Abstract
Porphyromonas gingivalis is a causative agent in the onset and progression of periodontal disease. This study aims to investigate the effects of quercetin, a natural plant product, on P. gingivalis virulence properties including gingipain, haemagglutinin and biofilm formation. Antimicrobial effects and morphological changes of quercetin on P. gingivalis were detected. The effects of quercetin on gingipains activities and hemolytic, hemagglutination activities were evaluated using chromogenic peptides and sheep erythrocytes. The biofilm biomass and metabolism with different concentrations of quercetin were assessed by the crystal violet and MTT assay. The structures and thickness of the biofilms were observed by confocal laser scanning microscopy. Bacterial cell surface properties including cell surface hydrophobicity and aggregation were also evaluated. The mRNA expression of virulence and iron/heme utilization was assessed using real time-PCR. Quercetin exhibited antimicrobial effects and damaged the cell structure. Quercetin can inhibit gingipains, hemolytic, hemagglutination activities and biofilm formation at sub-MIC concentrations. Molecular docking analysis further indicated that quercetin can interact with gingipains. The biofilm became sparser and thinner after quercetin treatment. Quercetin also modulate cell surface hydrophobicity and aggregation. Expression of the genes tested was down-regulated in the presence of quercetin. In conclusion, our study demonstrated that quercetin inhibited various virulence factors of P. gingivalis.
Similar content being viewed by others
Introduction
Periodontal disease is a common chronic inflammatory disease that characterized swelling and bleeding of the gums clinically, and leading to the progressive destruction of tooth-supporting tissues including the gingiva, alveolar bone, periodontal ligament, and cementum. Severe periodontitis can cause periodontal pockets formation, bone resorption, and eventually lead to tooth loss1. It has been estimated that nearly 50% of human population worldwide are affected by mild to moderate periodontitis and approximately 11% are severe forms2. In addition to the oral local sequelae, evidence has accumulated to suggest that periodontal disease is also related to a risk factor for systemic complications such as cardiovascular diseases, stroke, diabetes, arthritis, and preterm low birth weight for more than a decade3,4.
Two major etiological factors of periodontal disease are specific bacterial species called periodontopathogens colonizing subgingival sites, and subsequent host inflammatory and immune responses to these periodontopathogens5. Although over 700 microbial species have been identified in subgingival plaque, Porphyromonas gingivalis, a black-pigmented, rod-shaped, and Gram-negative anaerobic bacterium, is a keystone pathogen for periodontal disease6.
P. gingivalis possesses various potential virulence factors including gingipain proteases, haemagglutinin, fimbriae, capsule, lipopolysacharides and major outer-membrane proteins to evade the host immune defense system and destroy host connective tissues7,8. One major virulence factor is gingipain proteases consist of lysine-specific protease (Lys-gingipain (Kgp)) and arginine-specific protease (Arg-gingipain (Rgp)). Kgp is encoded by kgp gene and Rgp is further subdivided into RgpA and RgpB, encoded by rgpA and rgpB genes, respectively9,10. Hemagglutinin allowed P. gingivalis to adhere to host cells, which is an initial step in bacterial infection process. It facilitates the acquisition of heme through erythrocyte binding, causes lysis and aggregation of erythrocytes11. Besides, P. gingivalis is a late colonizer of subgingival plaque biofilm, and adheres to plaque-colonizing organisms including Streptococcus gordonii and Fusobacterium nucleatum12. A biofilm is a community of bacteria that attached to the surface and in a self-produced extracellular polymeric substances matrix consisting of protein, polysaccharides and nucleic acid13.
Therefore, the inhibition of P. gingivalis virulent effects may impede progression of periodontitis, prevent and control periodontal diseases. In the last decade, natural polyphenols have been studied since they have various biological functions such as antimicrobial, anti-inflammation, anti-oxidation, and anti-cancer. The largest class of polyphenols is flavonoids which have two aromatic rings linked with through three carbon atoms that form an oxygenated heterocycle. Quercetin (3,3′,4′,5,7-pentahydroxyflavone) is the most abundant flavonoid that exists in various vegetables and fruits including apples, tea, onions, red wine, red grapes, berries and tomatoes14,15. Quercetin has been reported to be effective against Gram-positive and Gram-negative bacteria, such as Staphylococcus aureus, Escherichia coli and Pseudomonas aeruginosa16,17,18. However, to our knowledge there are few studies focused on the influence of quercetin on P. gingivalis virulence factors. Therefore, in the present study, our aim was to determine the effect of quercetin on P. gingivalis virulent pathogenicity with respect to periodontal disease.
Results
Antimicrobial activity of quercetin against P. gingivalis planktonic
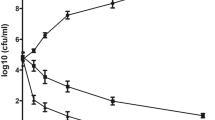
The antibacterial activity of quercetin against P. gingivalis was determined with a broth microdilution assay. Results of the susceptibility assay of P. gingivalis planktonic cultures to quercetin are shown in Fig. 1a,b. The MIC and the MBC values of quercetin were 200 and 400 μM, respectively.
Antibacterial activity and morphological changes of P. gingivalis. (a) MIC and MBC values of quercetin against planktonic P. gingivalis. 107 CFU/mL P. gingivalis suspension was added in a flat-bottomed 96-well microplate at final concentrations of 0–1600 μM quercetin and incubated under anaerobic conditions at 37 °C. (b) The number of colonies on blood agar. An aliquot of 10 μL cell suspension from each well was taken from above 96-well microplate, and bacterial clones were counted on the blood agar plates after incubation for 3–5 days. (c–f) TEM images of P. gingivalis treated with different concentration of quercetin. P. gingivalis cells with different concentrations of quercetin cultured at 37 °C for 4 h. The cell pellets were fixed with 2.5% glutaraldehyde at 4 °C, exposed to 2% osmium tetraoxide for 2 h, dehydrated in a series of ethanol and dried in acetone. Then the samples were embedded in resin blocks, cut into ultrathin sections, and stained with uranyl acetate and lead citrate. (c) 0 μM, (d) 100 μM, (e) 200 μM, (f) 400 μM, Bar = 200 nm.
Observation of morphological changes
The morphological changes of P. gingivalis treated with different concentrations of quercetin was observed by TEM. The cell membranes remained clearly intact in the control group (Fig. 1c). The structures of bacteria cells differ with final concentrations of 100, 200, and 400 μM quercetin (Fig. 1d–f). We saw considerably damage and discontinuity of cell membrane and the cell structures were damaged with increasing severity as concentration of quercetin increased. Thus, quercetin caused integrity loss of the cell membrane.
Effect of quercetin on gingipain activities
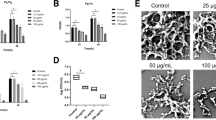
The activities of Rgp and Kgp influenced by quercetin at sub-MIC concentrations were measured. Quercetin inhibited Rgp and Kgp activities in dose-dependent manners significantly. The inhibitory effects within 1 h are also shown in Fig. 2a,c. The inhibition rates on Kgp activities decreased from 90.33% at 12.5 μM to 43.97% at 100 μM, as shown in Fig. 2b. The inhibition rates on Rgp activities decreased from 62.36% at 12.5 μM to 3.64% at 100 μM, as shown in Fig. 2d.
Effect of quercetin on Arg-gingipain (Rgp) and Lys-gingipain (Kgp) activity. P. gingivalis culture was harvested and suspended in PBS to OD660nm of 2 for Rgp activity and of 1 for Kgp activity. The P. gingivalis cells were incubated in PBS with or without quercetin at 37 °C in the dark for 3 h. Activity of Rgp (a,b) and Kgp (c,d) indicated as the hydrolysis of the specific chromogenic substrates (BAPNA and ALNA) was detected at every 2 min for 1 h by measuring the absorbance at 405 nm. The inhibition of substrate degradation was also measured as a function of time (b,d). Bars denoted by (****) indicate significant difference at p < 0.0001 by one-way analysis of variance (ANOVA) with Dunnett’s post hoc test.
Binding mode analysis
The substrate binding site of Kgp was reported to contain a catalytic triad including residues Cys477, His444 and Asp38819, which may form a charge-relay system for catalysis. Compared with Kgp, the catalytic triad of RgpB is Cys244, His211 and Glu15220. Quercetin was docked into active sites of Kgp and RgpB by using Glide21. The results showed that Quercetin could bind to the substrate binding site of Kgp and RgpB with favorable intermolecular interactions. In the predicted binding mode of quercetin to Kgp (Fig. 3a), the almost planar quercetin core accommodates the substrate binding site well by shape matching and forming two hydrogen bonds with residues Ala443O and Trp513N. Similarly, in the predicted binding mode of quercetin to RgpB (Fig. 3c), quercetin core also occupies the substrate binding site well and could form a hydrogen bond with residue Gly212N. When showing the surface of Kgp (Fig. 3b) and RgpB (Fig. 3d), it is observed that quercetin core could penetrate into the usually -Lys- and -Arg- fragments binding cavity of Kgp and RgpB respectively, which is beneficial for preventing the substrate from getting close and thus inhibiting the catalytic activity of the two enzymes.
Predicted binding modes of Quercetin in the binding pocket of Kgp (a,b, PDB ID: 6I9A) and RgpB (c,d, PDB ID: 1CVR). Quercetin is shown as pink sticks. Key residues around the binding pocket are displayed as sticks in (a) (Kgp, purple) and (c) (RgpB, blue). Hydrogen bonds are highlighted as red dashed lines, and water is depicted as small red balls. The surfaces of the substrate binding site in Kgp and RgpB are shown in (b) (purple) and (d) (blue), respectively.
Effect of quercetin on hemagglutination and hemolytic activity
Without adding quercetin, hemagglutination was observed up to a dilution of 1:16 for P. gingivalis. As shown in Fig. 4a, quercetin showed significantly reduced hemagglutination activity (fourfold for 100 μM and twofold for 50 μM, respectively). But quercetin at lower concentrations (12.5 and 25 μM) had a slight effect on hemagglutination.
Effect of quercetin on hemagglutination and hemolytic activity. (a) Overnight cultures of P. gingivalis with quercetin were harvested, and suspended in PBS. 100 μL of PBS was added to each well in a 96-well round-bottom microtiter plate, 100 μL of the bacterial cells was suspended in PBS, added to the first well and serially diluted (1:2 to 1:128). Sheep erythrocytes suspension (100 μL, 2% in PBS) and quercetin were added to each well, and the plate was incubated for 3 h. Hemagglutination activities were evaluated visually. (b) P. gingivalis cells were centrifuged, re-suspended to a final OD600nm of 1.5. The sheep erythrocytes at a concentration of 1% were mixed with an equal volume of bacterial cells with or without quercetin at 37 °C for 18 h. The hemolytic activity was determined at wavelength of 405 nm. Bars denoted by (****) indicate significant difference at p < 0.0001 by one-way analysis of variance (ANOVA) with Dunnett's post hoc test.
Because lysed erythrocytes by bacteria could release hemoglobin, the absorbance of 405 nm for red pigment was used to describe the hemolysis of erythrocytes. The effect of quercetin on the hemolysis of sheep erythrocytes by P. gingivalis was determined (Fig. 4b). The hemolytic activities of quercetin (12.5, 25, 50 and 100 μM) were 53.73 ± 4.67%, 49.54 ± 3.76%, 18.23 ± 1.94% and 13.45 ± 0.68%, respectively, compared to the control group. The results indicated that quercetin at sub-MIC concentrations had reduced capacities to lyse erythrocytes.
Effect of quercetin on biofilm formation
In addition to growth inhibition of P. gingivalis planktonic culture, quercetin also noticeably showed antimicrobial activity against P. gingivalis biofilms. The overall biomass of biofilms was quantified by the crystal violet assay with sub-MIC value of quercetin. As shown in Fig. 5a. P. gingivalis exhibited OD550nm values of 2.438 ± 0.241, whereas in the presence of quercetin, the values decreased from 1.817 ± 0.083 at 12.5 μM to 0.134 ± 0.022 at 100 μM.
Effect of quercetin on biofilm formation. (a) P. gingivalis suspension was grown in BHI supplemented with quercetin for 48 h at 37 °C in a 96-well polystyrene plates. After washing three times with sterile PBS, the adherent biofilms were incubated with methanol for 15 min followed by staining with 0.04% (w/v) crystal violet to determine overall biomass of the biofilm at wavelength of 550 nm. (b) The biofilm was formed as described for the above method. MTT (0.5 mg/mL) was added to detect the metabolic activity of the biofilms at wavelength of 590 nm. (c–g) CLSM images of P. gingivalis biofilm treated with quercetin. P. gingivalis biofilms were formed as described above assay for 48 h on glass coverslips with different concentrations of quercetin. Bacterial cells were washed three times with saline to remove unbound cells and stained with the LIVE/DEAD BacLigh Bacterial Viability Kit containing SYTO 9 dye and propidium iodide. (c) 0 μM, (d) 12.5 μM, (e) 25 μM, (f) 50 μM, (g) 100 μM, Bar = 50 μm. (h) Biofilm thickness. Bars denoted by (**), (***) and (****) indicate significant difference at p < 0.01, p < 0.001 and p < 0.0001, respectively by one-way analysis of variance (ANOVA) with Dunnett's post hoc test.
The metabolic activity of biofilms was quantified by the MTT assay with same concentrations of quercetin as above. The metabolic activity of biofilms exhibited OD590nm values with different concentrations of quercetin (12.5, 25, 50, and 100 μM) were 0.388 ± 0.015, 0.325 ± 0.036, 0.262 ± 0.015, and 0.055 ± 0.002, respectively, whereas in the absence of quercetin, the value was 0.456 ± 0.020. The results showed that quercetin decreased the biofilm metabolism compared to the control group and confirmed the crystal violet assay data (Fig. 5b).
Confocal laser scanning microscopy
To further determine the effect of quercetin on biofilm formation, the structures of the biofilms were observed by CLSM. Live bacteria showed fluorescent green, whereas dead bacteria were fluorescent red. In the absence of quercetin, the biofilm had a highly uniform distribution and dense layer structure (Fig. 5c). As quercetin concentration increased, the biofilm structures were highly dispersed and clearly sparse, and P. gingivalis cells tended to aggregate into distinct clusters easily discernable (Fig. 5d–g). In addition, the average thicknesses of biofilms were determined by CLSM (Fig. 5h). The thicknesses of quercetin-treated biofilms were 32.5 ± 0.1 μm at 12.5 μM, 25.6 ± 3.3 μm at 25 μM, 19.8 ± 1.3 μm at 50 μM and 13.3 ± 1.0 μm at 100 μM, respectively, which were thinner than the control group (44.2 ± 2.5 μm). These data confirmed that quercetin inhibited biofilm formation.
Effect of quercetin on hydrophobicity and aggregation
The hydrophobicity of the bacterial surfaces was determined by measuring the percentage of their adherence to hydrocarbons. The hydrophobicity assay revealed that quercetin increased P. gingivalis surface hydrophobicity in a dose-dependent manner as shown in Fig. 6a. The surface hydrophobicity rates with different concentrations of quercetin (12.5, 25, 50, and 100 μM) were 42.78 ± 1.09% (P = 0.044), 45.79 ± 2.09%, 58.10 ± 3.30%, and 68.95 ± 4.41%, respectively; which were substantially higher than that of the control group (36.06 ± 1.16%).
Effect of quercetin on hydrophobicity and aggregation. (a) Bacterial cells cultured in BHI with quercetin were washed and re-suspended in PUM buffer. 0.4 mL of hexadecane was added to 2 mL of the cell suspensions in tubes. After vortexed for 60 s and then incubated for 15 min at room temperature, the OD550nm of the aqueous phase was measured and the hydrophobic activity was calculated. (b) P. gingivalis were cultured anaerobically at 37 °C for 48 h in BHI broth, harvested and re-suspended in PBS to an OD600nm of 1.0. After 3 h, aggregation was calculated by measuring the decrease in OD600nm of each suspension at 37 °C. Bars denoted by (*), (**) and (****) indicate significant difference at p < 0.05, p < 0.01 and p < 0.0001, respectively by one-way analysis of variance (ANOVA) with Dunnett’s post hoc test.
We compared the aggregation of P. gingivalis with different concentrations of quercetin. As shown in Fig. 6b, the aggregation rate reached 18.35 ± 1.13% after 3 h incubation. There was a dose-dependent increase in bacterial aggregation with different concentrations of quercetin. With increasing concentrations of quercetin, the aggregation rate increased from 27.18 ± 1.38% at 12.5 μM to 73.49 ± 6.25% at 100 μM.
Effect of quercetin on gene expression
To gain insight into virulence factors and iron/heme utilization-related gene expression, real-time PCR analysis was used to quantify the effect of quercetin on P. gingivalis. As shown in Fig. 7, a significant and dose-dependent decrease expressions of all tested gene were observed. Among them, the virulence factor genes were including hagA, hagB, hem (involved in hemagglutination); kgp, rgpA, rgpB (involved in gingipain); ragA (immunodominant surface proteins), and vimA (virulence modulating gene A). Expression levels in virulence genes with quercetin were reduced ranging from 0.428- to 0.0884-fold at 50 μM and from 0.170-fold to 0.0340-fold at 100 μM. After investigating the effect of the quercetin on the expression of virulence factor genes, we also evaluated their effect on iron acquisition and metabolism (hmuR and ftn). At a concentration of 50 μM quercetin treatment, the expression of hmuR and ftn decreased 0.222-fold and 0.446-fold, respectively. More specifically, quercetin significantly inhibited the expression of both hmuR (0.0518-fold) and ftn (0.118-fold) at a concentration of 100 μM. The results indicate that quercetin inhibited the expression of virulence factors and iron/heme utilization-related genes in P. gingivalis.
Effect of quercetin on gene expression. Transcriptional levels of the virulence genes in P. gingivalis cultured in BHI supplemented with 50 and 100 μM quercetin were detected by real time-PCR. Bars denoted by (****) indicate significant difference at p < 0.0001 by one-way analysis of variance (ANOVA) with Dunnett's post hoc test.
Discussion
P. gingivalis which detected with greater frequency and at higher levels at periodontal sites is strongly associated with periodontal disease and has been considered as a major etiologic agent for periodontitis22,23. In the present study, we determined the effect of quercetin on gingipain activities, hemagglutination activity, hemolytic activity, biofilm formation, hydrophobicity and aggregation, which all contribute to P. gingivalis virulence factors. We also studied its effect on the expression of P. gingivalis various virulence factor genes by RT-PCR. Firstly, we detected the potential antibacterial activity of quercetin against planktonic P. gingivalis. The MIC and MBC studies showed the lowest concentrations to inhibit or hamper planktonic bacteria growth was 200 and 400 μM respectively, which indicate that quercetin have both bacteriostatic and bactericidal effects. Growth curve assay confirmed that quercetin did not affect the growth of P. gingivalis under the concentration of 200 μM (Fig. S1). Previous study found that flavonoids can generate hydrogen peroxide that damages membranes to inhibit or hamper bacterial activity24. Therefore, we investigate the morphological changes in cell structure by TEM and our results confirmed that quercetin damaged the cell membrane and lead to cell death. To the best of our knowledge, it is the first time to systematically investigate the effect of quercetin at sub-MIC concentration on P. gingivalis virulence and pathogenic properties.
Gingipains degrade a large variety of host proteins such as immunoglobulins, extracellular matrix proteins and bactericidal proteins25. Some of them are providing nutrients and growth factors like iron, peptides and amino acids; while others result in disrupt the host immune defense mechanisms by modulating cytokine functions, degrading immunoglobulin, complement proteins and cell membrane proteins26,27. Thus, based on the above factors, inhibition of bacterial gingipains by quercetin may potentiate its therapeutic effects in the therapy of periodontitis. Our data showed that without affecting bacteria growth, sub-MIC concentrations of quercetin could significantly inhibit gingipains activities of P. gingivalis cell (Fig. 2) and gingipain extracts (Fig. S2) which was consistent with previous study28. And molecular docking analysis were carried out to confirm that quercetin could accommodate the binding pocket of both Kgp and RgpB by well shape matching and favorable hydrogen bond and van der Waals interactions, suggesting that quercetin is capable to inhibit the bioactivity of Kgp or RgpB by occupying the catalytic binding site. These results suggested that quercetin may be beneficial to prevent or slow down periodontal tissue destruction by inhibiting the gingipain activity of P. gingivalis.
P. gingivalis is an obligate anaerobe that efficiently utilizes heme, transferrin, and hemoglobin as sources of iron (essential nutrition) for bacterial growth and can acquire heme on its cell surface, resulting in black-pigmented colonies on blood agar plates29. Unlike other microorganisms, P. gingivalis does not possess a siderophore scavenging system for iron uptake or the enzymes required for heme biosynthesis30. It employs alternative mechanisms to acquire heme directly from erythrocytes. The mechanism of hemin acquisition from erythrocytes is a defense mechanism of P. gingivalis against reactive oxygen species in the oral inflammatory microenvironment and a mechanism for iron storage31. The mechanism involves hemagglutination, hemolysis, binding, and degradation of the hemoglobin molecule32,33. To determine the effect of quercetin in heme acquisition, we choose hemagglutination and hemolysis assay as targets to investigate the potential inhibitory activities. Our results showed that quercetin inhibited hemagglutination and hemolytic activity. These activities were also related to hemagglutinin-adhesin domains of gingipains, which participate the maturation of hemoglobin-binding receptor protein and may modify erythrocyte surface molecules34. Gingipain deficient mutants resulted in decreased hemagglutination and hemolytic activities compared with the wild-type strain25,35. Hemagglutinin may not only mediate the adsorption and invasion of bacteria into host cells but also agglutinate and lyse erythrocytes to survive in vivo36. The development of compounds against P. gingivalis hemagglutinins could be a promising cytoprotective strategy to prevent the harmful effects of long-term bacterial infection30. Therefore, quercetin may be significant in controlling the pathogenic potential of P. gingivalis through interference with mechanisms involved in hemagglutination and hemolysis.
In most environments, bacteria exist mainly in the form of biofilms37. Developing biofilm communities is an adaptive strategy for the survival of bacteria in host environments38. The biofilm phenotype is physiologically and functionally distinct from the planktonic bacteria. Bacteria enclosed in the biofilm structure can tolerate antibiotics and antibacterial agents more than 100–1000 times higher than planktonic bacteria and protect bacteria from attack by the host immune system, which enables persistent infection38,39,40. P. gingivalis has the ability to form biofilm, matrix-enclosed structure in subgingival plaque. Thus, we studied the effects of quercetin on P. gingivalis biofilms by crystal violet and MTT assays. Crystal violet assay was used for quantification of biofilm biomass, while the MTT assay was utilized to evaluate the metabolic activities of viable bacteria in biofilms. We found that biofilm formation was significantly decreased by quercetin at sub-MIC concentrations in this study. Confocal imaging also confirmed that the biofilms were clearly thinner and sparser with increasing concentrations of quercetin. These results showed that quercetin show inhibitory effects on P. gingivalis biofilm formation.
Previous studies demonstrated that quercetin can alter the cell surface properties to inhibit biofilm formation41,42,43. Thus, we assessed the cell surface properties by cell surface hydrophobicity and aggregation assays. Cell surface hydrophobicity is an important attribute of bacteria that contributes to adhesion and biofilm formation, and is influenced by the growth medium, bacterial age, and bacterial surface structures44. Our results showed that quercetin increased cell surface hydrophobicity and may lead to the inhibition of biofilm, which was consistent with previous study45. During biofilm development, autoaggregation is a process through which a strain within the biofilm produces polymers to boost the integration of genetically identical strains46, and is also important for the bacterium to attach in the oral cavity, which are essential for the pathological process leading to periodontitis. Our results showed that quercetin increased aggregation but decreased biofilm formation. CLSM fluorescence imaging also confirmed that P. gingivalis cells tended to aggregate into distinct clusters easily discernable after adding quercetin. These results were in agreement with our previous study concerning the effect of resveratrol on F. nucleatum aggregation and biofilm formation46. Besides, P. gingivalis aggregation is associated with cell wall components and characteristics such as hydrophobicity47, which may be the reason of quercetin to influence bacterial aggregation.
In addition, real-time PCR analysis was performed to evaluate the effect of quercetin on the gene expressions involved in virulence of P. gingivalis. Our results showed that the expressions of all selected virulence genes were down-regulated in the presence of quercetin. Among them, rgpA, rgpB, kgp genes are involved in gingipain and hagA, hagB, hem genes are involved in hemagglutination, which are major virulences of P. gingivalis. rgpA gene is related with bacterial surface proteins, for example, fimbrillin and Kgp48. rgpB gene is responsible for the production of gingipains and important in the pathogenesis of periodontal disease49. The hagA, hagB, hem genes facilitate the acquisition of heme from erythrocytes and other host cells which is necessarily for bacterial growth50,51. HagA and HagB also involved in adhesion to human coronary artery endothelial cells and gingival epithelial cells51,52. RagA is immunodominant surface antigens found in the serum of patients with periodontal disease; and can be stimulated when P. gingivalis is exposed to cotinine, nicotine, and cigarette smoke extract53,54. The vimA (virulence modulating) gene plays a role in acetyl coenzyme A (acetyl-CoA) transfer and modulates lipid A and its associated proteins. It also has multifunctional properties including oxidative stress resistance, glycosylation and anchorage of several surface proteins, protein sorting and transport55,56. Previous studies found that a vimA deficient strain increased aggregation and changed membrane surface proteins57. The down-regulation of vimA gene expression caused by quercetin may result in increasing P. gingivalis aggregation, which was consistent with our results. But there was no change in the hydrophobicity or ability to form biofilm in vimA deficient strain which was inconsistent with our results. These different results might be due to different methods used to test hydrophobicity and biofilm formation. Besides, another explanation for this observation may be that the effects of quercetin on hydrophobicity and biofilm formation have no relationship with vimA gene. In our study, the genes involved in iron acquisition and metabolism (ftn and hmuR) were also down-regulated by quercetin. Ferritin, encoded by the ftn gene, is one of the intracellular iron-storage proteins and particularly important for P. gingivalis to survive under iron deprived conditions58. HmuR (heme/hemoglobin utilization receptor HmuR), which has homology with TonB-dependent outer membrane heme/hemoglobin receptors is utilized for hemin and hemoglobin binding and hemin transport59,60. Therefore, quercetin inhibited the expression of virulence factors and iron/heme utilization-related genes at the transcriptional level.
Previous studies have investigated antimicrobial-agents-containing mouth rinse for P. gingivalis in clinical studies61,62. The essential-oils mouth rinse group containing thymol could significant reduce the occurrence of P. gingivalis in saliva, and present reductions of pocket depth (PPD), plaque index (PI), and gingival index (GI) comparing baseline at 45 (T1) and 180 (T2) days after periodontal therapy61. Another mouth rinse containing Enteromorpha linza extract (consist of polyphenol) significantly inhibits P. gingivalis, and reduces plaque, improves the condition of gingival tissues, and reduces bleeding62. This study provides theoretical basis for application of quercetin, a natural polyphenol, in the clinical perspective.
In conclusion, our study demonstrated that quercetin inhibits virulence and physiological properties of P. gingivalis including gingipain activity, hemolytic activity, hemagglutination activity, biofilm formation, hydrophobicity, aggregation, and virulence gene expression. Our study provides new evidence that quercetin impair the pathogenicity of P. gingivalis and might be useful in treatment of periodontal disease. Further studies are required to better understand the molecular mechanism underlying of quercetin inhibition on P. gingivalis pathogenicity.
Materials and methods
Bacteria and culture conditions
Porphyromonas gingivalis ATCC 33277 was provided by Laboratory of Oral Microbiota and Systemic Diseases, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine and grown in Brain Heart Infusion Broth (BHI; Difco Laboratories, Sparks, MD, United States) supplemented with hemin (5 μg/mL), menadione (0.5 μg/mL) at 37 °C under anaerobic conditions (80% N2, 10% CO2, and 10% H2).
Bacteria antibiotic susceptibility assay
The drug susceptibility of planktonic cultures was determined by a broth microdilution assay. The quercetin (Sigma-Aldrich, St.Louis, MO, United States) was dissolved in dimethyl sulfoxide (DMSO, Sigma-Aldrich, St.Louis, MO, United States) and diluted at final concentrations of 0–1600 μM. A 20 μL quantity of P. gingivalis suspension (1 × 107 CFU/mL) was added in a flat-bottomed 96-well microplate and incubated under anaerobic conditions at 37 °C. A blank control and a vehicle control were also prepared. The minimal inhibitory concentration (MIC) referred to the lowest concentration of quercetin that inhibited visible bacterial growth. To determine the minimal bactericidal concentration (MBC) values, an aliquot of 10 μL cell suspension from each well was taken, and bacterial clones were counted after incubation for 3–5 days. The MBC was defined as to the lowest concentration at which no bacterial growth observed on the blood agar plates.
Transmission electron microscopy
Morphological changes in the cells were observed by transmission electron microscopy (TEM). The P. gingivalis cells with different concentrations of quercetin cultured at 37 °C for 4 h. The cell pellets were washed with PBS and fixed with 2.5% glutaraldehyde at 4 °C. Then, they were exposed to 2% osmium tetroxide for 2 h, dehydrated in a series of ethanol (30%, 50%, 70%, 85%, 95%, 100% and 100%) and dried in acetone solutions three times (50%, 100% and 100%) for 15 min each. Then the samples were embedded in resin blocks, cut into ultrathin (70-nm) sections, and stained with uranyl acetate and lead citrate. Each specimen was examined by TEM (FEI Talos L120C).
Gingipain activity assays
Gingipain activity was measured according to the previous methods with minor modifications63. Briefly, a 24 h P. gingivalis culture was harvested by centrifugation, washed, and suspended in PBS to an optical density at 660 nm of 2 for Rgp activity and of 1 for Kgp activity. P. gingivalis cells were incubated in PBS with or without quercetin, a specific substrate for Rgp (0.4 mM, N-α-benzoyl-dl-arginine-p-nitroanilide, BAPNA) or Kgp (0.4 mM, acetyl-lysine-p-nitroanilide, ALNA). The mixtures were incubated at 37 °C in the dark for 3 h. Activity of Rgp and Kgp indicated as the hydrolysis of the Rgp and Kgp-specific chromogenic substrates was detected at every 2 min for 1 h by measuring the absorbance at 405 nm. The assay was repeated three times independently to ensure reproducibility.
Molecular docking
Structures of Kgp (PDB: 6I9A) and RgpB (PDB: 1CVR) were retrieved from the Protein Data Bank. Proteins were prepared using the Protein Preparation Wizard in Maestro (Schrodinger, Inc., version 10.2). The crystal structures were minimized using the OPLS_2005 force field with the maximum root mean square deviation (RMSD) value of 0.3 Å. The ligands were prepared with the LigPrep module in Maestro, including adding hydrogen atoms, ionizing at a pH range from 5.0 to 9.0, and producing the corresponding low-energy 3D structure. The prepared compound was docked into the active site of Kgp/RgpB using Glide with default settings. The docked poses were ranked by Gscore, and the one with the lowest binding energy was selected for binding mode analysis.
Hemagglutination assays
The hemagglutination assays were performed as previously reported47. Briefly, overnight cultures of P. gingivalis with quercetin were harvested, centrifuged, and suspended in PBS. Then, 100 μL of PBS was added to each well of a 96-well round-bottom microtiter plate, 100 μL of the bacterial cells was added to the first well and serially diluted (1:2 to 1:128). At the same time, sheep erythrocytes were washed and re-suspended in PBS. Finally, sheep erythrocytes suspension (100 μL, 2% in PBS) and quercetin were added to each well, and the plate was incubated at room temperature for 3 h. Hemagglutination activities were evaluated visually. Assays were repeated three times independent experiments.
Hemolytic activity assays
Hemolytic activity was performed as previously reported with minor modification64. In brief, P. gingivalis cells were centrifuged, and re-suspended to a final OD600nm of 1.5 in PBS. Fresh sheep erythrocytes (Beiruite Bio-technology, Zhengzhou, China) were harvested by centrifugation from whole blood and washed with PBS until the supernatant did not contain hemoglobin pigment visibly. The sheep erythrocytes at a concentration of 1% were mixed with an equal volume of bacterial cells with or without quercetin at 37 °C for 18 h. The hemolytic activity was determined at wavelength of 405 nm. The means and standard deviations of three independent experiments were calculated.
Biofilm formation assays
Biofilm biomass was examined by the crystal violet staining method with minor modification13. P. gingivalis suspension was grown in BHI supplemented with quercetin for 48 h at 37 °C in a 96-well polystyrene plates. The culture supernatant was removed and washed three times with sterile PBS. Then, the adherent biofilms were incubated with methanol for 15 min followed by staining with 0.04% (w/v) crystal violet for 15 min. After washing with deionized water, 95% ethanol was added to detect. The OD values were recorded at wavelength of 550 nm.
The effect of quercetin on the viability of biofilm was determined using the 3-(4,5-dimethylthiazolyl-2)-2,5-diphenyltetrazoliumbromide (MTT) method65. The biofilm was formed as described for the above method. MTT (0.5 mg/mL) was added to detect the metabolic activity of the biofilms. The plates were cultured for 3 h in a dark place at 37 °C. Following incubation, MTT solution was gently aspirated from each well, and 100 μL of lysing solution was added to dissolve the formazan crystals. The absorbance of the solution was measured at a wavelength of 590 nm by microplate reader. All experiments were performed in triplicate with at least three replicates, and wells without cells were used as blank controls.
Biofilm structure observation
P. gingivalis biofilms were formed as described above assay for 48 h on glass coverslips with different concentrations of quercetin. Bacterial cells were washed three times with saline to remove unbound cells and stained with the LIVE/DEAD BacLight Bacterial Viability Kit (Molecular Probes Inc., Eugene, Oregon, United States). After staining for 30 min in the dark, CLSM (Leica TCS SP8; Leica Microsystems, Wetzlar, Germany) was applied to acquire images and measure the thicknesses of biofilm. The exciting laser intensity, background level, contrast and electronic zoom were fixed for each experiment. In each experiment, at least five random fields were recorded.
Hydrophobicity assays
The relative surface hydrophobicity was determined by measuring adherence to n-hexadecane66. Briefly, bacterial cells cultured with quercetin were washed in phosphate urea magnesium (PUM) buffer and re-suspended in the same buffer. Then, 0.4 mL of hexadecane (Sigma-Aldrich, St. Louis, MO, United States) was added to 2 mL of the cell suspensions in tubes. This mixture was vortexed for 60 s, and then incubated for 15 min at room temperature. The OD550nm of the aqueous phase was measured, and the hydrophobic activity was calculated using the formula: [(OD550nm before mixing) − (OD550nm after mixing)]/(OD550nm before mixing) × 100%. Independent experiments were repeated three times to verify the results.
Aggregation assays
Aggregation assays of P. gingivalis was performed as described previously with slight modification67. Briefly, P. gingivalis were cultured anaerobically in BHI broth, and harvested by centrifugation and re-suspended in PBS to an OD600nm of 1.0. After 3 h, aggregation was monitored by measuring the decrease in OD600nm of each suspension at 37 °C. The percentage of aggregation was calculated by the following equation: Aggregation rate = (ODInitial − OD3 h)/(ODInitial − ODBlank) × 100%. Bacterial aggregation assays were performed in triplicate independent experiments.
Quantitative analysis of gene expression by RT-PCR
The total RNA of P. gingivalis was extracted using Trizol reagent (Takara, Dalian, China), following the protocols provided by the manufacturer, and measured by Nanodrop 2000 to determine the RNA concentration. Reverse transcription was performed by cDNA synthesis kit (Takara, Dalian, China) to generate cDNA. Amounts of mRNA transcripts were measured by the Roche LightCycler 480 real-time PCR detection system (Roche, Basel, Switzerland). Reactions were performed with 20 μL of a mixture containing 10 μL of SYBR Premix (2 ×; Takara, Dalian, China), 1.6 μL of each primer, 3.4 μL of water and 5 μL of the cDNA template. The forward and reverse primer sequences are shown in Table 168,69. Real-time PCR conditions included 30 s at 95 °C; 10 s at 95 °C, 20 s at 60 °C and 15 s at 72 °C for 40 cycles. The fold changes in gene expression were using the ΔΔCt method. Each assay was performed with three independent RNA samples. The results correspond to three experiments independently.
Statistical analysis
All data were presented as means ± SD. Statistical analysis was performed with one-way analysis of variance (ANOVA) with Dunnett’s post hoc test. All statistical analyses were carried out using SPSS software (SPSS 15.0 software, United States). The difference was considered statistically significant at P < 0.05.
Data availability
All data generated or analyzed during this study are included in this published article.
References
Schmuch, J. et al. Extract from Rumex acetosa L. for prophylaxis of periodontitis: Inhibition of bacterial in vitro adhesion and of gingipains of Porphyromonas gingivalis by epicatechin-3-O-(4β→8)-epicatechin-3-O-gallate (procyanidin-B2-Di-gallate). PLoS ONE 10, e0120130. https://doi.org/10.1371/journal.pone.0120130 (2015).
Aliko, A. et al. Impact of Porphyromonas gingivalis peptidylarginine deiminase on bacterial biofilm formation, epithelial cell invasion, and epithelial cell transcriptional landscape. Sci. Rep. 8, 14144. https://doi.org/10.1038/s41598-018-32603-y (2018).
Thamban Chandrika, N. et al. Novel zafirlukast derivatives exhibit selective antibacterial activity against Porphyromonas gingivalis. Medchemcomm 10, 926–933. https://doi.org/10.1039/c9md00074g (2019).
Otomo-Corgel, J., Pucher, J. J., Rethman, M. P. & Reynolds, M. A. State of the science: Chronic periodontitis and systemic health. J. Evid. Base. Dent. Pract. 12, 20–28. https://doi.org/10.1016/S1532-3382(12)70006-4 (2012).
Liao, J., Zhao, L., Yoshioka, M., Hinode, D. & Grenier, D. Effects of Japanese traditional herbal medicines (Kampo) on growth and virulence properties of Porphyromonas gingivalis and viability of oral epithelial cells. Pharm. Biol. 51, 1538–1544. https://doi.org/10.3109/13880209.2013.801995 (2013).
Pham, T. A. V., Tran, T. T. P. & Luong, N. T. M. Antimicrobial effect of platelet-rich plasma against Porphyromonas gingivalis. Int. J. Dent. 2019, 7329103. https://doi.org/10.1155/2019/7329103 (2019).
Zhang, L. et al. Porphyromonas gingivalis lipopolysaccharide promotes T-helper 17 cell differentiation from human CD4+ naïve T cells via toll-like receptor-2 in vitro. Arch. Oral Biol. 107, 104483. https://doi.org/10.1016/j.archoralbio.2019.104483 (2019).
Jia, L. et al. Pathogenesis of important virulence factors of Porphyromonas gingivalis via toll-like receptors. Front. Cell. Infect. Microbiol. 9, 262. https://doi.org/10.3389/fcimb.2019.00262 (2019).
Liu, Y. et al. Infection of microglia with Porphyromonas gingivalis promotes cell migration and an inflammatory response through the gingipain-mediated activation of protease-activated receptor-2 in mice. Sci. Rep. 7, 11759. https://doi.org/10.1038/s41598-017-12173-1 (2017).
Sato, K. et al. Identification of a new membrane-associated protein that influences transport/maturation of gingipains and adhesins of Porphyromonas gingivalis. J. Biol. Chem. 280, 8668–8677. https://doi.org/10.1074/jbc.M413544200 (2005).
Sakai, E. et al. Construction of recombinant hemagglutinin derived from the gingipain-encoding gene of Porphyromonas gingivalis, identification of its target protein on erythrocytes, and inhibition of hemagglutination by an interdomain regional peptide. J. Bacteriol. 189, 3977–3986. https://doi.org/10.1128/JB.01691-06 (2007).
Lo, A. et al. FimR and FimS: Biofilm formation and gene expression in Porphyromonas gingivalis. J. Bacteriol. 192, 1332–1343. https://doi.org/10.1128/JB.01211-09 (2010).
He, Z., Huang, Z., Jiang, W. & Zhou, W. Antimicrobial activity of cinnamaldehyde on Streptococcus mutans Biofilms. Front. Microbiol. 10, 2241. https://doi.org/10.3389/fmicb.2019.02241 (2019).
Eseberri, I., Miranda, J., Lasa, A., Churruca, I. & Portillo, M. P. Doses of quercetin in the range of serum concentrations exert delipidating effects in 3T3-L1 preadipocytes by acting on different stages of adipogenesis, but not in mature adipocytes. Oxid. Med. Cell. Longev. 2015, 480943. https://doi.org/10.1155/2015/480943 (2015).
Casado-Díaz, A., Anter, J., Dorado, G. & Quesada-Gómez, J. M. Effects of quercetin, a natural phenolic compound, in the differentiation of human mesenchymal stem cells (MSC) into adipocytes and osteoblasts. J. Nutr. Biochem. 32, 151–162. https://doi.org/10.1016/j.jnutbio.2016.03.005 (2016).
Amin, M. U., Khurram, M., Khattak, B. & Khan, J. Antibiotic additive and synergistic action of rutin, morin and quercetin against methicillin resistant Staphylococcus aureus. BMC. Complement. Altern. Med. 15, 59. https://doi.org/10.1186/s12906-015-0580-0 (2015).
Ouyang, J. et al. Quercetin is an effective inhibitor of quorum sensing, biofilm formation and virulence factors in Pseudomonas aeruginosa. J. Appl. Microbiol. 120, 966–974. https://doi.org/10.1111/jam.13073 (2016).
Wang, S. et al. Bacteriostatic effect of quercetin as an antibiotic alternative in vivo and its antibacterial mechanism in vitro. J. Food Prot. 81, 68–78. https://doi.org/10.4315/0362-028X.JFP-17-214 (2019).
Guevara, T. et al. Structural determinants of inhibition of Porphyromonas gingivalis gingipain K by KYT-36, a potent, selective, and bioavailable peptidase inhibitor. Sci. Rep. 9, 4935. https://doi.org/10.1038/s41598-019-41354-3 (2019).
Halgren, T. A. et al. Glide: A new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J. Med. Chem. 47, 1750–1759. https://doi.org/10.1021/jm030644s (2004).
Eichinger, A. et al. Crystal structure of gingipain R: An Arg-specific bacterial cysteine proteinase with a caspase-like fold. EMBO J. 18, 5453–5462. https://doi.org/10.1093/emboj/18.20.5453 (1999).
Lewis, J. P., Dawson, J. A., Hannis, J. C., Muddiman, D. & Macrina, F. L. Hemoglobinase activity of the lysine gingipain protease (Kgp) of Porphyromonas gingivalis W83. J. Bacteriol. 181, 4905–4913 (1999).
Huck, O. et al. Akkermansia muciniphila reduces Porphyromonas gingivalis-induced inflammation and periodontal bone destruction. J. Clin. Periodontol. 47, 202–212. https://doi.org/10.1111/jcpe.13214 (2020).
Cushnie, T. P. & Lamb, A. J. Recent advances in understanding the antibacterial properties of flavonoids. Int. J. Antimicrob. Agents 38, 99–107. https://doi.org/10.1016/j.ijantimicag.2011.02.014 (2011).
Olczak, T., Dixon, D. W. & Genco, C. A. Binding specificity of the Porphyromonas gingivalis heme and hemoglobin receptor HmuR, gingipain K, and gingipain R1 for heme, porphyrins, and metalloporphyrins. J. Bacteriol. 183, 5599–5608. https://doi.org/10.1128/JB.183.19.5599-5608,2001 (2001).
Neyestani, T. R., Khalaji, N. & Gharavi, A. Black and green teas may have selective synergistic or antagonistic effects on certain antibiotics against Streptococcus pyogenes in vitro. J. Nutr. Environ. Med. 13, 258–266. https://doi.org/10.1080/13590840701703934 (2007).
Potempa, J., Banbula, A. & Travis, J. Role of bacterial proteinases in matrix destruction and modulation of host responses. Periodontology 2000(24), 153–192. https://doi.org/10.1034/j.1600-0757.2000.2240108.x (2000).
Kariu, T. et al. Inhibition of gingipains and Porphyromonas gingivalis growth and biofilm formation by prenyl flavonoids. J. Periodontal Res. 52, 89–96. https://doi.org/10.1111/jre.12372 (2017).
Hiratsuka, K., Kiyama-Kishikawa, M. & Abiko, Y. Hemin-binding protein 35 (HBP35) plays an important role in bacteria-mammalian cells interactions in Porphyromonas gingivalis. Microb. Pathog. 48, 116–123. https://doi.org/10.1016/j.micpath.2010.01.001 (2010).
Eltigani, S. A., Eltayeb, M. M., Ishihara, A. & Arima, J. Isolates from Monechma ciliatum seeds’ extract hampered Porphyromonas gingivalis hemagglutinins. J. Food Biochem. 43, e13029. https://doi.org/10.1111/jfbc.13029 (2019).
Sheets, S. M., Robles-Price, A. G., McKenzie, R. M., Casiano, C. A. & Fletcher, H. M. Gingipain-dependent interactions with the host are important for survival of Porphyromonas gingivalis. Front. Biosci. 13, 3215–3238. https://doi.org/10.2741/2922 (2008).
Dixon, D. R., Jeffrey, N. R., Dubey, V. S. & Leung, K. P. Antimicrobial peptide inhibition of Porphyromonas gingivalis 381-induced hemagglutination is improved with a synthetic decapeptide. Peptides 30, 2161–2167. https://doi.org/10.1016/j.peptides.2009.07.027 (2009).
Chu, L., Bramanti, T. E., Ebersole, J. L. & Holt, S. C. Hemolytic activity in the periodontopathogen Porphyromonas gingivalis: Kinetic of enzyme release and localization. Infect. Immunol. 59, 1932–1940 (1991).
Shi, Y. et al. Genetic analyses of proteolysis, hemoglobin binding, and hemagglutination of Porphyromonas gingivalis. J. Biol. Chem. 274, 17955–17960. https://doi.org/10.1074/jbc.274.25.17955 (1999).
Grenier, D. et al. Effect of inactivation of the Arg- and/or Lys-gingipain gene on selected virulence and physiological properties of Porphyromonas gingivalis. Infect. Immunol. 71, 4742–4748. https://doi.org/10.1128/iai.71.8.4742-4748.2003 (2003).
Kaizuka, K., Hosogi, Y., Hayakawa, M., Shibata, Y. & Abiko, Y. Human monoclonal antibody inhibits Porphyromonas gingivalis hemagglutinin activity. J. Periodontol. 74, 38–43. https://doi.org/10.1902/jop.2003.74.1.38 (2003).
Wood, T. K., Hong, S. H. & Ma, Q. Engineering biofilm formation and dispersal. Trends Biotechnol. 29, 87–94. https://doi.org/10.1016/j.tibtech.2010.11.001 (2011).
Macià, M. D., Rojo-Molinero, E. & Oliver, A. Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin. Microbiol. Infect. 20, 981–990. https://doi.org/10.1111/1469-0691.12651 (2012).
Kuang, X., Chen, V. & Xu, X. Novel approaches to the control of oral microbial biofilms. Biomed. Res. Int. 2018, 6498932. https://doi.org/10.1155/2018/6498932 (2018).
Ong, K. S., Mawang, C. I., Daniel-Jambun, D., Lim, Y. Y. & Lee, S. M. Current anti-biofilm strategies and potential of antioxidants in biofilm control. Expert Rev. Anti Infect. Ther. 16, 855–864. https://doi.org/10.1080/14787210.2018.1535898 (2018).
Vipin, C., Mujeeburahiman, M., Ashwini, P., Arun, A. B. & Rekha, P. D. Anti-biofilm and cytoprotective activities of quercetin against Pseudomonas aeruginosa isolates. Lett. Appl. Microbiol. 68, 464–471. https://doi.org/10.1111/lam.13129 (2019).
Vazquez-Armenta, F. J. et al. Quercetin reduces adhesion and inhibits biofilm development by Listeria monocytogenes by reducing the amount of extracellular proteins. Food Control 90, 266–273. https://doi.org/10.1016/j.foodcont.2018.02.041 (2018).
Hasan, S., Singh, K., Danisuddin, M., Verma, P. K. & Khan, A. U. Inhibition of major virulence pathways of Streptococcus mutans by quercitrin and deoxynojirimycin: A synergistic approach of infection control. PLoS ONE 9, e91736. https://doi.org/10.1371/journal.pone.0091736 (2014).
He, Z. et al. Role of the luxS gene in initial biofilm formation by Streptococcus mutans. J. Mol. Microbiol. Biotechnol. 25, 60–68. https://doi.org/10.1159/000371816 (2015).
Vazquez-Armenta, F. J. et al. Quercetin repressed the stress response factor (sigB) and virulence genes (prfA, actA, inlA, and inlC), lower the adhesion, and biofilm development of L. monocytogenes. Food Microbiol. 87, 103377. https://doi.org/10.1016/j.fm.2019.103377 (2020).
He, Z. et al. Anti-biofilm activities from resveratrol against Fusobacterium nucleatum. Front. Microbiol. 7, 1065. https://doi.org/10.3389/fmicb.2016.01065 (2016).
Slots, J. & Gibbons, R. J. Attachment of Bacteroides melaninogenicus subsp. asaccharolyticus to oral surfaces and its possible role in colonization of the mouth and of periodontal pockets. Infect. Immunol. 19, 254–264 (1978).
Zhang, R., Yang, J., Wu, J., Sun, W. B. & Liu, Y. Effect of deletion of the rgpA gene on selected virulence of Porphyromonas gingivalis. J. Dent. Sci. 11, 279–286. https://doi.org/10.1016/j.jds.2016.03.004 (2016).
Graziano, T. S. et al. Catecholamines promote the expression of virulence and oxidative stress genes in Porphyromonas gingivalis. J. Periodontal Res. 49, 660–669. https://doi.org/10.1111/jre.12148 (2014).
Lepine, G., Ellen, R. P. & Progulske-Fox, A. Construction and preliminary characterization of three hemagglutinin mutants of Porphyromonas gingivalis. Infect. Immunol. 64, 1467–1472 (1996).
Belanger, M., Kozarov, E., Song, H., Whitlock, J. & Progulske-Fox, A. Both the unique and repeat regions of the Porphyromonas gingivalis hemagglutin A are involved in adhesion and invasion of host cells. Anaerobe 18, 128–134. https://doi.org/10.1016/j.anaerobe.2011.10.005 (2012).
Song, H., Belanger, M., Whitlock, J., Kozarov, E. & Progulske-Fox, A. Hemagglutinin B is involved in the adherence of Porphyromonas gingivalis to human coronary artery endothelial cells. Infect. Immunol. 73, 7267–7273. https://doi.org/10.1128/IAI.73.11.7267-7273.2005 (2005).
Curtis, M. A., Slaney, J. M., Carman, R. J. & Johnson, N. W. Identification of the major surface protein antigens of Porphyromonas gingivalis using IgG antibody reactivity of periodontal case-control serum. Oral Microbiol. Immunol. 6, 321–326. https://doi.org/10.1111/j.1399-302x.1991.tb00502.x (1991).
Cogo, K. et al. Proteomic analysis of Porphyromonas gingivalis exposed to nicotine and cotinine. J. Periodontal Res. 47, 766–775. https://doi.org/10.1111/j.1600-0765.2012.01494.x (2012).
Aruni, A. W. et al. VimA-dependent modulation of acetyl coenzyme A levels and lipid A biosynthesis can alter virulence in Porphyromonas gingivalis. Infect. Immunol. 80, 550–564. https://doi.org/10.1128/IAI.06062-11 (2012).
Aruni, A. W., Robles, A. & Fletcher, H. M. VimA mediates multiple functions that control virulence in Porphyromonas gingivalis. Mol. Oral Microbiol. 28, 167–180. https://doi.org/10.1111/omi.12017 (2013).
Osbourne, D. O. et al. Role of vimA in cell surface biogenesis in Porphyromonas gingivalis. Microbiology 156, 2180–2193. https://doi.org/10.1099/mic.0.038331-0 (2010).
Ratnayake, D. B. et al. Ferritin from the obligate anaerobe Porphyromonas gingivalis: Purification, gene cloning and mutant studies. Microbiology 146, 1119–1127. https://doi.org/10.1099/00221287-146-5-1119 (2000).
Simpson, W., Olczak, T. & Genco, C. A. Characterization and expression of HmuR, a TonB-dependent hemoglobin receptor of Porphyromonas gingivalis. J. Bacteriol. 182, 5737–5748. https://doi.org/10.1128/jb.182.20.5737-5748.2000 (2000).
Simpson, W., Olczak, T. & Genco, C. A. Lysine-specific gingipain K and heme/hemoglobin receptor HmuR are involved in heme utilization in Porphyromonas gingivalis. Acta Biochim. Pol. 51, 253–262 (2004).
Cavalca Cortelli, S. et al. Clinical and microbiological effects of an essential-oil-containing mouth rinse applied in the “one-stage full-mouth disinfection” protocol—A randomized doubled-blinded preliminary study. Clin. Oral Investig. 13, 189–194. https://doi.org/10.1007/s00784-008-0219-3 (2008).
Cho, H. B. et al. Clinical and microbial evaluation of the effects on gingivitis of a mouth rinse containing an Enteromorpha linza extract. J. Med. Food 14, 1670–1676. https://doi.org/10.1089/jmf.2011.1710 (2011).
Sheets, S. M., Potempa, J., Travis, J., Casiano, C. A. & Fletcher, H. M. Gingipains from Porphyromonas gingivalis W83 induce cell adhesion molecule cleavage and apoptosis in endothelial cells. Infect. Immunol. 73, 1543–1552. https://doi.org/10.1128/IAI.73.3.1543-1552.2005 (2005).
Abdolhosseini, M., Nandula, S. R., Song, J., Hirt, H. & Gorr, S. U. Lysine substitutions convert a bacterial-agglutinating peptide into a bactericidal peptide that retains anti-lipopolysaccharide activity and low hemolytic activity. Peptides 35, 231–238. https://doi.org/10.1016/j.peptides.2012.03.017 (2012).
He, Z. et al. Use of the quorum sensing inhibitor furanone C-30 to interfere with biofilm formation by Streptococcus mutans and its luxS mutant strain. Int. J. Antimicrob. Agents 40, 30–35. https://doi.org/10.1016/j.ijantimicag.2012.03.016 (2012).
Lee, S. Y. & Lee, S. Y. Effects of sub-minimal inhibitory concentrations of antibiotics on the morphology and surface hydrophobicity of periodontopathic anaerobes. Anaerobe 55, 107–111. https://doi.org/10.1016/j.anaerobe.2018.11.005 (2019).
Tokuda, M., Karunakaran, T., Duncan, M., Hamada, N. & Kuramitsu, H. Role of Arg-gingipain A in virulence of Porphyromonas gingivalis. Infect. Immunol. 66, 1159–1166. https://doi.org/10.1128/IAI.66.3.1159-1166.1998 (1998).
Azelmat, J., Fournier-Larente, J. & Grenier, D. The anthraquinone rhein attenuates virulence gene expression in Porphyromonas gingivalis and exhibits synergistic antibacterial activity in association with metronidazole or natural compounds. Arch. Oral Biol. 60, 342–346. https://doi.org/10.1016/j.archoralbio.2014.11.006 (2015).
Zhou, W. et al. Melatonin receptor agonists as the “perioceutics” agents for periodontal disease through modulation of Porphyromonas gingivalis virulence and inflammatory response. PLoS ONE 11, e0166442. https://doi.org/10.1371/journal.pone.0166442 (2016).
Acknowledgements
We thank the Electron Microscopy center of Shanghai Institute of Precision Medicine, Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine, for their technical support and assistance on the electron microscopy. This work was supported by a grant from the National Natural Science Foundation of China (81971299/81670992) and was partly supported by a grant from Cross-disciplinary Research Fund of Shanghai Ninth People’s Hospital, Shanghai Jiao Tong University School of Medicine (JYJC202005).
Author information
Authors and Affiliations
Contributions
Z.Y.H. conducted the experiments and wrote the manuscript, X.Z. participated in the conception of the study and data analysis, Z.C.S., L.L. and H.S.C. conducted the experiments, S.L.L. supervised the study, W.Z. designed and supervised the study, reviewed the manuscript. All authors read and revised the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
He, Z., Zhang, X., Song, Z. et al. Quercetin inhibits virulence properties of Porphyromas gingivalis in periodontal disease. Sci Rep 10, 18313 (2020). https://doi.org/10.1038/s41598-020-74977-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-020-74977-y
- Springer Nature Limited
This article is cited by
-
Quercetin disrupts biofilm formation and attenuates virulence of Aeromonas hydrophila
Archives of Microbiology (2024)