Abstract
A large proportion of cases with chronic conditions including diabetes or pre-diabetes, hypertension and dyslipidemia remain undiagnosed. To include reproductive factors (RF) might be able to improve current screening guidelines by providing extra effectiveness. The objective is to study the relationships between RFs and chronic conditions’ biomarkers. A cross-sectional study was conducted. Demographics, RFs and metabolic biomarkers were collected. The relationship of the metabolic biomarkers were shown by correlation analysis. Principal component analysis (PCA) and autoencoder were compared by cross-validation. The better one was adopted to extract a single marker, the general chronic condition (GCC), to represent the body’s chronic conditions. Multivariate linear regression was performed to explore the relationship between GCC and RFs. In total, 1,656 postmenopausal females were included. A multi-layer autoencoder outperformed PCA in the dimensionality reduction performance. The extracted variable by autoencoder, GCC, was verified to be representative of three chronic conditions (AUC for patoglycemia, hypertension and dyslipidemia were 0.844, 0.824 and 0.805 respectively). Linear regression showed that earlier age at menarche (OR = 0.9976) and shorter reproductive life span (OR = 0.9895) were associated with higher GCC. Autoencoder performed well in the dimensionality reduction of clinical metabolic biomarkers. Due to high accessibility and effectiveness, RFs have potential to be included in screening tools for general chronic conditions and could enhance current screening guidelines.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Introduction
Type 2 diabetes mellitus (T2DM), hypertension and hyperlipidemia are chronic conditions that can result in severe complications1,2, including cardiovascular disease (CVD), the leading cause of death worldwide3,4. Unfortunately, a large number of patients with these conditions remain undiagnosed. Most updated literatures showed that in 2019 there are 50.1% (231.9 million) of diabetes patients still undiagnosed worldwide5. A large proportion of cases with hypertension6 and hyperlipidemia7 are also unware of their condition, particularly in low and middle income countries8.
Screening of those at risk of chronic conditions is of significance for both individuals and wider society, yet there are gaps in current practices. Early identification is generally based on commonly collected risk factors, such as age, gender, smoking status, body mass index (BMI) and family history. A number of societies and task forces have recommended various screening guidelines that consist of these risk factors9,10,11; however, there are growing concerns that such guidelines might be inadequate and inaccurate12,13,14,15. For example, the American Diabetes Association (ADA) and the US Preventive Services Task Force (USPSTF) guidelines have shown only a fair performance when externally validated12,13. Furthermore, a trial exploring the effectiveness of a population-based screening programme in the United Kingdom found that screening was not associated with a reduction in all-cause mortality over a median period of 9.6 years15. A number of commonly used screening functions have also been shown to be ineffective in population screening16.
To include novel or extra factors might help to identify high risk groups more accurately and has the potential to improve current screening guidelines for chronic conditions, in terms of both effectiveness and efficiency. Indeed, a growing body of studies has identified a strong relationship between women’s reproductive factors (RF) and chronic conditions. For example, early menarche has been found to be associated with an increased risk of T2DM17,18, obesity and insulin resistance19. Moreover, a retrospective study conducted in Europe showed that, after adjustment for confounding, early menopause and shorter reproductive life span was associated with T2DM20. A Japanese study also found a similar relationship regarding hypercholesterolemia21. Furthermore, the China Kadoorie Biobank study reported that Chinese women with late menopause (≥53 years) were 1.21 (95% CI: 1.03–1.42) times more likely to have T2DM than women with menopause at 46–52 years old (p < 0.0001)22. Another Chinese study also found that a higher number of live births was associated with hypertension and DM, and mediated by lifestyle and dyslipidemia23. Additional studies conducted in different regions and healthcare settings have shown similar results. It is also important to note that RFs are highly accessible in all medical settings with low cost, hence we hypothesized that RFs are associated with chronic conditions and, as novel factors, might be able to improve current screening guidelines to assess women’s risk of chronic conditions21.
The objective of the current study is to explore the relationship between RFs and chronic conditions in order to assess the application of RFs as preliminary screening tools for general chronic conditions in women, so as to allow for the early diagnosis and intervention. This is challenging as clinical biomarkers of chronic conditions consist of multiple parameters as dependent variable and it is difficult to clarify its relationship with RFs using standard statistical methods. Therefore, in order to investigate their association, we applied a machine learning based dimensionality reduction technique, autoencoder, to generalize one single marker to represent chronic conditions.
Methods
Participants
A cross-sectional study was conducted in the Gansu Province of China. Random stratified sampling was adopted to include participants who were under care in primary health service organizations. The sample size was assessed by the following formula24: \({\rm{N}}=\frac{{{\rm{Z}}}_{\frac{{\rm{\alpha }}}{2}}^{2}{\rm{P}}1-{\rm{P}}}{{{\rm{\varepsilon }}}^{2}}\). It was estimated that 384 participants ought to be enrolled from each of the 28 sample centres (accuracy = 95%, confidence = 95%). To allow for missing data, and the constitution of demographic factors, 12,000 participants were recruited, of which 11,115 completed the study.
Participants were eligible for inclusion if they were: (1) female; (2) postmenopausal; (3) no self-reported DM, hypertension or dyslipidemia.
Participants were excluded if they were: (1) diagnosed with secondary diabetes or secondary hypertension; (2) pregnant and lactating; (3) taking medicine that affects the metabolism of glucose and lipids within 3 months; and (4) diagnosed with type 1 diabetes; (5) Non-natural menopause.
Study design
From June to August 2016, seven investigators with a registered nursing practicing certificate administered a questionnaire, physical tests and biochemical tests to participants.
The questionnaire was designed based on related studies25,26 and modified according to pre-survey results in order to minimize bias. The investigators interviewed each participant face-to-face and completed the questionnaire accordingly. Five RFs were collected: age at menarche, age at menopause, reproductive life span, live births and abortion history.
The investigators performed five physical tests using standard instruments to measure height, weight, waist and hip circumference, heart rate, systolic blood pressure (SBP) and diastolic blood pressure (DBP). All tests were repeated three times and the average reading calculated. Body mass index (BMI) was calculated with weight (kg)/height2 (m2), and waist-to-hip ratio (WHR) was calculated using waist (cm)/hip (cm).
Three biochemical tests were performed, which included: 1) a fast blood-glucose test; 2) a blood lipids test; and 3) oral glucose tolerance test (OGTT). All laboratory assays were performed in accredited medical laboratories by the Chinese National Health Authority. Protocols were strictly adhered to. Total cholesterol (TC), total triglyceride (TG), high-density lipoprotein (HDL-C), low-density lipoprotein (LDL-C), fasting plasma glucose (FPG), OGTT 2 h plasma glucose (OGTT 2 h PG) were collected.
Ethical considerations
The ethics committee of the School of Public Health in Lanzhou University approved this study. All relevant ethical guidelines and regulations were strictly adhered to throughout. Informed consent was obtained from each participant whose data was included in the analyses.
Data analysis
First, a descriptive analysis was applied to summarize the biomarkers and RFs. Following that, since all the clinical biomarkers are continuous variables and are verified following normal (Gaussian) distribution (by Kolmogorov-Smirnov normality test), the relationship between included clinical biomarkers was explored using Pearson correlation analysis and hierarchical clustering analysis. Corresponding correlation plots were used to display the complex relationships between each of the two variables27. Through this method, we were able to demonstrate the redundancy of the clinical biomarkers. An autoencoder was then applied to generalize a single marker to represent 10 clinical biomarkers of the chronic conditions. Meanwhile, a more generic dimensionality reduction method, principle component analysis (PCA), was applied for comparison. Disease binary variables (positive or negative, represented by 1 or 0) of pathoglycemia, hypertension and dyslipidemia were determined from the continuous values of the original 10 clinical biomarkers according to the clinical ascertainment of these diseases. For both methods, using disease binary variables as labels and extracted single variable as the risk score to set different threshold, area under curve (AUC) with 95% confidence interval was calculated based on 10-fold cross-validation, in order to verify the representation power of the extracted variable. T-test was used to compare the representation power (AUC) of both methods.
An autoencoder is a data-driven neural network with an encoder, a bottleneck layer and a decoder combined. The encoder (also a multilayer network) can project the high-dimensional data onto a low-dimensional feature space at the output of the bottleneck layer, which can also be considered as a feature extraction of the input. The multilayer decoder network can then reconstruct the data from the coder layer reversely. Therefore, the network can be trained unsupervised with the same input and output by minimizing the mean square error (MSE) between them at the output of the network using a backpropagation algorithm28. The bottleneck layer is considered as a valid dimensionality reduction or feature extraction and then it can be used as a generalized marker to represent the input (the 10 biomarkers).
The structure of this autoencoder is presented in Fig. 1. Ten clinical biomarkers were used as the inputs and outputs to train the final autoencoder. All activation functions were set to be sigmoid functions for nonlinear transformation. Initially, considering the sample size and degree of freedom (decided by the number of parameters to be estimated), we set up a range for the number of layers and neurons. The specific numbers of layers and neurons were finally determined by greedy search, as well as other hyper-parameters, such as the learning rate and batch size.
After the better dimensionality reduction method was identified, it was used to extract the single biomarker of all cases. This single general marker is named as the general chronic condition (GCC) in this paper. Receiver operating characteristic curves (ROC) were plotted to show the capability of GCC to represent these three chronic conditions.
The relationship of GCC and RFs was then analyzed using multivariate linear regression. This confirmed whether the reproductive factors were associated with the GCC, and in other words, whether they are effective preliminary screening tools for chronic conditions. Missing data were handled using multiple imputation by chained equation (MICE) for 5 times and the results were pooled with Rubin’s rule29.
Statistical analysis was implemented on R 3.5.1. All significance tests were two-tailed and α = 0.05. PCA was implemented by prcomp function, and AUC was calculated by pROC package. The autoencoder was implemented on Python 3.5.4 using Tensorflow, which is an open-source software library for machine learning. Raw dataset and code can be found on Github. (https://github.com/dongdongdongdwn/Reproductive-factors-as-screening-tools-for-chronic-conditions-in-primary-care-using-a-machine-learn).
Results
Participant characteristics
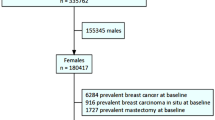
As shown in Fig. 2, 11,115 cases with valid data were included initially. According to our inclusion and exclusion criteria, 1,656 postmenopause women without self-report chronic conditions were retained for further analysis.
The clinical biomarkers collected from physical and biochemical tests are listed in Table 1 with respect to three age groups (years: 41–50, 51–65, >65). The mean values of the BMI were statistically different (p < 0.01) across the groups. A similar finding was also apparent for WHR, TC, fasting plasma glucose, OGTT 2 h plasma glucose, SBP and DBP. Table 2 describes the distribution of the 5 reproductive factors.
Correlation within clinical biomarkers
According to the correlation matrix and Hierarchical Clustering of the 10 clinical biomarkers (Fig. 3), none of the biomarkers were uncorrelated, which implies that complicated relationships and strong redundancy exist across the biomarkers, and dimensionality reduction could potentially extract a better representation.
Dimensionality reduction
Due to the internal correlation within the biomarkers, a dimensionality reduction method is reasonable to be used to extract representative features. Multilayer autoencoder and PCA were performed based on 10-fold cross validation and results were compared with a t-test. As shown in Table 3, AUCs of autoencoder were significantly higher (p value < 0.01) than the PCA for all chronic conditions (pathoglycemia, hypertension, dyslipidemia), indicating that autoencoder could produce a more representative variable to express the body’s metabolism.
Subsequently, autoencoder was applied to all cases, translated 10 biomarkers into a single general marker (GCC). As shown in Fig. 4, when distinguishing pathoglycemis, hypertension and dyslipidemia respectively, GCC showed good discrimination power with an AUC of more than 0.8 (AUC = 0.844, 0.824, 0.805).
Association between the reproductive factors and the GCC
Finally, the relationship between RFs and GCC was explored by multivariate linear regression. As illustrated in Table 4, after adjustment of age, GCC, as the dependent variable, was associated with age at menarche (p < 0.05) and reproductive life span (p < 0.01). It was also found that GCC was higher with early age at menarche (OR = 0.9976, 95%CI: 0.9961–0.9998) and shorter reproductive life span (OR = 0.9895, 95%CI: 0.9926–0.9864). No significant results were found as for age at menopause, live births and abortion history.
Discussion
This study explored the relationship between chronic conditions and reproductive factors and demonstrated that age at menarche and reproductive life span have potential to be incorporated into screening tools for general chronic conditions. The chronic conditions were generalized from relevant clinical biomarkers using one of the most advanced non-linear dimensionality reduction techniques in machine learning. Autoencoder outperformed a state-of-the-art dimensionality reduction method, PCA, and extracted a more discriminative general marker. To our knowledge, there is currently no similar study reported.
Metabolic syndrome (MS) is a traditional way to represent the chronic conditions, but had been given considerable doubt by both the American Diabetes Association and the European Association regarding its value as a CVD risk marker as too much critically important information was missing to warrant its designation as a “syndrome”30. Meanwhile, it has been shown that MS is insensitive to identifying some chronic metabolic diseases31. However, it is well know that these chronic conditions (i.e. pathoglycemia, hypertension and dyslipidemia) are characterized by metabolic disorder and show clustering on account of similar risk factors and correlative physiological mechanisms32,33. Hence, we used a machine learning based approach, autoencoder, to generate a representative marker to represent these chronic conditions.
Machine learning and artificial intelligence have become emerging techniques in health care for big data analysis34,35. Autoencoder was first introduced by Hinton in 2006 and has been verified to outperform traditional approaches for dimensionality reduction36 and gained increasing use as an application in medical studies37. In this study, the autoencoder was trained with more than 1,000 samples with 10 biomarkers and found to successfully extract one single marker, the GCC, to generalize the biomarkers for the chronic conditions. GCC was also shown to have the power to discriminate the chronic diseases (AUCs > 0.8). In comparison, the marker that was extracted by PCA was not discriminative enough (AUCs < 0.7). This could be interpreted by the nonlinear expressiveness of multilayer autoencoder36 which derives from its multiple hidden layers and nonlinear activation functions38. The improved nonlinear reconstruction of autoencoder over PCA has also been verified and explained by other researchers39,40. In addition, it is well known that a multilayer autoencoder requires more computation than PCA, however, due to the limited sample size (N = 1,656), these two methods did not show apparent difference in the computation time.
Via the multivariate linear regression, earlier age at menarche and shorter reproductive life span have been found to be associated with chronic conditions. In terms of age at menarche, our findings are in accordance with numbers of studies in which females with early age at menarche are at higher risk of chronic diseases. A recent study on Chinese elderly women (age = 70.39 ± 6.21) reported that women with metabolic syndrome had younger menarche age, higher gravidity and parity41. Additionally, in a multicenter case control study, Lecinana and his colleagues determined that very early exposure onset (age < 13) may do harm to body metabolism function42. Generally, adulthood adiposity is considered as potential mediator43. Apart from that, the association between reproductive life span and the risk of chronic conditions is also supported by relevant studies and could be interpreted by the protective effect of estrogen20. In terms of the age at menopause, currently there is not a uniform conclusion as some studies have not found any relationship44,45 whereas some have46,47. Mechanistic studies have demonstrated beneficial effects of estrogen on insulin secretion and glucose homeostasis. Meanwhile, some researchers believe the effect on TC is a result of a decrease in serum estradiol48 and a decrease in the activity of LDL-C receptors49. There is also an assumption that insulin resistance is associated with pregnancy and parturition50,51. However, after multivariable adjustment, we did not observe any such association. Besides, some specific reproductive conditions, such as polycystic ovary syndrome (PCOS), are associated with insulin resistance52 and secondary hyperandrogenism53. Lagana and his colleagues also found that insulin sensitizers could improve the PCOS symptoms54, which hints that additional RFs could benefit the chronic conditions screening. In sum, despite the fact that the relationship between RFs and chronic conditions could not be interpreted by a single factor, RFs do have strong associations with chronic conditions.
Low cost disease detection models are of great importance to reduce the health economic burden, and especially to benefit developing countries55. High accessibility and low cost are outstanding advantages of RFs. Furthermore, there are studies that have shown that the validity and reproducibility of self-reported RFs are good56. Therefore, RFs have potential as screening tools for chronic conditions and could improve current screening guidelines.
A number of important limitations need to be considered. First, this is a cross-sectional study and hence it cannot infer causality. Second, this study only tested the possibility that RFs can be incorporated into a screening tool and did not give the actual sensitivity and specificity of RFs to screen for chronic conditions. In terms of further research, a structured screening tool should be developed and externally validated. Third, although not included in the current study, uric acid and HbA1c are also crucial biomarkers for chronic conditions and it is important that future research takes them into account. Last, interpretability is always a key concern when applying machine learning to medical data analysis. Many advanced methods have been proposed to unfold the black box of neuron networks. In future study, we hope to focus on this specific question and explore the GCC more comprehensively.
To conclude, autoencoder performed well in the dimensionality reduction of clinical biomarkers, demonstrating its potential in further medical data process. Women with earlier age at menarche and shorter reproductive life span are more likely to suffer from chronic conditions. Due to high accessibility and effectiveness, RFs show potential to be included in preliminary screening tools for general chronic conditions in clinical practice and could enhance current screening guidelines.
Data availability
The original data is not currently available online but can be requested in machine-readable format from the corresponding author on reasonable request.
References
Rahelic, D. 7th Edition of Idf Diabetes Atlas–Call for Immediate Action. Lijec Vjesn 138, 57–58 (2016).
Innovative care for chronic health conditions. Rev Panam Salud Publica 12, 71–74 (2002).
Dixon, J. & Dewar, S. The NHS plan. BMJ 321, 315–316 (2000).
Koh, H. K., Blakey, C. R. & Roper, A. Y. Healthy People 2020: a report card on the health of the nation. JAMA 311, 2475–2476, https://doi.org/10.1001/jama.2014.6446 (2014).
Saeedi, P. et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas, 9 edition. Diabetes research and clinical practice 157, 107843, https://doi.org/10.1016/j.diabres.2019.107843 (2019).
Mozaffarian. Heart Disease and Stroke Statistics-2016 Update: A Report From the American Heart Association (vol 133, pg e38, 2016). Circulation 133, E599–E599, https://doi.org/10.1161/CIR.0000000000000409 (2016).
Koyama, A. K., Bali, V., Yermilov, I. & Legorreta, A. P. Identification of Undiagnosed Hyperlipidemia: Do Work Site Screening Programs Work? American Journal of Health Promotion 32, 971–978, https://doi.org/10.1177/0890117116671537 (2018).
Who, A. J. W. H. O. Global brief on hypertension. (2013).
Abid, A., Ahmad, S. & Waheed, A. Screening for Type II Diabetes Mellitus in the United States: The Present and the Future. Clinical Medicine Insights: Endocrinology and Diabetes 9, https://doi.org/10.4137/CMED.S38247 (2016).
Gillman Matthew, W., Caspard, H., Lane, K., Forman John, P. & Rifas-Shiman Sheryl, L. Diabetes and lipid screening among patients in primary care: A cohort study. BMC Health Services Research 8, 25, https://doi.org/10.1186/1472-6963-8-25 (2008).
Screening for Lipid Disorders in Adults: Recommendation Statement. Am. Fam. Physician 80, 1273–1274 (2009).
Kai Mckeever, B. et al. Receipt of Glucose Testing and Performance of Two US Diabetes Screening Guidelines, 2007–2012. PLoS ONE 10, e0125249, https://doi.org/10.1371/journal.pone.0125249.
Sheehy, A. M. et al. Analysis of guidelines for screening diabetes mellitus in an ambulatory population. Mayo Clinic proceedings 85, 27–35, https://doi.org/10.4065/mcp.2009.0289 (2010).
Ochoa, P. S. et al. Identification of Previously Undiagnosed Diabetes and Prediabetes in the Inpatient Setting Using Risk Factor and Hemoglobin A1C Screening. Annals of Pharmacotherapy 48, 1434–1439, https://doi.org/10.1177/1060028014547383 (2014).
Nwaneri, C., Bowen-Jones, D. & Cooper, H. Screening for type 2 diabetes and population mortality over 10 years. Lancet (London, England) 381, 901, https://doi.org/10.1016/S0140-6736(13)60665-0 (2013).
Toscano, C. M. et al. Cost-effectiveness of a national population-based screening program for type 2 diabetes: the Brazil experience. Diabetol Metab Syndr 7, 95, https://doi.org/10.1186/s13098-015-0090-8 (2015).
He, C. et al. Age at menarche and risk of type 2 diabetes: results from 2 large prospective cohort studies. Am J Epidemiol 171, 334–344, https://doi.org/10.1093/aje/kwp372 (2010).
Muka, T. et al. Age at natural menopause and risk of type 2 diabetes: a prospective cohort study. Diabetologia 60, 1951–1960, https://doi.org/10.1007/s00125-017-4346-8 (2017).
Adair, L. S. & Gordon-Larsen, P. Maturational timing and overweight prevalence in US adolescent girls. American journal of public health 91, 642, https://doi.org/10.2105/AJPH.91.4.642 (2001).
Brand, J. S. et al. Age at menopause, reproductive life span, and type 2 diabetes risk: results from the EPIC-InterAct study. Diabetes care 36, 1012, https://doi.org/10.2337/dc12-1020 (2013).
Lee, J. S. et al. Independent association between age at natural menopause and hypercholesterolemia, hypertension, and diabetes mellitus: Japan nurses’ health study. Journal of Japan Atherosclerosis Society 20, 161–169 (2013).
Wang, M. et al. Age at natural menopause and risk of diabetes in adult women: Findings from the China Kadoorie Biobank study in the Zhejiang area. Journal of Diabetes Investigation 9, 762–768, https://doi.org/10.1111/jdi.12775 (2018).
Xu, B., Chen, Y., Xiong, J. P., Lu, N. & Tan, X. Association of Female Reproductive Factors with Hypertension, Diabetes and LQTc in Chinese Women. Sci Rep, 7, https://doi.org/10.1038/srep42803 (2017).
Daniel, W. W. Biostatistics: a foundation for analysis in the health sciences. 9th ed. edn (John Wiley & Sons, 2009).
Feng, Y. et al. Prevalence of metabolic syndrome and its relation to body composition in a Chinese rural population. Obesity (Silver Spring) 14, 2089–2098, https://doi.org/10.1038/oby.2006.244 (2006).
Gu, D. et al. Prevalence of the metabolic syndrome and overweight among adults in China. Lancet 365, 1398–1405, https://doi.org/10.1016/S0140-6736(05)66375-1 (2005).
John A. RiceLes informations contenues dans cette page sont à usage strict de et ne doivent être utilisées ou copiées par un tiers.Powered by, and. Mathematical Statistics and Data Analysis, (Wadsworth, 1988).
Rumelhart, D. E., Hinton, G. E. & Williams, R. J. Learning representations by back-propagating errors. Nature 323, 533–536, https://doi.org/10.1038/323533a0 (1986).
Rubin, D. B. Multiple Imputation for Nonresponse in Surveys, https://doi.org/10.1002/9780470316696 (2008).
Kahn, R., Buse, J., Ferrannini, E. & Stern, M. The metabolic syndrome: time for a critical appraisal: joint statement from the American Diabetes Association and the European Association for the Study of Diabetes. Diabetes care 28, 2289–2304, https://doi.org/10.2337/diacare.28.9.2289 (2005).
DeBoer, M. D. & Gurka, M. J. Low sensitivity of the metabolic syndrome to identify adolescents with impaired glucose tolerance: An analysis of NHANES 1999-2010. Acta Veterinaria Scandinavica 13, 83, https://doi.org/10.1186/1475-2840-13-83 (2014).
Ford, E. S., Li, C. & Zhao, G. Prevalence and correlates of metabolic syndrome based on a harmonious definition among adults in the US. J Diabetes 2, 180–193, https://doi.org/10.1111/j.1753-0407.2010.00078.x (2010).
Beltran-Sanchez, H., Harhay, M. O., Harhay, M. M. & McElligott, S. Prevalence and trends of metabolic syndrome in the adult U.S. population, 1999–2010. J Am Coll Cardiol 62, 697–703, https://doi.org/10.1016/j.jacc.2013.05.064 (2013).
Obermeyer, Z. & Emanuel, E. J. Predicting the Future - Big Data, Machine Learning, and Clinical Medicine. N Engl J Med 375, 1216–1219 (2016).
Beam, A. L. & Kohane, I. S. Big Data and Machine Learning in Health Care. JAMA 319, 1317–1318, https://doi.org/10.1001/jama.2017.18391 (2018).
Hinton, G. E. & Salakhutdinov, R. R. Reducing the dimensionality of data with neural networks. Science 313, 504–507, https://doi.org/10.1126/science.1127647 (2006).
Majumdar, A., Gogna, A. & Ward, R. Semi-supervised Stacked Label Consistent Autoencoder for Reconstruction and Analysis of Biomedical Signals. IEEE Transactions on Biomedical Engineering PP, 1–1 (2016).
Plaut, E. From Principal Subspaces to Principal Components with Linear Autoencoders. (2018).
Wang, Y., Yao, H. & Zhao, S. Auto-encoder based dimensionality reduction. Neurocomputing 184, 232–242, https://doi.org/10.1016/j.neucom.2015.08.104 (2016).
Tan, C. & Eswaran, C. Reconstruction and recognition of face and digit images using autoencoders. Neural Comput & Applic 19, 1069–1079, https://doi.org/10.1007/s00521-010-0378-4 (2010).
Liu, M. et al. Association between reproductive variables and metabolic syndrome in chinese community elderly women. Archives of Gerontology & Geriatrics 63, 78–84 (2015).
Alonso, D. L. M. et al. Risk of ischemic stroke and lifetime estrogen exposure. Neurology 68, 33 (2007).
Lakshman, R. et al. Association between age at menarche and risk of diabetes in adults: results from the EPIC-Norfolk cohort study. Diabetologia 51, 781–786 (2008).
Mishra, G. D., Carrigan, G., Brown, W. J., Barnett, A. G. & Dobson, A. J. Short-term weight change and the incidence of diabetes in midlife: results from the Australian Longitudinal Study on Women’s Health. Diabetes care 30, 1418, https://doi.org/10.2337/dc06-2187 (2007).
Soriguer, S. F. et al. Type 2 diabetes mellitus and other cardiovascular risk factors are no more common during menopause: longitudinal study. Menopause 16, 817–821, https://doi.org/10.1097/GME.0b013e31819d4113 (2009).
Malacara, J. M., Huerta, R., Rivera, B., Esparza, S. & Fajardo, M. E. Menopause in normal and uncomplicated NIDDM women: physical and emotional symptoms and hormone profile. Maturitas 28, 35–45, https://doi.org/10.1016/S0378-5122(97)00051-0 (1997).
Cho, G. J. et al. Postmenopausal status according to years since menopause as an independent risk factor for the metabolic syndrome. Menopause-the Journal of the North American Menopause Society 15, 524–529 (2008).
Chakravarti, S. & Studd, J. Hormonal profiles after the menopause. British Medical Journal 2, 784–787 (1976).
Arca, M., Vega, G. L. & Grundy, S. M. Hypercholesterolemia in postmenopausal women: Metabolic defects and response to low-dose lovastatin. Jama 47, 87–88 (1994).
Gunderson, E. et al. Long-term plasma lipid changes associated with a first birth - The coronary artery risk development in young adults study. Am. J. Epidemiol. 159, 1028–1039, https://doi.org/10.1093/aje/kwh146 (2004).
Smith, D. E. et al. Longitudinal Changes in Adiposity Associated With Pregnancy: The CARDIA Study. JAMA 271, 1747–1751, https://doi.org/10.1001/jama.1994.03510460039030 (1994).
Laganà, A. S., Garzon, S., Casarin, J., Franchi, M. & Ghezzi, F. Inositol in Polycystic Ovary Syndrome: Restoring Fertility through a Pathophysiology-Based Approach. Trends in Endocrinology and Metabolism 29, 768–780, https://doi.org/10.1016/j.tem.2018.09.001 (2018).
Laganà, A. S., Vitale, S. G., Noventa, M. & Vitagliano, A. Current Management of Polycystic Ovary Syndrome: From Bench to Bedside. International Journal of Endocrinology 2018, https://doi.org/10.1155/2018/7234543 (2018).
Laganà, A. S. et al. Evidence-Based and Patient-Oriented Inositol Treatment in Polycystic Ovary Syndrome: Changing the Perspective of the Disease. International journal of endocrinology and metabolism 15, e43695, https://doi.org/10.5812/ijem.43695 (2017).
Bayati, M., Bhaskar, S. & Montanari, A. Statistical analysis of a low cost method for multiple disease prediction. Stat Methods Med Res 27, 2312–2328, https://doi.org/10.1177/0962280216680242 (2018).
Den Tonkelaar, I. Validity and reproducibility of self-reported age at menopause in women participating in the DOM-project. Maturitas 27, 117–123, https://doi.org/10.1016/S0378-5122(97)01122-5 (1997).
Acknowledgements
We are grateful for the efforts of all Research Assistants in the Chinese Academy of Science and the University of Hong Kong. Many thanks for your unremitting help and support. We would also like to express our appreciation to the patients in the primary care setting who consented to participate in this research. This study is supported by National Key R&D Program of the Ministry of Science and Technology of China (Grant No. 2016YFC1301602), National Natural Science Foundation of China (Grant Nos. 81871447 and 81661168015) and Shenzhen Science and Technology Innovation Commission (Grant No. JCYJ20160608153506088). The funding agencies played no role in the overall process of the research.
Author information
Authors and Affiliations
Contributions
J.L. and S.T. were involved in the whole process of this study, including study design, data collection, data process and results interpretation. S.T. and W.D. took charge of data analysis, model construction and testing, as well as the drafting of paper. K.C. and X.L. were responsible for the supervision of laboratory tests and also helped to revise the paper. L.B. contributed to the interpretation of the results and the revision of the paper. Prof. J.L. was the academic lead on this study and finalized the manuscript. All the authors have approved the final version of the publication, agree with the submission and accept responsibility for the paper’s validity.
Corresponding author
Ethics declarations
Competing interests
All the authors declare no competing interests. All have agreed to the submission to this journal and can confirm that the manuscript is not currently under submission in any other journal.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tian, S., Dong, W., Chan, K.L. et al. Screening for chronic conditions with reproductive factors using a machine learning based approach. Sci Rep 10, 2848 (2020). https://doi.org/10.1038/s41598-020-59825-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-59825-3
- Springer Nature Limited