such as sugar and galactose, can be converted into one of these intermediates. Intermediates can then be used directly. Metatranscriptomic analysis revealed pyruvate metabolism in yeasts (genera Pichia, Schizosaccharomyces, Saccharomyces and Zygosaccharomyces) and lactic acid bacteria (genus Lactobacillus), which represented two stages in the production of flavor components, with the genera Schizosaccharomyces and Lactobacillus serving as the core functional microorganisms36. Specific expression of genes in metabolism also requires subsequent omics analysis.
In summary, this study is the first to reveal the differences between different sorghum-fermented CRWs and the microorganisms that are related to the production of the main flavor compounds. In addition, we can also deduce that tannins can affect the microbial structure, degrading themselves to produce more ester. Through the association of flavor substances and microbial fermentation process analysis, we can deduce the fermentation of the core microbiota. On the other hand, we target to produce important aromas and health ingredient strains. This provides a theoretical basis for the brewing of CRW with sorghum.
Materials and Methods
Sample collection
Samples were collected in Zhangjiakou city in the Hebei Province in April 2017. The fermentation was performed at a constant temperate of 28 °C and over a period of 12 days. Forty-two samples from different fermentation days were collected. Each sample was divided into two parts: one part was kept at 4 °C immediately after collection for flavor testing, and the other was transferred to sterile bottles and stored at −80 °C for DNA extraction.
Determination of acid, reducing sugar, alcohol and flavor compounds
The DNS method was adopted to determine the reducing sugar content using glucose as reference standard. A 1.0 mL liquid sample was ten times diluted. Then, 1.0 mL of the dilution was shaken well with 1.0 mL of DNS solution and put in a boiling water bath for 5 min. When it cooled to room temperature, the mixture was diluted by water to a final volume of 10.0 mL, and its absorbance at 540 nm was read37. The alcohol content and acidity were measured according to the standard of GB/T13662-2008 and used to evaluate wine quality.
The flavor compounds and organic acids in the samples were analyzed by HS-SPME/GC-MS and HPLC, respectively. For HS-SPME-GC-MS analysis, each wine sample (8 mL) was placed in a 15 mL SPME glass vial together with 2.5 g of sodium chloride. The vial was tightly capped, and the extraction head (50/30 μm DVB/CAR/PDMS) was inserted. The vial was then placed in a water bath with ultrasonic waves for 45 min at 50 °C. After extraction, the fiber was introduced into the injection port of the GC-MS system (at 230 °C for 5 min), and the analytes from the fiber were thermally desorbed. Identification was carried out using a Shimadzu-QP2010 Plus-GCMS. Each concentrated fraction was analyzed on a DB-wax column (30 m × 0.25 mm i.d., 0.25 µm film thickness). The carrier gas, helium, was circulated at 1 mL/min in split-flow mode, and the split ratio was 50/1. The oven temperature program was as follows: 35 °C for 4 min; 5 °C/min ramp to 150 °C and holding for 2 min; and 3 °C/min ramp to 210 °C. The injector and detector temperature was 230 °C, and the ion source temperature was set 200 °C. The ion energy for electron impact (EI) was always 70 eV. The chromatograms were recorded by monitoring the total ion currents in the 30–350 mass range. Semi-quantification of the volatile compounds was performed using 2-octanol as the internal standard16. For HPLC analysis, each wine sample (5 mL) was placed in a tube, centrifuged at 10,000 r/min for 15 min, and then filtered through a 0.45 mm microporous membrane. Chromatographic conditions were based on the method described by Ye et al.38. The determination of organic acids was carried out by HPLC. The separations were carried out on an Agilent 1260 Infinity II equipped with a 250 mm × 4.6 mm and a 5 μm welch ultimate XB-C18 column. The column temperature was set at 30 °C. A mixture of phosphate buffer (0.01 mol/L (NH4)2HPO4), adjusted with phosphoric acid solution to pH 3.0, was used as the mobile phase with a flow rate of 0.8 mL/min. The detection wavelength was 215 nm.
DNA extraction, PCR amplification and Illumina MiSeq sequencing
Microbial DNA was extracted from the samples using the E.Z.N.A.® soil DNA kit (Omega Bio-Tek, Norcross, GA, U.S.A.) according to the manufacturer’s protocols. The final DNA concentration and purification were determined by NanoDrop 2000 UV-vis spectrophotometer (Thermo Scientific, Wilmington, USA), and DNA quality was checked by 1% agarose gel electrophoresis. The V3-V4 hypervariable regions of the bacterial 16S rRNA gene were amplified by polymerase chain reactions (95 °C for 5 min; 25 cycles of 95 °C for 30 s, 55 °C for 30 s and 72 °C for 40 s; and a final extension at 72 °C for 10 min) with primers 338 F (5′-ACTCCTACGGGAGGCAGCAG-3′) and 806 R (5′-GGACTACHVGGGTWTCTAAT-3′)39 by a thermocycler PCR system (GeneAmp 9700, ABI, USA). For fungi ITS1 region was amplified by PCR (95 °C for 2 min; 30 cycles at 95 °C for 30 s, 61 °C for 30 s, and 72 °C for 45 s; and a final extension at 72 °C for 10 min) with the ITS1F (5′-AxxxCTTGGTCATTTAGAGGAAGTAA-3′) and ITS2 (5′-BGCTGCGTTCTTCATCGATGC-3′)40 primers. PCR products were purified according to a previous study41. Purified amplicons were pooled in equimolar ratios and paired-end sequenced (2 × 300) on an Illumina MiSeq platform (Illumina, San Diego, USA) according to standard protocols by Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China).
Processing of sequencing data
Raw fastq files were demultiplexed, quality filtered by Trimmomatic and merged by FLASH with the following criteria: (i) The reads were truncated at any site receiving an average quality score <20 over a 50 bp sliding window. (ii) Primers were matched exactly, allowing 2 nucleotide mismatches, and the reads containing ambiguous bases were removed. (iii) Sequences whose overlap was longer than 10 bp were merged according to their overlapping sequence42.
Operational taxonomic units (OTUs) were clustered with a 97% similarity cutoff using UPARSE (version 7.1 http://drive5.com/uparse/), and chimeric sequences were identified and removed using UCHIME. The taxonomy of each 16 S rRNA gene sequence was analyzed by an RDP Classifier algorithm (http://rdp.cme.msu.edu/) against the Silva (SSU123) 16 S rRNA database using a confidence threshold of 70%. The taxonomy of each ITS gene sequwence was analyzed by Unite (Release 6.0 http://unite.ut.ee/index.php)43.
Alpha rarefaction was performed in QIIME (version 1.7.0) using the Chao1 estimates of species abundance44, while observed species estimation of the amount of unique OTUs found in each sample and Shannon index45 were calculated. Cluster analysis was preceded by NMDS46. For the QIIME calculation, the beta diversity of the unweighted and weighted UniFrac distances was used for the UPGMA clustering and principal coordinate analysis47. To identify the differences in the bacterial communities between the two groups, similarities were analyzed using the Bray-Curtis dissimilarity distance matrices.
The metagenomic functional composition prediction analysis for all biofilm samples from 16 S data in the latest Kyoto Encyclopedia of Genes and Genomes (KEGG) database was performed using PICRUSt pipeline as described by Langille et al.48.
To associate the microbiota and flavour compounds, the significance of correlations between the microbial genera and flavour were tested(pearson correlation). The p values were adjusted by FDR using Benjamini–Hochberg method, the cutoff of adjusted p value was set as 0.05. The association network was constructed by using all significant associations, and it was displayed using R language programming.
References
Xu, E. et al. Characterization of Volatile Flavor Compounds in Chinese Rice Wine Fermented from Enzymatic Extruded Rice. Journal of Food Science. 80, 1476–1489 (2015).
Fu, J. Introduction to yellow rice wine in China from ancient time to present day. Liquor-Making Science & Technology. 113–116 (2011).
Fan, W. & Qian, M. Characterization of aroma compounds of chinese “Wuliangye” and “Jiannanchun” liquors by aroma extract dilution analysis. J Agric Food Chem. 54, 2695–2704 (2006).
Xiong, R. Preliminary study on the selection criteria of raw materials for the Chinese Rice Wine (Zhejiang A&F University, Zhejiang, Linan, 2015).
Chen, S., Luo, T., Xu, Y., Fan, W. & Zhao, G. Effects of yeast strains and raw materials on β-phonylethanol production in Chinese rice wines. China Brewing. 28, 23–26 (2009).
Cui, L. Biosynthetic Pathways and Steps of Pyrazine Compounds in Maotai-flavor Liquor. Liquor Making. 34, 39–40 (2007).
Zhang, CL. The relationship between quality, microorganism and aroma components of Luzhou Laojiao Daqu. (Jiangnan Universty. Jiangsu, Wuxi, 2012).
Fujii, T. et al. Molecular cloning, sequence analysis, and expression of the yeast alcohol acetyltransferase gene. Applied & Environmental Microbiology. 60, 2786–2792 (1994).
Beaumont, M. Flavouring composition prepared by fermentation with Bacillus spp. Int. J. Food Microbiol. 75, 189–196 (2002).
Wei, Z., Tabrizi, M., Robert, N. M. J. & Zhong, H. Daqu - A Traditional Chinese Liquor Fermentation Starter. Journal of the Institute of Brewing. 117, 82–90 (2011).
De, F., Genovese, A., Ferranti, P., Gilbert, J. & Ercolini, D. Metatranscriptomics reveals temperature-driven functional changes in microbiome impacting cheese maturation rate. Scientific Reports. 6, 2045–2322 (2016).
Liu, S. P. et al. Bacterial succession and the dynamics of volatile compounds during the fermentation of Chinese rice wine from Shaoxing region. World J Microbiol Biotechnol. 31, 1907–1921 (2015).
Wang, P. X. et al. Changes in flavour characteristics and bacterial diversity during the traditional fermentation of Chinese rice wines from Shaoxing region. Food Control. 44, 58–63 (2014).
Lv, Y. et al. Characterization of microbial communities in Chinese rice wine collected at Yichang city and Suzhou city in China. Journal of Microbiology & Biotechnology. 27, 1409–1418 (2017).
Guo, X., Hu, P., Xu, Y. & Zhao, G. Research on the Volatile Flavoring Substances in Yellow Rice Wine. Liquor-making Science & Technology. 5 (2004).
Cui, Y., Cao, X., Wang, C. & Zhou, G. Research progress of 4-Vinylguaiacol and 4-Ethylguaiacol in fermentation industry. China Brewing. 28, 14–17 (2009).
Mason, D. & Braunwald, E. The Effects of Nitroglycerin and Amyl Nitrite on Arteriolar and Venous Tone in the Human Forearm. Circulation. 32, 755 (1965).
Luo, T., Fan, W. & Xu, Y. Characterization of volatile and semi-volatile compounds in Chinese rice wines by headspace solid phase microextraction followed by gas. Journal of the Institute of Brewing. 114, 172–179 (2008).
Yu, L., Ding, F. & Ye, H. Analysis of characteristic flavour compounds in Chinese rice wines and representative fungi in wheat Qu samples from different regions. Journal of the Institute of Brewing. 118, 114–119 (2012).
Lv, X. C. et al. Bacterial community dynamics during the traditional brewing of Wuyi Hong Qu glutinous rice wine as determined by culture-independent methods. Food Control. 34, 300–306 (2013).
Jiménez, N., Curiel, J., Reverón, I., Rivas, B. & Muñoz, R. Uncovering the Lactobacillus plantarum WCFS1 Gallate Decarboxylase Involved in Tannin Degradation. Appl Environ Microbiol. 79, 4253–4263 (2013).
Chowdhury, S., Khanna, S., Verma, S. & Tripathi, A. Molecular diversity of tannic acid degrading bacteria isolated from tannery soil. Journal of Applied Microbiology. 97, 1210–1219 (2004).
Kasai, D., Masai, E., Miyauchi, K., Katayama, Y. & Fukuda, M. Characterization of the Gallate Dioxygenase Gene: Three Distinct Ring Cleavage Dioxygenases Are Involved in Syringate Degradation by Sphingomonas paucimobilis SYK-6†. Journal of Bacteriology. 187, 5067–5074 (2005).
Nogales, J. et al. Unravelling the gallic acid degradation pathway in bacteria: the gal cluster from Pseudomonas putida. Molecular Microbiology. 79, 359–374 (2011).
Luan, J. Research on the Flavor in Rice Wine. China Brewing 21, 21–24 (2002).
Li, H., et al. Integrated Electromicrobial Conversion of CO2 to Higher Alcohols. Science. 335 (2012).
Chen, H., Wu, Z. & Xie, G. Detection of Organic Acids in Rancid Yellow Rice Wine in Storage. Liquor-Making Science & Technology. 2, 78–81 (2017).
Liu, J. J. et al. Research and Application on L-Malic Acid. Chian Food Additives. 3, 53–56 (2003).
Feng, A., Zhao, W., Bai, W. & Pang, J. Determination of organic acids in different rice wine. China Brewing. 29, 144–146 (2010).
Erikax, B. & Bernardoa, L. Characterization of Cladosporium Rot in Grapevines,a Problem of Growing Importance in Chile. Plant Disease. 92, 1635–1642 (2008).
Takada, T., Kurakawa, T., Tsuji, H. & Nomoto, K. Fusicatenibacter saccharivorans gen. nov., sp. nov., isolated from human faeces. International Journal of Systematic & Evolutionary Microbiology. 63, 3691–3696 (2013).
Xia, R. & Che, B. Determination of Isoamyl nitrite by gas chromatography. Chinese Journal of Medicine. 41, 48–48 (2006).
Palleroni, N. & Bradbury, J. Stenotrophomonas, a new bacterial genus for Xanthomonas maltophilia (Hugh 1980) Swings et al. 1983. Int J Syst Bacteriol. 43 (1993).
Pontesucre, A. ABC transporters in microorganisms: research, innovation and value as targets against drug resistance. (ed. Ponte-Sucre, A) Ch. 2, 35–36 (Caister Academic, 2009).
Ter Beek, J., Guskov, A. & Slotboom, D. J. Structural diversity of ABC transporters. Journal of General Physiology. 143, 419–435 (2014).
Song, Z., Du, H., Zhang, Y. & Xu, Y. Unraveling Core Functional Microbiota in Traditional Solid-State Fermentation by High-Throughput Amplicons and Metatranscriptomics Sequencing. Frontiers in Microbiology. 8 (2017).
Guan, B., Ding, Y., Xie, L. & Long, Y. The Modification of the DNS Method for the Determination of Reducing Sugar. Journal of Wuxi University of Light Industry. 18 (1999).
Ye, F., Chen, X. & Ni, X. Study of Analytical Method for Organic acids in Yellow Rice Wines by High Performance Liquid Chromatography. Liquor Making. 40, 80–85 (2013).
Zhang, X. L., Tian, X. Q., Ma, L. Y. & Yang, Q. Biodiversity of the Symbiotic Bacteria Associated with Toxic Marine Dinoflagellate Alexandrium tamarense. Journal of Biosciences & Medicines. 3, 23–28 (2015).
Buee, M. et al. 454 Pyrosequencing analyses of forest soils reveal an unexpectedly high fungal diversity. New Phytologist. 184, 449–456 (2009).
Ren, G., Ren, W., Teng, Y. & Li, Z. Evident bacterial community changes but only slight degradation when polluted with pyrene in a red soil. Frontiers in Microbiology. 6 (2015).
Magoč, T. & Salzberg, S. FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963 (2011).
Kõljalg, U. et al. Towards a unified paradigm for sequence-based identification of fungi. Molecular Ecology. 22, 5271–5277 (2013).
Caporaso, J. G. et al. QIIME allows analysis of high-throughput community sequencing data. Nature Methods. 7, 335–336 (2010).
Kemp, P. & Aller, J. Bacterial diversity in aquatic and other environments: what 16 S rDNA libraries can tell us. Fems Microbiology Ecology. 47, 161–177 (2004).
Werley, H., Devine, E., Zorn, C., Ryan, P. & Westra, B. The NMDS: Abstraction tool for standardized, comparable, essential data. American Journal of Public Health. 81, 421–426 (1991).
Lozupone, C., Lladser, M., Knights, D., Stombaugh, J. & Knight, R. UniFrac: an effective distance metric for microbial community comparison. Isme Journal. 5 (2011).
Langille, M. G. I. et al. Predictive functional profiling of microbial communities using 16 S rRNA marker gene sequences. Nature Biotechnology. 31, 814–821 (2013).
Acknowledgements
This study was supported by National Key R&D Program of China(2016YFD0400500) and National Natural Science Foundation of China (No. 31400161). We are also grateful to all wine producers that collaborate with this study.
Author information
Authors and Affiliations
Contributions
J.L.X., X.L. and Q.R. designed the research; X.L. and Z.P.L. analyzed the results; H.J.W. and Z.W.W. performed the experiments; J.L.X. and F.P.Z. participated in writing the manuscript and revision; Q.R. supervised the research.
Corresponding author
Ethics declarations
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Xu, J., Wu, H., Wang, Z. et al. Microbial dynamics and metabolite changes in Chinese Rice Wine fermentation from sorghum with different tannin content. Sci Rep 8, 4639 (2018). https://doi.org/10.1038/s41598-018-23013-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-018-23013-1
- Springer Nature Limited
This article is cited by
-
The initial composition and structure of microbial community determined the yield and quality of Baijiu during the spontaneous fermentation
International Microbiology (2023)
-
Microbial succession and exploration of higher alcohols-producing core bacteria in northern Huangjiu fermentation
AMB Express (2022)
-
Metabolite-based cell sorting workflow for identifying microbes producing carbonyls in tobacco leaves
Applied Microbiology and Biotechnology (2022)
-
Global cocoa fermentation microbiome: revealing new taxa and microbial functions by next generation sequencing technologies
World Journal of Microbiology and Biotechnology (2021)
-
Bacterial succession and the dynamics of flavor compounds in the Huangjiu fermented from corn
Archives of Microbiology (2020)