Abstract
The highly conserved plant microRNA, miR156, affects plant development, metabolite composition, and stress response. Our previous research revealed the role of miR156 in abiotic stress response in Medicago sativa exerted by downregulating SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE transcription factors. Here we investigated the involvement and possible mechanism of action of the miR156/SPL module in flooding tolerance in alfalfa. For that, we used miR156 overexpressing, SPL13RNAi, flood-tolerant (AAC-Trueman) and -sensitive (AC-Caribou) alfalfa cultivars exposed to flooding. We also used Arabidopsis ABA insensitive (abi1-2, abi5-8) mutants and transgenic lines with either overexpressed (KIN10-OX1, KIN10-OX2) or silenced (KIN10RNAi-1, KIN10RNAi-2) catalytic subunit of SnRK1 to investigate a possible role of ABA and SnRK1 in regulating miR156 expression under flooding. Physiological analysis, hormone profiling and global transcriptome changes revealed a role for miR156/SPL module in flooding tolerance. We also identified nine novel alfalfa SPLs (SPL1, SPL1a, SPL2a, SPL7, SPL7a, SPL8, SPL13a, SPL14, SPL16) responsive to flooding. Our results also showed a possible ABA-dependent SnRK1 upregulation to enhance miR156 expression, resulting in downregulation of SPL4, SPL7a, SPL8, SPL9, SPL13, and SPL13a. We conclude that these effects induce flooding adaptive responses in alfalfa and modulate stress physiology by affecting the transcriptome, ABA metabolites and secondary metabolism.
Similar content being viewed by others
Introduction
Climate change is expected to increase the mean annual temperature and precipitation, which are correlated with flooding events1,2,3, affecting crop quality and yield4,5. Plants use escape or adaptive mechanisms to cope with flooding stress (FS). For example, flood-escaping rice genotypes elongate plant height by increasing stem cell size6 maintaining leaf gas exchange. SUBMERGENCE1A (SUB1A), SNORKEL1 (SK1) and SK2 were identified in Arabidopsis thaliana through their sequence similarity to rice homologs and were found to play a major role in flooding response7,8. Unlike plants employing the energy demanding flood-escape strategy, flood-adapting plants, such as wheat, reduce their energy metabolism, remaining submerged, and revive after the stress has ceased to exist9.
Regulating energy metabolism is important, especially under stress during which a reduced photosynthesis rate decreases glucose levels, despite the increased demand for carbon skeletons to biosynthesize specialized metabolites to scavenge Reactive Oxygen Species. Reduced energy metabolism (REM) associated with decreased oxygen availability and reduced photosynthesis assimilation during FS has been reported in different studies10,11,12. REM is perceived by a sucrose non-fermenting-related protein kinase, SnRK113,14, which is associated with a hexokinase1 (HXK1)15. The heterotrimeric protein kinase SnRK1 is catalyzed by the α subunits KIN10 and KIN1116 and regulated by the β and γ subunits in Arabidopsis17. SnRK1 expression is also triggered in Arabidopsis by the stress-related hormone abscisic acid11,18. The activation of SnRK1 regulates metabolic stress response and development in Arabidopsis14.
Previous studies showed that overexpression of microRNA156 in Medicago sativa (alfalfa) increased shoot branching, delayed flowering, reduced stem length19,20,21, and played a positive role in abiotic stress tolerance22,23,24,25,26. Considering these miR156 functions, we hypothesised that miR156 might play a role in flooding tolerance and its expression might be regulated by SnRK1. Non-protein-coding RNA sequencing revealed multiple differentially expressed microRNAs in response to flooding in poplar and maize27,28. In maize, Zhang et al.28 identified over 100 differentially expressed microRNAs upon FS of which miR159, miR395, and miR474 were increased while others (miR166, miR167, miR171, miR396, miR399) were decreased. Besides screening for differentially expressed microRNAs, it is important to validate microRNAs and their target genes by modulating their expression and investigating the plant’s response to FS. In plants, miR156 down regulates SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE (SPL) genes29. So far, at least seven SPLs (SPL2, SPL3, SPL4, SPL6, SPL9, SPL12, SPL13) have been identified as direct targets of miR156 in alfalfa30. In the current study, we investigated the role of miR156/SPL module in flooding tolerance using hormone and transcriptomic profiling combined with physiological responses of miR156OE and SPL RNAi alfalfa plants. We also investigated a possible crosstalk between miR156/SPL and ABA-dependent SnRK1 using ABA insensitive (abi1-2 and abi5-8) Arabidopsis mutants along with KIN10 overexpressing and RNAi silenced Arabidopsis plants under ABA and low sugar treatments.
Material and methods
Genetic material and flooding stress
We performed one field and two greenhouse flooding experiments at the Agriculture and Agri-Food Canada (AAFC) Research Centre in Kentville, Nova Scotia (45°03′46.38″ N, 64°29′10.26″ W) and AAFC Research Center in London, Ontario, respectively. For the field experiment we used wild type (WT), empty-vector alfalfa genotypes, a local cultivar (AC-Caribou), FS-tolerant (AAC-Trueman), miR156OE (A8a, miR156-A8, A16, A11, A11a, A17), miR156-regulated SPL6RNAi (SPL6-405, SPL6-425, SPL6-428), and miR156-regulated SPL13RNAi (SPL13R-2, SPL13R-5, SPL13R-6) genotypes as described in supplementary Figs. S1 and 2. The transgenic plants were generated previously in our lab20,23. The nucleic acid sequences were obtained from either M. truncatula or M. sativa, depending on the availability of gene sequences in the public domain. The sequences of miR156 and SPLs are those of M. sativa. Where gene sequences were not known for M. sativa, we used primers designed based on M. truncatula sequences to amplify their homologues M. sativa (primers used in qRT-PCR are indicated in supplementary Table S1). Based on field phenotypic responses, forage yield, and transcript analysis, genotypes used in the greenhouse experiment were reduced to WT, AC-Caribou, AAC-Trueman, SPL13R-5, SPL13R-6 and miR156-A8. One-month-old alfalfa plants were subjected to FS in the greenhouse by submerging eight pots of same genotype per tray with water level up to soil surface for two weeks while control plants were well-drained (WD). Greenhouse growth conditions were kept as described in Feyissa et al.24.
Measurement of photosynthesis-related parameters
Midday photosynthesis assimilation rates and dark-adapted chlorophyll fluorescence (Fv/Fm) were measured in newly growing unshaded leaves using LI-6400XT portable photosynthesis meter coupled with a Fluorescence System (LI-COR Biosciences, USA). To determine photosynthesis efficiency, photosynthesis assimilation responses across a gradient of CO2 levels (0–2000 ppm) (A/Ci) were measured and the maximum rate of rubisco carboxylase activity (Vcmax) and maximum photosynthesis electron transport rate (Jmax) were calculated using the R statistical software31.
Hormone profiling
Shoot samples were lyophilized with a Labconco freeze drier system (Kansas, USA) at − 50 °C for three days, and ~ 50 mg of dried samples were used for hormone profiling. Phytohormone analysis was performed at the National Research Council of Canada, Saskatoon, SK on a fee-for-service basis. UPLC/ESI–MS/MS of Waters ACQUITY UPLC system, coupled to a Waters Micromass Quattro Premier XE quadrupole tandem mass spectrometer via a Z-spray interface was used. Quantification of phytohormones was performed using deuterium labeled internal standards32.
Total monomeric anthocyanin and polyphenol determination
Flash-frozen shoots were used to determine monomeric anthocyanin (TMA) employing a pH deferential extraction method as described in Lee et al.33 and Cheok et al.34. TMA were expressed as mg cyanidin-3-0-glucoside (CG) equivalent.
RNA extraction for qRT-PCR and RNAseq
Top shoot tip leaves (~ 50 mg) were collected in Precellys lysing tubes, flash frozen with liquid nitrogen, homogenized by a PowerLyzer 24 and total RNA was extracted using QIAGEN RNeasy Plant mini kit. Total RNA was treated with Ambion TURBO DNA-free DNase followed by iScript cDNA synthesis for qRT-PCR using the CFX96 Real-Time PCR detection system (Bio-Rad). Specifically, we mixed 2 µL(200 ng) cDNA, 1 µL forward and reverse gene-specific primers (10 µM each) (Table S1), 5 µL SsoFast EvaGreen Supermixes, and 2 µL of nuclease-free water. PCR amplification was performed using: cDNA denaturation at 95 °C for 30 s followed by 40 cycles of 95 °C for 10 s, 58 °C for 30 s and 72 °C for 30 s followed by a melting curve that ran from 65 to 95 °C with a gradual increment of 0.5 °C per 5 s. All reactions were performed with three technical and four biological replicates. Transcript levels were analysed relative to acetyl-CoA carboxylase (ACC1) and ACTIN housekeeping genes.
Total RNA quality was checked using a BioRad Bioanalyzer before RNAseq analysis. Stranded mRNA library was prepared with NEBNext followed by sequencing with Illumina NovaSeq6000 with 100 bp fragment pair end reads at Genome Quebec, Montreal as a fee-for-service. Raw RNA sequencing reads can be accessed from the National Center for Biotechnology Information, NCBI, BioProject PRJNA596791.
RNAseq and pathway analysis
RNAseq data was analyzed according to Trapnell et al.35 on Biocluster with a Linux interface. M. truncatula Mt4.0 V2 (http://www.medicagogenome.org/http://www.medicagogenome.org/downloads) was used as a reference genome due to lack of publicly available alfalfa genome sequence. With the recent tetraploid alfalfa genome report36, we searched for the gene IDs of the novel MsSPLs (Table S2). The alfalfa nucleotide sequences of the novel MsSPLs were locally aligned to the newly reported tetraploid alfalfa genome sequence. Gene IDs with 300–680 nucleotide sequences, depending on the MsSPL, and more than 95% identity were considered as a potential candidate from the annotated alfalfa General Feature Format, .gff, file36. To identify gene expression patterns and module identification, R-software V3.5.2-based weighted gene co-expression network analysis (WGCNA) was conducted using ‘BiocManager’ package37. Pathway analysis was done using MapMan V3.6 (https://mapman.gabipd.org/) with a M. truncatula reference genome, and manual incorporation of the differentially expressed genes (DEG) into phenylpropanoid and ABA biosynthesis pathways.
5′RACE-based miR156 cleavage site identification
To understand whether miR156 cleaves the newly identified SPLs (SPL7a, 8 and 13a) that were silenced under FS, 5′RACE (FirstChoice RLM-RACE) was performed using manufacturer’s—provided two forward primers with designed gene-specific inner (GSI) and outer (GSO) reverse primers (Table S1). miR156 overexpressing (A17) alfalfa genotype was used in this study. The PCR products were cloned into a pJET1.2 (Clone JET PCR cloning kit), transformed into E. coli TOP10 cells by heat shock, and the purified plasmids (GeneJET Plasmid Miniprep kit) were sequenced.
Investigating protein–protein interaction between SnRK1 and miR156 biogenesis genes
To investigate the role of SnRK1 in miR156 biogenesis, we tested for interaction between SnRK1 (Medtr1g034030.1) and miR156-biogenesis proteins DICER-LIKE (DCL) (Medtr3g102270.2) and finger protein SERRATE (SE) (Medtr8g043980) using the yeast-two-hybrid system (Y2H). We used ProQuest Two-Hybrid System (Invitrogen) system. A detailed description of making constructs and conducting Y2H assay is provided in Table S3.
Investigating SnRK1 regulation by sugar and ABA in Arabidopsis
To understand whether the expression of SnRK1 is regulated through ABA signalling pathway, low-sugar and ABA, ABA-insensitive Arabidopsis mutants (abi1-2 and abi5-8) were planted in ½ MS media supplemented with T1-control (44 mM sucrose, no ABA), T2 (22 mM sucrose, no ABA), or T3 (22 mM sucrose, 1 µM ABA). Arabidopsis seeds were surface sterilized with 10% bleach for 15 min, rinsed 5 times with sterile water, and plated on the designated treatment plates. Tissue culture plates were stratified at 4 °C for three days and placed at low-light intensity (10 µmol/m2s) to reduce photosynthesis-mediated sugar supplement. Treatments were replicated three times placing all genotypes together in each plate per replicate for one week (each genotype contained 50 seedlings). Subsequently, tissues were collected consistently at 2 PM, to rule out variation due to circadian rhythm and an optimized 4-h ABA response, and transcript levels of SnRK1-related genes (Fig. 5) were determined.
Investigating SnRK1 regulation by miR156 in alfalfa
To investigate whether the expression of SnRK1 was upstream of miR156 upon low sugar and ABA treatment in alfalfa, rooted cuttings of miR156OE (A17) and WT alfalfa were exposed to either T1-control (15 g/L sucrose, no ABA) or T2 (no sucrose, 100 µM ABA) under FS and kept plants at low-light intensity (25 µmol/m2s). One-week-old seedlings were treated with ABA and tissues were collected after 4 h treatment (at 2 PM), and transcript levels of SnRK1-related genes (Fig. 5) were determined.
Investigating miR156 expression dependence on SnRK1 in Arabidopsis
Arabidopsis seedlings with increased (KIN10-OX1, KIN10-OX2) and silenced (KIN10RNAi-1, KIN10RNAi-2) expression of the catalytic subunit KIN10 were used to understand whether miR156 expression is dependent on activation of SnRK1 during ABA treatment and sugar starvation. These plants were kindly provided by Dr Filip Rolland, KU, Leuven13. The treatment arrangement is similar to the one used for the ‘investigation of SnRK1 expression by sugar and ABA’ except for genotype differences. Subsequently, miR156 expression levels were determined (Fig. 5).
Data analysis
Physiological and hormonal data were first checked for normal distribution using a Shapiro–Wilk test in the R-software environment 3.5.2, followed by ANOVA and post hoc Tukey multiple comparison tests. Pair-wise comparison was performed using student t-test when necessary.
Results
To investigate the role of miR156 in flooding tolerance in alfalfa, we compared miR156OE, SPL6RNAi, SPL13RNAi, WT, and empty-vector plants combined with flooding-sensitive (AC-Caribou) and flooding-tolerant (AAC-Trueman) cultivars. The plants were initially characterized for flooding response using physiological parameters and expression of select flooding-responsive genes (Figs. S1, 3; Table S4). Based on these results, we narrowed the focus to miR156-A8, SPL13RNAi-5, SPL13RNAi-6, AC-Caribou, AAC-Trueman, and WT and repeated the experiment in the greenhouse.
miR156/SPL module mediates physiological responses of alfalfa during flooding
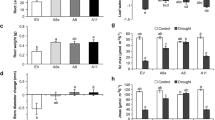
Flooding negatively affects photosynthesis and respiration38, so we set out to determine whether miR156 regulates this response. Alfalfa plants were subjected to flooding stress (FS) and well-drained conditions (WD) for two weeks (Figs. 1A, S1). Leaf yellowing was observed in all genotypes under FS, but was most severe in AC-Caribou and WT (Fig. 1B) which also had reduced photosynthesis rates than other genotypes (Fig. 1D). The flooding tolerant cultivar, AAC-Trueman, showed increased red colouration in the stems under FS (Fig. 1C). Under WD, there were no significant differences between genotypes in photosynthesis rate (Figs. 1D, S1). Under FS dark-adapted, chlorophyll fluorescence (Fv/Fm), often used as a stress tolerance indicator39,40, was maintained at a higher level in AAC-Trueman, miR156-A8, and SPL13RNAi plants relative to WT (Fig. 1E). To further understand the photosynthesis efficiency, the maximum rubisco carboxylase activity, Vcmax, and maximum photosynthesis electron transport, Jmax were determined (Fig. 1F,G). The data showed a similar pattern to the measurements of photosynthesis rate and Fv/Fm ratio observed in Fig. 1D,E. SPL13RNAi, miR156-A8, and AAC-Trueman maintained a higher level of Vcmax and Jmax except for AC-Caribou which showed comparable levels despite FS (Fig. 1F,G).
Physiological responses of flood-stressed and control plants. One-month alfalfa plants growing under (A) control, and (B) flood-stressed conditions, (C) stem colour development in AAC-Trueman plants upon flood stress, (D) photosynthesis assimilation rate in µ mol CO2 m−2 leaf area S−1, (E) chlorophyll fluorescence response, Fv/Fm, (F) Vcmax, the maximum rate of rubisco carboxylase activity, (G) Jmax, maximum photosynthesis electron transport rate. Values are sample means ± SE, n = 8 individual plants. A significant difference from ANOVA was followed by Post hoc Tukey multiple comparisons test and indicated with different letters.
ABA-metabolites are increased in miR156OE alfalfa plants under flooding
Plant hormones, such as ethylene, mediate rice and Arabidopsis responses to FS by inducing SUB1A and ABA5,7,41,42. The role of ABA-catabolite, phaseic acid (PA), was also demonstrated in Arabidopsis response to drought, where it enhances ABA-like PYRABACTIN RESISTANCE/PYR-LIKE/REGULATORY COMPONENTS OF ABA RECEPTORS-based (PYR/PYL/RCAR) signal perception and PA-specific responses41,43. We investigated the hormone profiles of FS- and WD-treated alfalfa genotypes to understand hormonal changes. Total ABA metabolites, which comprises of ABA, PA, ABA-glucose ester (ABAGE), and another four ABA derivatives, were increased in FS plants relative to their WD by 1.5- (AAC-Trueman) to 2.5 –fold (miR156-A8), while a 30% reduction was observed in WT (Fig. 2A). Specifically, ABA-metabolites from glucosyl esterification and oxidation reactions, ABAGE and PA, respectively, contributed a significant portion of the total ABA metabolite abundance under FS, second only to ABA (Fig. 2B–D). For instance, while PA concentration was not changed between FS and WD WT, an average fold-increase of 2.95- (AAC-Trueman) to 3.75 (SPL13RNAi-6) was observed in the tolerant genotypes (Fig. 2C). The results suggest an ABA-dependent response mechanism in flood tolerant alfalfa plants (Fig. 2).
UPLC/ESI–MS/MS-based hormone profiling in flood stressed and control alfalfa genotypes. (A) The abundance of total abscisic acid, ABA, metabolites (B) abscisic acid glucose ester, ABAGE, (C) phaseic acid, PA, (D) ABA, (E) indole acetic acid-aspartic acid, IAA-Asp, (F) total auxin, (G) gibberellic acid GA19, (H) total gibberellic acid GA, and (I) total cytokinin in ng per gram leaf dry weight. Total ABA metabolites in ‘A’ comprises ABA, Dihydrophaseic acid (DPA), ABAGE (Abscisic acid glucose ester), Phaseic acid (PA), 7′-Hydroxy-abscisic acid (7′OH-ABA), neo-Phaseic acid (neo-PA), and trans-Abscisic acid (t-ABA). Total auxin in ‘F’ comprises Indole-3-acetic acid (IAA) and N-(Indole-3-yl-acetyl)-aspartic acid (IAA-Asp). Total gibberellic acid in ‘H’ comprises gibberellin 8, 9, 19, 20, 24, 29, 34, 44, 51, and 53. Total cytokinin in ‘I’ comprises (trans) Zeatin-O-glucoside (t-ZOG), (cis) Zeatin-O-glucoside (c-ZOG), (cis) Zeatin riboside (c-ZR), Dihydrozeatin riboside (dhZR), Isopentenyladenosine (iPR), Values are sample means ± SE, n = 3 individual plants as biological replicates. A significant difference from ANOVA was followed by Post hoc Tukey multiple comparisons test and indicated with different letters.
Developing adventitious roots and shoot elongation are regulated by auxin, ethylene, and gibberellin, and are used as a flood escape mechanisms by some plants44. There was a significant decrease in IAA-Asp and total gibberellin levels in response to FS (Fig. 2E–H, Table S5). Variable reductions in vegetative growth (Fig. 1A) were suggested to correlate with reduced cytokinin levels45, a correlation observed in all alfalfa genotypes in this study (Fig. 2I). The detailed hormone profile is provided in Table S6.
miR156 regulates secondary metabolism pathways to improve flooding tolerance
To investigate whether the miR156 gene regulatory network is involved in flooding tolerance, determined RNA expression levels of the flooding responsive SNORKEL18, as well as miR156 and miR156-regulated SPL4 and SPL13 genes20,30 in miR156OE, SPL13RNAi, WT, AC-Caribou and AAC-Trueman plants (Fig. S3). Transcript levels of SNORKEL1 and miR156 were increased in miR156OE plants, whereas those of SPL4 and SPL13 were decreased upon FS (Fig. S3). Based on these results, we decided to investigate changes in global transcriptome using miR156-A8, SPL13RNAi-6, AC-Caribou, AAC-Trueman, and WT alfalfa.
Transcript profiles comparing FS and WD revealed that over 60% of DEG were decreased in FS WT, whereas the differences between upregulated and downregulated genes were minimal (51% increased and 49% decreased) in FS miR156-A8 (Fig. 3A). But AC-Caribou, SPL13R-6, and AAC-Trueman reduced 53, 56 and 59% of DEG, respectively. To further identify genes contributing to alfalfa flooding tolerance, we compared DEG from FS WT to those from AC-Caribou, AAC-Trueman, SPL13RNAi, and miR156-A8 FS (Fig. 3B). Genotype-specific and commonly shared DEG were observed. Interestingly, two up-regulated and 11 down-regulated genes were commonly shared by the flood-tolerant AAC-Trueman, miR156-A8 and SPL13RNAi-6 genotypes. The two up-regulated transcripts code for Gly-Asp-Ser-Leu (GDSL)-like lipase/acyl hydrolase (Medtr8g087870) and a reticuline oxidase-like protein (Medtr2g031560). On the other hand, five of the 11 commonly down-regulated transcripts under FS code for carbonic anhydrase (Medtr0219s0070), galactinol-raffinose galactosyltransferase (Medtr7g091880), AP2 domain class transcription factor (Medtr3g098580), PAR1 protein (Medtr1g101120), and sieve element occlusion protein (Medtr1g074990).
Differentially expressed genes and their associated function upon flood stress in alfalfa. (A) Differentially expressed genes, DEG, in each genotype comparing flooding and well-drained control alfalfa plants, (B) Venn diagram illustrating commonality and uniqueness in DEG obtained by comparing flood-stressed WT to AC-Caribou, AAC-Trueman, SPL13RNAi-6 and miR156-A8 genotypes independently upon flooding, (C) Venn diagram illustrating commonly shared and unique DEG obtained by comparing flood-stressed and their respective well-drained control counterparts plants of each genotypes, (D) functional distribution of DEG generated by comparing flood-stressed WT to AC-Caribou, AAC-Trueman, SPL13RNAi-6 and miR156-A8 genotypes. Upper and lower panel numbers in ‘B’ and ‘C’ indicate increased and decreased DEG, respectively, compared to WT flood stress or the same genotype under control conditions respectively. Novaseq 6000-based RNAseq analysis was performed for three biological replicates for each treatment condition and genotype. Venn diagram was constructed using an online tool http://www.interactivenn.net/ from DEG.
GDSL-like lipase/acyl hydrolase contributes to plant response to abiotic and biotic stresses and to seed development by modulating lipid metabolism46,47,48,49. A comparison of WT with the other genotypes under FS identified two other major GDSL-related genes, including GDSL-like lipase/acyl hydrolase and Pmr5/Cas1p GDSL/SGNH-like acyl-esterase (Fig. S4A). Of these genes, one of three in SPL13RNAi-6, 29 of 33 in miR156-A8, two of six in AC-Caribou and nine of 14 in AAC-Trueman were increased under flooding compared to FS WT (Fig. S4A). The second commonly increased transcript in all three genotypes (miR156-A8, SPL13RNAi-6 and AAC-Trueman) codes for the reticuline oxidase-like protein which binds flavin adenine dinucleotide (FAD) that serves as hydrogen acceptor in a bi-covalent manner, and possesses an oxidoreductase activity in (S)-scoulerine biosynthesis50. The (S)-scoulerine alkaloid biosynthesis pathway is downstream of tyrosine required for the biosynthesis of important alkaloids such as berberine51. Transcripts of this gene were increased 1.7-fold in FS AAC-Trueman relative to its WD while other genotypes (WT, AC-Caribou, miR156-A8, and SPL13R-6) showed a reduced 0.22 to 0.53-fold change compared to their respective WD (Fig. S4B). Despite this reduction, reticuline oxidase-like protein transcripts were increased 1.6 (miR156-A8) to sixfold higher (AAC-Trueman) relative to FS WT (Fig. S4B).
To understand transcript plasticity of genotypes in response to flooding, we compared the transcript profiles of WD and FS plants of each genotype and compared their similarity. We identified 1071 upregulated and 1624 downregulated DEG shared by all genotypes (Fig. 3C). In addition, 89, 3764, 337, 471, and 437 upregulated and 148, 993, 199, 327, and 497 downregulated genotype-specific DEG were detected in WT, SPL13RNAi-6, miR156-A8, AC-Caribou, and AAC-Trueman genotypes, respectively (Fig. 3C). Among the transcripts differentially expressed in all genotypes upon FS, ABA biosynthesis, SnRK1, and phenylpropanoid pathway genes were upregulated (Fig. S4C,D, Table S7, 8) while SPLs had lower transcript levels (Fig. 4B). The number and distribution of different molecular function-associated DEG between FS WT and all other genotypes code primarily for protein post-translational modification, miscellaneous UDP glucosyl and glucoronyl transferases, RNA regulation of transcription, signalling receptor kinase, biotic and abiotic stress, transport, secondary metabolism, plant hormone metabolism, development and cell wall cellulose synthase (Fig. 3D).
Phylogenetic analysis and identification of new SPLs in alfalfa. (A) Amino acid coding sequence-based phylogenetic analysis of SPLs, (B) Differentially expressed SPLs between control and flood-stressed alfalfa plants, (C) Validation of the consistently regulated SPLs from RNAseq using qRT-PCR, (D) The conserved SQUAMOSA PROMOTER-BINDING PROTEIN, SBP, domain in the newly identified SPLs containing two zinc-finger binding domains (Zn 1 and 2) and nuclear localization signal (NLS), (E) Complementarity of SPL7a, 8 and 13a to that of matured miR156 sequence along with 5′ RLM RACE determined cleavage sites. SPLs boxed with red line in ‘A’ and underlined in ‘B’ are newly identified SPLs while boxed with black lines were previously identified (Aung et al., 2015b; Gao et al., 2016). Arrows in ‘B’ and ‘C’ indicates commonly reduced SPLs during FS. Phylogenetic analysis is done using clustal omega online tool (https://www.ebi.ac.uk/Tools/msa/clustalo/) followed by FigTree v.1.4.2 free software. The ‘*’ in ‘C’ indicates significance level at p < 0.05 when compared between control and flooding transcript levels. In silico amino acid sequence in ‘D’ is analysed using http://weblogo.berkeley.edu/logo.cgi. In ‘D’ zinc-finger binding domains (Zn 1 and 2) and nuclear localization signals (NLS) are indicated with red and green boxes, respectively. Arrows in ‘D’ indicates cleavage site by miR156.
Novel SPLs regulated by miR156 in alfalfa
Previous reports revealed seven SPLs (SPL2, SPL3, SPL4, SPL6, SPL9, SPL12, and SPL13) were regulated by miR156 in alfalfa20,30. Our observation that miR156 expression was enhanced under FS (Fig. S3D) prompted us to investigate if miR156-regulated SPLs contribute to alfalfa’s response to FS. RNAseq analysis followed by transcript annotation of five alfalfa genotypes exposed to FS showed that 15 SPLs were affected by FS relative to their WD counterparts (Fig. 4 A,B). We retrieved amino acid coding nucleotide sequences from a database (http://www.medicagogenome.org/) for the new SPLs along with previously used sequences30 for phylogenetic analysis for Medicago truncatula, Glycine max, and Arabidopsis thaliana (Table S9). Besides, the previously known seven SPLs, nine new SPLs (SPL1, SPL1a, SPL2a, SPL7, SPL7a, SPL8, SPL13a, SPL14 and SPL16) were identified in this study from database search and amino acid sequence alignment (Fig. 4A). The naming of the new SPLs is based on the closely related known SPLs from the phylogenetic tree in the clade. Phylogenetic analysis grouped the SPLs into eight clades that have over 75% coding sequence similarity. Clade I (SPL1, SPL1a, SPL14), clade II (SPL2, SPL3, SPL4, SPL7a), clade III (SPL6), clade IV (SPL7), clade V (SPL8), clade VI (SPL9), clade VII (SPL2a, SPL12), and clade VIII (SPL13, SPL13a, SPL16) (Fig. 4A). Of these SPLs, SPL4, SPL7a, SPL8, SPL9, SPL13, SPL13a were downregulated under FS compared to WD in all genotypes, SPL3 was downregulated only in miR156-A8, while SPL12 and SPL16 were not consistently downregulated in all the genotypes (Fig. 4B). The transcriptomic data were validated by qRT-PCR (Fig. 4C) by amplifying each SPL (Table S1).
The newly identified SPLs were further analyzed for their conserved SBP domain, nuclear localization signal, and presence of miR156 binding nucleotide sequences. In silico amino acid sequence analysis (http://weblogo.berkeley.edu/logo.cgi) of the newly identified SPLs revealed they all contained the conserved SBP domain (Fig. 4E). Using an amino acid sequence-based online tool (http://nls-mapper.iab.keio.ac.jp/cgi-bin/NLS_Mapper_form.cgi), nuclear localization signals were also detected in the newly identified SPLs (Fig. S4).
To understand whether the newly identified SPLs are regulated by miR156, we first checked for complementarity to mature miR156 ‘ATGCTCTCTCTCTTCTGTCA’ in a 5′ to 3′ orientation and found different proportions of miR156 matching sequence in SPLs ranging from 13/20 in SPL1 and SPL7 to 19/20 in SPL2a, SPL7a and SPL13a (Fig. S5). Of the different proportions of miR156 matching sequence to SPLs, SPL7a, SPL8 and SPL13a possessed 19/20, 18/20 (in two fragments) and 19/20 nucleotide matches to miR156, respectively (Fig. 4D). miR156 cleavage sites in transcripts of SPLs silenced under flooding (SPL7a, SPL8 and SPL13a) were determined using 5′-RACE. The sequencing results revealed SPL7a, SPL8 and SPL13a were cleaved by miR156 upstream of the complementary target sequence (Fig. 4D). These results revealed that miR156 downregulates SPL4, SPL7a, SPL8, SPL9, SPL13 and SPL13a in alfalfa in response to FS.
Flooding enhances SnRK1 expression in an ABA-dependent manner
The protein kinase SnRK1 was reported to be elevated in Arabidopsis that undergo a REM response due to abiotic stress13,14. We observed an upregulation of SnRK1 (Medtr1g034030) and its regulatory β subunit (Medtr5g098510 and Medtr2g095290) in alfalfa under FS (Fig. S4C). The catalytic α subunit KIN11 (Medtr6g048250 and Medtr6g012990) was also increased consistent with SnRK1 expression in two of the genotypes (AC-Caribou and SPL13RNAi-6) (Fig. S4C). Considering the elevated levels of ABA-metabolites in AAC-Trueman, miR156-A8 and SPL13RNAi-6 (Fig. 2) upon FS, we investigated the ABA signaling pathway and ABA-responsive elements in these plants. The ABA signaling PYL9/PYR1 receptors as well as ABI2 and ABA-responsive element ABRE were upregulated, whereas ABI1 was downregulated under FS (Figs. 2, S4D). Given an enhanced level of SnRK1 and ABA and findings from the literature showing SnRK1’s central role in sugar and ABA signalling 18,52, we investigated whether the expression of SnRK1 is ABA-dependent.
Noticing the upregulation of SnRK1, ABA-signaling elements (Table S7, Fig. S4D) and ABA (Fig. 2) in alfalfa under FS, we determined the transcript levels of KIN10, KIN11 and DARK INDUCED genes (DIN). This was done in ABA insensitive Arabidopsis mutants (abi1-2, and abi5-8)53,54, due to the lack of similar mutants in alfalfa. The ABA-insensitive mutant and WT Arabidopsis seedlings were treated with either 22 mM sucrose and 1 µM ABA or 22 mM sucrose alone under low light growth conditions. Expression level of KIN11 was reduced in abi5-8 while both KIN10 and KIN11 were increased in abi1-2 plants during ABA treatment, where only one of the calcium and protein binding elements (ABI1) is silenced (Fig. 5A,B). We also determined whether the expression of dark-induced and multiple stress responsive DIN genes (DIN1, DIN6 and DIN10)13 were affected by ABA treatment. We found that the expression of DIN10 was increased with reduced sugar level in abi1-2 mutants while reduced in abi5-8 mutants with ABA application (Fig. 5C). This result shows DIN10 expression is ABA-dependent and regulated by ABI5 (Fig. 5C). On the other hand, RNA transcript levels of DIN1 and DIN6 were increased in the presence of ABA despite a reduced level of ABI1 and ABI5 (Fig. 5D,E).
ABA-dependent expression of SnRK1 may regulate miR156. SnRK1 catalytic subunits (A) KIN10, (B) KIN11, relative transcript levels of DARK INDUCED, DIN, (C) DIN10, (D) DIN6, (E) DIN1 genes in Arabidopsis, (F) relative transcript levels of SnRK1 and SnRK2 along with SnRK1 regulatory and catalytic subunits in response to 100 µM ABA treatment, (G) transcript levels of SnRK1 upon 100 µM ABA treatment relative to their counterpart control alfalfa plants, (H) relative transcript levels of miR156 in KIN10 overexpressing and RNAi silenced Arabidopsis plants, (I) Yeast-two-hybrid (Y2H) assay for the interaction between SnRK1 and miR156 biogenesis genes. n = 50 biological replicates of Arabidopsis in ‘A’ to ‘E’ and ‘H’ while three alfalfa plants in ‘F’ and ‘G’. Arabidopsis specific three housekeeping genes (elongation factor-alpha, tubulin, and actin) are used for relative transcript analysis relative to WT in ‘A’ to ‘E’ and ‘H’ while alfalfa specific elongation factor, actin and ubiquitin10 are used in ‘F’ and ‘G’. Values are means with ± SE. In ‘I’, protein-coding whole sequences of the genes were amplified using gene-specific primers with the addition of ‘CACC’ in the forward primer, for directional cloning (Table S1). The interaction between the pEXP32/Krev1 (rat Krev1) with pEXP22/RalGDS-wt (ras association domain of RalGDS) is used as positive control while for the negative control pEXP32/Krev1 with the mutated ras association domain of RalGDS, pEXP22/RalGDS-m2, was used according to the user’s manual.
To determine whether the transcription of SnRK1 in alfalfa is affected by miR156 expression level, we exposed one-month old rooted cuttings of alfalfa plants (miR156OE and WT) to FS for one week followed by treatment with of 100 µM ABA for 4 h. Then the transcript levels of SnRK1 and its catalytic α (KIN10 and KIN11) and regulatory (β and γ) subunits, and SnRK2 were analysed. Under control conditions, only SnRK2 was significantly increased in miR156OE plants whereas the other regulatory and catalytic subunits were not affected relative to WT (Fig. 5F). Interestingly, under 100 µM ABA treatment, the expression levels of all the catalytic, regulatory subunits, SnRK1 and SnRK2 were not significantly different from those of WT (Fig. 5F). Both WT and miR56OE genotypes had higher SnRK1 expression under 100 µM ABA treatment compared to their counter part controls indicating the ABA-dependent SnRK1 activation is not dependent on miR156 (Fig. 5G).
Does SnRK1 regulate miR156?
To understand whether SnRK1 regulates miR156 during flooding response, we used Arabidopsis plants with altered KIN10 expression (KIN10-OX-1, KIN10-OX-2, KIN10RNAi-1, KIN10RNAi-2)13 treated with 3 µM ABA, and determined miR156 transcript level. The transcript level of miR156 was significantly higher in KIN10-OX while KIN10RNAi plants showed a comparable (KIN10RNAi-2) or lower (KIN10RNAi-1) levels relative to WT (Fig. 5H). Interestingly, under ABA treatment, miR156 expression level stayed significantly higher at least in one KIN10-OX (KIN10-OX-2) but lower in KIN10RNAi plants when compared to WT (Fig. 5H).
DICER-LIKE 1 (DCL1), SERRATE (SE) and HYPONASTIC LEAVES (HYL1) are critical for the biogenesis of microRNAs55. Our observation that increased KIN10 expression was correlated with upregulation of miR156 (Fig. 5H) while overexpression of miR156 did not affect KIN10 (Fig. 5F) prompted us to investigate whether SnRK1 regulates miR156 biogenesis in alfalfa. A recent report showed that inactivation of SnRK2 kinases causing reduced phosphorylation of SE and HYL1 led to a decrease in microRNA biogenesis under stress conditions56. We used a yeast-two-hybrid assay to investigate the in vivo pairwise protein–protein interactions between SnRK1 with DCL1 and SE. Apparently, no protein–protein interaction was detected between SnRK1 and either DCL1 or SE (Fig. 5I).
miR156/SPL module enhances flooding adaptive mechanisms
The transcriptomic profile of SPL13RNAi plants shows an increase in genes coding for trehalose-6-phosphate synthase (TPS) and trehalose-6-phosphate phosphatase (TPP) (Fig. 6A). The reduction in photosynthesis assimilation rate after two weeks of FS (Figs. 1C, 6B, Table S10) may cause lower level of fructose and glucose, which in flooding tolerant plants is compensated for by increased levels of TPS and TPP to enhance sucrose hydrolysis. Such REM is sensed by SnRK1 in Arabidopsis to coordinate metabolic responses57. Likewise, levels of SnRK1 along with its catalytic and regulatory subunits are increased in alfalfa during FS (Figs. 5G, S3C). Moreover, induction of the phenylpropanoid pathway is observed in flood-tolerant SPL13RNAi plants to scavenge ROS (Fig. 6C, Table S8). We previously reported an interplay between miR156-SPL13 and WD40-1F-DFR in alfalfa to enhance biosynthesis of abiotic stress-alleviating anthocyanin through the phenylpropanoid pathway24. Consistent with these findings, we detected an increase in total anthocyanin monomers under FS (Fig. 6C).
miR156-based regulation of SPL13 enhances photosynthesis, phenylpropanoid, and ABA biosynthesis in response to flooding. (A) Illustration of reduced Glycolysis and TCA cycle with an enhanced phenylpropanoid pathway in flood-stressed SPL13RNAi plants based on differentially expressed genes, (B) MapMan-based pathway analysis illustrating Photosystem I and II associated increased transcript abundance along with electron transport chain in miR156-A8 plants compared to WT plants during FS, (C) Total monomeric anthocyanin in Gallic acid equivalents, (D) enrichment of ABA biosynthesis in SPL13RNAi plants upon flooding. n = 3 biological replicates. Pathways in ‘A’ and ‘C’ were manually generated based on the KEGG pathway database (https://www.genome.jp/kegg/pathway.html)70 with permission and previous reports on glycolysis, phenylpropanoid, and carotenoid biosynthesis pathway. DEG corresponding to construct the phenylpropanoid and glycolysis pathway in ‘A’ are presented in Tables S6 and S7, respectively. Values in ‘C’ are mean with ± SE.
In the current study, we detected an increase in the level of ABA metabolites (PA, ABAGE) during FS in flood-tolerant alfalfa genotypes (AAC-Trueman, miR156-A8 and SPL13RNAi-6) (Fig. 2). The increase in accumulation of ABA metabolites under FS is regulated at the transcript level based on results of global transcriptomic-derived pathway analysis in SPL13RNAi plants (Fig. 6D) and other flood-tolerant genotypes (Table S7).
Discussion
miR156 regulates physiological processes during flooding stress
Plants survive abiotic stress by employing adaptation or escape strategies (individually or in combination) to maintain essential physiological processes required for plant growth and development. Altering root architecture (among others) has been reported for plants adapted to growth under flooding58, and is a mechanism by which some plants can regulate response to flooding stress. For example, in Alternanthera philoxeroides flooding induces the formation of adventitious roots that help the plant absorb oxygen from ambient water, thus alleviating the deleterious effects of this stress59. In alfalfa, plants overexpressiong miR156E had enhanced drought stress, and showed thicker, denser and more adventitious roots compared to WT control23. Our results showed that miR156OE alfalfa plants maintained an active photosynthesis activity during FS as expressed in photosynthesis electron transport and maximum rate of carboxylase activity. This came with a higher dark-adapted chlorophyll florescence, Fv/Fm, and ultimately a stable photosynthesis assimilation rate. Interestingly, SPL13RNAi and the flooding-tolerant genotype, AAC-Trueman, phenocopied the response of miR156-A8. The reduced levels of Fv/Fm and photosynthesis assimilation rate, but with no effect on Vcmax and Jmax in AC-Caribou under FS, suggests a viable Ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) enzyme but its activity may be hindered by the scarcity of CO2 under stress. Understanding the maintenance of Vcmax and Jmax in AC-Caribou under FS would require studies on the Rubisco enzymatic activity.
Phaseic acid-dependent regulation of flooding tolerance in alfalfa
We investigated the role of phytohormones in alfalfa’s response to FS and found an increased level of ABA-metabolites under FS in tolerant genotypes (AAC-Trueman, miR156-A8 and SPL13RNAi-6), whereas WT had reduced levels. Of the different ABA-metabolites involved in signalling, phaseic acid (PA), was recently investigated for its role in plant adaptive plasticity41. The report showed how PA could have an ABA-like effect, and PA-specific responses during FS. In our study, we also detected a significant increase in the amount of PA in alfalfa genotypes of SPL13RNAi, miR156OE and AAC-Trueman under FS relative to WD counterparts.
Gibberellic acid, GA, is involved in internode elongation resulting in taller plants benefiting underwater-submerged plants60. The increase in plant height under the influence of GA comes with the cost of energy dissipation that competes with other physiological processes. In this regard, WT enhanced the GA abundance and increased plant height while miR156OE and SPL13RNAi reduced their GA levels under FS. This follows the typical miR156OE alfalfa phenotype with increased number of branches but reduced shoot height19. It seems that miR156OE and SPL13RNAi plants channel the assimilated carbon into the biosynthesis of stress mitigating metabolites, such as anthocyanin. Increasing plant height might be an important strategy in fully underwater-submerged plants facilitating gas exchange, but our experiment involved FS only up to the soil surface. It remains to be investigated whether the hormone profile will change in these alfalfa genotypes if plants were completely submerged under water.
Genotype-specific enhancement of secondary metabolism and photosynthesis
Under FS, plants have a reduced availability of O2 and CO2, which are important for protein biosynthesis and energy production10,12. On the other hand, gene expression followed by translation is an energy consuming process61. In our study, the flood-tolerant miR156-A8 plants maintained a comparative level of DEG between FS and WD while others showed mainly reduction under FS.
Besides the proportion of DEG affected by FS, identifying FS-responsive genes and their association/network via pathway enrichment analysis is important to understand FS-regulation mechanism. The increased expression levels of photosynthesis-related genes in SPL13RNAi and flood-tolerant AAC-Trueman genotypes during FS indicates the maintenance of physiological processes. This is in line with the observed maintenance of Vcmax and Jmax that ultimately resulted in a relatively higher photosynthesis assimilation rate during FS. FS induces the production of ROS (as part of the photosynthesis process) resulting in a negative feed-back that damages cellular integrity and enzymatic reactions62. Flood-tolerant plants, on the other hand, retain ROS levels to maintain normal physiological processes. Plant use of secondary metabolites to scavenge ROS is well documented63,64. The observed enrichment of secondary metabolism, specifically those of the phenylpropanoid pathway, suggests that flood-tolerant genotypes of alfalfa used a similar strategy to mitigate FS. Pathway enrichment analysis showed enhancement of cell wall metabolism, ROS-scavenging ascorbates and glutathione associated transcripts. This agrees with the transcript profile of SPL13RNAi plants, where Flavanone 3 hydroxylase (F3H), Flavanone 3′5′hydroxylase (F3′5H) and Dihydroflavonol 4-reductase (DFR) were upregulated in accordance with the enhanced level of total monomeric anthocyanin during FS. Analyzing the network of global transcriptome changes to metabolome under FS will provide a better understanding of alfalfa’s response to FS.
Identification of novel SPLs in alfalfa
A previous study identified seven SPLs (SPL2, SPL3, SPL4, SPL6, SPL9, SPL12 and SPL13) that are regulated by miR156 in alfalfa20,30. Here, RNAseq followed by gene ontology analysis revealed 16 SPLs in alfalfa, of which nine are novel sequences. SPLs are transcription factors that regulate the expression of downstream genes having diverse functions in plants65. Identifying new SPLs should shed light on novel molecular factors that control various aspects of alfalfa growth and development. The nine newly identified SPLs (SPL1, SPL1a, SPL2a, SPL7, SPL7a, SPL8, SPL13a, SPL14 and SPL16) can now be subjected to functional characterization in alfalfa.
Based on their sequence similarities, the previously identified seven SPLs and the nine SPLs reported in this study were organized into eight clades with those of Medicago truncatula, Glycine max and Arabidopsis thaliana in the phylogenetic tree. The newly identified SPLs are distributed into six out of the eight clades. Understanding the clade distribution of the newly identified SPLs along with the other SPLs from different plant species could provide information regarding their putative function considering their similarity in coding sequences. For example, clade I assigned SPLs, such as AtSPL1, are important for thermo-tolerance at the reproductive stage, redundantly with AtSPL1266. The newly identified SPL1, SPL1a and SPL14 may also have a role in thermos-tolerance, but this should be validated using gene silencing and overexpressing alfalfa plants under heat stress. Similarly, SPL7a may have similar function to that of SPL2, SPL3, SPL4, in alfalfa and AtSPL3, AtSPL4 and AtSPL5 in Arabidopsis. In line with this, SPLs from clade II (SPL7a and SPL4) were silenced under FS in the current study.
miR156-regulated SPLs are involved in alfalfa flooding response
Identification of SPLs affected by FS is important to understand the role of miR156/SPL gene network in alfalfa’s response to this stress. In this study, we identified three new SPLs (SPL7a, SPL8 and SPL13a) and five of the previously known (SPL3, SPL4, SPL9, SPL12, SPL13) to be downregulated in miR156-A8 plants exposed to flooding. SPL13 is one of the six SPLs (SPL4, SPL7a, SPL8, SPL9, SPL13, SPL13a) that were downregulated in all alfalfa genotypes upon FS, and is also silenced by miR156. Similar to miR156OE, SPL13RNAi plants withstood FS as manifested by their ability to maintain their physiological activities. It remains to be investigated whether other SPLs silenced by FS are directly involved in flooding response, and whether the effects of different SPLs are redundant or additive.
ABA-dependent SnRK1 potentially regulates miR156 for flooding tolerance
In our study, flooding caused an increase in ABA metabolites and upregulation of ABA biosynthesis genes. Due to the conserved ABA signalling pathway in plants41 we investigated the exogenous ABA application effect on SnRK1 using ABI insensitive Arabidopsis mutant seedlings (abi1-2, abi5-8) (due to the lack of similar mutants in alfalfa). The results showed a decrease in KIN11 (one of the catalytic subunits of SnRK1) expression due to the combined effect of ABA and reduced sugar concentration. KIN11 was more significantly reduced in abi5-8 than in abi1-2 mutant seedlings. This could be explained if the other calcium binding ABI (ABI2) is still active in the abi1-2 mutant, complementing the defective ABI1. As KIN11 is affected by ABA and low sugar availability in Arabidopsis13,14,18, we investigated whether this response depends on the level of miR156 expression in alfalfa. SnRK1 was induced in both WT and miR156OE alfalfa genotypes in response to ABA treatment, with the highest level in WT. Under ABA treatment, the expression levels of the catalytic (KIN11), regulatory (β and γ), SnRK1 and SnRK2 were comparable between the genotypes under control condition, except for SnRK2 which was significantly higher in miR156OE. Interestingly, under ABA treatment, the expression of SnRK1-associated genes (SnRK2, KIN11, SnRK1-γ subunit, SnRK1-β2 subunit) in miR156OE was similar to that in WT. This suggests the expression of SnRK1 may not depend on the level of miR156 expression, but SnRK1 may act upstream of miR156.
Due to the highly conserved SnRK function in plants13, and the lack of alfalfa mutants with altered KIN10 expression, we determined whether the expression of miR156 depends on SnRK1 using Arabidopsis with silenced (via RNAi) or overexpressed KIN10. When the plants were subjected to ABA, expression levels of miR156 was significantly higher in KIN10 overexpressing plants compared to WT or KIN10RNAi plants under control or ABA treatment conditions. The observed ABA-dependent expression of SnRK1 catalytic subunits (KIN10 and KIN11) with KIN10 overexpression resulting in higher miR156 levels suggests that SnRK1 may upregulate miR156 in Arabidopsis. Despite the evidence we showed in Arabidopsis, investigating the direct effect of ABA-dependent SnRK1 on miR156 using transgenic alfalfa genotypes with altered expression of SnRK1 is still indispensable. To understand how SnRK1 could increase the expression of miR156 in vivo, it is necessary to investigate possible protein–protein interaction between SnRK1 and miR156 biogenesis proteins (such as DCL1, SE and HYL1). In our hands, we did not detect a pairwise protein–protein interaction between SnRK1 and DCL1 and SE. This could be associated with the possibility that more than two protein components may be required for interaction to occur, as reported among three cellulose synthase catalytic subunits (irregular xylem 1, 3, and 5) of CesA family for the proper assembly of the cellulose synthesizing complex67, or that SnRK1 may interact with other miR156 biogenesis proteins in alfalfa. Consistent with a possible role for SnRK1 in miR156 biogenesis, inactivation of the redundant SnRK2 kinases under stress inhibits the biosynthesis of miR156 and other microRNAs involved in stress tolerance56.
Conclusion
Of the different phytohormones involved in abiotic stress response, ABA’s role has been well documented68. In our study, we found an enhanced level of ABA in flooding tolerant genotypes. ABA-dependent stress tolerance in plants involves regulating genes that control various plant functions, including REM that triggers protein kinase SnRK1. In our study we found an enhanced level of SnRK1 in alfalfa under FS, and that expression of its α subunits KIN10 and KIN11 was ABA-dependent. Sensing REM associated with reduced photosynthesis dictates the metabolite dynamics, considering that the major carbon skeleton of ROS scavenging secondary metabolites, such as phenylpropanoids, is derived from sugar. In our study, a reallocation of resources from energy metabolism into secondary metabolism (anthocyanin) was observed at the transcriptomic and metabolic levels. Similar to the drought stress inducing anthocyanin through regulating miR156/SPL13-WD40-1/DFR24, there was an enhanced level of miR156 upon flooding in alfalfa dictating an enhanced anthocyanin biosynthesis to scavenge ROS. Enhanced levels of miR156 and miR157 were reported in alfalfa upon drought stress69, suggesting a close functional overlap between miR156 and miR157. Whereas our investigation was focussed on the role of miR156 in flooding tolerance, we cannot rule out a redundant or additive role for miR157 in response to this stress.
We propose that the reduced but continuous photosynthesis assimilation in flooding-tolerant genotypes maintains the carbon skeleton demand to produce secondary metabolism. An enhanced level of SnRK1 governed by ABA and REM may enhance the miR156 expression to regulate the downstream genes in alfalfa. Subsequently, the enhanced level of miR156 upon FS silences three newly identified SPLs (SPL7a, SPL8, SPL13a) and three previously identified SPLs (SPL4, SPL9, SPL13) to regulate downstream genes and physiological functions. Based on our present results and others in the literature, we propose a model through which the response to FS is regulated in alfalfa via miR156 (Fig. 7).
Proposed model for flooding tolerance in alfalfa. In flooding-tolerant cultivars, ABA is induced upon FS triggering PYL/PYR9 signaling molecules. This signaling pathway induces ABI2 and ABI5 while reducing the phosphorylated PP2C for increased expression of ABA-responsive elements, ABRE. ABA also induces a REM protein kinase SnRK1 along with its α and β subunits (KIN10 and KIN11) through ABI5. The increased expression of SnRK1 may trigger miR156 and channels resources towards the phenylpropanoid pathway. Induced miR156 expression silences SPL13, to mediate anthocyanin biosynthesis by its interaction with DFR24, and other SPLs (SPL4, 7a, 8, 9 and 13a). The increased abundance of ABA metabolites antagonistically affects gibberellic acid, cytokinin and auxin hormones. To exploit the newly identified SPLs (1, 1a, 2a, 7, 7a, 8, 13a, 14 and 16) in alfalfa breeding, it will be necessary to investigate their functional roles individually and in combination in response to FS and other physiological responses.
Abbreviations
- ABAGE:
-
ABA-glucose ester
- Abi:
-
ABA insensitive
- ACC1:
-
Acetyl-CoA carboxylase
- DCL:
-
DICER-LIKE
- DEG:
-
Differentially expressed genes
- DFR :
-
Dihydroflavonol 4-reductase
- DIN :
-
DARK INDUCED
- F3H :
-
Flavanone 3 hydroxylase
- F3′5H :
-
Flavanone 3′5′hydroxylase
- FS:
-
Flooding stress
- GA:
-
Gibberellic acid
- GDSL:
-
Gly-Asp-Ser-Leu (GDSL)-like lipase/acyl hydrolase
- GSI:
-
Gene-specific inner reverse primer
- GSO:
-
Gene-specific outer reverse primer
- HXK1:
-
Hexokinase1
- HYL1 :
-
HYPONASTIC LEAVES
- J max :
-
Maximum photosynthesis electron transport rate
- KIN10 and KIN11 :
-
Catalytic subunits of non-fermenting-related protein kinase1
- miR:
-
MicroRNA
- NLS:
-
Nuclear localization signals
- PA:
-
Phaseic acid
- PYL:
-
PYR-Like
- PYR:
-
ABA-like PYRABACTIN RESISTANCE
- 5′-RACE:
-
Rapid amplification of cDNA ends
- RCAR:
-
REGULATORY COMPONENTS OF ABA RECEPTORS
- REM:
-
Reduced energy metabolism
- ROS:
-
Reactive oxygen species
- Rubisco:
-
Ribulose-1,5-bisphosphate carboxylase/oxygenase
- SBP:
-
SQUAMOSA-PROMOTER BINDING PROTEIN
- SE:
-
SERRATE
- SK1 :
-
SNORKEL1
- SnRK:
-
Non-fermenting-related protein kinase
- SPL :
-
SQUAMOSA-PROMOTER BINDING PROTEIN-LIKE
- SUB1A :
-
SUBMERGENCE1A
- TCA:
-
Tricarboxylic acid
- TMA:
-
Total monomeric anthocyanin
- TPP :
-
Trehalose-6-phosphate phosphatase
- TPS :
-
Trehalose-6-phosphate synthase
- UPLC-MS:
-
Ultra-performance liquid chromatography-mass spectrometry
- V cmax :
-
Maximum rate of rubisco carboxylase activity
- WD:
-
Well-drained
- WGCNA:
-
Weighted gene co-expression network analysis
- WT:
-
Wild type
- Y2H:
-
Yeast-two-hybrid system
References
Alexander, L. V. et al. Global observed changes in daily climate extremes of temperature and precipitation. J. Geophys. Res. 111(D05), 109 (2006).
Brown, S. et al. What are the implications of sea-level rise for a 1.5, 2 and 3 °C rise in global mean temperatures in the Ganges-Brahmaputra-Meghna and other vulnerable deltas?. Reg. Environ. Change 18(6), 1829–1842 (2018).
Rogelj, J. et al. Scenarios towards limiting global mean temperature increase below 1.5 °C. Nat. Clim. Change 8(4), 325–332 (2018).
Bailey-Serres, J., Lee, S. C. & Brinton, E. Waterproofing crops: Effective flooding survival strategies. Plant Physiol. 160(4), 1698 (2012).
Yeung, E. et al. A stress recovery signaling network for enhanced flooding tolerance in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 115(26), E6085–E6094 (2018).
Dubois, V., Moritz, T. & García-Martínez, J. L. Comparison of the role of gibberellins and ethylene in response to submergence of two lowland rice cultivars Senia and Bomba. J. Plant Physiol. 168(3), 233–241 (2011).
Xu, K. et al. Sub1A is an ethylene-response-factor-like gene that confers submergence tolerance to rice. Nature 442(7103), 705–708 (2006).
Hattori, Y. et al. The ethylene response factors SNORKEL1 and SNORKEL2 allow rice to adapt to deep water. Nature 460(7258), 1026–1030 (2009).
Herzog, M. et al. Physiology, gene expression, and metabolome of two wheat cultivars with contrasting submergence tolerance. Plant Cell Environ. 41(7), 1632–1644 (2018).
Branco-Price, C., Kaiser, K. A., Jang, C. J. H., Larive, C. K. & Bailey-Serres, J. Selective mRNA translation coordinates energetic and metabolic adjustments to cellular oxygen deprivation and reoxygenation in Arabidopsis thaliana. Plant J. 56(5), 743–755 (2008).
Gupta, K. J., Zabalza, A. & Van-Dongen, J. T. Regulation of respiration when the oxygen availability changes. Physiol. Plant. 137(4), 383–391 (2009).
Mustroph, A. et al. Profiling translatomes of discrete cell populations resolves altered cellular priorities during hypoxia In Arabidopsis. Proc. Natl. Acad. Sci. USA 106(44), 18843–18848 (2009).
Baena-González, E., Rolland, F., Thevelein, J. M. & Sheen, J. A central integrator of transcription networks in plant stress and energy signalling. Nature 448, 938 (2007).
Ramon, M. et al. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell 31(7), 1614–1632 (2009).
Moore, B. et al. Role of the Arabidopsis Glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300(5617), 332–336 (2003).
Fragoso, S. et al. SnRK1 isoforms AKIN10 And AKIN11 are differentially regulated in arabidopsis plants under phosphate starvation. Plant Physiol. 149(4), 1906–1916 (2009).
Wurzinger, B., Nukarinen, E., Nägele, T., Weckwerth, W. & Teige, M. The SnRK1 kinase as central mediator of energy signaling between different organelles. Plant Physiol. 176(2), 1085–1094 (2018).
Jossier, M. et al. SnRK1 (SNF1-Related Kinase 1) has a central role in sugar and ABA signalling in Arabidopsis thaliana. Plant J. 59(2), 316–328 (2009).
Aung, B. et al. Ectopic expression of LjmiR156 delays flowering, enhances shoot branching, and improves forage quality in alfalfa. Plant Biotechnol. Rep. 9(6), 379–393 (2015).
Aung, B. et al. microRNA156 as a promising tool for alfalfa improvement. Plant Biotechnol. J. 13(6), 779–790 (2015).
Aung, B., Gruber, M. Y. & Hannoufa, A. The microRNA156 system: A tool in plant biotechnology. Biocatal. Agric. Biotechnol. 4(4), 432–442 (2015).
Arshad, M., Gruber, M. Y., Wall, K. & Hannoufa, A. An insight into microRNA156 role in salinity stress responses of alfalfa. Front. Plant Sci. 8, 356 (2017).
Arshad, M., Feyissa, B., Amyot, L., Aung, B. & Hannoufa, A. microRNA156 improves drought stress tolerance in alfalfa (Medicago sativa) By Silencing SPL13. Plant Sci. 258, 122–136 (2017).
Feyissa, B. A., Arshad, M., Gruber, M. Y., Kohalmi, S. E. & Hannoufa, A. The interplay between miR156/SPL13 And DFR/WD40–1 regulate drought tolerance in alfalfa. BMC Plant Biol. 19(1), 434 (2019).
Matthews, C., Arshad, M. & Hannoufa, A. Alfalfa response to heat stress is modulated by microRNA156. Physiol. Plant. 165(4), 830–842 (2019).
Feyissa, B. A., Renaud, J., Nasrollahi, V., Kohalmi, S. E. & Hannoufa, A. Transcriptome-IPMS analysis reveals a tissue-dependent miR156/SPL13 regulatory mechanism in alfalfa drought tolerance. BMC Genom. 21(1), 721 (2020).
Lu, S., Sun, Y.-H. & Chiang, V. L. Stress-responsive microRNAs in populus. Plant J. 55(1), 131–151 (2008).
Zhang, Z. et al. Submergence-responsive microRNAs are potentially involved in the regulation of morphological and metabolic adaptations in maize root cells. Ann. Bot. 102(4), 509–519 (2008).
Wang, J.-W., Schwab, R., Czech, B., Mica, E. & Weigel, D. Dual effects of miR156-targeted SPL genes and CYP78A5/KLUH on plastochron length and organ size in Arabidopsis thaliana. Plant Cell 20(5), 1231–1243 (2008).
Gao, R., Austin, R. S., Amyot, L. & Hannoufa, A. Comparative transcriptome investigation of global gene expression changes caused by miR156 overexpression IN Medicago sativa. BMC Genom. 17(1), 658 (2016).
Duursma, R. A. Plantecophys—an R package for analysing and modelling leaf gas exchange data. PLoS ONE 10(11), e0143346 (2015).
Lulsdorf, M. et al. Androgenesis-inducing stress treatments change phytohormone levels in anthers of three legume species (Fabaceae). Plant Cell Rep. 31(7), 1255–1267 (2012).
Lee, J., Durst, R. & Wrolstad, R. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 88(5), 1269–1278 (2005).
Cheok, C. Y., Chin, N. L., Yusof, Y. A., Talib, R. A. & Law, C. L. Optimization of total monomeric anthocyanin (TMA) and total phenolic content (TPC) extractions from mangosteen (Garcinia mangostana Linn.) hull using ultrasonic treatments. Ind. Crops Prod. 50, 1–7 (2013).
Trapnell, C. et al. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat. Protoc. 7, 562 (2012).
Chen, H. et al. Allele-aware chromosome-level genome assembly and efficient transgene-free genome editing for the autotetraploid cultivated alfalfa. Nat. Commun. 11(1), 2494 (2020).
Langfelder, P. & Horvath, S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform. 9(1), 559 (2008).
Caudle, K. L. & Maricle, B. R. Effects of flooding on photosynthesis, chlorophyll fluorescence, and oxygen stress in plants of varying flooding tolerance. Trans. Kansas Acad. Sci. 115, 5–18 (2012).
Sharma, D. K., Andersen, S. B., Ottosen, C.-O. & Rosenqvist, E. Wheat cultivars selected for high Fv/Fm under heat stress maintain high photosynthesis, total chlorophyll, stomatal conductance transpiration and dry matter. Physiol. Plant. 153(2), 284–298 (2015).
Su, L., Dai, Z., Li, S. & Xin, H. A novel system for evaluating drought-cold tolerance of grapevines using chlorophyll fluorescence. BMC Plant Biol. 15(1), 82 (2015).
Weng, J.-K., Ye, M., Li, B. & Noel, J. P. Co-evolution of hormone metabolism and signaling networks expands plant adaptive plasticity. Cell 166(4), 881–893 (2016).
Fukao, T., Barrera-Figueroa, B. E., Juntawong, P. & Peña-Castro, J. M. Submergence and waterlogging stress in plants: A review highlighting research opportunities and understudied aspects. Front. Plant Sci. 10, 340 (2019).
Rodriguez, P. L. Abscisic acid catabolism generates phaseic acid, a molecule able to activate a subset of ABA receptors. Mol. Plant 9(11), 1448–1450 (2016).
Voesenek, L. A. C. J. & Bailey-Serres, J. Flood adaptive traits and processes: An overview. New Phytol. 206(1), 57–73 (2015).
Werner, T., Motyka, V., Strnad, M. & Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 98(18), 10487–10492 (2001).
Akoh, C. C., Lee, G.-C., Liaw, Y.-C., Huang, T.-H. & Shaw, J.-F. GDSL family of serine esterases/lipases. Prog. Lipid Res. 43(6), 534–552 (2004).
Oh, I. S. et al. Secretome analysis reveals an arabidopsis lipase involved in defense against Alternaria brassicicola. Plant Cell 17(10), 2832–2847 (2005).
Hong, J. K. et al. Function of a novel GDSL-type pepper lipase gene, CaGLIP1, in disease susceptibility and abiotic stress tolerance. Planta 227(3), 539–558 (2008).
Tan, X. et al. Characterization and expression of A GDSL-like lipase gene from Brassica napus in Nicotiana benthamiana. Protein J. 33(1), 18–23 (2014).
Sato, F. et al. Metabolic engineering of plant alkaloid biosynthesis. Proc. Natl. Acad. Sci. USA 98(1), 367–372 (2001).
Chou, W.-M. & Kutchan, T. M. Enzymatic oxidations in the biosynthesis of complex alkaloids. Plant J. 15(3), 289–300 (1998).
Rodrigues, A. et al. ABI1 and PP2CA phosphatases are negative regulators of Snf1-related protein kinase1 signaling in Arabidopsis. Plant Cell 25(10), 3871–3884 (2013).
Rubio, S. et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 150(3), 1345–1355 (2009).
Zou, Y. et al. miR172b controls the transition to autotrophic development inhibited by ABA in Arabidopsis. PLoS ONE 8(5), e64770 (2013).
Yu, Y., Jia, T. & Chen, X. The ‘how’ and ‘where’ of plant microRNAs. New Phytol. 216(4), 1002–1017 (2017).
Yan, J. et al. The SnRK2 kinases modulate miRNA accumulation in Arabidopsis. PLoS Genet. 13(4), e1006753 (2017).
Ramon, M. et al. Default activation and nuclear translocation of the plant cellular energy sensor SnRK1 regulate metabolic stress responses and development. Plant Cell 31(7), 1614–1632 (2019).
Sauter, M. J. Root responses to flooding. Curr. Opin. Plant Biol. 16(3), 282–286 (2013).
Ayi, Q. et al. Oxygen absorption by adventitious roots promotes the survival of completely submerged terrestrial plants. Ann. Bot. 118(4), 675–683 (2016).
Ayano, M. et al. Gibberellin biosynthesis and signal transduction is essential for internode elongation in deep water rice. Plant Cell Environ. 37(10), 2313–2324 (2014).
Lindqvist, L. M., Tandoc, K., Topisirovic, I. & Furic, L. Cross-talk between protein synthesis, energy metabolism and autophagy in cancer. Curr. Opin. Genet. Dev. 48, 104–111 (2018).
Zhang, Y. et al. Different reactive oxygen species scavenging properties of flavonoids determine their abilities to extend the shelf life of tomato. Plant Physiol. 169(3), 1568–1583 (2015).
Chen, Y. et al. Plant morphology, physiological characteristics, accumulation of secondary metabolites and antioxidant activities of Prunella vulgaris L. under UV solar exclusion. Biol. Res. 52(1), 17 (2019).
Ayenew, B. et al. Metabolite profiling and transcript analysis reveal specificities in the response of a berry derived cell culture to abiotic stresses. Front. Plant Sci. 6, 728 (2015).
Preston, J. C., Jorgensen, S. A., Orozco, R. & Hileman, L. C. Paralogous SQUAMOSA PROMOTER BINDING PROTEIN-LIKE (SPL) genes differentially regulate leaf initiation and reproductive phase change in petunia. Planta 243(2), 429–440 (2016).
Chao, L.-M. et al. Arabidopsis transcription factors SPL1 And SPL12 confer plant thermotolerance at reproductive stage. Mol. Plant 10(5), 735–748 (2017).
Taylor, N. G., Howells, R. M., Huttly, A. K., Vickers, K. & Turner, S. R. Interactions among three distinct cesa croteins essential for cellulose synthesis. Proc. Natl. Acad. Sci. USA 100(3), 1450–1455 (2003).
Vishwakarma, K. et al. Abscisic acid signaling and abiotic stress tolerance in plants: A review on current knowledge and future prospects. Front. Plant Sci. 8, 161–161 (2017).
Li, Y. et al. Identification of drought-responsive microRNAs from roots and leaves of alfalfa by high-throughput sequencing. Genes (Basel) 8(4), 119 (2017).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl. Acids Res. 28(1), 27–30 (2000).
Acknowledgements
We acknowledge Dr F. Rolland, KU, Leuven for providing seeds of Arabidopsis KIN10 overexpression and silenced plants. Seeds of ABA insensitive mutant lines were provided by R. Thapa from Dr. Y. Cui lab at AAFC, London. We also thank R. Thom for her technical support. We are grateful to Dr. M. Gruber for critical reading and editing of the manuscript.
Author information
Authors and Affiliations
Contributions
B.A.F. developed materials and performed the experiments at AAFC, London, Ontario; L.A. assisted in data collection; V.N. assisted in new alfalfa genome annotation; Y.P. performed the experiments at AAFC, Kentville, Nova Scotia, Canada; Y.P., S.E.K. and A.H. supervised the project; B.A.F., Y.P. and A.H. designed the research; B.A.F. drafted manuscript; B.A.F., L.A., Y.P., S.E.K., and A.H. edited and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Feyissa, B.A., Amyot, L., Nasrollahi, V. et al. Involvement of the miR156/SPL module in flooding response in Medicago sativa. Sci Rep 11, 3243 (2021). https://doi.org/10.1038/s41598-021-82450-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-82450-7
- Springer Nature Limited
This article is cited by
-
Screening of microRNAs and target genes involved in Sclerotinia sclerotiorum (Lib.) infection in Brassica napus L.
BMC Plant Biology (2023)
-
Identification and regulatory network analysis of SPL family transcription factors in Populus euphratica Oliv. heteromorphic leaves
Scientific Reports (2022)
-
Investigation of proteins’ interaction network and the expression pattern of genes involved in the ABA biogenesis and antioxidant system under methanol spray in drought-stressed rapeseed
3 Biotech (2022)
-
Deciphering the role of SPL12 and AGL6 from a genetic module that functions in nodulation and root regeneration in Medicago sativa
Plant Molecular Biology (2022)
-
Identification and Characterization of SRS Genes in Phaseolus vulgaris Genome and Their Responses Under Salt Stress
Biochemical Genetics (2022)