Abstract
LncRNAs and tumor microenvironment (TME) exert an important effect in antitumor immunity. Nonetheless, the role of m6A-related lncRNA clustering patterns in prognosis, TME and immunotherapy of cervical cancer (CC) remains unknown. Here, based on 7 m6A-related prognostic lncRNAs obtained from TCGA-CC dataset, two m6AlncRNA clustering patterns were determined. m6AlncRNA clusterA was characterized by immune cell infiltrates and immune activation. m6AlncRNA clusterB was characterized by enrichment of immune evasion and tumorigenic activation pathways as well as survival and clinical stage disadvantage. Then, principal component analysis algorithms were used to construct m6AlncRNAscore based on prognostic differentially expressed genes between two m6AlncRNA clusters to quantify m6AlncRNA clustering patterns. m6AlncRNAscore was an independent prognostic protective factor. Higher Th2 and Treg cells and enrichment of immunosuppressive pathways were observed in the low-m6AlncRNAscore group, with poorer survival. High-m6AlncRNAscore was characterized by increased infiltration of activated CD8 T cell, enrichment of immune activation pathways, lower IL-10 and TGF-beta1 levels, and higher immunophenscore values, indicating inflamed TME and better anti-tumor immunotherapy efficacy. Quantitative Real-Time Polymerase Chain Reaction was used for detection of m6A-related prognostic lncRNAs. Collectively, we identified two m6AlncRNA clustering patterns which play a nonnegligible role in the prognosis, TME heterogeneity and immunotherapy of CC patients.
Similar content being viewed by others
Introduction
Cervical cancer (CC) is the fourth leading cause of cancer deaths in women globally1. In China, 59,000 women died of CC in 2020, accounting for about 17% of all CC deaths worldwide in the same year2. High mortality of CC is associated with recurrence and an advanced stage at diagnosis. Patients with recurrent and/or advanced CC have limited treatment options and poor prognosis, with a 5-year survival probability of 17%3. Immunotherapy represented by immune checkpoint inhibitors (ICIs) provides a promising perspective for cancer treatment. However, the overall response rate of ICIs was only 10–25% in previous clinical trials for CC4. The mechanisms behind the poor response of ICIs deserve further investigation. Increasing evidence indicates that tumor microenvironment (TME) not only influences tumor cell growth and metastasis, but also strongly affects tumor immune escape and immunotherapy efficacy5,6. Therefore, the heterogeneity and complexity of TME and novel biomarkers associated with TME should be further explored to predict immunotherapeutic response and provide new therapeutic targets for tumors.
N6-methyladenosine (m6A) modification, the most common epigenetic modification in eukaryotic messenger RNAs and long non-coding RNAs (lncRNAs), plays a crucial role in RNA processing, splicing, degradation, and translation, thereby affecting cell self-renewal, differentiation, tumorigenesis, and tumor progression7,8,9. m6A modification is a dynamic reversible process regulated by methyltransferases, binding proteins, and demethylases10. Methyltransferases are composed of METTL3/14/16, ZC3H13, RBM15, WTAP, VIRMA, and RBM15B, and catalyze the RNA methylation modification process10,11,12. Demethylases include FTO12 and ALKBH3/512, and mediate the RNA methylation removal process. Binding proteins consist of YTHDF1/2/3, YTHDC1/2, HNRNPA2B1, LRPPRC, FMR1, TRMT112, ZCCHC4, NUDT21, CPSF6, SETD2, SRSF3, SRSF10, XRN1, NXF1, PRRC2A, IGF2BP1/2/3, IGFBP3, and RBMX, exerting a vital role in carcinogenesis, invasion, and metastasis by combining with m6A motif10,11,12.
Although over 85% of the human genome is transcribed, less than 3% of the transcripts encode protein, and the remaining transcripts mainly are non-coding RNAs13. LncRNAs, more than 200 nucleotides in length, constitute the largest group of ncRNAs and play a key role in transcriptional and post-transcriptional regulation14. It has been reported that lncRNAs exert an important effect on oncogenesis, metastasis, TME, and tumor immune escape and might be potential therapeutic targets for cancer14,15,16,17,18. However, the relationship between m6A-related lncRNA clustering patterns and TME immune infiltration remains unclear.
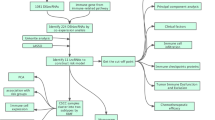
In this study, we established a scoring system, m6AlncRNAscore, to quantify the m6AlncRNA clustering patterns in individual patients with CC. Further, we explored the independent prognostic value of m6AlncRNAscore in the overall survival (OS), progression-free survival (PFS), and disease-specific survival (DSS), and the potential predictive role in immunotherapy efficacy. Additionally, we explored the correlation of m6AlncRNA clustering patterns with clinicopathologic characteristics, TME immune infiltration, and somatic mutation, as well as the potential mechanisms in CC. Finally, we validated the expression of 4 m6A-related prognostic lncRNAs in tumor samples and normal tissues.
Materials and methods
Data acquisition
The RNA sequencing and somatic mutation data were downloaded from the Cancer Genome Atlas (TCGA, https://portal.gdc.cancer.gov/) database. The immunophenscore (IPS) data were downloaded from the Cancer Immunome Atlas (TCIA, https://tcia.at/) database. The clinical information was downloaded from UCSC Xena (https://xenabrowser.net/). The sequencing data of 306 CC samples and 3 normal tissues were downloaded. The clinical information was summarized in Supplementary Table 1. Patients with OS less than or equal to 30 days were excluded, leaving the remaining 273 CC patients were enrolled into the further survival analysis. All methods were performed in accordance with the relevant guidelines and regulations.
Identification of m6A-related lncRNAs
The transcriptome sequencing genes were divided into mRNA genes and lncRNA genes according to the human genome annotation data. Expression levels of 34 m6A regulators were extracted from the mRNA data. Pearson correlation coefficient was then used to assess the correlation between m6A regulators and lncRNAs. The lncRNAs with absolute correlation coefficient > 0.40 and P value < 0.001 were regarded as m6A-related lncRNAs. Next, univariate Cox regression analysis was applied to determine lncRNAs associated with prognosis. The m6A-related lncRNAs with P value < 0.05 were considered as m6A-related prognostic lncRNAs.
Consensus clustering analysis
The “ConsensusClusterPlus” package (1000 iterations) was utilized to divide patients into different clustering patterns, referred to as m6AlncRNA clusters, based on the expression levels of m6A-related prognostic lncRNAs. According to the expression of prognosis-associated DEGs between different m6AlncRNA clusters, patients were again classified into different clustering subtypes, termed as gene clusters.
Gene set variation analysis (GSVA)
To explore the difference of biological process activity between different subgroups, we conducted GSVA enrichment analysis by using the “GSVA” package. The “c2.cp.kegg.v7.4.symbols” gene sets were downloaded from MSigDB database for running GSVA analysis. Adjusted P value < 0.05 was regarded as statistically significant.
Estimation of TME immune cell infiltration
The single-sample gene-set enrichment analysis (ssGSEA) algorithm was used to quantify the relative infiltration levels of TME immune cells. The gene set for marking 23 immune cell types was acquired from the published study19,20. The ssGSEA score was applied to represent the relative abundance of each infiltrating immune cell in each patient. Moreover, ESTIMATE algorithm was utilized to quantify the stromal and immune components for each patient.
Identification of DEGs and KEGG pathway enrichment analysis
The “limma” package was utilized to determine differentially expressed genes (DEGs) between different m6AlncRNA clusters. The significance criterion for determining DEGs was set as adjusted P value < 0.001. The “clusterProfiler” package was employed to perform KEGG enrichment analysis for the DEGs to explore their potential biological behaviors.
Generation of m6AlncRNAscore
To further investigate the role of m6AlncRNA clustering patterns in CC, we constructed a scoring system, namely m6AlncRNAscore, for individual patients based on the prognosis-associated DEGs between different m6AlncRNA clusters. The procedures for m6AlncRNAscore establishment were as follows: first, we extracted prognosis-associated DEGs by univariate Cox regression analysis; second, principal component analysis (PCA) was used to construct m6AlncRNAscore based on each prognostic DEG expression for each sample. The advantage of this method is that the score is focused on the largest well-correlated (or anti-correlated) gene block in the set, while the contribution weight from genes that are not tracked with other set is reduced. Similar to GGI establishment20,21, the m6AlncRNAscore formula was as follows:
where n is the total number of prognosis-associated DEGs, and i is the expression of the ith prognostic DEG.
Somatic mutation analysis
The “maftools” package was used to analyze the somatic mutation data of patients. Tumor mutation burden (TMB), mutations per million bases, was calculated for each patient. Then, we compared TMB between different m6AlncRNAscore groups.
Prediction of response to ICIs
TCIA database provides immune profiles and antigenomes for 20 solid tumors including CC. IPS ranges from 0 to 10, represents tumor immunogenicity. The larger the IPS value, the stronger the immunogenicity. It has been validated that IPS could predict the response of tumor patients to ICIs19,20,21,22,23.
Drug sensitivity prediction
We predicted the chemotherapeutic drug sensitivity based on the Genomics of Drug Sensitivity in Cancer (GDSC) database (https://www.cancerrxgene.org/). oncoPredict package was used to estimate the half-maximal inhibitory concentration (IC50).
Construction of ceRNA network
We firstly obtained the miRNAs interacting with the m6A-related prognostic lncRNAs by co-expression method. The miRNAs with absolute correlation coefficient < -0.20 and P value < 0.001 were regarded as related miRNAs. Then, we predicted the miRNA target genes (mRNA) by miRanda, miRDB, miRTarBase and TargetScan software. When all four kinds of software consider this gene as the target gene of miRNA, we regard this gene as the final target gene. The lncRNA-miRNA and miRNA-mRNA regulatory relationships were integrated to construct the competing endogenous RNA (ceRNA) network using Cytoscape software.
Sample collection
We totally collected 14 cervical tissue specimens, including 6 cervical cancer samples and 6 healthy controls in the Gynecology Department of Cancer Hospital Affiliated to Shanxi Medical University. Ethical approval was obtained from the Science Research Ethics Committee of Cancer Hospital Affiliated to Shanxi Medical University (No: SJJ202105). Informed consent and approval were provided by all participants. 6 patients with cervical cancer were newly diagnosed FIGO stage I/II patients without receiving any treatment.
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated from 12 samples using RNA TRIzol reagent (Tiangen Biotech Co., Ltd., Beijing, China, #DP451). cDNA synthesis was conducted with PrimeScriptTM RT Master Mix (Takara Biomedical Technology Co., Ltd., Beijing, China, #RR036Q). Real-time PCR was then performed with TB Green Premix Ex Taq (Takara Biomedical Technology Co., Ltd., Beijing, China, #RR820A). Relative expression of lncRNAs were normalized to GAPDH and calculated by 2-ΔΔCt method. Primers sequences are listed in Supplementary Table 2.
Statistical analysis
All statistical analyses were done in R version 4.0.4. Pearson correlation test was employed for assessing the relationship between m6A regulators and lncRNAs. Wilcoxon rank sum test was applied to compare the quantitative data such as m6A-related prognostic lncRNAs, immune cell infiltration, and m6AlncRNAscore between groups. Kaplan–Meier method was utilized to draw survival curves, and log-rank test was performed to compare the survival difference between groups. The predictive accuracy of m6AlncRNAscore was evaluated using the receiver operating characteristic (ROC) curve and area under curve (AUC). Multivariable Cox regression model was applied to ascertain the independent prognostic factors of CC. According to the association between m6AlncRNAscore, TMB, and OS, we used the “survminer” package to find the optimal cutoff values of m6AlncRNAscore and TMB, respectively. Patients were then divided into different groups according to the optimal cutoff value. Unless otherwise specified, a two-sided P value < 0.05 was considered statistically significant.
Results
Identification of m6A-related lncRNAs
Pearson correlation analysis was used to assess the relationship between 14,086 lncRNAs and 34 m6A regulators. Total 112 lncRNAs with absolute correlation coefficient > 0.40 and P value < 0.001 were considered as m6A-related lncRNAs. Univariate Cox regression analysis was used to explore the prognostic roles of m6A-related lncRNAs. Of the 112 m6A-related lncRNAs, 7 were associated with the OS (Table 1). These results indicated that the 7 m6A-related lncRNAs, including AC024270.4, AC008124.1, AL109811.2, AC015922.2, AC099850.4, AC025176.1, and RPP38-DT, might be potential prognostic biomarkers of CC, termed as m6A-related prognostic lncRNAs.
Expression profiles of m6A-related prognostic lncRNAs
To explore the potential biological function of m6A-related lncRNAs in the occurrence of CC, we compared the expression profiles of 7 m6A-related prognostic lncRNAs between CC samples and normal tissues. Notably, the tumor samples showed significantly lower expression levels of AC024270.4, AC008124.1, AL109811.2, and AC015922.2, but higher levels of AC099850.4, AC025176.1, and RPP38-DT, compared with the normal samples (Supplementary Fig. 1). These findings suggested that the 7 m6A-related prognostic lncRNAs might possess important biological roles in the development of CC.
Consensus clustering patterns of m6A-related prognostic lncRNAs
The “ConsensusClusterPlus” package, using the 7 m6A-related prognostic lncRNAs, was utilized to explore the molecular subtypes of patients. According to the cumulative distribution function (CDF), the area under the CDF curve, the tracking plot from k = 2 to 9 (Supplementary Fig. 2a-c), and the number of cases in any cluster cannot be too small, the k = 2 was identified as the cluster number in our study to divide patients into two different m6A-related lncRNA clustering patterns (Fig. 1a), including 209 cases in m6AlncRNA clusterA and 64 cases in m6AlncRNA clusterB. m6AlncRNA clusterA had a notably better outcome compared with clusterB (Fig. 1b). In addition, the heatmap revealed that m6AlncRNA clusterB was preferentially related to a high FIGO stage (Fig. 1c).
Differences in OS, clinicopathological and biological features, and TME characteristics between two different m6AlncRNA clustering patterns constructed based on 7 m6A-related prognostic lncRNAs. (A) Consensus clustering matrix for k = 2. (B) Kaplan–Meier curves of OS between m6AlncRNA clusterA and m6AlncRNA clusterB. (C) Heatmap and clinicopathological features of two m6AlncRNA clusters (*, P < 0.05). (D) Heatmap and the activation states of biological pathways in two different m6AlncRNA clustering patterns. (E) The abundance of each TME infiltrating cell in two m6AlncRNA clustering patterns (*, P < 0.05; **, P < 0.01; ***, P < 0.001). (F) Immune score, (G) ESTIMATE score, and (H) stromal score in two m6AlncRNA clusters.
TME immune infiltration characteristics of different m6AlncRNA clustering patterns
GSVA was applied to explore the biological behaviors between different m6AlncRNA clustering patterns. m6AlncRNA clusterA presented enrichment pathways related to oxidative phosphorylation, cardiac muscle contraction, histidine catabolism, and arachidonic acid metabolism (Fig. 1d). m6AlncRNA clusterB was significantly enriched in immune evasion, stromal, and tumorigenic activation pathways such as TGF beta signaling pathway, ubiquitin mediated proteolysis, focal adhesion, and pathways in cancer. Subsequently, we further compared TME cell infiltrates between two m6AlncRNA clusters. ClusterA showed higher infiltration levels of multiple immune cells such as activated B cell and activated CD8 T cell than clusterB (Fig. 1e). The TME cell-infiltrating characteristic of clusterA was consist with its matching survival advantage. As expected, clusterA exhibited higher immune score (Fig. 1f) and ESTIMATE score (Fig. 1g), suggesting that clusterA had a significantly higher immune cell content and lower tumor purity. However, no significant difference of stromal score was displayed between two clusters (Fig. 1h). These results indicated that the two distinct m6AlncRNA clustering patterns had markedly different TME.
Generation of m6AlncRNA genes and KEGG pathway enrichment analysis
To further explore the potential biological behaviors of each m6AlncRNA clustering pattern, we determined 786 DEGs between two m6AlncRNA clusters using the limma package, named as m6AlncRNA genes. Then, we used the clusterProfiler package to perform KEGG enrichment analysis for the DEGs. Figure 2a showed the pathways with significant enrichment. To our surprise, these genes presented enrichment of pathways associated with PD-L1 expression and PD-1 checkpoint pathway and infection-related pathways such as viral carcinogenesis and Epstein-Barr virus infection. Afterward, we utilized univariate Cox regression analysis to explore the effect of DEGs on the survival of patients. Among the 786 genes, 140 were positively or negatively related to the OS with P value < 0.05, regarded as m6AlncRNA prognostic genes (Supplementary Table 3).
KEGG pathway analysis of DEGs between two m6AlncRNA clustering patterns and differences in OS and clinicopathological characteristics between two gene clusters constructed based on m6AlncRNA prognostic genes. (A) KEGG pathway analysis for DEGs between two m6AlncRNA clusters. (B) Consensus clustering matrix of m6AlncRNA prognostic genes for k = 2. (C) Kaplan–Meier curves of OS between gene clusterA and gene clusterB. (D) Heatmap and clinicopathological characteristics of two gene clusters (***, P < 0.001).
Consensus clustering of m6AlncRNA prognostic genes
To further assess the regulation mechanism of m6AlncRNA clustering pattern in CC, we subsequently performed consensus clustering analysis based on the 140 prognostic DEGs so as to divide patients. The consensus clustering of the 140 m6AlncRNA prognostic genes classified patients into two different genomic subtypes, considered as gene clusterA (n = 224) and gene clusterB (n = 49), respectively (Supplementary Fig. 3a-c and Fig. 2b). We found that 4 out of the 7 m6A-related prognostic lncRNAs showed significantly different expression levels in the two gene clusters (Supplementary Fig. 4a). Gene clusterB had significantly better prognosis than gene clusterA (Fig. 2c). Moreover, the heatmap showed gene clusterA was preferentially associated with m6AlncRNA clusterB (Fig. 2d).
Generation of m6AlncRNAscore and prognostic value
To reveal the role of 140 prognostic DEGs in CC, we used PCA to construct a scoring system to quantify the m6AlncRNA clustering pattern in each patient, termed as m6AlncRNAscore. We then divided patients into the high-m6AlncRNAscore group (n = 147) and the low-m6AlncRNAscore group (n = 126) according to the cutoff value -0.85 determined by the survminer package. Of the 7 m6A-related prognostic lncRNAs, 6 displayed significantly different levels between two different m6AlncRNAscore groups (Supplementary Fig. 4b). A better prognosis was observed in the high-m6AlncRNAscore subgroup (Fig. 3a). The alluvial diagram showed the corresponding relationship between m6AlncRNA cluster grouping, gene cluster grouping, m6AlncRNAscore grouping, and survival outcomes (Fig. 3b). The matching rates of m6AlncRNA clusterA with high-m6AlncRNAscore and m6AlncRNA clusterB with low-m6AlncRNAscore were 67.5% and 90.6%, respectively. To assess the accuracy of m6AlncRNAscore in predicting the OS, we performed ROC analysis and found that the 3-year AUC value was 0.708, implying that m6AlncRNAscore had a good prognostic discrimination performance (Fig. 3c). Subsequently, we compared the m6AlncRNAscore between different clustering subtypes. The m6AlncRNAscore in m6AlncRNA clusterA, as expected, was dramatically higher than that in m6AlncRNA clusterB (Fig. 3d). Similarly, gene cluster B had significantly higher m6AlncRNAscore compared with gene cluster A (Fig. 3e). Further stratified survival analysis results showed that the OS time in the low-m6AlncRNAscore group was dramatically shorter compared with the high-m6AlncRNAscore group, no matter for patients with grade1/2, grade3/4, age ≤ 60 years, age > 60 years, stage I/II, or stage III/IV (Fig. 3f-k).
Construction of m6AlncRNAscore and prognostic value. (A) Kaplan–Meier curves of OS between the high- and low-m6AlncRNAscore groups. (B) Alluvial diagram showing the changes of m6AlncRNA clusters, gene clusters, m6AlncRNAscore, and survival state. (C) The 3-year ROC curve of m6AlncRNAscore in the OS. (D) Comparison of m6AlncRNAscore between two m6AlncRNA clusters. (E) Comparison of m6AlncRNAscore in two gene clusters. Kaplan–Meier curves of OS between the high- and low-m6AlncRNAscore groups in (F) grade1/2 patients, (G) grade3/4 patients, (H) patients with age ≤ 60 years, (I) patients with age > 60 years, (J) stage I/II patients, and (K) stage III/IV patients.
Independent prognostic value of m6AlncRNAscore in the prognosis of CC
As shown in Fig. 4a, the univariate Cox analysis results showed that m6AlncRNAscore, age, and FIGO stage were significantly related to the OS of CC patients. Subsequent multivariate Cox analysis results displayed that age was an independent risky factor (HR = 2.157, P value = 0.015), but m6AlncRNAscore was an independent protective factor (HR = 0.918, P value < 0.001) for the OS of CC patients (Fig. 4b).
Considering the significance of PFS and DSS in tumor prognosis, we further validated the prognostic value of m6AlncRNAscore in the PFS and DSS. In the univariate analysis, high-m6AlncRNAscore was significantly associated with better PFS (Fig. 4c) and DSS (Fig. 4d). Moreover, further multivariate Cox regression analysis results showed that m6AlncRNAscore was not only an independent prognostic factor for the PFS (Fig. 4e), but also an independent prognostic factor for the DSS (Fig. 4f). Our results strongly indicated that m6AlncRNAscore had good prognostic value in CC.
TME immune infiltration characteristics of different m6AlncRNAscore groups
To verify the biological behaviors of m6AlncRNA clustering patterns in TME, we performed GSVA and ssGSEA analyses in two m6AlncRNAscore groups. The high-m6AlncRNAscore group was characterized by enrichment of hallmark pathways such as oxidative phosphorylation and cardiac muscle contraction (Fig. 5a) and infiltration of activated CD8 T cell, CD56dim natural killer cell, and monocyte (Fig. 5b). The low-m6AlncRNAscore group was characterized by enrichment of immunosuppressive, stromal, and carcinogenic activation pathways such as wnt signaling pathway, TGF beta signaling pathway, MAPK signaling pathway, ERBB signaling pathway, focal adhesion, extracellular matrix (ECM)-receptor interaction, and pathways in cancer (Fig. 5a). Besides, the low-m6AlncRNAscore group was rich in T helper 2 (Th2) and regulatory T (Treg) cells, two types of tumor immunosuppressive T cells (Fig. 5b). We then explored the expression profiles of immunosuppressive factors IL-10 (Fig. 5c) and TGF-beta1 (Fig. 5d) and found that their levels in the low-m6AlncRNAscore group were significantly higher. The above results indicated again that m6A-related lncRNA clustering patterns played a vital role in shaping TME landscape.
TME cell infiltration characteristics in the high- and low-m6AlncRNAscore groups. (A) Heatmap and the activation states of biological pathways in two m6AlncRNAscore groups. (B) The abundance of each TME infiltrating cell in two m6AlncRNAscore groups (*, P < 0.05; **, P < 0.01; ***, P < 0.001). The expression levels of (C) IL-10 and (D) TGF-beta1 in two m6AlncRNAscore groups.
Clinical and somatic mutation characteristics of different m6AlncRNAscore groups
As expected, the m6AlncRNAscore was significantly higher in patients with stage I/II than those with stage III/IV (Fig. 6a). However, no significant m6AlncRNAscore difference was observed in different age or grade subgroups. TMB quantification analysis results showed that the low-m6AlncRNAscore group presented no significant TBM difference in relative to the high-m6AlncRNAscore group (Fig. 6b). Next, the survminer package was applied to classify patients with information of somatic mutation and survival into the high-TMB group (n = 28) and the low-TMB group (n = 226) according to the cutoff value 6.32. A better prognostic tendency was observed in the high TMB group, while no significant difference was displayed between the high- and low-TMB groups (Fig. 6c). Moreover, we found that patients with low-m6AlncRNAscore and low-TMB had the worst prognosis, and the prediction power of m6AlncRNAscore was not disturbed by TMB during the first 5 years of follow-up (Fig. 6d).
Clinical and somatic mutation characteristics in the high- and low-m6AlncRNAscore groups. (A) Comparison of m6AlncRNAscore between patients with stage I/II and patients with stage III/VI. (B) Comparison of TMB between the high- and low-m6AlncRNAscore groups. (C) Kaplan–Meier curves of OS in high- and low-TMB groups. (D) Survival analyses for subgroup patients stratified by m6AlncRNAscore and TMB using Kaplan–Meier curves.
Patient’s response to ICIs in different m6AlncRNAscore groups
It has been reported that IPS values could predict the response of patients to ICIs. Whether patients received anti-CTLA-4 (Fig. 7a), anti-PD-L1 (Fig. 7b) or anti-CTLA-4 and anti-PD-L1 combination treatments (Fig. 7c), the IPS values of the high-m6AlncRNAscore group were dramatically higher compared with the low-m6AlncRNAscore group, suggesting that the corresponding ICI therapy responses in the high-m6AlncRNAscore group were significantly better than those of the low-m6AlncRNAscore group. These results indicated that patients with high-m6AlncRNAscore were more likely to benefit from ICIs.
Drug sensitivity prediction in different m6AlncRNAscore groups
Then, we used the GDSC database to predict the valid drugs of high- and low-m6AlncRNAscore groups. Supplementary Fig. 5 showed that the CC patients in the high-m6AlncRNAscore group sensitively responded to 12 drugs (AZD3759, BI-2536, CDK95038, Dasatinib, ERK2440, Erlotinib, Gefitinib, Ibrutinib, NU7441, Osimertinib, Sapitinib, and UMI-77). The therapy responses to 18 drugs (Afuresertib, Axitinib, AZD6482, AZD8055, Dactolisib, GNE-317, GSK269962A, Ipatasertib, Leflunomide, MK-2206, Navitoclax, Nilotinib, OSI-027, Oxaliplatin, Palbociclib, PF-4708671, Ribociclib, and SB505124) in the low-m6AlncRNAscore group were significantly better than those of the high-m6AlncRNAscore group.
Construction of the ceRNA network of the 7 m6A-related prognostic lncRNAs
To further identify the mechanism of the 7 m6A-related prognostic lncRNAs in CC patients, we constructed the lncRNAs-miRNAs-mRNAs ceRNA network. First, we obtained 30 miRNAs by the co-expression method. Then, miRanda, miRDB, miRTarBase and TargetScan software were used to identify 166 mRNA. Furthermore, we constructed and visualized the ceRNA network by incorporating 7 m6A-related prognostic lncRNAs, 30 miRNAs, and 166 mRNA (Supplementary Fig. 6).
Validation of the expression levels of four m6A-related lncRNAs in CC samples
qRT-PCR assay was used to detect the expression of AC024270.4, AC008124.1, AC025176.1 and RPP38-DT in 6 tumor tissues and 6 normal samples. As shown in Supplementary Fig. 7, compared with normal tissues, cervical cancer tissues had higher AC024270.4 and AC008124.1, but lower AC025176.1. There was no difference in AC024270.4 expression between tumor samples and normal samples.
Discussion
Increasing evidence indicates that TME plays an indispensable role in tumor immune escape and immunotherapy efficacy5,6. Therefore, identifying the role and potential regulatory mechanisms of m6A-related lncRNA clustering patterns in survival prediction and immune infiltration will deepen our understanding of tumor immune escape and enrich the effective population for cancer immunotherapy.
Here, we revealed two distinct m6AlncRNA clustering patterns based on 7 m6A-related prognostic lncRNAs. m6AlncRNA clusterA was characterized by low tumor purity and high infiltration level of immune cells, such as activated B cell and activated CD8 T cell, which are key effectors of anti-tumor immunity24,25. Further, m6AlncRNA clusterA were mainly involved in immune activation pathways such as oxidative phosphorylation, cardiac muscle contraction, and arachidonic acid metabolism. Inhibition of oxidative phosphorylation alone limits the proliferation of T cells exposed to persistent antigen and promotes T cell exhaustion by upregulating genes associated with T cell exhaustion26. Therefore, we speculated that the enriched oxidative phosphorylation signaling pathway in m6AlncRNA clusterA might promote the self-renewal of T cells, thereby enhancing antitumor immunity. Mediators released from arachidonic acid metabolic pathway play vital roles in maintaining the immune system normal function27,28. Cardiac muscle contraction pathway has been reported to be associated with autoimmune diseases characterized by abnormally activated immune response29,30. However, m6AlncRNA clusterB was characterized by enrichment of immune evasion and tumorigenic activation pathways such as TGF beta signaling pathway, ubiquitin mediated proteolysis, and pathways in cancer. Existing studies imply that TGF beta signaling inhibits not only the innate immunity but also the adaptive immune system, leading to tumor immune evasion and poor response to ICIs31,32. Thus, TGF beta signaling pathway is a potential tumor therapeutic target worthy of in-depth study33. Ubiquitin mediated proteolysis is involved in multiple biological processes including immune regulation and inflammatory response34. Melanoma patients with high level HECTD2, the E3 ubiquitin ligase involved in ubiquitin mediated proteolysis, had worse antitumor immunity and worse outcome of ICI treatment than those with low level HECTD234. Consistent with TME immune infiltration characterizations, m6AlncRNA clusterA had better clinical outcomes in relative to m6AlncRNA clusterB, which was preferentially related to a higher FIGO stage.
Further, we explored the transcriptome difference between two m6AlncRNA clustering patterns. These DEGs were significantly linked to PD-L1 expression and PD-1 checkpoint pathway and immune-related infection pathways. PD-1/PD-L1 axis negatively regulates T cell activation by inhibiting Ras-Raf-MEK-ERK35. These findings demonstrated again that the m6AlncRNA clustering pattern dissimilarity was associated with tumor immunity difference. Moreover, we classified patients into two different genomic subtypes and two distinct m6AlncRNAscore groups based on the prognostic DEGs. Gene clusterA with poor prognosis was preferentially associated with m6AlncRNA clusterB with poor prognosis. To our surprise, up to 72.9% of patients had m6AlncRNA cluster grouping consistent with m6AlncRNAscore grouping. By integrated analyses, we found that the m6AlncRNAscore was a reliable and independent prognostic protective biomarker for the OS, PFS, and DSS of CC patients. No matter in the overall or in stratified survival analysis, patients with high-m6AlncRNAscore had better prognosis than low-m6AlncRNAscore. Previous studies have reported that high-TMB predicts a better clinical outcome and a higher ICI response rate in some tumors36,37. In our study, patients with high-TMB had longer OS than patients with low TMB, while no significant difference was observed. Although it could not be considered that the long-term prediction ability of m6AlncRNAscore was not affected by TMB due to the small sample size, we could determine that its prediction ability within 5 years was unaffected by TMB. Our findings indicated that m6AlncRNA clustering patterns might affect tumor immune escape by regulating TME, and finally affected the prognosis of patients.
Similar to GSVA results of m6AlncRNA clusters, the high-m6AlncRNAscore group was enriched in immune-inflamed pathways such as oxidative phosphorylation and cardiac muscle contraction, while the low-m6AlncRNAscore group was significantly related to immunosuppressive pathways such as wnt signaling pathway, TGF beta signaling pathway, and MAPK signaling pathway. As discussed earlier in this study, oxidative phosphorylation signaling and cardiac muscle contraction signaling are involved in activated immune response in humans26,38,39. TGF beta signaling31,32,33,34,40, Wnt signaling41, and MAPK signaling42 have been reported to promote tumor immune escape and limit antitumor immune response. Besides, the abundance of activated CD8 T cell and CD56dim natural killer cell was higher in the high-m6AlncRNAscore group. Consistent with the enrichment of immunosuppressive pathways, the fractions of Th2, Treg, IL-10, and TGF-beta1 were higher in patients with low-m6AlncRNAscore. Th2 and Treg are tumor immunosuppressive cells. Researchers have already found that patients with cervical cancer express higher Th2 and Treg than women with normal cervix43. Treg, a major barrier to effective anti-tumor immunotherapy, promotes tumor immune escape by production of immunosuppressiv cytokines such as IL-10 and TGF-beta44,45. TGF-beta in turn promotes the expansion of Treg44,45. It has been reported recently that m6A-related lncRNAs are novel prognostic biomarkers in lung cancer and breast cancer and are associated with TME46,47. Therefore, m6AlncRNAscore dissimilarity was significantly associated with TME difference. These findings could provide novel insights for cancer immunotherapy, that is, targeting m6A-related lncRNAs or m6A-related lncRNA relevant genes to reverse adverse TME, and then developing new immunotherapeutic drugs.
IPS comprehensively represents tumor immunogenicity and has been verified to predict the ICI treatment efficacy19,20,21,22,23. In our study, the high-m6AlncRNAscore group had higher IPS values than the low-m6AlncRNAscore group regardless of ICI therapy regimen, indicating that patients with high-m6AlncRNAscore might be more likely to benefit from ICIs. Additionally, IL-10 and TGF-beta might be potential new targets for anti-tumor immunotherapy in patients with high-m6AlncRNAscore, deserving further study. Furthermore, drug sensitivity analysis results found that the high-m6AlncRNAscore group had different sensitive drugs compared with the high-m6AlncRNAscore group. These findings demonstrated that m6AlncRNA clustering patterns could affect the therapeutic efficacy of ICIs.
Previous research on the 7 m6A-related lncRNAs was few. AC099850.4 was reported to be in relation to the prognosis of ovarian cancer through lncRNA-miRNA-mRNA competing triplets48. In our study, we also constructed the ceRNA network, which could help us better understand the mechanisms of the 7 m6A-related lncRNAs in CC. Consistent with the lower expression levels of AC024270.4 and AC008124.1 in cervical cancer tissues in relative to normal tissues in TCGA database, our qRT-PCR results displayed that AC024270.4 and AC008124.1 had significantly lower levels in tumor samples than normal tissues. The level of AC025176.1 in tumor samples was remarkably higher compared with normal tissues both in our experiment and TCGA analysis results. However, the expression of RPP38-DT in cervical cancer tissues was higher in TCGA database, but lower in our experiment in comparison with normal controls. The expression difference of RPP38-DT may be associated with the small sample size in TCGA-cervical cancer database with only three controls and our experiment with 6 pairs of case–control samples.
One limitation of our study was that our results were not validated in another database. Another limitation was that the full mechanisms of the 7 m6A-related lncRNAs in CC remained unclear. More research is needed in the future.
In summary, we firstly revealed the important regulation role of m6A-related lncRNA clustering patterns on prognosis, TME, and ICI therapeutic efficacy in CC. The difference of m6A-related lncRNA clustering patterns was a vital factor leading to the heterogeneity and complexity of individual TME. The systematic evaluation of m6A-related lncRNA clustering patterns will help improve our understanding of TME immune infiltration characteristics and might provide novel potential approaches for immunotherapy response prediction and patient prognostic stratification in CC.
Data availability
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (http://www.cancer.gov/tcga), TCIA database (https://tcia.at/)", and UCSC Xena database (https://xenabrowser.net/).
References
Sung, H. et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J. Clin. 71(3), 209–249 (2021).
Cao, W., Chen, H. D., Yu, Y. W., Li, N. & Chen, W. Q. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin. Med. J (Engl) 134(7), 783–791 (2021).
Marret, G., Borcoman, E. & Le Tourneau, C. Pembrolizumab for the treatment of cervical cancer. Expert Opin. Biol. Ther. 19(9), 871–877 (2019).
Otter, S. J., Chatterjee, J., Stewart, A. J. & Michael, A. The role of biomarkers for the prediction of response to checkpoint immunotherapy and the rationale for the use of checkpoint immunotherapy in cervical cancer. Clin. Oncol (R Coll Radiol) 31(12), 834–843 (2019).
Arneth, B. Tumor microenvironment. Medicina (Kaunas). 31(12), 834–843 (2019).
Wu, T. & Dai, Y. Tumor microenvironment and therapeutic response. Cancer Lett. 387, 61–68 (2017).
He, L. et al. Functions of N6-methyladenosine and its role in cancer. Mol. Cancer 18(1), 176 (2019).
Zhou, Z. et al. Mechanism of RNA modification N6-methyladenosine in human cancer. Mol. Cancer. 19(1), 104 (2020).
Li, M., Zha, X. & Wang, S. The role of N6-methyladenosine mRNA in the tumor microenvironment. Biochim. Biophys. Acta. Rev. Cancer. 1875(2), 188522 (2021).
Yang, Y., Hsu, P. J., Chen, Y. S. & Yang, Y. G. Dynamic transcriptomic m(6)A decoration: Writers, erasers, readers and functions in RNA metabolism. Cell Res. 28(6), 616–624 (2018).
Chen, X. Y., Zhang, J. & Zhu, J. S. The role of m(6)A RNA methylation in human cancer. Mol. Cancer. 18(1), 103 (2019).
Xu, J. et al. The identification of critical m6A RNA methylation regulators as malignant prognosis factors in prostate adenocarcinoma. Front. Genet. 11, 602485 (2020).
Hangauer, M. J., Vaughn, I. W. & McManus, M. T. Pervasive transcription of the human genome produces thousands of previously unidentified long intergenic noncoding RNAs. PLoS Genet. 9(6), e1003569 (2013).
Yang, G., Lu, X. & Yuan, L. LncRNA: A link between RNA and cancer. Biochim. Biophys. Acta. 1839(11), 1097–1109 (2014).
Li, J., Meng, H., Bai, Y. & Wang, K. Regulation of lncRNA and its role in cancer metastasis. Oncol. Res. 23(5), 205–217 (2016).
Luo, Y. et al. Long Non-coding RNAs: Emerging Roles in the Immunosuppressive Tumor Microenvironment. Front. Oncol. 10, 48 (2020).
Chang, L. et al. Roles of long noncoding RNAs on tumor immune escape by regulating immune cells differentiation and function. Am. J. Cancer Res. 11(6), 2369–2385 (2021).
Kumar, M. M. & Goyal, R. LncRNA as a therapeutic target for angiogenesis. Curr. Top Med. Chem. 17(15), 1750–1757 (2017).
Charoentong, P. et al. Pan-cancer immunogenomic analyses reveal genotype-immunophenotype relationships and predictors of response to checkpoint blockade. Cell Rep. 18(1), 248–262 (2017).
Barbie, D. A. et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature 462(7269), 108–112 (2009).
Sotiriou, C. et al. Gene expression profiling in breast cancer: Understanding the molecular basis of histologic grade to improve prognosis. J. Natl. Cancer Inst. 98(4), 262–272 (2006).
Zeng, D. et al. Tumor microenvironment characterization in gastric cancer identifies prognostic and immunotherapeutically relevant gene signatures. Cancer Immunol. Res. 7(5), 737–750 (2019).
Yi, M. et al. Immune signature-based risk stratification and prediction of immune checkpoint inhibitor’s efficacy for lung adenocarcinoma. Cancer Immunol. Immunother. 70(6), 1705–1719 (2021).
Reina-Campos, M., Scharping, N. E. & Goldrath, A. W. CD8(+) T cell metabolism in infection and cancer. Nat. Rev. Immunol. 21(11), 718–738 (2021).
Tokunaga, R. et al. B cell and B cell-related pathways for novel cancer treatments. Cancer Treat. Rev. 73, 10–19 (2019).
Vardhana, S. A. et al. Impaired mitochondrial oxidative phosphorylation limits the self-renewal of T cells exposed to persistent antigen. Nat. Immunol. 21(9), 1022–1033 (2020).
Hanna, V. S. & Hafez, E. A. A. Synopsis of arachidonic acid metabolism: A review. J. Adv. Res. 11, 23–32 (2018).
Grover, M., Behl, T., Bungau, S. & Aleya, L. Potential therapeutic effect of Chrysopogon zizanioides (Vetiver) as an anti-inflammatory agent. Environ. Sci. Pollut. Res. Int. 28(13), 15597–15606 (2021).
Wu, R. et al. Identification of hub genes in rheumatoid arthritis through an integrated bioinformatics approach. J. Orthop. Surg. Res. 16(1), 458 (2021).
Jiang, X. H. et al. Effect of grilled nux vomica on differential rna expression profile of gastrocnemius muscle and tolllike receptor 4 (TLR-4)/nuclear factor kappa B (NF-kappaB) Signaling in experimental autoimmune myasthenia gravis rats. Med. Sci. Monit. 26, e919150 (2020).
Batlle, E. & Massague, J. Transforming growth factor-beta signaling in immunity and cancer. Immunity 50(4), 924–940 (2019).
Chung, J. Y. et al. TGF-beta signaling: from tissue fibrosis to tumor microenvironment. Int. J. Mol. Sci. 22(14), 7575 (2021).
Yang, Y. et al. The role of TGF-β signaling pathways in cancer and its potential as a therapeutic target. Evid. Based. Complement Alternat. Med. 2021, 6675208 (2021).
Ciechanover, A., Orian, A. & Schwartz, A. L. Ubiquitin-mediated proteolysis: Biological regulation via destruction. BioEssays 22(5), 442–451 (2000).
Ottina, E. et al. E3 ubiquitin ligase HECTD2 mediates melanoma progression and immune evasion. Oncogene 5(230), 46 (2012).
Patsoukis, N. et al. Selective effects of PD-1 on Akt and Ras pathways regulate molecular components of the cell cycle and inhibit T cell proliferation. Sci. Signal. 10, 573141 (2020).
Kang, K., Xie, F., Mao, J., Bai, Y. & Wang, X. Significance of tumor mutation burden in immune infiltration and prognosis in cutaneous melanoma. Front. Oncol. 15(9), 1409–1424 (2020).
Sholl, L. M. et al. The promises and challenges of tumor mutation burden as an immunotherapy biomarker: A perspective from the international association for the study of lung cancer pathology committee. J. Thorac. Oncol. 16(1), 1–9 (2021).
Wu, R. et al. Identification of hub genes in rheumatoid arthritis through an integrated bioinformatics approach. J. Orthop. Surg. Res. 26, e919150-919151 (2020).
Jiang, X. H. et al. Effect of grilled Nux vomica on differential RNA expression profile of gastrocnemius muscle and toll-like receptor 4 (TLR-4)/nuclear factor kappa B (NF-κB) signaling in experimental autoimmune myasthenia gravis rats. Med. Sci. Monit. 40(37), 5567–5578 (2021).
Patel, S., Alam, A., Pant, R. & Chattopadhyay, S. Wnt signaling and its significance within the tumor microenvironment: Novel therapeutic insights. Front. Immunol. 10, 2872 (2019).
Li, P. et al. GC-derived EVs enriched with MicroRNA-675-3p contribute to the MAPK/PD-L1-mediated tumor immune escape by targeting CXXC4. Mol. Ther. Nucleic Acids. 22, 615–626 (2020).
Peghini, B. C. et al. Local cytokine profiles of patients with cervical intraepithelial and invasive neoplasia. Hum. Immunol. 73(9), 920–926 (2012).
Horii, M. & Matsushita, T. Regulatory B cells and T cell regulation in cancer. J. Mol. Biol. 433(1), 166685 (2021).
Paluskievicz, C. M. et al. T regulatory cells and priming the suppressive tumor microenvironment. Front. Immunol. 10, 2453 (2019).
Zhao, J., Lin, X., Zhuang, J. & He, F. Relationships of N6-Methyladenosine-related long non-coding RNAs with tumor immune microenvironment and clinical prognosis in lung adenocarcinoma. Front. Genet. 12, 714697 (2021).
Zhong, X. et al. Identification of N6-Methyladenosine-related LncRNAs for predicting overall survival and clustering of a potentially novel molecular subtype of breast cancer. Front. Oncol. 11, 742944 (2021).
Zhao, J. et al. Identification of potential prognostic competing triplets in high-grade serous ovarian cancer. Front. Genet. 11, 607722 (2021).
Acknowledgements
This work was supported by grants from the Shanxi Provincial Natural Science Foundation of China (No: 20210302124420).
Author information
Authors and Affiliations
Contributions
H.X.J. designed and drafted the manuscript. H.X.J. and J.H.W. collected and analyzed the data. H.X.J. collected samples and performed experiments. J.T.W., M.T.C., and S.H.H. revised the manuscript. All authors contributed to the article and approved the submitted version.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jia, H., Cao, M., Hao, S. et al. Prediction of prognosis, immune infiltration and immunotherapy response with N6-methyladenosine-related lncRNA clustering patterns in cervical cancer. Sci Rep 12, 17256 (2022). https://doi.org/10.1038/s41598-022-20162-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-022-20162-2
- Springer Nature Limited
This article is cited by
-
Identification of N6-Methyladenosine-Associated lncRNAs and Analysis of Prognostic Signature in Breast Cancer
Biochemical Genetics (2024)