Abstract
Recent therapeutic advances in breast cancer (BC) have improved survival outcomes; however, the prognosis for patients with bone metastasis (BM) remains poor. Hence, novel clinical biomarkers are needed to accurately predict BC BM as well as to promote personalized medicine. Here, we discovered a novel biomarker, TOR1B, for BM in BC patients via analysis of BC gene expression data and clinical information downloaded from open public databases. In cancer cells, we found high expression levels of TOR1B in the nucleus and endoplasmic reticulum. Regarding gene expression, the level of TOR1B was significantly upregulated in BC patients with BM (p < 0.05), and the result was externally validated. In addition, gene expression clearly demonstrated two distinct types of prognoses in ER- and PR-positive patients. In multivariate regression, the gene could be an independent predictor of BM in BC patients, i.e., a low expression level of TOR1B was associated with delayed metastasis to bone in BC patients (HR, 0.28; 95% CI 0.094–0.84). Conclusively, TOR1B might be a useful biomarker for predicting BM; specifically, patients with ER- and PR-positive subtypes would benefit from the clinical use of this promising prognostic biomarker.
Similar content being viewed by others
Introduction
Bone metastasis (BM) is the most devastating condition in patients with advanced breast cancer (BC), occurring in approximately 70% of patients, predominantly those with luminal A or estrogen/progesterone receptor‐positive (ER + /PR +) subtypes1,2,3. It seriously affects quality of life and survival outcomes through skeletal-related events (SREs), including bone pain, tumor-induced fracture, spinal cord compression, hypercalcemia and other complications4,5. The overall five-year survival rate for patients with BM is 22.8%, whereas for BC patients without BM, it is greater than 95%6,7. Although recent therapeutic advances in BC, such as targeted drugs and immunotherapy, have improved survival8, it is still unknown why some patients develop BM while others do not, even if they possess the disease phenomena. In most cases, metastatic lesions enter into a latent state that can last months, years, or even decades before forming a clinically detectable macrometastasis1,9. Numerous molecular markers may critically regulate the tumor microenvironment during the latency period, and the detection of tumors that are more prone to metastasis, particularly in bone, is vital1,10,11. Therefore, early detection of biomarkers may be crucial for the implementation of targeted therapeutic measures, eventually reducing the risk of skeletal complications and improving survival and quality of life.
A number of serum and tissue biomarkers have been extensively studied for the early diagnosis or prognosis of BM in BC patients. Integrin alpha 5 (ITGA5) is a mediator of breast-to-bone metastasis and a therapeutic target for treating osteolytic lesions in BC patients12. However, clinical trials have repeatedly failed to demonstrate the therapeutic benefits of integrin inhibitors in cancer patients13. Serum markers of bone formation (N-terminal propeptide of type-1 collagen [P1NP]) and bone resorption [C-telopeptide of type-1 collagen (CTX) and pyridinoline cross-linked carboxy-terminal telopeptide of type-1 collagen (1-CTP)] seem to have good prognostic value for bone-specific recurrence. Nevertheless, those markers have several limitations, such as poor sensitivity, small event numbers and lack of independent predictive ability9. Dedicator of cytokinesis protein 4 (DOCK4) is a promising prognostic biomarker for the early prediction of skeletal recurrence in BC. However, the DOCK4 expression level does not significantly influence overall survival in BC patients14. Other biomarkers, such as ICAM-1, cadherin-11, osteoactivin, bone sialoprotein, CCN3, IL-11, CCL2, CITED2, CXCR4, CTGF, OPN, CX3CR1, TWIST1, adrenomedullin and Enpp1, have also been identified as potential prognostic markers in BC with BM patients7,15,16. However, these biomarkers cannot be rationally used in clinical practice17. Therefore, additional novel useful biomarkers need to be identified and validated for the early prediction of BM in BC patients.
A previous study revealed that the Torsin/cofactor system has a functional relationship with nuclear envelope/nuclear pore complex (NPC) biogenesis18. Rodriguez-Bravo et al. molecularly established that NPC deregulation directly contributes to the progression of lethal prostate cancer and suggested a novel therapeutic target for prostate cancer19. A recent study characterized transcriptomic complexity with a personalized genomics approach in prostate cancer and identified TOR1A as the master regulator20. TOR1B and TOR1A are colocalized in the cell compartments retaining the same protein complex, demonstrating similar related functions21. A recent study discovered a higher level of TOR1B autoantigen in patients with pancreatic diseases, including pancreatic cancer22. However, the role of TOR1B in BC has not yet been determined. To investigate the relationship between TOR1B and BC BM, the present study aimed to identify the location of the protein in the cell and determine whether TOR1B has any prognostic value in BC BM, particularly in ER + BC, which is the most prone to developing BM.
Results
Subcellular localization and expression of TOR1B
The subcellular localization of TOR1B was predicted using the COMPARTMENTS database by considering multiple information sources, i.e., database annotations, automatic text mining of the biomedical literature and sequence-based predictions regarding different cell types. The TOR1B protein product was identified with the highest confidence (confidence level 5) in the nucleus and endoplasmic reticulum and moderate confidence (confidence level 4) in the extracellular space (p value). The lowest confidence (confidence level 1) was in the cytosol, cytoskeleton, mitochondrion, peroxisome, endosome, lysosome and Golgi apparatus (Fig. 1A). TOR1B protein expression was projected using subcellular Atlas from the Human Protein Atlas database, and it was highly expressed in the nucleus and endoplasmic reticulum (Fig. 1B).
The localization of TOR1B at the cellular level. Subcellular locations from COMPARTMENTS (A) and the Protein Atlas (B). The confidence of each association is signified by stars, where ★★★★★ is the highest confidence and ★☆☆☆☆ is the lowest. All Image data shown were generated and downloaded with searching the queries from COMPARTMENTS (compartments.jensenlab.org) or Human Protein Atlas (www.proteinatlas.org, Protein Atlas version 22.0).
TOR1B expression was upregulated in BC patients with BM
The mRNA expression of TOR1B was evaluated between metastatic and nonmetastatic BC patients in the E-MTAB-365 and GSE2034 datasets. The upregulation of TOR1B was highly significant in patients with BM (p = 8e-04 and p = 4.5e-05, metastatic vs. nonmetastatic BC, E-MTAB-365 and GSE2034 datasets, respectively) (Fig. 2). These results indicate that higher TOR1B expression at the transcriptional level may be associated with BM in BC patients.
TOR1B expression and its correlation with clinicopathological features of BC
This study included 425 patients with BC from the E-MTAB-365 dataset and 286 patients from the GSE2034 dataset. The association of TOR1B expression with clinical features is shown in Table 1. Based on the median expression of TOR1B, patients in the E-MTAB-365 dataset were classified into groups: high expression (n = 212) and low expression (n = 213). The chi-square test was performed to study the association of clinicopathological features of BC patients with TOR1B expression (high and low). The results showed that the age and tumor grade of BC patients were not significantly correlated with TOR1B expression (p > 0.05). Regarding hormone receptor status, the presence or absence of HER2 was significantly associated with TOR1B expression (p = 0.008). Furthermore, TP53 mutation was highly correlated with TOR1B upregulation (p = 0.007). Remarkably, the CIT classification, TNM stage and BM of BC patients were significantly associated with TOR1B expression (p = 5.00E-04, p = 0.023 and p = 0.001, respectively). The clinicopathological characteristics of the BC patients are presented in Table 1.
BM predictive ability of TOR1B in BC patients
The sensitivity and specificity of TOR1B in predicting BM were evaluated from the area under the curve (AUC) of receiver operating characteristic (ROC) curve analysis. The AUC of TOR1B showed significant predictive capability for BM in BC patients in both the E-MTAB-365 and GSE2034 datasets (AUC = 0.627, p < 0.001 and AUC = 0.663, p < 0.001, respectively) (Fig. 3).
High expression level of TOR1B was associated with poor outcomes in BM
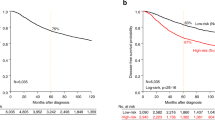
To study the prognosis of BC patients with respect to TOR1B upregulation, we generated a Kaplan‒Meier plot, and the results showed that patients with upregulated TOR1B expression underwent earlier BM than patients with low TOR1B expression (Fig. 4). We further studied TOR1B expression in patients with BM using various clinicopathological features, i.e., age, presence or absence of hormone receptors, etc., to identify which patients were the most prone to develop earlier metastasis. The results demonstrated that TOR1B upregulation was significantly associated with earlier BM in ER + (E-MTAB-365, p = 0.0159 and GSE2034, p = 0.00182) and PR + (E-MTAB-365, p = 0.00742) BC, and patients who were younger than 55 years were more vulnerable (E-MTAB-365, p = 0.0182) to early metastasis (Fig. 5). Furthermore, to identify the prognostic predictor in BC patients, we performed a multivariate Cox proportional hazard regression analysis of TOR1B with other clinical information. The results revealed that TOR1B was an independent predictor of BM in BC patients (p = 0.024) (Fig. 6).
Significant association of TOR1B expression molecular markers and clinical information. Kaplan‒Meier curve based on the expression of TOR1B in ER-positive breast cancer patients from the E-MTAB-356 (A) and GSE2034 (B) datasets, PR-positive breast cancer patients (C) and patients aged more than 55 years (D) from the E-MTAB-356 dataset. Middle-aged groups of patients. The p value was calculated by log-rank tests.
TOR1B interaction network analysis
To study the interaction network of TOR1B with other proteins and genes, we performed a molecular network analysis by introducing TOR1B into the GeneMANIA and STRING databases. TOR1B was closely associated with other proteins, such as NEDD8, TOR1A1P2, DYNLT1, TOR1A, TNFRSF25, PRPF4B, WDR11, STON2, TNFRSF10A and COPS4 (Fig. 7). In addition, among the various Homo sapiens genes, TOR1B was related to the TOR1A, TOR1A1P2, TOR1A1P1, TOR2A, TOR3A, TOR4A, BUB1, GIMAP2, GAPVD1, LIPC, GPR107, FBXO6, FUBP3, UBE201, LAGE3, EN2, FAM32A, SPTLO1, ERP44 and TOPORS genes (Fig. 3).
The interaction networks of TOR1B with other genes and proteins. (A) Interaction networks of TOR1B with other genes were generated using GeneMANIA. (B) Interaction networks of TOR1B with other genes were generated using STRING. All Image data shown were generated and downloaded with searching the queries from GeneMANIA (genemania.org) or STRING (string-db.org).
Discussion
Bone is the most frequent and common site for metastatic growth in BC patients. Once cancer cells have metastasized to the bone, they may have devastating impacts on cancer patients. There are several treatment approaches, such as hormone therapy, chemotherapy and bisphosphonate administration, while systemic therapies, including surgery, external radiation therapy and bone-targeted therapies, are used to treat BM23; however, a curative drug or treatment method has not yet been developed. Currently, multiple biomarkers have been identified for the prognostication of BM in BC patients14,24,25, which has improved therapeutic management; however, there is an unmet need to explore novel prognostic and predictive biomarkers. In this study, we identified and demonstrated TOR1B as a potential prognostic and predictive biomarker for BM in BC patients. TOR1B is an ATPase found primarily in the ER and nuclear envelope, and the protein product of the gene may act as a molecular chaperone assisting in the proper folding of secreted and/or membrane proteins and maintaining membrane integrity (https://www.proteinatlas.org/ENSG00000136816-TOR1B)26. Aggressive tumor microenvironments generated from diverse oncogenic, transcriptional and metabolic abnormalities disrupt ER homeostasis, leading to the aberrant activation of ER stress sensors, and the pathways of their downstream signaling may act as key drivers of tumor growth and metastasis, particularly failure to respond to therapies such as chemotherapy and targeted immunotherapy26. Therefore, enhancement of ER integrity and homeostasis by modulating TOR1B may provide novel therapeutic insight into metastatic BC patients. The protein is moderately available in the cytosol, which is also reflected in our study, and is highly similar to the neighboring gene TOR1A, which impairs the TGFβ signaling pathway in dystonia27. There are different variants or splices available for TOR1B that may have diverse functions, i.e., mutation of the protein (S38L variant) has a direct impact on lung squamous cell carcinoma (https://activedriverdb.org/mutation/show/NM_014506/38/L). Surprisingly, only a few functions of the protein have been discovered in which the crucial metabolic and immune functions are completely unknown yet (https://www.proteinatlas.org/ENSG00000136816-TOR1B/immune+cell).
Currently established BC prognostic markers, such as lymph node metastasis, primary tumor size, grade, etc., have several limitations13,14,28. Therefore, many women are overtreated and suffer severe side effects, as only 40% of patients develop recurrence and ultimately die due to metastasis, while 80% of them receive adjuvant chemotherapy28. The gene expression of primary and metastatic tumors has shown a similar pattern29,30, indicating that the differentially expressed genes between highly metastatic cells and their parental cells may only represent alterations caused by microenvironmental stimuli rather than their own gene expression pattern. Therefore, changes in differentially expressed genes in primary tumors are suggested as a powerful predictor that more accurately predicts the metastatic risk for individual BC patients24,28,31,32,33. In our previous study, we demonstrated 70 genes (92 probes) in late recurrence in primary BC patients, particularly those with lymph node-positive disease33. In the present study, we found overexpression of a single TOR1B gene in BC patients with BM. We robustly validated it in an external dataset. Based on high and low expression levels, the gene efficiently separated two subtypes of patients, those who developed BM and those who did not. Indeed, BC patients with high expression levels of TOR1B had early metastasis to bone compared to those with low expression levels of the gene. This finding is consistent with the previous study of Wang et al., who showed that patients with higher expression levels of the SRXN1 and TOR1B genes were not suitable for radiotherapy (https://doi.org/10.21203/rs.3.rs-1935488/v1). In fact, the gene can be used to predict a worse prognosis for BM in ER- and PR-positive BC patients, which is the distinct finding of our study, as TOR1B expression is only observed in overall BC without subtype specificities (https://www.proteinatlas.org/ ENSG00000136816-TOR1B/immune + cell). In addition, patients younger than 55 years of age were more prone to develop metastasis. This is also supported by the finding that increasing age at diagnosis is inversely associated with distant metastasis to bone and viscera34. The sensitivity and specificity of the prognostic marker showed satisfactory predictive capabilities (AUC 0.627 and 0.663) for BM in BC patients. This finding supports the meta-analysis of Zhao et al.by showing similar predictive ability in different studies of BC diagnostic accuracy35.
In both gene and protein interaction networks, the TOR1B gene has shown very close and strong associations with TOR1A, which impairs TGFβ signaling and generates multiorgan inflammation36,37. In humans, TGF-β acts as a metabolic driver in the tumor microenvironment to promote bone and lung metastasis via different mechanisms38,39,40. Therefore, exploration of the effect of TOR1B on TGF-β or other functional proteins might provide new therapeutic insight into BC metastasis.
In conclusion, we discovered TOR1B as a promising biomarker for predicting BM in BC patients, implying its prognostic value in the era of personalized medicine. We suggest that TOR1B might be used as a molecular diagnostic tool, particularly in ER + /PR + BC patients who are most prone to develop BM. Thus, we believe that TOR1B might be a helpful tool for orthopedic oncologists.
Methods
Subcellular localization of TOR1B
Subcellular localization of TOR1B was predicted and visualized by using the COMPARTMENTS database (http://compartments.jensenlab.org)41. All data were searched and filtered by selecting gene types from Homo sapiens, and the genes were further collected from different databases using unique gene symbols.
Protein expression of TOR1B by immunohistochemistry
Protein expression levels of TOR1B were identified using the subcellular Atlas from the Human Protein Atlas database (https://www.proteinatlas.org/)42. Gene symbols were used to search for information from the database. Immunohistochemistry (IHC) staining of TOR1B in the endoplasmic reticulum and nucleus was assessed in the subcellular Atlas.
Gene expression profiles, clinical data and analysis
Gene expression profiles and clinical information of BC patients were downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/) and ArrayExpress (https://www.ebi.ac.uk/arrayexpress/). The robust multiarray average algorithm (RMA) was used to normalize the raw data to the median via Cluster 3.0. The GSE2034 and E-MTAB-365 datasets comprised 286 and 425 patients, respectively, and were taken as the training and validation cohorts. The probe set identifiers were transformed into gene symbols. The gene expression levels were normalized to the median center and were classified into two expression groups. Patients with TOR1B expression levels higher than the median were considered the high expression group, and patients with TOR1B expression levels less than or equal to the median were the low expression group. These groups were used for further analyses.
TOR1B expression and its correlation with clinical information
A total of 711 patients were selected from the GSE2034 and E-MTAB-365 datasets to observe the mRNA expression levels of TOR1B among BM BC patients. Based on mRNA expression levels, the Kaplan‒Meier method was used to compare the survival between BC patients who developed BM and those who did not develop BM in total as well as those with ER- and PR-positive BC. Chi-square and log-rank tests were used to assess patient survival and metastasis.
TOR1B gene‒gene and protein‒protein interaction network analysis
Interactions between the TOR1B gene and other genes were analyzed using the GeneMANIA database (https://genemania.org/)43. The gene interaction networks were built based on a functional association of genes present in the datasets, including protein and genetic interactions, signaling pathways, coexpression, colocalization and protein domain similarities. The analytical parameters were set according to the default indicators. Protein‒protein interaction (PPI) network analysis was performed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (http://www.string-db.org/)44. The PPI network of TOR1B was investigated in STRING’s Homo sapiens database. TOR1B protein networks were established based on the following eight criteria: experimental evidence and curated databases, gene neighborhood, gene fusion, gene co-occurrence, text mining, co-expression and protein homology.
Statistical analysis
Kaplan‒Meier curves, chi-square tests and log-rank tests were performed using the R programming language (www.r-project.org). The Wilcoxon signed-rank test was used to compare the clusters between the two groups. The predictive capability of the biomarker was evaluated by multivariate analysis. The sensitivity and specificity of the prognostic biomarker were assessed by AUC. p < 0.05 was considered to be statistically significant.
Data availability
All the data analyzed in this study are publicly available and can be downloaded from the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=gse2034 for the dataset GSE2034) and ArrayExpress (www.ebi.ac.uk/biostudies/arrayexpress/studies/E-MTAB-365 for the dataset E-MTAB-365).
References
Salvador, F., Llorente, A. & Gomis, R. R. From latency to overt bone metastasis in breast cancer: Potential for treatment and prevention. J. Pathol. 249, 6–18. https://doi.org/10.1002/path.5292 (2019).
Monteran, L. et al. Bone metastasis is associated with acquisition of mesenchymal phenotype and immune suppression in a model of spontaneous breast cancer metastasis. Sci. Rep. 10, 13838. https://doi.org/10.1038/s41598-020-70788-3 (2020).
Schroder, J. et al. Treatment and pattern of bone metastases in 1094 patients with advanced breast cancer: Results from the prospective German tumour registry breast cancer cohort study. Eur. J. Cancer 79, 139–148. https://doi.org/10.1016/j.ejca.2017.03.031 (2017).
Martin, M. et al. Bone-related complications and quality of life in advanced breast cancer: Results from a randomized phase III trial of denosumab versus zoledronic acid. Clin. Cancer Res. 18, 4841–4849. https://doi.org/10.1158/1078-0432.CCR-11-3310 (2012).
Fornetti, J., Welm, A. L. & Stewart, S. A. Understanding the bone in cancer metastasis. J. Bone Miner Res. 33, 2099–2113. https://doi.org/10.1002/jbmr.3618 (2018).
Song, X., Wei, C. & Li, X. The signaling pathways associated with breast cancer bone metastasis. Front. Oncol. 12, 855609. https://doi.org/10.3389/fonc.2022.855609 (2022).
Awolaran, O., Brooks, S. A. & Lavender, V. Breast cancer osteomimicry and its role in bone specific metastasis; An integrative, systematic review of preclinical evidence. Breast 30, 156–171. https://doi.org/10.1016/j.breast.2016.09.017 (2016).
Duan, Z. & Luo, Y. Targeting macrophages in cancer immunotherapy. Signal Transduct. Target Ther. 6, 127. https://doi.org/10.1038/s41392-021-00506-6 (2021).
Brown, J. et al. Associations between serum bone biomarkers in early breast cancer and development of bone metastasis: Results from the AZURE (BIG01/04) trial. J. Natl. Cancer Inst. 110, 871–879. https://doi.org/10.1093/jnci/djx280 (2018).
Roato, I. & Ferracini, R. Cancer stem cells, bone and tumor microenvironment: Key players in bone metastases. Cancers (Basel). https://doi.org/10.3390/cancers10020056 (2018).
Ren, Q. et al. Gene expression predicts dormant metastatic breast cancer cell phenotype. Breast Cancer Res. 24, 10. https://doi.org/10.1186/s13058-022-01503-5 (2022).
Pantano, F. et al. Integrin alpha5 in human breast cancer is a mediator of bone metastasis and a therapeutic target for the treatment of osteolytic lesions. Oncogene 40, 1284–1299. https://doi.org/10.1038/s41388-020-01603-6 (2021).
Hamidi, H. & Ivaska, J. Every step of the way: Integrins in cancer progression and metastasis. Nat. Rev. Cancer 18, 533–548. https://doi.org/10.1038/s41568-018-0038-z (2018).
Westbrook, J. A. et al. Identification and validation of DOCK4 as a potential biomarker for risk of bone metastasis development in patients with early breast cancer. J. Pathol. 247, 381–391. https://doi.org/10.1002/path.5197 (2019).
Li, X. Q., Zhang, R., Lu, H., Yue, X. M. & Huang, Y. F. extracellular vesicle-packaged CDH11 and ITGA5 induce the premetastatic niche for bone colonization of breast cancer cells. Cancer Res. 82, 1560–1574. https://doi.org/10.1158/0008-5472.CAN-21-1331 (2022).
Huang, Y. H. et al. Expression pattern and prognostic impact of glycoprotein non-metastatic B (GPNMB) in triple-negative breast cancer. Sci. Rep. 11, 12171. https://doi.org/10.1038/s41598-021-91588-3 (2021).
Boshuizen, J. & Peeper, D. S. Rational cancer treatment combinations: An urgent clinical need. Mol. Cell 78, 1002–1018. https://doi.org/10.1016/j.molcel.2020.05.031 (2020).
Laudermilch, E. et al. Dissecting torsin/cofactor function at the nuclear envelope: A genetic study. Mol. Biol. Cell 27, 3964–3971. https://doi.org/10.1091/mbc.E16-07-0511 (2016).
Rodriguez-Bravo, V. et al. Nuclear pores promote lethal prostate cancer by increasing POM121-driven E2F1, MYC and AR nuclear import. Cell 174, 1200-1215 e1220. https://doi.org/10.1016/j.cell.2018.07.015 (2018).
Iacobas, S. & Iacobas, D. A. A personalized genomics approach of the prostate cancer. Cells https://doi.org/10.3390/cells10071644 (2021).
Hewett, J. W. et al. TorsinB–perinuclear location and association with torsinA. J. Neurochem. 89, 1186–1194. https://doi.org/10.1111/j.1471-4159.2004.02404.x (2004).
Ghassem-Zadeh, S. et al. Novel autoantibody signatures in sera of patients with pancreatic cancer, chronic pancreatitis and autoimmune pancreatitis: a protein microarray profiling approach. Int. J. Mol. Sci. https://doi.org/10.3390/ijms21072403 (2020).
Choi, J. Y. Treatment of bone metastasis with bone-targeting radiopharmaceuticals. Nucl. Med. Mol. Imaging 52, 200–207. https://doi.org/10.1007/s13139-017-0509-2 (2018).
Savci-Heijink, C. D., Halfwerk, H., Koster, J. & van de Vijver, M. J. A novel gene expression signature for bone metastasis in breast carcinomas. Breast Cancer Res. Treat 156, 249–259. https://doi.org/10.1007/s10549-016-3741-z (2016).
Westbrook, J. A. et al. CAPG and GIPC1: Breast cancer biomarkers for bone metastasis development and treatment. J. Natl. Cancer Inst. https://doi.org/10.1093/jnci/djv360 (2016).
Chen, X. & Cubillos-Ruiz, J. R. Endoplasmic reticulum stress signals in the tumour and its microenvironment. Nat. Rev. Cancer 21, 71–88. https://doi.org/10.1038/s41568-020-00312-2 (2021).
Koh, Y. H., Rehfeld, K. & Ganetzky, B. A Drosophila model of early onset torsion dystonia suggests impairment in TGF-beta signaling. Hum. Mol. Genet. 13, 2019–2030. https://doi.org/10.1093/hmg/ddh208 (2004).
Weigelt, B., Peterse, J. L. & Van’t Veer, L. J. Breast cancer metastasis: Markers and models. Nat. Rev. Cancer 5, 591–602. https://doi.org/10.1038/nrc1670 (2005).
Weigelt, B. et al. Gene expression profiles of primary breast tumors maintained in distant metastases. Proc. Natl. Acad. Sci. U. S. A. 100, 15901–15905. https://doi.org/10.1073/pnas.2634067100 (2003).
Ramaswamy, S., Ross, K. N., Lander, E. S. & Golub, T. R. A molecular signature of metastasis in primary solid tumors. Nat Genet 33, 49–54. https://doi.org/10.1038/ng1060 (2003).
Kang, Y. et al. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell 3, 537–549 (2003).
Priedigkeit, N. et al. Exome-capture RNA sequencing of decade-old breast cancers and matched decalcified bone metastases. JCI Insight. https://doi.org/10.1172/jci.insight.95703 (2017).
Akter, S. et al. Prognostic value of a 92-probe signature in breast cancer. Oncotarget 6, 15662–15680. https://doi.org/10.18632/oncotarget.3525 (2015).
Purushotham, A. et al. Age at diagnosis and distant metastasis in breast cancer–a surprising inverse relationship. Eur. J. Cancer 50, 1697–1705. https://doi.org/10.1016/j.ejca.2014.04.002 (2014).
Zhao, F. et al. Comparing biomarkers for predicting pathological responses to neoadjuvant therapy in HER2-positive breast cancer: A systematic review and meta-analysis. Front. Oncol. 11, 731148. https://doi.org/10.3389/fonc.2021.731148 (2021).
Kulkarni, A. B. et al. Transforming growth factor beta 1 null mutation in mice causes excessive inflammatory response and early death. Proc. Natl. Acad. Sci. U.S.A. 90, 770–774. https://doi.org/10.1073/pnas.90.2.770 (1993).
Shull, M. M. et al. Targeted disruption of the mouse transforming growth factor-beta 1 gene results in multifocal inflammatory disease. Nature 359, 693–699. https://doi.org/10.1038/359693a0 (1992).
Drabsch, Y. & ten Dijke, P. TGF-beta signaling in breast cancer cell invasion and bone metastasis. J. Mammary Gland Biol. Neoplasia 16, 97–108. https://doi.org/10.1007/s10911-011-9217-1 (2011).
Zhang, L. et al. EZH2 engages TGFbeta signaling to promote breast cancer bone metastasis via integrin beta1-FAK activation. Nat. Commun. 13, 2543. https://doi.org/10.1038/s41467-022-30105-0 (2022).
Chiou, W. C. et al. alpha-viniferin and epsilon-viniferin Inhibited TGF-beta1-Induced epithelial-mesenchymal transition, migration and invasion in lung cancer cells through downregulation of vimentin expression. Nutrients https://doi.org/10.3390/nu14112294 (2022).
Binder, J. X. et al. COMPARTMENTS: Unification and visualization of protein subcellular localization evidence. Database 1–9, 2014. https://doi.org/10.1093/database/bau012 (2014).
Uhlén, M. et al. Tissue-based map of the human proteome. Science 347, 12604191–12604199. https://doi.org/10.1126/science.1260419 (2015).
Warde-Farley, D. et al. The GeneMANIA prediction server: Biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 38, W214-220. https://doi.org/10.1093/nar/gkq537 (2010).
Hoffmann, R. & Valencia, A. A gene network for navigating the literature. Nat. Genet. 36, 664. https://doi.org/10.1038/ng0704-664 (2004).
Acknowledgements
This study was supported by National Research Foundation of Korea (NRF) grants funded by the Korean government (MEST) (Grant No: NRF-2018R1A6A1A03025124 and NRF-2020H1D3A1A04080389 to S.S.K. and NRF-2020R1I1A1A01069013 to T.G.C.
Author information
Authors and Affiliations
Contributions
M.N.N. and S.Ak. conceived and designed the study; M.N.N., S.Ak. and T.G.C. analyzed and interpreted the data; H.A., S.An., S.H. and Y.S., were involved in the relevant literature search and primary draft writing; H.A., M.N.N., S.Ak. and T.G.C. wrote and improved the manuscript. S.S.K., T.G.C., I.K., J.H., M.N.N. and S.Ak. reviewed and edited the manuscript. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nguyen, M.N., Akter, S., Akhter, H. et al. TOR1B: a predictor of bone metastasis in breast cancer patients. Sci Rep 13, 1495 (2023). https://doi.org/10.1038/s41598-023-28140-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-023-28140-y
- Springer Nature Limited
This article is cited by
-
YTHDF2 governs muscle size through a targeted modulation of proteostasis
Nature Communications (2024)
-
Prognostic implications of TOR1B expression across cancer types: a focus on basal-like breast cancer and cellular adaptations to hypoxia
Journal of Cancer Research and Clinical Oncology (2024)