Abstract
Dinoflagellates of the Symbiodiniaceae family encompass diverse symbionts that are critical to corals and other species living in coral reefs. It is well known that sexual reproduction enhances adaptive evolution in changing environments. Although genes related to meiotic functions were reported in Symbiodiniaceae, cytological evidence of meiosis and fertilisation are however yet to be observed in these taxa. Using transcriptome and genome data from 21 Symbiodiniaceae isolates, we studied genes that encode proteins associated with distinct stages of meiosis and syngamy. We report the absence of genes that encode main components of the synaptonemal complex (SC), a protein structure that mediates homologous chromosomal pairing and class I crossovers. This result suggests an independent loss of canonical SCs in the alveolates, that also includes the SC-lacking ciliates. We hypothesise that this loss was due in part to permanently condensed chromosomes and repeat-rich sequences in Symbiodiniaceae (and other dinoflagellates) which favoured the SC-independent class II crossover pathway. Our results reveal novel insights into evolution of the meiotic molecular machinery in the ecologically important Symbiodiniaceae and in other eukaryotes.
Similar content being viewed by others
Introduction
Sex is part of the life cycle of nearly all eukaryotes and has most likely been so since the last eukaryotic common ancestor1. Even lineages that were traditionally thought to be asexual, such as the Amoebozoa, possess the molecular machinery required for sex2. Dinoflagellates, a group of flagellated, mostly marine phytoplankton, are no exception. From the deeper-branching species Gymnodinium catenatum to the more recently-diverging Alexandrium minutum, syngamy and meiosis have been observed cytologically and through mating-type experiments3. The family Symbiodiniaceae, a lineage that branches between these two4, has been suggested to be sexual based on an early life-cycle description5. Some species of Symbiodiniaceae are symbionts, associated with a wide range of coral reef organisms, including cnidarians, molluscs, and foraminifera. Importantly, the dissociation of Symbiodiniaceae from reef-building corals under environmental stress (i.e., coral bleaching) can lead to coral death and eventual collapse of coral reefs6. A thorough understanding of the molecular mechanisms that underpin the reproduction of Symbiodiniaceae will elucidate the selective forces acting on this trait and adaptation of these ecologically important taxa.
Incongruence between the phylogenies of multiple isoenzymes and the internal transcribed spacer (ITS) regions7, and between the phylogenies of organellar and nuclear gene markers8 suggests that Symbiodiniaceae undergo hybridisation, in addition to clonal propagation. The identification of many meiotic toolkit genes in four diverse Symbiodiniaceae species (Symbiodinium microadriaticum, Breviolum minutum, Cladocopium goreaui, and Fugacium kawagutii) supports the notion that Symbiodiniaceae may be sexual9,10. Symbiodiniaceae cells grow as motile, flagellated cells (mastigotes) under light and divide in the dark as coccoid cells11. Symbiodiniaceae are believed to be isogamous5, but it is difficult to confirm through direct observation if these cell divisions are mitotic or meiotic. If sex occurs during the dark part of the life cycle as found in A. minutum3, one may assume that dinoflagellates including Symbiodiniaceae can respond to selection pressure by producing genetic variation through sexual recombination12. Some genes have a preferred set of codons due to variable abundance of distinct tRNAs; this codon usage bias affects the efficiency of gene expression. In mammals, codon usage bias was hypothesised to be a result of GC-biased gene conversion driven by meiotic recombination13. A higher rate of recombination does not necessarily improve the efficacy of selection, or of the removal of deleterious mutations14. Nevertheless, a strong codon usage bias likely arose via positive selection, and would inform us about processes that have been favoured in the evolution of Symbiodiniaceae.
One of the hallmarks of meiosis is the formation of the synaptonemal complex (SC), a proteinaceous scaffolding that juxtaposes homologous chromosomes, mediating their synapsis (i.e., the pairing of chromosomes for potential crossovers) during prophase I. The conventional SC consists of tripartite proteinaceous elements in parallel: a central element and two outer lateral elements, like two ladders attached side-to-side, with rungs of transversal filament proteins holding the elements together. Loops of sister chromatids are tethered to each lateral element. The process of synapsis “zips” down the scaffolding, with crossovers occurring at recombination nodules. Unsynapsed regions of the lateral elements are referred to as axial elements (see Loidl15 for a detailed diagram). These axial elements comprise homologous pairing protein 1 (Hop1) and reductional division protein 1 (Red1), whereas pachytene checkpoint protein 2 (Pch2) prevents chromosome segregation when synapsis and recombination are defective16. Formation of the SC is triggered by the synapsis initiation complex proteins, more commonly known as the ZMM proteins: (a) the transversal filament protein Zip1 (not to be confused with zinc transporter Zip1) attaches the central element to the pair of lateral elements; (b) Zip2, Zip3, and Zip4 mediate protein-protein interaction; (c) Mer3, a DNA helicase unwinds double-stranded DNA; and (d) the Msh4-Msh5 heterodimer binds to Holliday junction17. Metazoan equivalents of these proteins are named synaptonemal complex proteins (SYCP) and synaptonemal complex central element (SYCE) proteins. Genes encoding the SC and ZMM proteins are meiosis-specific and have been used as indicators of sex in diverse eukaryotes2.
Some genes, although not strictly in meiosis-specific pathways, are also relevant to meiosis. For example, postmeiotic segregation increased homologs 1 and 2 (PMS1 and PMS2) are part of the DNA mismatch repair system during both mitosis and meiosis. They compete to heterodimerise with MutL homolog 1 (MLH1), which is then assembled into the MutL-MutS heteroduplex that aids in degradation of DNA strands. Genes involved in syngamy (i.e., gamete fusion during fertilisation) are also relevant to sexual reproduction. Cell membranes fuse first (plasmogamy), followed by nuclear fusion (karyogamy); genes associated with these processes include HAP2 (haploid-disrupting 2) and gamete expressed protein 1 (GEX1). Thought to be an ancestral gene in all eukaryotes, HAP2 encodes the transmembrane protein Hap2-GCS1 that inserts into the target membrane using a hydrophobic fusion loop18. GEX1 gene products are nuclear envelope proteins involved in karyogamy19.
To attain a comprehensive overview of sex and reproduction in Symbiodiniaceae, here we used available genome and transcriptome data from broadly sampled taxa to investigate the presence of genes that encode functions critical to different stages of meiosis and syngamy, and their associated protein complexes. We also investigated codon usage in nuclear genes from each isolate, and compared the predicted functions between genes with strong codon usage preference and those under neutral selection.
Results
Canonical synaptonemal complex is absent in Symbiodiniaceae
The datasets of 21 isolates of Symbiodiniaceae used in this study are shown in Table 1. We found that meiosis-specific genes involved in the formation of the synaptonemal complex (SC) were largely missing from these microalgae. We did not recover genes encoding Hop1, Red1, Pch2, Zip1, Zip2, Zip3, and Zip4 (Fig. 1). However, we identified some of the genes that encode the ZMM proteins: the DNA helicase Mer3 and the Holliday junction heterodimer Msh4-Msh5. We also found most of the representative genes involved in homologous recombination, i.e., HOP2, MND1, DMC1, RAD51A and ATR. This result suggests that Symbiodiniaceae is capable of producing genetically diverse gametes in the absence of a canonical SC. This pattern of non-SC mediated meiotic crossovers may also hold for other dinoflagellates; Gymnodinium pseudopalustre Schiller, Amphidinium cryophilum, Alexandrium tamarense (basionym Gonyaulax tamarensis), Alexandrium minutum, and several species of Tovelliaceae (basionym Woloszynska) have all been observed to lack obvious SC or SC-like structures3,20,21,22. The lack of SC proteins in dinoflagellates was previously described23, but the studied taxa remain unspecified, and the genetic resources from dinoflagellates were very limited at that time, with no genome-scale data available. Table 2 shows the loss of canonical SCs reported in eukaryote lineages.
Sex-associated gene inventory of the 21 Symbiodiniaceae isolates analysed in this study, using Polarella glacialis isolates as outgroups. For Cladocopium goreaui MI SCF055, Polarella glacialis CCMP1383, and Polarella glacialis CCMP2088, both transcriptomes and predicted proteins were searched, with the latter versions marked with asterisks. Symbiodinium tridacnidorumH denotes proteins predicted from the hybrid genome assembly; Symbiodinium tridacnidorum denotes proteins predicted from the short-read only assembly. BUSCO completion includes both complete and fragmented orthologs. The tree topology shown on the left is based on LaJeunesse et al.69; branch lengths are not to scale. Distinct processes or protein complexes attributed to meiosis and fertilisation are shown at the top, with pie charts beneath showing genes associated with each process/complex. Genes in boldface are meiosis-specific. Genes under “double-stranded breaks” refer to those involved in inducing double-stranded DNA breaks while those under “homologous recombination” are involved in repairing said breaks. Presence of a gene is represented by coloured sections of a pie chart, with different colours assigned to different genera (e.g., Cladocopium goreaui has MSH5 and MER3 from the ZMM proteins). Absence of the genes HOP1, RED1, and PCH2, which are essential in the formation of the SC, is shown against a grey background.
Genes playing major roles in meiosis and syngamy are present
We identified the gamete fusogen gene, HAP2, in the transcriptomes of Cladocopium sp. SM (also known as isolate WSY) and Cladocopium sp. C1 MMETSP1367, and in the genomes of Symbiodinium natans and Symbiodinium tridacnidorum (both high-quality hybrid-reads assemblies24). This suggests that some of the cells observed to be attached together, i.e., “large tetrads” described in early microscopic observations may indeed be fertilisation. It is unsurprising that HAP2 occurs in Symbiodiniaceae, as this gene has been observed in other taxa in the Alveolata (to which dinoflagellates also belong): the ciliate Tetrahymena thermophila (UniProtKB: HAP2_TETTH, “evidence at protein level”) and in the apicomplexan Plasmodium berghei (UniProtKB: HAP2_PLABA, “evidence at transcript level”). We also recovered HAP2 candidates in other dinoflagellates, namely Polarella glacialis CCMP1383, Prorocentrum minutum, Gymnodinium catenatum, Noctiluca scintillans, and Oxyrrhis marina; its presence in the latter two species has been independently confirmed by Hofstatter and Lahr25. See Supplementary Fig. S1 online for a phylogeny showing the clustering of alveolate Hap2 sequences. Although we did not find clear evidence of HAP2 among the predicted gene models from all six available symbiodiniacean genomes26, we recovered fragments of this gene in the transcriptomes of S. natans and S. tridacnidorum. The presence of HAP2 remains to be more thoroughly investigated as more high-quality genomes become available to guide gene prediction methods.
We did not recover the karyogamy gene GEX1 in all the isolates studied here, but its absence does not necessarily mean Symbiodiniaceae do not undergo nuclear fusion. The malaria-causing parasite Plasmodium falciparum, the ciliate Tetrahymena thermophila, the diatom Thalassiosira pseudonana, and the blight-causing oomycete Phytophthora infestans also appear to be missing GEX1, but gamete fusion is well-established in these cases.
Symbiodiniaceae appear to have a reduced set of cohesin complex genes. We found only genes encoding the main component of the cohesin complex, the heterodimer Smc1-Smc3, which forms a proteinaceous ring around sister chromosomes or sister chromatids. We did not find REC8, SMC5, or SMC6. Because REC8 plays a role in anaphase I (separating homologous chromosomes) and anaphase II (separating sister chromatids), this raises the possibility that Symbiodiniaceae may not go through the same mechanism of chromosome/chromatid separation in canonical meiosis. In canonical meiosis, the Smc5-Smc6 heterodimer (encoded by SMC5 and SMC6) recruits Smc1-Smc3 to double-strand DNA breaks. It appears that in Symbiodiniaceae, Smc5-Smc6 has been replaced by another protein complex, or the recruitment process does not occur.
We found candidate genes for the mismatch repair proteins Pms1 and Pms2 in Symbiodiniaceae; these sequences were clustered as a single family (see Supplementary Fig. S2 online). In comparison, the opisthokont counterparts are sufficiently distinct as separate subfamilies (see PANTHER family tree PTHR10073). Searching against the KEGG ortholog HMM database (KOfam), these candidate proteins in Symbiodiniaceae shared higher sequence similarity to “DNA mismatch repair protein PMS2” (KOfam ID: K10858) than to “DNA mismatch repair protein PMS1” (KOfam ID: K10864); we thus annotated them as Pms2 here. Since Pms1 and Pms2 play the same role in DNA mismatch repair, it is unlikely that the absence of either one dramatically affects meiosis in Symbiodiniaceae.
Other genes that we did not recover in Symbiodiniaceae or P. glacialis are DNA2 and REC114 from the set of genes that induce double-strand DNA breaks, SLX4 from the crossover I pathway, MMS4 (also known as EME1) from the crossover II pathway, and MSH3 from the set of mismatch-correction genes. These genes are not meiosis-specific, and not critical to their implicated processes. As is the case for PMS1 and PMS2, we do not expect their absence to impact meiosis in Symbiodiniaceae.
Our observations of meiosis-specific and meiosis-related genes for B. minutum and S. microadriaticum are broadly consistent with earlier results by Chi et al.9, except for the absence of two genes in this study: HOP2 in B. minutum and PMS1 in S. microadriaticum based on the revised gene models. The HOP2-encoding sequence identified in B. minutum by Chi et al.9 failed to align to any putative protein homologs from SwissProt. The PMS1-encoding sequence identified in S. microadriaticum by Chi et al.9 aligned poorly to homologs from model organisms (mouse, human, Dictyostelium discoideum, Arabidopsis thaliana, yeasts S. cerevisiae and S. pombe) with a mismatch of 67% over 146 parsimony-informative sites. These results suggest that the previously identified genes are highly fragmented, or false positives. Liu et al.10 searched for meiosis-associated genes in C. goreaui and F. kawagutii, and found many more candidates than Chi et al.9. These likely include false positives, especially among genes that share high sequence identity such as the different MSHs and SMCs; for instance, a candidate protein for SLX1 may have been misannotated as SLX4. To make a stronger case for gene presence, we constructed single-gene phylogeny trees as described in Chi et al.9. Genes reported by Liu et al.10 to be present in S. microadriaticum, B. minutum, C. goreaui, and F. kawagutii but completely absent here based on revised gene models from Chen et al.26 are: HOP1, ZIP1, REC8, SMC5, SMC6, RAD17, SLX4, MMS4, and MSH3.
We only identified nine sex-associated genes in the F. kawagutii predicted gene set, but Morse27 found nine additional genes (MND1, MLH1, MSH2, MSH3, MSH4, MSH5, RAD51A, SGS1, and SMC3) using one-way BLAST searches from the F. kawagutii transcriptome data. Our putative candidates for the additional nine that Morse27 found did not fit our criteria (i.e., BLASTP hits with e-value < 10−3 and mutual coverage \(\ge \)25% against query sequences) and were thus considered absent.
For isolates for which genome data are not available (Table 1), absence of genes (based on searches using only transcriptome data) may represent false negatives. For instance, expression level of these genes may have been too low or likely not expressed under the conditions for which the transcript data were generated. This is likely the case for Cladocopium sp. Md and Cladocopium sp. C3k, in which completeness of transcriptome data based on Benchmarking Universal Single-Copy Orthologs (BUSCO) was 42.1% and 22.2% respectively (Fig. 1).
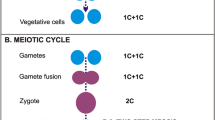
We present the hypothetical sexual stages of the life cycle in Symbiodiniaceae in Fig. 2. During plasmogamy (Fig. 2a), Hap2 allows the “minus” mating type gamete to insert a loop into an opposing gamete28. The mechanism for karyogamy remains unclear because we did not recover GEX1. Meiosis begins in a diploid cell (Fig. 2b), following which Spo11 makes double-strand breaks in DNA (Fig. 2c). In canonical meiosis, the SC forms during synapsis and then degrades at the end of prophase I; this does not occur in Symbiodiniaceae (Fig. 2d,e,f). A pair of MRN complexes, composed of Rad50, Mre11, and Atm, tethers the broken ends of DNA strands together29. The Smc1-Smc3 heterodimer keeps sister chromosomes together, and the Hop2-Mnd1 complex then binds to DNA strands and searches for homologous chromosomes (Fig. 2d)30. Rad51 and Dmc1 assemble on double-strand breaks; Hop2-Mnd1 interacts with Rad51 and Dmc1, allowing single-strand invasion31. Crossover II then occurs: double Holliday junctions are resolved with the help of endonucleases Mus81 and Slx1, and helicases Sgs1 and Mer3 (Fig. 2e). The Msh4-Msh5 heterodimer, along with Mlh1 and Mlh3, keeps homologous chromosomes together. Finally, the Msh2-Msh6 complex recognises base-base mismatches in DNA (Fig. 2f), activated by Pms2 and Mlh1. Exo1 then excises the incorrect bases, and the DNA strands are further repaired32. At the end of prophase I, we presume the rest of meiosis proceeds similarly to other dinoflagellates (Fig. 2g); chromosomes stay condensed and are segregated via spindles that form outside the nucleus33.
Hypothetical sexual stages of the life cycle in Symbiodiniaceae (ploidy n), showing all the relevant meiotic toolkit proteins. The missing complexes and processes, inferred from absence of genes, are shown against a yellow backdrop. The stages shown are (a) plasmogamy, (b) completion of gamete fusion, (c) formation of DNA double-strand break, (d) pairing of homologous chromosomes, (e) crossover II, (f) base-base mismatch correction, and (g) the end of meiosis, in which “tetrad” of daughter cells divides into four gametes. Stages (c) through (f) occur during prophase I of meiosis. The canonical synaptonemal complex forms around stage (d) until the end of prophase I. The “n” represents the haploid set of chromosomes. See text for detail.
Are sex-associated genes under selection pressure?
We found that most sex-associated genes in Symbiodiniaceae have no codon usage preference, but trend towards “neutrality” (Table 3; see Supplementary Fig. S3, Supplementary Fig. S4 and Supplementary Fig. S5 online). This is similar to the meiotic genes in the yeast Schizosaccharomyces pombe, which showed an unbiased codon usage, in contrast to the highly biased ribosomal proteins87. From a total of 287 sex-associated genes found in this study, only seven (six MER3s and one MND1) show evidence of putative biased codon usage (i.e., distance of >25 units away from the diagonal line of neutrality plots in Supplementary Fig. S4 online). In addition, MER3 in Symbiodiniaceae is highly diverged when compared to model organisms (e.g., see branch lengths in Supplementary Fig. S6 online), suggesting non-neutral evolution.
To determine which gene functions were undergoing selection, we annotated all the predicted gene models using Gene Ontology (GO) terms (Table 3). Genes with a strong codon preference were significantly enriched for the reductive pentose-phosphate cycle (GO:0019253), i.e., photosynthesis via C3 carbon fixation. Genes under neutral selection were enriched in several RNA-associated processes: RNA-dependent DNA biosynthetic process (GO:0006278), RNA-mediated transposition (GO:0032197), and RNA phosphodiester bond hydrolysis (GO:0090501 and GO:0090502). If taken together with another enriched process DNA integration (GO: 0015074), genes under neutral selection appear to be largely involved in the propagation of transposable elements. See Supplementary Table S1 online for the top five enriched GO terms for each isolate.
Most isolates, including P. glacialis showed a similar trend of codon usage: overall GC-content of 50–58% with several hundred coding sequences (CDSs) exhibiting strong codon usage bias; see Supplementary Fig. S3, Supplementary Fig. S4 and Supplementary Fig. S5 online for codon usage trends for each isolate. Assuming that Symbiodiniaceae have a large effective population and that the effect of GC-biased gene conversion is small, most CDSs appear to be under non-neutral selection. The slight variation in codon usage observed for each isolate is attributed to the synonymous third codon position. For this analysis, we focused on Cladocopium goreaui as a representative of all isolates. Figure 3a shows the effective number of codons of CDSs in C. goreaui versus the GC-content of the synonymous third codon position (GC3s). The slope for the trend line for C. goreaui is 0.13 (i.e. <1.0), indicating that most of these CDSs may have been under selection for elevated GC-content in third codon positions. As shown in Fig. 3a, 790 CDSs show a distance of >25 units below the expected curve, indicating strong codon-usage preference; 74% of all CDSs in C. goreaui have >50% GC-content in all three codon positions (Fig. 3b). Figure 3c shows the multi-variate correspondence plot of relative synonymous codon usage, with each of the two axes representing the relative inertia that explains variation of the observed codon usage. Axis 1 explains 19.6% of variation in codon usage of all C. goreaui genes based on differences in GC3s.
Codon usage trends of Cladocopium goreaui. Coding sequences (CDSs) were plotted according to their properties. (a) Effective number of codons (in y-axis) plotted against GC-content of synonymous third codon position (GC3s; in x-axis), in which the black curve represents the expected number of codons used under neutral selection. We consider CDSs at a distance >25 units below the expected curve to have a strong codon-usage preference. (b) Plot of neutrality, in which the diagonal line y = x represents neutrality. Genes closer to this line are under neutral selection. The trend line is shown in red; the percentage refers to the proportion of CDSs in each quadrant of GC-content. (c) Multi-variate correspondence analysis of relative synonymous codon usage, in which the x and y axes represent relative inertia. CDSs with GC3s-content below 50% and above 80% are highlighted in darker shades of blue.
Discussion
Our results, based on the analysis of genome-scale data from a broadly sampled set of taxa, provide strong evidence for the lack of canonical SCs in Symbiodiniaceae. The lack of SCs has also been reported in other dinoflagellates. In species that have well-characterised sexual life cycles, such as in Gymnodinium pseudopalustre, chromosomes were “paired at a distance”20, suggesting that SCs are not needed for synapsis. One exception is the stretched-out chromosomes observed to form axial-loop structures in Prorocentrum micans during meiosis34, but they lacked the characteristic ladder-like organisation. These structures may represent dinoflagellate-specific synaptonemal complexes, although further confirmation is needed (Marie-Odile Soyer-Gobillard, personal communication, 26 August 2019). Such cytological observations do not exist for Symbiodiniaceae. Ciliates, which are basal alveolates, lack SCs in some lineages, while retaining “residual SC structures” in others (Table 2). Apicomplexans including Plasmodium falciparum are basal to dinoflagellates, but P. falciparum has canonical SCs and a near-complete meiotic gene inventory35. Therefore, our results suggest an independent loss of canonical SCs in dinoflagellates.
In canonical meiosis, SCs mediate class I crossovers. This pathway, favoured in Arabidopsis and mammals, involves ZMM proteins and MutL homologs that ensure crossovers happen at a distance from one another on a chromosome, resulting in a phenomenon known as interference. The tension produced from these distant points of crossovers allow the paired chromosomes to separate properly into daughter cells36. In contrast, the class II pathway recruits the Mus81-Mms4 complex, among others, to repair double-strand breaks at random positions37. Class II crossovers can also occur on SCs, usually near pericentric heterochromatin38. The highly repetitive sequences near the centromere are known to produce complex recombination intermediates that are thought to be better resolved by the class II pathway39.
In the SC-lacking S. pombe and ciliates, crossovers occur predominantly via the class II pathway40,41. We recovered MUS81 and SGS1 in Symbiodiniaceae, which encode the crossover junction endonuclease Mus81 and the DNA helicase Sgs1, respectively. These two proteins are essential for meiotic crossover in the SC-lacking ciliate Tetrahymena thermophila41. Therefore, we propose that Symbiodiniaceae also undergo class II crossovers in lieu of SC-mediated crossovers. To verify this class II crossover bias, we suggest comparing frequencies of different classes via immunostaining39.
The loss of SCs in dinoflagellates may be explained by two conditions. First, repeat sequences make up a substantial proportion (20–40%) of Symbiodiniaceae genomes24,42, and an even higher proportion (68%) in the genome of Polarella glacialis43, a closely related, earlier-diverging sister lineage. Second, dinoflagellates including Symbiodiniaceae have permanently condensed chromosomes. Although genome data from other more-anciently diverged dinoflagellates remain lacking, these conditions represent key idiosyncratic genome features in dinoflagellates44. We speculate that the tension provided by class I crossovers is not necessary for chromosome separation and therefore, SCs became expendable. We hypothesise that this loss started with the loss of function of ZIP1 which encodes the transversal filaments (the axial elements are encoded by HOP1 and RED1, which are still present in SC-lacking S. pombe45). Our hypotheses cannot explain the loss of SCs seen in some ciliates however, because they possess canonical chromosome architecture.
Talbert and Henikoff46 theorise that cell size increase in dinoflagellates during the Eocene due to warming temperatures led to genome size expansion. Together with the gain of dinoflagellate viral nucleoproteins, they propose that novel ways of packaging chromosomes arose in dinoflagellates. Extending this study to more basal dinoflagellates and alveolates will help reveal if this loss coincides with the appearance of permanently condensed chromosomes and an increase in repetitive elements47.
Diatoms also lack genes encoding canonical SCs; SC-like structures observed in some species suggest that other unidentified proteins may have replaced the SC48. Both cytological and in silico analysis demonstrated the lack of SCs in the fungus Ustilago maydis49,50. Similarly, SCs are completely absent in the fission yeast S. pombe; they instead produce single-lined linear elements. On the other hand, spermatogenesis in male fruit flies involves interlocked homologous chromosomes called bivalents that are physically sequestered into pockets of the prophase nucleus, leading to achiasmate meiosis51. Without cytological evidence, we cannot rule out the possibility that there may be such unidentified SC substitutes in Symbiodiniaceae. We also cannot rule out a parasexual process such as that in the fungus Candida albicans, in which a fusion of haploids undergoes recombination and chromosome loss until it is haploid again52. Many of the genes in the meiotic toolkit also play alternative roles in non-meiotic processes; for instance, MND1 that repairs double-strand breaks in meiosis is also involved in telomere maintenance53. Therefore, the presence of the toolkit genes alone cannot prove an organism is sexual.
Identification of the HAP2 gene using our approach suggests the existence of Symbiodiniaceae gametes. Questions about other characteristics, however, remain: are these gametes isogamous mastigotes as previous research suggests, and what are their mating types? Immunostaining of the Hap2 protein at different life cycle stages may prove useful in answering these questions.
The codon usage trends seen here corroborate results from earlier studies54,55, and agree with general codon usage in dinoflagellates. The Symbiodiniaceae and dinoflagellates studied thus far have an overall GC-content >50% in their coding regions, with the third codon GC-content varying more than the first or second, and the variation of codon bias within each species or isolate being correlated with the GC-content of the synonymous third codon position (GC3s)56. Hypotheses that may explain these findings are: (a) Symbiodiniaceae underwent selection pressure whereby high GC3s was favoured, or (b) as one would expect in mammals, many genes underwent concerted evolution which led to GC-biased gene conversion57. In any case, meiotic recombination likely drives these codon usage trends14, supporting the hypothesis that Symbiodiniaceae are sexual. Our results also suggest that these species undergo non-canonical meiosis using the class II crossover pathway that bypasses SC formation.
Methods
Dataset
We collected transcriptomes used in González-Pech et al.55. Original sources are listed in Table 1. Incomplete ORFs were removed from transcriptomes, then translated using EMBOSS transeq58. Proteins encoded by plastid and mitochondrial genes were removed using BLASTP59 searches against the RefSeq database (mitochondrion and plastid; release 75) following González-Pech et al.55. For the six species Symbiodinium microadriaticum, Symbiodinium tridacnidorum, Breviolum minutum, Fugacium kawagutii, Cladocopium sp. C92, and Cladocopium goreui, we used predicted gene models from Chen et al.26. To assess the completeness of each dataset, we searched for the alveolata_odb10 BUSCO60 genes using BLASTP (e-value < 10–5).
Searching for homologous sequences
The relevant protein sequences available in the SwissProt database (release 2019_05) were used as queries to search against our dataset for putative homologs. Accession numbers of queries are listed in Supplementary Table S2 online. For genes that were very divergent, Symbiodinium microadriaticum61 and apicomplexan sequences from TrEMBL were used as queries. We chose a total of 42 genes, a combination of genes analysed in Chi et al.9 and Hofstatter et al.2. Genes that were peripheral players (e.g., RAD52, which mediates RAD51 in homologous recombinational repair), or genes that did not have close homologs outside of metazoans (e.g., BRCA1, BRCA2) were excluded from our analysis. Hits with e-value < 10−3 and mutual coverage \(\ge \)25% were selected as candidates.
Validation of candidate sequences
Identified homologs were further verified using a phylogenetic approach. Here, candidate sequences from each gene were first aligned to the query sequences, other SwissProt homologs, and homologs from closely-related taxa using MAFFT v762 with the --auto setting. Alignments were trimmed using either trimAL v1.263 with the -automated1 algorithm or BMGE v1.1264 with gap cut-off -g 0.4. Single-gene phylogenetic trees were constructed using IQ-TREE ModelFinder Plus using ultrafast bootstrap of 1000 replicates65,66,67. Putative homologs were verified if (a) they branched together with other dinoflagellates or alveolates, and (b) had branch lengths similar to those of queries. These sequences were then annotated by KofamScan68 using the KOfam eukaryotic database. The KOfam annotation of these genes are listed in Supplementary Table S3 online. The predicted gene models and protein sequences are available at https://doi.org/10.14264/uql.2020.483. Many Mnd1 candidates from the BLASTP step did not have corresponding KOfam hits to Mnd1, but this may be because Symbiodiniaceae sequences are very divergent.
Codon usage trends
The effective number of codons, GC3s, and GC-content of each codon position for each CDS in the dataset were obtained using CodonW v1.3 (http://codonw.sourceforge.net/culong.html) and EMBOSS cusp58. Relative synonymous codon usage was used for correspondence analysis. Plots were generated using the R package ggpubr version 0.2.2 (https://cran.r-project.org/package=ggpubr). For transcriptome-alone isolates, we assumed 1 transcript = 1 CDS, as we did not have information on gene splicing from these data.
Gene Ontology (GO) enrichment analysis
We performed GO enrichment analysis independently for each of the seven species: S. microadriaticum, S. tridacnidorum, S. natans, B. minutum, F. kawagutii, C. goreaui, and Cladocopium sp. C92 (Supplementary Table S1 online). For each species, we first used the predicted proteins as query and searched against the SwissProt database (release 2019_11; BLASTP, E ≤ 10−5) for putative homologs. Each protein sequence was then annotated based on the top SwissProt hit, and the associated GO terms to this hit were recovered using two in-house Python scripts (https://github.com/TimothyStephens/Annotate_GOterms_from_BLAST), following UniProt-GOA mapping (release 2019_05). Using the R package topGO version 2.36.0 (https://bioconductor.org/packages/topGO/), the enrichment analysis for GO terms associated with Biological Process was conducted using Fisher’s exact test. Independently for each species, a comparison was conducted for (a) proteins coded by genes exhibiting strong codon preference, and for (b) proteins coded by genes under neutral selection, each against all predicted proteins in the corresponding genome as background. Using combined, predicted proteins from all seven species, a comparison was also conducted independently for (a) proteins coded by genes exhibiting a strong codon preference, and for (b) proteins coded by genes under neutral selection, each against all predicted proteins from the seven genomes as background; these results are shown in Table 3. In total, our approach yielded 126,979 GO-annotated proteins, 2,400 proteins coded by genes exhibiting strong codon preference, and 47,422 proteins coded by genes under neutral selection (i.e., at a distance <5% from the diagonal line of y = x in Fig. 3b).
Data availability
All gene models and protein sequences used in this study are available at https://doi.org/10.14264/uql.2020.483, except for the predicted protein sequences of Polarella glacialis and Symbiodinium tridacnidorum (hybrid genome assemblies) that were obtained from Stephens et al.43 and González-Pech et al.24 respectively.
References
Speijer, D., Lukeš, J. & Eliáš, M. Sex is a ubiquitous, ancient, and inherent attribute of eukaryotic life. Proc. Natl. Acad. Sci. USA 112, 8827–8834 (2015).
Hofstatter, P. G., Brown, M. W. & Lahr, D. J. G. Comparative genomics supports sex and meiosis in diverse amoebozoa. Genome Biol. Evol. 10, 3118–3128 (2018).
Figueroa, R. I., Dapena, C., Bravo, I. & Cuadrado, A. The hidden sexuality of Alexandrium minutum: An example of overlooked sex in dinoflagellates. PLoS One 10, e0142667 (2015).
Price, D. C. & Bhattacharya, D. Robust Dinoflagellata phylogeny inferred from public transcriptome databases. J. Phycol. 53, 725–729 (2017).
Freudenthal, H. D. Symbiodinium gen. nov. and Symbiodinium microadriaticum sp. nov., a zooxanthella: taxonomy, life cycle, and morphology. J. Protozool. 9, 45–52 (1962).
Hughes, T. P. et al. Global warming transforms coral reef assemblages. Nature 556, 492–496 (2018).
LaJeunesse, T. C. Investigating the biodiversity, ecology, and phylogeny of endosymbiotic dinoflagellates in the genus Symbiodinium using the ITS region: In search of a “species” level marker. J. Phycol. 37, 866–880 (2001).
Brian, J. I., Davy, S. K. & Wilkinson, S. P. Multi-gene incongruence consistent with hybridisation in Cladocopium (Symbiodiniaceae), an ecologically important genus of coral reef symbionts. PeerJ 7, e7178 (2019).
Chi, J., Parrow, M. W. & Dunthorn, M. Cryptic sex in Symbiodinium (Alveolata, Dinoflagellata) is supported by an inventory of meiotic genes. J. Eukaryot. Microbiol. 61, 322–327 (2014).
Liu, H. et al. Symbiodinium genomes reveal adaptive evolution of functions related to coral-dinoflagellate symbiosis. Commun. Biol. 1, 95 (2018).
Fitt, W. K. & Trench, R. K. The relation of diel patterns of cell division to diel patterns of motility in the symbiotic dinoflagellate Symbiodinium microadriaticum Freudenthal in culture. New Phytol. 94, 421–432 (1983).
Hadany, L. & Comeron, J. M. Why are sex and recombination so common? Ann. N. Y. Acad. Sci. 1133, 26–43 (2008).
Pouyet, F., Mouchiroud, D., Duret, L. & Sémon, M. Recombination, meiotic expression and human codon usage. eLife 6, e27344 (2017).
Webster, M. T. & Hurst, L. D. Direct and indirect consequences of meiotic recombination: implications for genome evolution. Trends Genet. 28, 101–109 (2012).
Loidl, J. Conservation and variability of meiosis across the eukaryotes. Annu. Rev. Genet. 50, 293–316 (2016).
San-Segundo, P. A. & Roeder, G. S. Pch2 links chromatin silencing to meiotic checkpoint control. Cell 97, 313–324 (1999).
Lynn, A., Soucek, R. & Börner, G. V. ZMM proteins during meiosis: crossover artists at work. Chromosome Res. 15, 591–605 (2007).
Fédry, J. et al. The ancient gamete fusogen Hap2 is a eukaryotic class II fusion protein. Cell 168, 904–915 (2017).
Ning, J. et al. Comparative genomics in Chlamydomonas and Plasmodium identifies an ancient nuclear envelope protein family essential for sexual reproduction in protists, fungi, plants, and vertebrates. Genes Dev. 27, 1198–1215 (2013).
von Stosch, H. A. Observations on vegetative reproduction and sexual life cycles of two freshwater dinoflagellates, Gymnodinium pseudopalustre Schiller and Woloszynskia apiculata sp. nov. Br. Phycol. J. 8, 105–134 (1973).
Wilcox, L. W., Wedemayer, G. J. & Graham, L. E. Amphidinium cryophilum sp. nov. (Dinophyceae) a new freshwater dinoflagellate. II. Ultrastructure. J. Phycol. 18, 18–30 (1982).
Lindberg, K., Moestrup, Ø. & Daugbjerg, N. Studies on woloszynskioid dinoflagellates I: Woloszynskia coronata re-examined using light and electron microscopy and partial LSU rDNA sequences, with description of Tovellia gen. nov. and Jadwigia gen. nov. (Tovelliaceae fam. nov.). Phycologia 44, 416–440 (2005).
Grishaeva, T. M. & Bogdanov, Y. F. Conservation and variability of synaptonemal complex proteins in phylogenesis of eukaryotes. Int. J. Evol. Biol., 856230 (2014).
González-Pech, R. A. et al. Structural rearrangements drive extensive genome divergence between symbiotic and free-living Symbiodinium. bioRxiv, 783902 (2019).
Hofstatter, P. G. & Lahr, D. J. G. All eukaryotes are sexual, unless proven otherwise. BioEssays 41, 1800246 (2019).
Chen, Y., González-Pech, R. A., Stephens, T. G., Bhattacharya, D. & Chan, C. X. Evidence that inconsistent gene prediction can mislead analysis of dinoflagellate genomes. J. Phycol. 56, 6–10 (2020).
Morse, D. A transcriptome-based perspective of meiosis in dinoflagellates. Protist 170, 397–403 (2019).
Baquero, E., Fedry, J., Legrand, P., Krey, T. & Rey, F. A. Species-specific functional regions of the green alga gamete fusion protein HAP2 revealed by structural studies. Structure 27, 113–124 (2019).
Alsbeih, G. MRE11A gene mutations responsible for the rare ataxia telangiectasia-like disorder in Human Genetic Diseases (ed. Plaseska-Karanfilska, D.) 79–90 (InTech Open, 2011).
Zhao, W. et al. Mechanistic insights into the role of Hop2-Mnd1 in meiotic homologous DNA pairing. Nucleic Acids Res. 42, 906–917 (2014).
Vignard, J. et al. The interplay of RecA-related proteins and the MND1-HOP2 complex during meiosis in Arabidopsis thaliana. PLoS Genet. 3, e176 (2007).
Wei, K., Kucherlapati, R. & Edelmann, W. Mouse models for human DNA mismatch-repair gene defects. Trends Mol. Med. 8, 346–353 (2002).
Fukuda, Y. & Suzaki, T. Unusual features of dinokaryon, the enigmatic nucleus of dinoflagellates in Marine Protists: Diversity and Dynamics (eds Ohtsuka, S. et al.) 23–45 (Springer Japan, 2015).
Soyer-Gobillard, M. O., Bhaud, Y. & Hilaire, D. New data on mating in an autotrophic dinoflagellate, Prorocentrum micans Ehrenberg. Vie Milieu 52, 167–175 (2002).
Lee, A. H., Symington, L. S. & Fidock, D. A. DNA repair mechanisms and their biological roles in the malaria parasite Plasmodium falciparum. Microbiol. Mol. Biol. Rev. 78, 469–486 (2014).
Nambiar, M., Chuang, Y. C. & Smith, G. R. Distributing meiotic crossovers for optimal fertility and evolution. DNA Repair 81, 102648 (2019).
Higgins, J. D., Buckling, E. F., Franklin, F. C. H. & Jones, G. H. Expression and functional analysis of AtMUS81 in Arabidopsis meiosis reveals a role in the second pathway of crossing-over. Plant J. 54, 152–162 (2008).
Demirci, S. et al. Distribution, position and genomic characteristics of crossovers in tomato recombinant inbred lines derived from an interspecific cross between Solanum lycopersicum and Solanum pimpinellifolium. Plant J. 89, 554–564 (2017).
Anderson, L. K. et al. Combined fluorescent and electron microscopic imaging unveils the specific properties of two classes of meiotic crossovers. Proc. Natl. Acad. Sci. USA 111, 13415–13420 (2014).
Chi, J., Mahé, F., Loidl, J., Logsdon, J. & Dunthorn, M. Meiosis gene inventory of four ciliates reveals the prevalence of a synaptonemal complex-independent crossover pathway. Mol. Biol. Evol. 31, 660–672 (2013).
Lukaszewicz, A., Howard-Till, R. A. & Loidl, J. Mus81 nuclease and Sgs1 helicase are essential for meiotic recombination in a protist lacking a synaptonemal complex. Nucleic Acids Res. 41, 9296–9309 (2013).
González-Pech, R. A. et al. Genomes of Symbiodiniaceae reveal extensive sequence divergence but conserved functions at family and genus levels. bioRxiv, 800482 (2019).
Stephens, T. G. et al. Genomes of the dinoflagellate Polarella glacialis encode tandemly repeated single-exon genes with adaptive functions. BMC Biology 18, 56 (2020).
Lin, S. Genomic understanding of dinoflagellates. Res. Microbiol. 162, 551–569 (2011).
Lorenz, A. S. pombe meiotic linear elements contain proteins related to synaptonemal complex components. J. Cell Sci. 117, 3343–3351 (2004).
Talbert, P. B. & Henikoff, S. Chromatin: packaging without nucleosomes. Curr. Biol. 22, R1040–R1043 (2012).
Gornik, S. G., Hu, I., Lassadi, I. & Waller, R. F. The biochemistry and evolution of the dinoflagellate nucleus. Microorganisms 7, 245 (2019).
Patil, S. et al. Identification of the meiotic toolkit in diatoms and exploration of meiosis-specific SPO11 and RAD51 homologs in the sexual species Pseudo-nitzschia multistriata and Seminavis robusta. BMC Genomics 16, 930 (2015).
Fletcher, H. L. A search for synaptonemal complexes in Ustilago maydis. J. Cell Sci. 50, 171–180 (1981).
Donaldson, M. E. & Saville, B. J. Bioinformatic identification of Ustilago maydis meiosis genes. Fungal Genet. Biol. 45, S47–S53 (2008).
Vazquez, J., Belmont, A. S. & Sedat, J. W. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12, 1473–1483 (2002).
Bennett, R. J. & Johnson, A. D. Completion of a parasexual cycle in Candida albicans by induced chromosome loss in tetraploid strains. EMBO J. 22, 2505–2515 (2003).
Maciver, S. K. Ancestral eukaryotes reproduced asexually, facilitated by polyploidy: a hypothesis. BioEssays 41, 1900152 (2019).
Bayer, T. et al. Symbiodinium transcriptomes: genome insights into the dinoflagellate symbionts of reef-building corals. PLoS One 7, e35269 (2012).
González-Pech, R. A., Ragan, M. A. & Chan, C. X. Signatures of adaptation and symbiosis in genomes and transcriptomes of Symbiodinium. Sci. Rep. 7, 15021 (2017).
Williams, E., Place, A. & Bachvaroff, T. Transcriptome analysis of core dinoflagellates reveals a universal bias towards “GC” rich codons. Mar. Drugs 15, 125 (2017).
Duret, L. & Galtier, N. Biased gene conversion and the evolution of mammalian genomic landscapes. Annu. Rev. Genomics Hum. Genet. 10, 285–311 (2009).
Rice, P., Longden, I. & Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 16, 276–277 (2000).
Altschul, S. F., Gish, W., Miller, W., Myers, E. W. & Lipman, D. J. Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990).
Simão, F. A., Waterhouse, R. M., Ioannidis, P., Kriventseva, E. V. & Zdobnov, E. M. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015).
Aranda, M. et al. Genomes of coral dinoflagellate symbionts highlight evolutionary adaptations conducive to a symbiotic lifestyle. Sci. Rep. 6, 39734 (2016).
Katoh, K. & Standley, D. M. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013).
Capella-Gutiérrez, S., Silla-Martínez, J. M. & Gabaldón, T. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25, 1972–1973 (2009).
Criscuolo, A. & Gribaldo, S. BMGE (Block Mapping and Gathering with Entropy): a new software for selection of phylogenetic informative regions from multiple sequence alignments. BMC Evol. Biol. 10, 210 (2010).
Hoang, D. T., Chernomor, O., von Haeseler, A., Minh, B. Q. & Vinh, L. S. UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522 (2018).
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K. F., von Haeseler, A. & Jermiin, L. S. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Meth. 14, 587–589 (2017).
Nguyen, L. T., Schmidt, H. A., von Haeseler, A. & Minh, B. Q. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015).
Aramaki, T. et al. KofamKOALA: KEGG ortholog assignment based on profile HMM and adaptive score threshold. Bioinformatics 36, 2251–2252 (2020).
LaJeunesse, T. C. et al. Systematic revision of Symbiodiniaceae highlights the antiquity and diversity of coral endosymbionts. Curr. Biol. 28, 2570–2580 (2018).
Davies, S. W., Marchetti, A., Ries, J. B. & Castillo, K. D. Thermal and pCO2 stress elicit divergent transcriptomic responses in a resilient coral. Front. Mar. Sci. 3, 112 (2016).
Ladner, J. T., Barshis, D. J. & Palumbi, S. R. Protein evolution in two co-occurring types of Symbiodinium: an exploration into the genetic basis of thermal tolerance in Symbiodinium clade D. BMC Evol. Biol. 12, 217 (2012).
González-Pech, R. A., Vargas, S., Francis, W. R. & Wörheide, G. Transcriptomic resilience of the Montipora digitata holobiont to low pH. Front. Mar. Sci. 4, 403 (2017).
Levin, R. A. et al. Sex, scavengers, and chaperones: transcriptome secrets of divergent Symbiodinium thermal tolerances. Mol. Biol. Evol. 33, 2201–2215 (2016).
Parkinson, J. E. et al. Gene expression variation resolves species and individual strains among coral-associated dinoflagellates within the genus Symbiodinium. Genome Biol. Evol. 8, 665–680 (2016).
Xiang, T., Nelson, W., Rodriguez, J., Tolleter, D. & Grossman, A. R. Symbiodinium transcriptome and global responses of cells to immediate changes in light intensity when grown under autotrophic or mixotrophic conditions. Plant J. 82, 67–80 (2015).
Shoguchi, E. et al. Draft assembly of the Symbiodinium minutum nuclear genome reveals dinoflagellate gene structure. Curr. Biol. 23, 1399–1408 (2013).
Shoguchi, E. et al. Two divergent Symbiodinium genomes reveal conservation of a gene cluster for sunscreen biosynthesis and recently lost genes. BMC Genomics 19, 458 (2018).
Loidl, J. & Scherthan, H. Organization and pairing of meiotic chromosomes in the ciliate Tetrahymena thermophila. J. Cell Sci. 117, 5791–5801 (2004).
Round, F. E., Crawford, R. M. & Mann, D. G. The Diatoms: Biology & Morphology of the Genera (Cambridge University Press, 1991).
Mann, D. G. & Stickle, A. J. Meiosis, nuclear cyclosis, and auxospore formation in Navicula sensu stricto (Bacillariophyta). Br. Phycol. J. 24, 167–181 (1989).
Manton, I., Kowallik, K. & von Stosch, H. A. Observations on the fine structure and development of the spindle at mitosis and meiosis in a marine centric diatom (Lithodesmium undulatum). II. The early meiotic stages in male gametogenesis. J. Cell Sci. 5, 271–298 (1969).
Holloman, W. K., Schirawski, J. & Holliday, R. The homologous recombination system of Ustilago maydis. Fungal Genet. Biol. 45, S31–S39 (2008).
Kohli, J. & Bähler, J. Homologous recombination in fission yeast: absence of crossover interference and synaptonemal complex. Experientia 50, 295–306 (1994).
Egel-Mitani, M., Olson, L. W. & Egel, R. Meiosis in Aspergillus nidulans: another example for lacking synaptonemal complexes in the absence of crossover interference. Hereditas 97, 179–187 (1982).
Rasmussen, S. W. Ultrastructural studies of spermatogenesis in Drosophila melanogaster meigen. Z. Zellforsch. Mikrosk. Anat. 140, 125–144 (1973).
Gilboa, L. & Lehmann, R. How different is Venus from Mars? The genetics of germ-line stem cells in Drosophila females and males. Development 131, 4895–4905 (2004).
Hiraoka, Y, Kawamata K., Haraguchi T. & Chikashige, Y. Codon usage bias is correlated with gene expression levels in the fission yeast Schizosaccharomyces pombe. Genes Cells 14, 499-509 (2009).
Acknowledgements
S.S. and Y.C. are supported by the University of Queensland Research Training Scholarship. This project was supported by the Australian Research Council grant DP190102474 awarded to C.X.C. and D.B., and the computational resources of the National Computational Infrastructure (NCI) National Facility systems through the NCI Merit Allocation Scheme (Project d85) awarded to C.X.C. D.B. was supported by a NIFA-USDA Hatch grant (NJ01170). We thank Timothy Stephens for providing analytic tools, and Raúl González-Pech for his insights into interpreting the results.
Author information
Authors and Affiliations
Contributions
S.S. and C.X.C. conceived the study; Y.C. provided in-house scripts and computational pipelines; S.S. and Y.C. conducted all computational analyses; S.S. prepared all figures, tables, and the first draft of the manuscript; S.S., C.X.C. and D.B. interpreted the results; all authors reviewed, commented on and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Shah, S., Chen, Y., Bhattacharya, D. et al. Sex in Symbiodiniaceae dinoflagellates: genomic evidence for independent loss of the canonical synaptonemal complex. Sci Rep 10, 9792 (2020). https://doi.org/10.1038/s41598-020-66429-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-66429-4
- Springer Nature Limited
This article is cited by
-
New insights on the role of nitrogen in the resistance to environmental stress in an endosymbiotic dinoflagellate
Environmental Science and Pollution Research (2023)
-
A practical reference for studying meiosis in the model ciliate Tetrahymena thermophila
Marine Life Science & Technology (2022)
-
Comparison of 15 dinoflagellate genomes reveals extensive sequence and structural divergence in family Symbiodiniaceae and genus Symbiodinium
BMC Biology (2021)
-
Direct evidence of sex and a hypothesis about meiosis in Symbiodiniaceae
Scientific Reports (2021)