Abstract
To investigate the effects of temperature on Brassica napus (canola) resistance to Leptosphaeria maculans (LM), the causal agent of blackleg disease, metabolic profiles of LM infected resistant (R) and susceptible (S) canola cultivars at 21 °C and 28 °C were analyzed. Metabolites were detected in cotyledons of R and S plants at 48- and 120-h post-inoculation with LM using UPLC-QTOF/MS. The mock-inoculated plants were used as controls. Some of the resistance-related specific pathways, including lipid metabolism, amino acid metabolism, carbohydrate metabolism, and aminoacyl-tRNA biosynthesis, were down-regulated in S plants but up-regulated in R plants at 21 °C. However, some of these pathways were down-regulated in R plants at 28 °C. Amino acid metabolism, lipid metabolism, alkaloid biosynthesis, phenylpropanoid biosynthesis, and flavonoid biosynthesis were the pathways linked to combined heat and pathogen stresses. By using network analysis and enrichment analysis, these pathways were identified as important. The pathways of carotenoid biosynthesis, pyrimidine metabolism, and lysine biosynthesis were identified as unique mechanisms related to heat stress and may be associated with the breakdown of resistance against the pathogen. The increased susceptibility of R plants at 28 °C resulted in the down-regulation of signal transduction pathway components and compromised signaling, particularly during the later stages of infection. Deactivating LM-specific signaling networks in R plants under heat stress may result in compatible responses and deduction in signaling metabolites, highlighting global warming challenges in crop disease control.
Similar content being viewed by others
Introduction
Blackleg or phoma stem canker is a significant disease of canola (Brassica napus) and many other cruciferous species worldwide. Blackleg epidemics frequently occur in canola crops across Europe, Canada, and Australia1,2. Leptosphaeria maculans (Desm.) Ces. & De Not. (LM) and L. biglobosa are two closely related species that can cause blackleg disease on canola, previously addressed as aggressive (Tox+, A-group) and non-aggressive (less aggressive, Tox0, B-group), respectively1,3,4,5. The two species exist in most canola-growing regions1,4. In Iran, few studies have been carried out on canola blackleg. The blackleg disease occurs typically in canola-growing areas with humidity sufficient for disease development. Blackleg exists in 30 regions and 10 provinces in Iran3. Like Poland, China, and Lithuania, L. biglobosa is the dominant species and causes minor damage across canola growing regions in Iran3.
The hemibiotrophic fungus LM produces ascospores acting as primary inoculum while conidia produced on infected leaves act as secondary inoculum. Hyphae from germinated ascospores or conidia enter the plant through the stomata or wounds5. The fungus grows between the cells and into the stem after the primary infection of the leaf. The intercellular growth is systemic, biotrophic, and visually symptomless6. Finally, the fungus invades the stem cortical tissues in the necrotrophic phase, where the blackened cankers are developed after killing the invaded tissues. The blackleg disease can also spread by infected seeds when the fungus infects the seed pods of B. napus, but this is far less common4.
The most cost-effective and reliable management approach for blackleg is the use of resistant cultivars7. Both qualitative (monogenic) and quantitative (polygenic) resistances against LM in Brassica species have been reported. Quantitative resistance is conferred by several genes, associated with genomic regions or QTL (quantitative trait loci) and expressed in the adult stage8; however, qualitative resistance is mediated by single resistance genes and is effective in protecting plants during the leaf infection stage7. Twenty-two major genes for resistance to LM have been identified in Brassica species (B. rapa, B. nigra, B. oleracea, B. napus, and B. juncea lines), among all only five genes (LepR3, Rlm2, Rlm4, Rlm7, and Rlm9) have been cloned, all of which encode membrane-bound cell surface–localized receptor proteins used to detect apoplastic elicitors9.
The effects of temperature on host plant resistance responses have been reported in several pathosystems8. Formerly, it has been reported that temperature affects effectiveness of both quantitative and R gene-mediated qualitative resistances against LM6,8,9,10,11. The temperature sensitivity for solanaceous resistance is attributable to the NLR sensor and not to its downstream signaling components in potatoes, which is not yet known in other plants, including Brassicaceae12. Identifying thermostable resistance is essential for understanding the mechanisms of resistance and for better managing the disease under elevated temperatures11,13. Rlm1 and Rlm6-mediated resistance to LM are temperature-sensitive6,14. When cotyledons of canola cv. Quinta were inoculated with LM isolate A2; the plant response was resistant at 18 °C but susceptible at 27 °C14. The interaction between Quinta and the isolate A2 was characterized as a specific Rlm1/AvrLm1 interaction15,16. Results of Huang et al. (2006) under controlled environment experiments indicated that Rlm6-mediated resistance in DarmorMX (carrying Rlm6) against LM was not effective at 25 °C6. Investigations of effects of temperature on effectiveness of R gene-mediated resistance (e.g. Rlm4) in different genetic backgrounds showed that combining R gene with quantitative resistance can maintain stability of resistance at elevated temperature10. Results of determining temperature sensitivity and stability of R gene-mediated resistance at 28 °C by using four cultivars (Topas-Rlm1, Topas-Rlm2, Topas-Rlm3, and Topas-Rlm4) under four temperature regimes (15/10, 20/15, 25/20, and 30/25 °C) showed that lesion size is highly dependent on temperature; however, the influence of temperature on R-mediated resistance appears to depend on the specific cultivar harboring the R gene17.
Different 'omics' have been utilized to study how plants react to biotic or abiotic stress conditions. These tools have provided unique opportunities to examine the study of genomics and transcriptomics in the host and the pathogen18. However, metabolomics, defined as the comprehensive analysis of metabolites in a biological specimen, and proteomics, compared to the other two branches, are still lagging. Despite the vast diversity of existing metabolites, few of them have been found in host–pathogen interactions with the Brassicaceae family18,19. Most studies have focused on specific target groups of metabolites, such as glycosinolates, phenylpropanoids, and a small number of phytoalexins20.
Metabolites produced in leaves of B. napus and B. rapa inoculated with either different races of the biotroph Albugo candida or sprayed with CuCl2 were determined by Pedras et al.19. This investigation established consistent phytoalexin (spirobrassinin, cyclobrassinin, and rutalexin) and phytoanticipin (indolyl-3-acetonitrile, arvelexin, caulilexin C, and 4-methoxyglucobrassicin) production and six new phenylpropanoids and two new flavonoids in canola19. Abdel-Farid et al. used NMR-based metabolomics to study B. rapa's reactions to infection by L. maculans, Aspergillus niger and F. oxysporum. F. oxysporum-infected plants accumulated more phenylpropanoids (sinapoyl-, feruloyl, and 5-hydroxyferuloyl malate), flavonoids (kaempferol and quercetin), and fumaric acid than plants infected with the other two species20. Huang et al. transposed A. thahliana reduce the synapine composition. The transformation led to a change in derivatives of sinapoyl, quercetin, salicylic acid, and indolyl glucosinolates. Transgenic plants were susceptible to fungal infection, and it was found that these metabolites play an important role in protecting the Brassicaceae plants21. Then, Botanga et al. inoculated A. thahliana C01 plants with Aternaria brassicola and found approximately half of the identified metabolites were affected by the inoculation. Due to the infection by A. brassicola, several phytohormones, oxylipins, amino acids, synaptics, ligands, and hundreds of potential biomarkers were affected22. However, very few studies have been done to evaluate metabolic mechanisms of plant defense against infection by the fungal pathogen LM in the Brassicaceae family, especially canola. Currently, there is a lack of metabolomics investigations on canola plants subjected to concurrent exposure to high temperature and LM infection. With the aim to improve our knowledge on plant-fungus interactions challenged with high temperatures, the canola cultivars, Excel (carrying the resistance gene Rlm7, resistant) and Drakkar (without any resistance genes, susceptible) were inoculated with the LM isolate ME24 under the optimum temperatures (21/16 °C) and elevated temperatures (28/23 °C), the changes in metabolites at different time intervals after inoculation were investigated.
The resistance gene Rlm7 in B. napus is a valuable source of resistance for controlling blackleg disease on canola2,23,24,25. The Rlm7 gene was more durable than other resistance genes (e.g., Rlm1) for control of LM because it is still used in commercial cultivars to prevent blackleg epidemics, even though it has been used commercially for more than 15 years2,23. The major goal of this study was to investigate the effects of temperature on Rlm7 at the metabolite level to understand the metabolic changes in canola plants under various biotic and abiotic stresses. Unraveling the complex plant-pathogen interactions under biotic and abiotic stresses can lead to better understanding of the molecular and physiological basis of host plant resistance to fungi in response to global warming.
Results
Disease progress on different cultivars under different temperatures
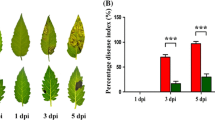
Daily assessment of symptoms showed that disease severity statistically differed between cultivars Excel (resistant, carrying Rlm7) and Drakkar (susceptible, lacking any R genes) at 21 °C and 28 °C. While large lesions developed on Drakkar at 14 days post inoculation (dpi) at both temperatures, the symptoms differed between temperatures on Excel (Fig. 1a). No visible symptoms were observed on Excel at 21 °C up to 10 dpi and only small dark lesions restricted to the inoculation sites were observed at 14 dpi. However, large lesions were observed on Excel at 28 °C at 14 dpi, suggesting the breakdown of resistance. Assessment of the lesion area of 30 plants for these symptoms in the areas of lesions measured using ImageJ software showed that on the susceptible cultivar Drakkar the lesion size dropped half at 28 °C compared to those at 21 °C. On the other hand, the size of lesions on the resistant cultivar Excel increased three times at 28 °C compared to those at 21 °C (Fig. 1b).
(a) The symptoms on cotyledons of cultivars Excel (resistant, carrying Rlm7) and Drakkar (susceptible, lacking any R genes) at 14 dpi. Plants were inoculated with Leptosphaeria maculans isolate ME24 at 21 °C and 28 °C. (b) disease severity on cultivars seedling Excel and Drakkar at 21 °C (optimum temperatures) and 28 °C (Elevated temperatures). Each bar represents the mean of disease severity (on a 0–1 ratio scale) 30 leaf samples and the error bars are the standard error of the mean (SEM).
Metabolic profiles of the B. napus-L. maculans interactions
Out of 9966 and 11,652 peaks in positive and negative ionization modes, respectively, 3319 and 7096 metabolites in positive and negative ionization modes were filtered based on a > 30,000 measured mean abundance and used for further multivariate analysis. To obtain candidate metabolites, the selected parent ions were collected using online databases with multivariate analysis. In the positive ionization mode, 2758 metabolites were discovered, while in the negative ionization mode, 6400 metabolites were putatively identified based on matches to publicly accessible databases including METLIN, KEGG, Pubchem, and the Plant Metabolic Network (PMN). In the positive ionization mode, 1369 metabolites at 48 h post inoculation (hpi) and 928 metabolites at 120 hpi and in negative ionization modes, 1988 metabolites at 48 hpi and 1703 metabolites at 120 hpi showed significant (p < 0.05) changes between temperatures, cultivars, and/or their interactions. The Supplementary files (Supplementary Tables S1 and S2) provide tables that include information on the annotation and mass error of metabolites, retention time, as well as all transitions and their intensities.
Univariate and multivariate analysis of the metabolite profile
The score plots obtained from PCA analysis consistently exhibited clustering among the biological replicates for each treatment, indicating a high level of repeatability across various treatments (Figs. 2 and 3; Supplementary Figs. S1–S8). The metabolic profiles of R and S plants were evaluated by creating loading plots to assess the overall differences. To validate the findings of PCA using a more robust pattern recognition technique, we conducted a supervised PLS-DA analysis. The score plots and corresponding VIP (Variable of Importance in Projection) plots derived from the models were built for the metabolic profiles of the canola cotyledon tissues. The results of PLS-DA analyses of the positive and negative ion modes showed that there were differences between cultivars and temperatures in score plots based on the first couple of components (Figs. 2 and 3; Supplementary Figs. S1–S8). The PLS-DA model is believed to be reliable when Q2 > 0.5 and suitable cross-validation is obtained for R2 > Q2. In this analysis, a Q2 > 0.736 and R2 > 0.983 indicated good predictive power. The samples of mock and LM inoculated at both temperatures and at different time points were separated bycomponent 1 only (not component 2) (Figs. 2 and 3). For instance, component 1 measured 22.2% and 21.3% of variations among the samples of Excel inoculated with LM vs mock inoculation, at 48 hpi, at 21 °C and 28 °C, posetive ion mode respectively. Our VIP analysis from PLS-DA analysis revealed the top 15 metabolites of negative ion mode for B. naupus–L. maculans pathosystem which showed better sensitivity in terms of peak intensity and numbers (Figs. 2 and 3; Supplementary Figs. S9–S16). The enrichment analysis module performs metabolite set enrichment analysis (MSEA) on several libraries containing ~ 6300 groups of metabolite sets. We selected chemical structures as a metabolite set library, which includes: 1. super-class (39 super chemical class metabolite sets or lipid sets), 2. main-class (617 main chemical class metabolite sets or lipid sets), and 3. sub-class (1250 sub-chemical class metabolite sets or lipid sets).The results of enrichment analysis for chemical structure classification of the key metabolites in the B. naupus-L. maculans pathosystem showed that metabolites are placed in 9 supper classes (Fig. 4a and Supplementary Table S3), in 22 main classes (Fig. 4b and Supplementary Table S4), and 39 subclasses (Fig. 4c and Supplementary Table S5).
PCA and PLS-DA analysis; (a) Principal component analysis PCA score plot showing distinct relationship between samples and differences in resistant cultivar inoculated with the pathogen LM (RP) vs resistant cultivar with mock inoculation (RM) at 48 hpi/21 °C in negative ionization mode with the MetaboAnalystR 6.0 software package along the x-axis (PC1) and within groups along the y-axis (PC2). PC1 and PC2 respectively summarize 29.8% and 27.1% of the observed variance in the metabolites, green and red shaded areas are the 95% confidence regions of each group; (b) Scores plot of metabolite features in PLS-DA (partial least squares-discriminant analysis) model representing covariance between component 1 (25.8%) at x-axis and component 2 (26.5%) at y-axis; (c) Variable of Importance in Projection (VIP) score plots derived from PLS-DA analysis, a higher VIP score (> 2.0) for metabolites represent higher contribution in the PLS-DA model.
PCA and PLS-DA analysis; (a) principal component analysis PCA score plot showing distinct relationship between samples and differences in susceptible cultivar inoculated with the pathogen LM (SP) vs susceptible cultivar with mock inoculation (SM) at 48 hpi/28 °C in negative ionization mode with the MetaboAnalystR 6.0 software package along the x-axis (PC1) and within groups along the y-axis (PC2). PC1 and PC2 respectively summarize 38.2% and 25.7% of the observed variance in metabolites, green and red shaded areas are the 95% confidence regions of each group; (b) Scores plot of metabolite features in PLS-DA (partial least squares-discriminant analysis) model representing covariance between component 1 (25.1%) at x-axis and component 2 (18.8%) at y-axis; (c) Variable of Importance in Projection (VIP) score plots derived from PLS-DA analysis, a higher VIP score (> 2.0) for metabolites represent higher contribution in the PLS-DA model.
Enrichment analysis for chemical structure classification of the key metabolic pathways in the Brassica napus–Leptosphaeria maculans pathosystem with the MetaboAnalystR 6.0 software package. (a) Supper class, (b) Main class, (c) Subclass. The color is based on the p-value (darker colors indicate more significance) and the size of each circle is based on the enrichment ratio (the larger indicates a higher ratio). The top 25 crucial chemical groups were studied in each class. The details of the enrichment analysis results for the identified metabolites in each class were presented in supplementary tables (Tables S3, S4, S5).
Fold change (FC ≥ 2.0) analysis was conducted to identify up- and down-regulated discriminant annotated metabolites by univariate feature analysis in Metaboanalyst 6.0. Fold change (FC ≥ 2.0) analysis in positive ionization in resistant cultivar inoculated with the pathogen LM (RP) vs. resistant cultivar with mock inoculation (RM) at 48 hpi /21 °C suggested 95 metabolites significantly up- and 56 down-regulated metabolite (Fig. 5a). In negative ionization, in the RP vs. RM at 48 hpi /21 °C suggested 129 metabolites significantly up- and 142 down-regulated metabolite. Fold change (FC ≥ 2.0) analysis in positive ionization in RP vs. RM at 48 hpi /28 °C suggested 310 metabolites significantly up- and 109 down-regulated metabolite. In negative ionization, in RP vs. RM at 28 °C/48 hpi suggested 366 metabolites significantly up- and 315 down-regulated metabolite (Fig. 5b). In RP vs. RM at 48hpi/21 °C under VIP ≥ 2, in positive mode, 81 features showed significant differences, 38 up-regulated and 53 down-regulated. In negative mode, RP vs. RM at 48hpi/21 °C, with VIP ≥ 2, 171 features showed significant differences, 76 up-regulated and 95 down-regulated. In elevated temperature, RP vs. RM at 48hpi/28 °C with VIP ≥ 2, in positive mode, 72 features showed significant differences, 42 up-regulated and 30 down-regulated. In negative mode, RP vs. RM at 48hpi /28 °C, in VIP ≥ 2, in positive mode, 164 features showed significant differences, 76 up-regulated and 88 down-regulated.
Univariate feature analysis in Metaboanalyst 6.0. fold change (FC threshold ≥ 2) analysis of metabolite in (a) resistant cultivar inoculated with the pathogen LM (RP) vs resistant cultivar with mock inoculation (RM) at 48 hpi/21 °C in positive ionization and (b) RP vs RM at 48 hpi/28 °C in negative ionization.
Effects of temperature on metabolic pathways related to L. maculans infection
Only 128 out of 1000 significant metabolites at 21 °C and 28 °C (FDR < 0.05) were recognized in 66 Arabidopsis pathways using the database of Arabidopsis metabolic pathways from KEGG or HMDB as a reference for a global test algorithm (Table 1 and Supplementary Table S6). Most of the 54 and 63 pathways identified in the positive and negative ionization modes were common. By comparing pathways impacted in LM-infected plants of R (resistant) and S (susceptible) cultivars at 21 °C, several pathways down-regulated in S cultivar were up-regulated in the R cultivar at both periods (2 and 5 dpi) and interestingly these pathways were among those down-regulated in R cultivars at 28 °C. In this case, the pathways were classified as either pathogen stress-related or host resistance-related. These pathways included alpha-Linolenic acid metabolism, nicotinate and nicotinamide metabolism, galactose metabolism, histidine metabolism, aminoacyl tRNA biosynthesis, porphyrin and chlorophyll metabolism, fatty acid elongation in mitochondria, fatty acid metabolism, and fatty acid biosynthesis. Some pathways were down-regulated at 21 °C and at the same time up-regulated at 28 °C (Table 1). They could be classified as heat stress-specific related pathways. Lysine biosynthesis, carotenoid biosynthesis, and pyrimidine metabolism were grouped in this category. On the other hand, following pathogen inoculation at 21 °C some pathways were up-regulated in the R cultivar at all-time points and remained up-regulated at 28 °C. Consequently, these pathways that were related to both pathogen and heat stresses were categorized as general or non-specific stress-related pathways (Table 1). Given the fact that a metabolite could be a component of different pathways and influence multiple biological processes, as a result, detecting plant metabolic networks becomes a challenging task. To solve this problem and simplify the process, the standardized overview of pathways was achieved by adapting the KEGG classification for canola pathways (Figs. 6 and 7). Phenylalanine metabolism, tryptophan metabolism, alpha-Linolenic acid metabolism, beta-Alanine metabolism, isoquinoline alkaloid biosynthesis, pantothenate and CoA biosynthesis, and tyrosine metabolism had the most effect on the B. napus-L. maculans interaction (Figs. 6 and 7). Also, according to the results of network analysis based on the KEGG global metabolic network with the MetaboAnalystR 6.0 software package, 15 significant pathways were identified (Table 2).
Overview of pathways in the Brassica napus–Leptosphaeria maculans pathosystem. (a) Metabolic pathway analysis (MetPA) from positive ionization mode with the MetaboAnalystR 6.0 software package. All the matched pathways are displayed as circles. The color and size of each circle are based on p-value (darker colors indicate more significant changes of metabolites in the corresponding pathway) and pathway impact value (the larger size the more impact), respectively. The most impacted pathways are annotated. 1: Phenylalanine metabolism, 2: Tryptophan metabolism, 3: alpha-Linolenic acid metabolism, 4: beta-Alanine metabolism, 5: Isoquinoline alkaloid biosynthesis, 6: Tyrosine metabolism, 7: Pantothenate and CoA biosynthesis, 8: Sphingolipid metabolism, 9: Glyoxylate and dicarboxylate metabolism, 10: Limonene and pinene degradation. (b) Metabolite sets enrichment overview. The horizontal bar (white-to-red) reflects increasing statistical significance.
Overview of pathways in Brassica napus–Leptosphaeria maculans pathosystem. (a) Metabolic pathway analysis (MetPA) from negative ionization mode with the MetaboAnalystR 6.0 software package. All the matched pathways are displayed as circles. The color and size of each circle are based on p-value (darker colors indicate more significant changes of metabolites in the corresponding pathway) and pathway impact value (the larger the value, the more impact), respectively. The most impacted pathways are annotated. 1: Alanine, aspartate, and glutamate metabolism, 2: Phenylalanine metabolism, 3: Tyrosine metabolism, 4: Tryptophan metabolism, 5: alpha-Linolenic acid metabolism, 6: Riboflavin metabolism, 7: Isoquinoline alkaloid biosynthesis, 8: Stilbenoid, diarylheptanoid, and gingerol biosynthesis, 9: Glyoxylate and dicarboxylate metabolism, 10: Citrate cycle (TCA cycle), 11: Indole alkaloid biosynthesis, 12: Phenylpropanoid biosynthesis, 13: Porphyrin and chlorophyll metabolism, 14: Pantothenate and CoA biosynthesis. (b) Metabolite sets enrichment overview. The horizontal bar (white-to-red) reflects increasing statistical significance.
Discussion
Elevated temperature is the primary abiotic stress for plants under climate change. With increased temperatures due to global warming, future plant disease epidemics are anticipated to be more frequent and more severe. In nature, temperature-sensitive resistance in plants has been regularly reported10,11,12,13,26. Metabolic pathways down/up-regulated under pathogen attack to reinforce the defense mechanisms at elevated temperatures can neutralize defense through altering these regulations, may also be exploited to engineer new crop cultivars and to understand how temperature affects plant innate immunity and discover elite genes that confer disease resistance at elevated temperature27. Nevertheless, little is known about the molecular mechanisms of plant defense against pathogen infections and how climate change influences them26,27. To better comprehend the physiological responses arbitrated by the resistance gene Rlm7 when confronted with the pathogen L. maculans (LM) under incompatible and compatible interactions, a comparative metabolomic study was done. Our study is the first to examine the effectiveness of the resistance gene Rlm7 at elevated temperatures using metabolomic techniques.
Large blackleg lesions were observed on the susceptible cultivar Drakkar at both 21 °C and 28 °C. However, the lesion size at 28 °C was half of that at 21 °C. This may be due to reduced growth of the pathogen and drop of humidity at 28 °C which may have slowed down the spread of the lesion. No pycnidia were observed on cotyledons of Drakkar at 28 °C may also be due to low humidity at 28 °C since low humidity is hostile for pathogen sporulation. Small dark lesions around the inoculation areas were observed on the resistance cultivar Excel at 21 °C, while large lesions were observed at 28 °C. The size of lesions on Excel increased three times at 28 °C in comparison with those at 21 °C, suggesting the Rlm7 gene-mediated resistance in Excel is sensitive to temperature. This is consistent with a recent study of Rlm7 in the Topas introgression line (Topas-Rlm7) which showed that the Rlm7 is temperature-sensitive in Topas-Rlm713. Previous studies showed that the genetic background plays a crucial role in single Rlm gene-mediated resistance10,17,23, this study reconfirmed that the Rlm7 resistance gene specific for LM in the Excel cultivar was temperature-sensitive. Similarly, other studies suggest that Rlm7 is the next best candidate resistance gene to be introduced into the canola-host gene pool23,24,25. The Rlm9, Rlm4, and Rlm7 encode wall-associated kinase-like (WAKL) proteins28,29. Cloning these three genes has shown them to be allelic variants of one another. WAKLs are a newly emerging class of Avr-responsive, ETI-inducing R genes.
The findings of this study demonstrate that metabolic profiling is a highly effective approach for investigating the underlying mechanisms of the pathogenesis of the hemibiotrophic pathogen LM and the mechanisms of host resistance in canola. Our results showed that LM could cause up-regulation of a range of metabolic pathways. Some metabolites associated with different pathways were likely to be involved in canola resistance to LM infection. Results of our study also indicate a hierarchical structure in the control of multi-dimensional metabolic networks responding to numerous simultaneous stressors, such as pathogen (biotic) and heat (abiotic). The observed activation of metabolic pathways may indicate the rapid inhibition of the fungal infection, as shown in other plant-pathogen interactions30. The results of metabolic network analysis showed three classes of pathways: resistance-related specific pathways, non-specific stress-related pathways, and heat stress-related specific pathways.
Resistance-related specific pathways
Metabolic pathways for fatty acids, alpha-linolenic acid, nicotinate and nicotinamide, galactose, histidine, porphyrin and chlorophyll, fatty acid elongation in mitochondria, and aminoacyl tRNA biosynthesis were considered as host resistance-related specific pathways or pathogen stress-related specific pathways. Fatty acids (FA) metabolic pathways have significant roles in plant defense against pathogens. Previous studies on FAs reveal that they play passive roles in plant defense, serving as biosynthetic precursors for cuticular components or the phytohormone jasmonic acid. Studies of FA metabolic mutants demonstrated an active signaling role for the cuticle in plant defense31.
Our study showed that alpha-linolenic acid metabolism has significant effects in both positive and negative ionization modes (Figs. 6 and 7). Alpha-linolenic acid, as a plant hormone, responds to plant infection by pathogens, cooperates with other plant hormones to stimulate growth, adapt to stresses, and regulate metabolic pathways32. Its application enhances the activity of various antioxidant enzymes, including sodium dismutase, catalase, ascorbate peroxidase, NADH peroxidase, and glutathione reductase33. It is speculated that oxidation regulation of unsaturated fatty acids can be used for their long-distance transport in plants34. The metabolisms of 12-OPDA, 13(S)-HPOT, and alpha-linolenic acids were up-regulated in the LM-canola interaction at 120 hpi in the R cultivar under 21 °C. The 12-OPDA is a precursor for synthesizing jasmonic acid (JA), the increasing JA at 120 hpi is probably in response to the fungal necrotrophic stage. The 12-Oxo-9(Z)-dodecenoic acid strongly inhibited mycelial growth and spore germination of eukaryotic microbes35. Oxylipins have several functions including direct antimicrobial effect, stimulation of plant defense gene expression, and regulation of plant cell death36.
The abundance of palmitic acid (hexadecanoic acid) a precursor for JA37, was significantly reduced in both R and S plants inoculated with the pathogen LM compared to their control at 48 hpi at 21 °C. This reduction is probably related to suppressing the production of JA and increasing the expression of salicylic acid (SA) synthesis. The increase of palmitic acid at 120 hpi suggests higher demands for the production of JA. Palmitic acid also affects the pathways of fatty acid biosynthesis, fatty acid elongation in mitochondria, and fatty acid metabolism. The study by Šašek et al. revealed that the expression of multiple marker genes linked to different plant hormones was elevated in resistant and susceptible B. napus plants infected with LM38. Additionally, the study revealed that the expression of genes related to the ET pathway increased at later stages of the infection (8–10 dpi), while genes related to SA were induced earlier (4–6 dpi)38. Similarly, the study by Lowe et al. (2014) demonstrated that the marker genes for SA and ABA were up-regulated in B. napus inoculated with LM at 7 and 14 dpi39. Haddadi et al. provided a more detailed picture of the global transcriptome that is active in B. napus during cotyledon infection by LM. The gene expression profile associated with different plant hormones revealed the importance of SA at the earlier stages of infection (until 4 dpi) and the up-regulation of genes related to the JA pathway at the later stages (6–8 dpi)40. SA is more effective in defending against biotrophic and hemibiotrophic pathogens, while ET/JA signaling is more capable in resisting against necrotrophic pathogens and herbivorous insects. Gene-for-gene interaction studies in the B. napus–L. maculans pathosystem showed early induction of SA/JA responsive factors, indicating the importance of those genes in the incompatible interaction. The early activation of SA-responsive factors in resistant B. napus genotypes guarantees effective slowdown of the pathogen infection due to the reinforcement of the defense throughout the plant via the associated SAR (systemic acquired resistance). Each hormone has certain responsive factors and signaling pathways, where the responsive pathways of different hormones also have different potential connections, building up an integrated and systemic signaling network to cope with various challenges41. SA–JA relationship is not always antagonistic. Tamaoki et al. explained that the defense system is activated by both SA and JA signaling interaction during the induction of defense response42. There may be at least a partly shared signal transduction pathway used for signaling both JA and SA.
The increased activity of nicotinate and nicotinamide metabolic pathway in the R cultivar compared to the control at 48 hpi under 21 °C (48 hpi/21 °C) might be considered as an indication of a greater supply of these two vital precursors for the generation of coenzymes, NAD + and NADP +, which are crucial for redox reactions and electron transportation from one reaction to another. These coenzymes are essential for many metabolic pathways including the TCA cycle, glycolysis, fatty acid biosynthesis, pentose phosphate cycle, and many other metabolic pathways. Among these redox carriers, NAD is the keystone of cellular oxidations along catabolism and is essential for plant growth and development43. In addition to its redox role, there is compelling evidence that NAD is a signal molecule controlling crucial functions in primary and secondary carbon metabolism. Studies using integrative omics approaches combined with molecular plant pathology have shown that manipulating NAD biosynthesis and recycling led to an alteration of metabolite pools and developmental processes and changes in resistance to various pathogens43. NAD levels could be used as a potential target to improve tolerance to biotic stress and crop improvement.
The increased histidine metabolism in the R cultivar compared to control at 48 hpi/21 °C suggesting the importance of histidine in host resistance since histidine biosynthesis pathway is integrated with several metabolic pathways, including tryptophan. Tryptophan plays a significant role in regulating plant development and defense responses and is the precursor for indoleacetic acid, a plant hormone necessary for cell expansion44. Liquid fertilizer originated from a yeast cell extract containing histidine exerted inhibitory effects on the development of bacterial wilt disease in tobacco plants caused by the soil-borne bacterial pathogen Ralstonia solanacearum45.
An increase in the abundance of galactose metabolism at 48 hpi compared to control in the R cultivar at 21 °C showed the importance of the central carbohydrate metabolism as one of the key sources for protecting cells from stresses46. Galactose has a defensive function and is involved in synthesizing hemicellulose47. Sugars play a crucial role in the interaction between plants and pathogens, serving as a vital energy source for defense and functioning as signaling molecules for the regulation of defense genes48. The concept of “Sweet Immunity” and sugar-enhanced defense has emerged due to the potential involvement of certain sugars in plant immunity47,48,49. Our network analysis and Enrichment analysis also identified galactose metabolism as an important pathway. The elevated levels of aminoacyl-tRNA biosynthesis in the R cultivar compared to control at 48 hpi/21 °C support previous study that the function of aminoacyl-tRNA synthesis is to precisely match amino acids with tRNAs containing the corresponding anticodon50.
Non-specific stress-related pathways
The network analysis and enrichment analysis identified the importance of phenylalanine metabolism, tryptophan metabolism, beta-Alanine metabolism, isoquinoline alkaloid biosynthesis, tyrosine metabolism, pantothenate and CoA biosynthesis, sphingolipid metabolism, glyoxylate, and dicarboxylate metabolism as general or non-specific stress-related pathways. Phenylalanine metabolism and tyrosine metabolism as non-specific stress-related pathways were increased in all treatments compared to control at 21 °C and in resistance cultivar at 48 hpi under the elevated temperature regime. Tyrosine metabolism was up-regulated in soybean at 1 and 2 dpi in response to R. solani, which is consistent with our findings51,52. Tyrosine can undergo catabolism, resulting in the production of diverse secondary metabolites such as tocopherols, tocotrienols, and tyramine. These metabolites serve as precursors for the synthesis of hydroxycinnamic acid amides that are linked to the cell wall53. Phenylalanine, tyrosine, and tryptophan, as precursors for a wide variety of secondary metabolites, play a critical role in plant defense against biotic stressors and resistance to pathogen infections54. It has been hypothesized that tyrosine decarboxylated precursors (tyramine and dopamine) are used in the synthesis of isoquinolines in plants due to the up-regulation of the isoquinoline alkaloids pathway. Historically, the physiological activity of these metabolites has a role in plant defense systems, either by a direct toxic impact or by suppressing certain processes of invading pathogens55,56.
Pantothenate (vitamin B5) is the precursor to coenzyme A (CoA), which in turn, plays an essential role in fatty acid and pyruvate metabolism57. The main function of vitamins is to act as a cofactor in diverse metabolic pathways, facilitate the production of essential compounds for plants, induce resistance against pathogens, directly promote plant growth, and participate in energy conservation in the plant from stored compounds57. This is supported by the results that sphingolipid metabolism was increased at 120 hpi compared to control in the R cultivar under both optimum/elevated temperature regimes. Sphingolipids could act as second messengers with effects on cellular homeostasis.
Heat stress-related specific pathways
Carotenoid biosynthesis, pyrimidine metabolism, and lysine biosynthesis were classified as heat stress-specific related pathways. This is supported by the results that carotenoid biosynthesis pathway in pathogen-inoculated plants of the resistant cultivar compared to those of mock up-regulated at 120 hpi/28 °C but down-regulated at 120 hpi/21 °C. The functional metabolism of carotenoids encompasses various processes, such as the stability of membrane lipid bilayers, the elimination of free radicals induced by reactive oxygen species, and the safeguarding of membrane lipid integrity58,59. The plant hormones abscisic acid and strigolactones as well as other apocarotenoids that are involved in different developmental processes and stress responses, are all synthesized from carotenoids60. Quan et al. revealed that glutamic acid and poly-γ-glutamic acid boosted the heat resistance of Chinese cabbage (Brassica rapa L. ssp. Pekinensis) by refining carotenoid biosynthesis, photosynthesis, and ROS Signaling61. Exogenous application of biostimulants, such as carotenoids, can alleviate the damage caused by elevated temperatures in canola and improve growth parameters and physiological characteristics.
Nucleotide metabolism is a crucial component of plant metabolism and affects many metabolic pathways. Specifically, pyrimidine nucleotides participate directly in nucleic acid synthesis52,62. The observed up-regulation of the pyrimidine metabolism pathway at 28 °C, with the increase in this pathway more pronounced in the S cultivar than in the R cultivar, suggests that genes of the pyrimidine catabolic pathway are important in plant abiotic stress responses. The role of pyrimidine catabolism in stress tolerance may be related to the production of proline biosynthesis precursors62. In plants, a complete characterization of this pathway is lacking63,64. The accumulation of purine and pyrimidine bases in R cultivars at 48 hpi/28 °C might be interpreted as the plant's reaction to mixed pathogen and heat stress. Furthermore, pyrimidine derivatives have broad-spectrum biological activities, including antibacterial, antifungal, insecticidal, herbicidal, and antiviral activities65.
The lysine biosynthesis was increased at 48 and 120 hpi compared to the control in both R and S cultivars under elevated temperatures, which suggests the importance of lysine biosynthesis in plant heat stress response. Lysine is an important precursor for the synthesis of glutamate, which is an essential final energy source for the production of carbohydrates, lipids, peptides, and secondary metabolites. Lysine levels increase under abiotic stress, such as salt, water, heavy metals, and nutritive stress66. Endogenous lysine metabolism or its exogenous application helps reduce abiotic stress67. It has also been suggested that the jasmonate signaling pathway and tryptophan metabolism through stress responses are induced by lysine metabolism68.
Global metabolic network
KEGG mapping of the global metabolic network displayed enriched biosynthetic pathways for the biosynthesis of phenylalanine, tyrosine, tryptophan, pantothenate and CoA, sphingolipid, valine, leucine, isoleucine, arginine, proline, caffeine, biotin, glycan, starch and sucrose, galactose, fructose and mannose, ubiquinone and terpenoids-quinones, as well as selenocompounds (Figs. 6 and 7). A significant number of these pathways are related to amino acid metabolism, which serves as a common source for secondary metabolites such as phenolics and alkaloids69. Enrichment of amino acid metabolism is essential for influencing metabolic processes after pathogen contact. Pantothenic acid, a precursor for some cofactors, is crucial for regulating enzyme activity and hence influencing plant development via metabolic pathways. Selenocompounds protect plants from a variety of abiotic stresses70. Metabolites that enhance the ubiqinone pathway can act as electron transporters in the process of photosynthesis71. Caffeine, classified as a xanthine alkaloid, may protect the young and soft tissues from infections caused by pathogens72. Glycans function as signaling molecules in stress and plant defense responses, as well as being involved in the energy metabolism of a plant73. In summary, the enrichment analysis showed a significant presence of metabolite hits in many pathways. The prevalence of these hits suggests that plant responses are influenced in terms of defense, resistance, and/or survival.
Network analysis provides a clear snapshot of the pathways in which the annotated metabolites are engaged, enhancing our knowledge of their significance in these processes74. Network analysis of different pathways in canola cotyledons in response to L. maculans infection identified 15 significant pathways (Table 2) that are mostly associated with the production of amino acids and subsequently carbohydrates. Cardoso et al. performed a meta-analysis on a wide range of species and various stress circumstances such as water deficit, cold, salt, and nitrogen deprivation stress75. Their findings emphasized that amino acid metabolism plays a vital role in plant stress adaptation. Amino acids are not the only source of carbon and nitrogen for different pathways but also good stress signal messengers that aid the entire plant in acclimating to stress conditions75.
The specific outcomes of disease severity when a plant is simultaneously challenged by an invading pathogen and heat stress, depend on many factors, including host and pathogen genetics. Most studies have described negatively impacted host resistance under elevated temperatures, while others noticed positive or neutral effects of elevated temperatures on host resistance. Development of strategies to protect food security under global warming will require an understanding of how plants deal, both positively and negatively, with simultaneous abiotic and biotic stresses76. Our results obtained from B. napus–L. maculans (LM) interaction suggest that the elevated temperatures might negatively affect the resistance to LM. However, this work was done using canola cotyledons and the effects of temperature on resistance to LM might be different at various canola growth stages. There is a need to further investigate the effects of temperature on the effectiveness of Rlm7 at different canola growth stages. One of the most effective approaches to developing crops that are adaptable to climate change is to understand the diverse plant immune mechanisms impacted by temperature and identify key genes that can enhance disease resistance through genome editing techniques. This study identified important alterations in a diversity of metabolite classes, suggesting an infection-related reprogramming of particular pathways. Our results indicate the importance of hierarchical regulation of multidimensional metabolic networks in the face of several concurrent challenges, such as pathogen and heat stress. To identify other mechanisms in physiological processes under biotic and abiotic stresses, further research is needed using RNA sequencing, proteomics, and metabolomics with other platforms.
Conclusion
This work profiles the metabolisms of canola cotyledons at the initial infection stage (biotrophic stage) and the necrotrophic stage, in both compatible and incompatible (with the resistance gene Rlm7) interactions at normal (21 °C) and elevated (28 °C) temperatures using UPLC-QTOF/MS. Resistance-related specific pathways, heat stress-related specific pathways, and pathways associated with combined heat and pathogen stresses have been identified. The identified pathways associated with the breakdown of resistance at elevated temperatures, such as carotenoid biosynthesis, pyrimidine metabolism, and lysine biosynthesis will play an important role in developing climate-resilient crops by using effective techniques to manipulate these pathways. However, to fully understand the multidimensional nature of plant-pathogen interactions and produce disease-resistant cultivars that are resilient to climate change, there is a need for future research investigating a wider range of environmental conditions.
Materials and methods
Plant growth and infection conditions
Two canola cultivars (Drakkar—susceptible without any resistance genes and Excel- resistant carrying the resistance gene Rlm7) and the LM isolate EM24 carrying three known effectors (AvrLm1, AvrLm6, and AvrLm7), were used in this study. Before planting, seeds were distributed in a 9-cm-diameter sterile Petri dish with one layer of saturated filter paper. The petri dish was placed in an incubator at 20 °C ± 1 with 45% relative humidity, for two days. The germinated seeds were sown in plastic pots containing pasteurized soil (1:3 mixture of peat and soil). Pots were maintained in a growth chamber under controlled environmental conditions at two different temperatures: 21 °C (light)/16 °C (dark) and 28 °C (light)/23 °C (dark), with 75% relative humidity and 16 h photoperiod with 13,000 lx light intensity at canopy level.
After 14 days, the cotyledons of individual seedlings were wounded, using a sterile needle at each of the four lobes. Each of the wounded sites was inoculated with a 10 μl drop of conidial suspension or sterile distilled water (control). Spore suspensions of L. maculans were prepared from cultures grown on V8-agar. The fungal inoculum was prepared according to Chen and Fernando77. The concentration of spores was diluted to a final spore concentration of 1 × 107 spores mL−1. Each treatment included three biological replicates (each replicate consists of five pots with five plants per pot). The treatments were arranged in a completely randomized design. At 48, 120 h post-inoculation (hpi), 300 mg of whole cotyledon tissues per time point, corresponding to 3 cotyledons (3 cotyledons from 3 plants with one cotyledon from each plant), were randomly collected for metabolomics analysis from pathogen- and mock-inoculated plants. The leaf samples were shock-frozen in liquid nitrogen and stored at − 80 °C until processing. The rest of the seedlings were kept under the same growth conditions and disease progression was monitored.
Disease severity assessment
After sampling cotyledon tissues for metabolite analysis, the remaining plants were kept in the growth chamber for disease symptom evaluation up to 14 dpi in 3-day intervals by rating the infected cotyledons on a 0 to 9 scale (where 0 = no symptom and 9 = tissue collapse and appearance of conidia)78. The 0–9 original ordinal score of disease severity data was converted to a ratio scale normalized to the 0–1 range using the following equation:
where y = blackleg severity on a ratio scale, i to j are the number of infected canola plants scored between 0 and 9, respectively according to their disease severities and n is the total number of plants scored. The normalized blackleg severity (y) values over time were used to construct a disease progress curve. The disease severity over time was subsequently reduced to a single summary variable by calculating the area under the disease progress curve (AUDPC) based on the following formulae:
where i is the number of individual observations, n is the total number of observations, t is time after inoculation (days) and y is the normalized blackleg severity. At 14 dpi, the lesions were digitally imaged and the areas of lesions were measured using ImageJ software79.
Metabolite extraction and UPLC-QTOF/MS analysis
To prepare 200 ml of extraction mixture, 250 µL of 80% methanol–water was added to 40 mg freeze-dried samples that were already grounded into a fine powder using an electric mill. After the extraction solvent was added, the samples were sonicated for 15 min and subsequently centrifuged at a speed of 14,000 rpm for 10 min. Finally, supernatants were transferred to new 1.5 mL micro-tubes after filtration with a 0.2 µm micropore PTFF membrane54. For each sample, 5 µL was injected into an ultra-high performance liquid chromatography system (Thermo Dionex Ultimate 3000 LC) connected to a quadrupole time of flight detector (Bruker Compact™ QTOF-MS) with a heated-electrospray ionization source (ESI). Chromatographic separation was performed as described in Tortosa et al. using a binary gradient solvent mode consisting of 0.1% formic acid in water (solvent A) and acetonitrile (solvent B). The following gradient was used: 3% B (0–3 min), from 3 to 25% B (3–10 min), from 25 to 80% B (10–18 min), from 80 to 100% B (18–22 min), and to hold 100% B until 24 min. The flow rate was established at 0.3 ml/min and the column temperature was controlled at 35 °C80.
MS data were acquired using an acquisition rate of 2 Hz over the mass range of 50–1200 m/z. Both polarities (±) ESI mode were used under the following specific conditions: gas flow 8 l/min; nebulizer pressure 38 psi; dry gas 7 l/min; dry temperature 220 °C. Capillary and endplate offsets were set to 4500 and 500 V, respectively. To monitor the performance of data acquisition, the run sequence was started with 3 blanks (methanol, the solvent used in sample extraction), and a standard compound (triphenyl phosphate for positive ionization mode and chloramphenicol for negative ionization mode) followed by every 10 samples to ensure instrument drift was minimal81.
Metabolite identification and quantification
Total ion chromatograms generated by LC/MS were imported straight into Data Analysis (Bruker) and then exported as mzXML using CompassXport (Bruker). Peaks were aligned and identified using the ProfileAnalysis 2.1 program82. The peaks were aligned using a 10-s, 8-mDa frame and the "split buckets with multiple features" setting. Buckets were created using the molecular characteristics option, which takes into consideration the retention duration, m/z isotopic pattern, and the charge state of the probable compounds. The investigation focused on metabolites with an average abundance larger than 30,000. The intensity of peaks that exhibit a larger change on RT was added by hand to the bucket table. Metabolites reported in commercial databases including METLIN, KEGG, Pubchem, HMDB, and Plant Metabolic Network (PMN) were used for tentative identification using accurate mass (accurate mass error (AME) = 5 ppm).
Statistical analysis
Multivariate analysis including principal components analysis (PCA) and Partial least squares discriminant analysis (PLS-DA) was carried out using the online software Metaboanalyst 6.0 (2023). The data from each day under different temperatures were uploaded separately for analysis. The data was Pareto scaled to reduce the influence of intense peaks while emphasizing weaker peaks that may have more biological relevance. The corresponding loadings of intense peaks were reduced and loadings from weak peaks were increased due to Pareto scaling. PLS-DA is a supervised approach that aims to differentiate between classes (Y) in highly complex data sets, despite within-class variability in the observed variables (X), in other words, to find the metabolic differences between the inoculated groups and mock groups. PLS-DA models were cross-validated using R2 and Q2 parameters. The quality evaluation (Q2) and R-squared (R2) statistics give a quantitative measure of consistency between the anticipated and original data and provide estimates of the predictive power of the model, as well. The PLS-DA model utilizing the first principal component of VIP (variable importance in the projection) values paired with univariate analysis (Fold change) (P ˂ 0.05) to detect differentially expressed metabolites. By computation of log twofold change (log 2 FC (inoculated/mock)) for each metabolite in both genotypes (resistant and susceptible) and based on VIP˃2, metabolites linked to resistance or susceptibility or heat stress were identified. In many studies, a VIP value > 2 is a suitable threshold for feature selection; however, this cut-off depends on the number of variables employed83. To identify the metabolites responsible for discrimination among the metabolomic profiles, the variable importance in the projection (VIP) score was used to select features with the most significant contribution in a PLS-DA model. VIP scores are a weighted sum of PLS weights for each variable and measure the contribution of each predictor variable to the model. Further, the VIP statistic summarizes the importance of the metabolites in differentiating the time points in multivariate space. Metabolites exhibiting high VIP scores (≥ 2) are the more influential variables.
Metabolic pathway analysis
Metabolite set enrichment analysis (MSEA) using metabolites significantly altered was conducted to identify the altered metabolic pathways (metabolite sets) associated with phenotypes of interest. Metabolomics Pathway Analysis (MetPA) was used for the analysis and visualization of metabolomics data within the biological context of metabolic pathways according to KEGG and HMDB metabolic databases. Arabidopsis metabolic pathway databases as references were used for pathway analysis. The final result is a ranking of the relative importance of pathways in the KEGG and HMDB databases together with their associated p-values from enrichment analysis82,84. Network analysis based on the KEGG global metabolic network according to the KEGG and HMDB metabolic database was also done85. The eenrichment analysis, pathway analysis, and network analysis were done using MetaboAnalystR 6.0 software package.
Ethical approval
The study was conducted in accordance with relevant guidelines and legislation.
Data availability
Data are available from the corresponding author upon reasonable request. The datasets used and/or analyzed during the current study are also available from the corresponding author on reasonable request or downloadable from Google Drive. All LC-Qtof MS files in mzXML format have been converted to two archive files (positive and negative ionization modes). The addresses of links: Positive mode ionization (https://drive.google.com/file/d/1RXcMzDDkRy7QCCx_UrtK93FZq-PGmkQv/view?usp=drive_link). Negative mode ionization (https://drive.google.com/file/d/1Pz_yC6pBFnmt23qo3UksYRp4Ph-W8gKc/view?usp=drive_link).
References
Fitt, B.D.L., Brun, H., Barbetti, M.J. & Rimmer, S.R. World-wide importance of phoma stem canker (Leptosphaeria maculans and L. biglobosa) on oilseed rape (Brassica napus). Eur. J. Plant Pathol. 114, 3–15 (2006).
Van de Wouw, A. P. et al. A new set of international Leptosphaeria maculans isolates as a resource for elucidation of the basis and evolution of blackleg disease on Brassica napus. Plant Pathol. 73, 170–185 (2024).
Zamanmirabadi, A., Hemmati, R., Dolatabadian, A. & Batley, J. Current progress in studying blackleg disease (Leptosphaeria maculans and L. biglobosa) of canola in Iran: Where do we stand now? Plant Pathol. 71, 239–250 (2022).
West, J. S., Kharbanda, P. D., Narbetti, M. J. & Fitt, B. D. L. Epidemiology and management of Leptosphaeria maculans (phoma stem canker) on oilseed rape in Australia Canada and Europe. Plant Pathol. 50, 10–27 (2001).
Huang, Y. J., Toscano-Underwood, C., Fitt, B. D. L., Hu, X. J. & Hall, A. M. Effects of temperature on ascospore germination and penetration of oilseed rape (Brassica napus) leaves by A- or B-group Leptosphaeria maculans (phoma stem canker). Plant Pathol. 52, 245–255 (2003).
Huang, Y. J. et al. Temperature and leaf wetness duration affect phenotypic expression of Rlm6-mediated resistance to Leptosphaeria maculans in Brassica napus. New Phytol. 170, 129–141 (2006).
Kutcher, H. R. et al. Blackleg disease of canola mitigated by resistant cultivars and four-year crop rotations in western Canada. Can. J. Plant Pathol. 35, 209–221 (2013).
Raman, H., Raman, R. & Larkan, N. Genetic dissection of blackleg resistance loci in rapeseed (Brassica napus L.); plant breeding from laboratories to fields. InTech. 4, 86–120 (2013).
Borhan, M. H., Van de Wouw, A. P. & Larkan, N. J. Molecular interactions between Leptosphaeria maculans and Brassica Species. Annu. Rev. Phytopathol. 60, 237–257 (2022).
Noel, K. et al. Influence of elevated temperatures on resistance against phoma stem canker in oilseed rape. Front. Plant Sci. 13, 785804 (2022).
Huang, Y. J. et al. Identification of environmentally stable QTL for resistance against Leptosphaeria maculans in oilseed rape (Brassica napus). Theor. Appl. Gen. 129, 169–180 (2016).
Richard, M. M., Knip, M., Aalders, T., Beijaert, M. S. & Takken, F. L. Unlike many disease resistances, Rx1-mediated immunity to potato virus X is not compromised at elevated temperatures. Front. Genet. 11, 417 (2020).
Noel, K. et al. Transcriptomics of temperature-sensitive R gene-mediated resistance identifies a WAKL10 protein interaction. Sci. Rep. 14, 5023 (2024).
Badawy, H. M. A., Kakau, J. & Hoppe, H. H. Temperature and ageing of host tissue affect the interactions between different oilseed rape cultivars and pathotype groups of Leptosphaeria maculans. J. Phytopathol. 134, 255–263 (1992).
Ansan-Melayah, D., Balesdent, M. H., Buée, M. & Rouxel, T. Genetic characterization of AvrLm1, the first avirulence gene of Leptosphaeria maculans. Phytopathol. 85, 1525–1529 (1995).
Balesdent, M. H. et al. Genetic control and host range of avirulence toward Brassica napus cultivars Quinta and Jet Neuf in Leptosphaeria maculans. Phytopathol 91, 70–76 (2001).
Fernando, D., Rong, L. & Borhan, H. Temperature effects on lesion development and on R-genes when infected by Leptosphaeria maculans causing blackleg disease of canola. Phytopathol. 105, 43 (2015).
Kaur, B. et al. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: Applications, challenges, and prospects. Plants 10, 1989 (2021).
Pedras, M. S. C., Zheng, Q. A., Gadagi, R. S. & Rimmer, S. R. Phytoalexins and polar metabolites from the oilseeds canola and rapeseed: Differential metabolic responses to the biotroph Albugo candida and to abiotic stress. Phytochem. 69, 894–910 (2008).
Abdel-Farid, I. B. et al. Fungal infection-induced metabolites in Brassica rapa. Plant Sci. 176, 608–615 (2009).
Huang, J. et al. Pleiotropic changes in Arabidopsis f5h and sct mutants revealed by large-scale gene expression and metabolite analysis. Planta 230, 1057–1069 (2009).
Botanga, C. J. et al. Metabolite profiling of Arabidopsis inoculated with Alternaria brassicicola reveals that ascorbate reduces disease severity. Mol. Plant Microbe Interact. 25, 1628–1638 (2012).
Huang, Y. J., Mitrousia, G. K., Sidique, S. N. M., Qi, A. & Fitt, B. D. Combining R gene and quantitative resistance increases effectiveness of cultivar resistance against Leptosphaeria maculans in Brassica napus in different environments. PLoS One 13, 0197752 (2018).
Mitrousia, G. K., Huang, Y. J., Qi, A., Sidique, S. N. M. & Fitt, B. D. L. Effectiveness of Rlm7 resistance against Leptosphaeria maculans (phoma stem canker) in UK winter oilseed rape cultivars. Plant Pathol. 67, 1339–1353 (2018).
Padmathilake, K. R. E. & Fernando, W. G. D. Less virulent Leptosphaeria biglobosa immunizes the canola plant to resist highly virulent L. maculans, the blackleg pathogen. Plants 11, 996 (2022).
Desaint, H. et al. Fight hard or die trying: When plants face pathogens under heat stress. New Phytol. 229, 712–734 (2021).
Son, S. & Park, S. R. Climate change impedes plant immunity mechanisms. Front. Plant Sci. 13, 1032820 (2022).
Larkan, N. J., Ma, L., Haddadi, P., Buchwaldt, M. & Parkin, I. A. P. The Brassica napus wall-associated kinase-like (WAKL) gene Rlm9 provides race-specific blackleg resistance. Plant J. 104, 892–900 (2020).
Haddadi, P. et al. Brassica napus genes Rlm4 and Rlm7, conferring resistance to Leptosphaeria maculans, are alleles of the Rlm9 wall-associated kinase-like resistance locus. Plant Biotechol. J. 20, 1229 (2022).
Sade, D. et al. Comparative metabolomics and transcriptomics of plant response to Tomato yellow leaf curl virus infection in resistant and susceptible tomato cultivars. Metabolomics 11, 81–97 (2015).
Kachroo, A. & Kachroo, P. Fatty acid-derived signals in plant defense. Annu. Rev. Phytopathol. 47, 153–176 (2009).
Siedow, J. N. Plant lipoxygenase: Structure and function. Annu. Rev. Plant Biol. 42, 145–188 (1991).
Pietryczuk, A. & Czerpak, R. Effect of traumatic acid on antioxidant activity in Chlorella vulgaris (Chlorophyceae). Plant Growth Regul. 65, 279–286 (2011).
Li, L., Li, C., Lee, G. I. & Howe, G. A. Distinct roles for jasmonate synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. 99, 6416–6421 (2002).
Prost, I. et al. Evaluation of the antimicrobial activities of plant oxylipins supports their involvement in defense against pathogens. Plant Physiol. 139, 1902–1913 (2005).
Raffaele, S. et al. A MYB transcription factor regulates very-long-chain fatty acid biosynthesis for activation of the hypersensitive cell death response in Arabidopsis. Plant Cell 20, 752–767 (2008).
Farmer, E. E., Weber, H. & Vollenweider, S. Fatty acid signaling in Arabidopsis. Planta 206, 167–174 (1998).
Šašek, V. et al. Recognition of avirulence gene AvrLm1 from hemibiotrophic ascomycete Leptosphaeria maculans triggers salicylic acid and ethylene signaling in Brassica napus. Mol. Plant Microbe Interact. 25, 1238–1250 (2012).
Lowe, R. G. et al. Genomes and transcriptomes of partners in plant-fungal-interactions between canola (Brassica napus) and two Leptosphaeria species. PloS One 9, e103098 (2014).
Haddadi, P., Ma, L., Wang, H. & Borhan, M. H. Genome-wide transcriptomic analyses provide insights into the lifestyle transition and effector repertoire of Leptosphaeria maculans during the colonization of Brassica napus seedlings. Mol. Plant Pathol. 17, 1196–1210 (2016).
Yang, C. & Fernando, W. D. Hormonal responses to susceptible, intermediate, and resistant interactions in the Brassica napus–Leptosphaeria maculans pathosystem. Int. J. Mol. Sci. 22, 4714 (2021).
Tamaoki, D. et al. Jasmonic acid and salicylic acid activate a common defense system in rice. Plant Signal. Behav. 8, 24260 (2013).
Gakière, B., Fernie, A. R. & Pétriacq, P. More to NAD+ than meets the eye: A regulator of metabolic pools and gene expression in Arabidopsis. Free Radic. Biol. Med. 122, 86–95 (2018).
Stepansky, A. & Leustek, T. Histidine biosynthesis in plants. Amino Acids 30, 127–142 (2006).
Seo, S. et al. L-Histidine induces resistance in plants to the bacterial pathogen Ralstonia solanacearum partially through the activation of ethylene signaling. Plant Cell Physiol. 57, 1932–1942 (2016).
Nokhrina, K., Ray, H., Bock, C. & Georges, F. Metabolomic shifts in Brassica napus lines with enhanced BnPLC2 expression impact their response to low temperature stress and plant pathogens. GM Crops Food 5, 120–131 (2014).
Bolouri Moghaddam, M. R. & Van den Ende, W. Sweet immunity in the plant circadian regulatory network. J. Exp. Bot. 64, 1439–1449 (2013).
Bolton, M. D. Primary metabolism and plant defense fuel for the fire. Mol. Plant Microbe Interact. 22, 487–497 (2009).
Trouvelot, S. et al. Carbohydrates in plant immunity and plant protection: Roles and potential application as foliar sprays. Front. Plant Sci. 5, 592 (2014).
Ibba, M. & Söll, D. Aminoacyl-tRNA synthesis. Annu. Rev. Biochem. 69, 617–650 (2000).
Zeier, J. New insights into the regulation of plant immunity by amino acid metabolic pathways. Plant Cell Environ. 36, 2085–2103 (2013).
Aliferis, K. A., Faubert, D. & Jabaji, S. A metabolic profiling strategy for the dissection of plant defense against fungal pathogens. PLoS One 9, 111930 (2014).
Tzin, V. & Galili, G. New insights into the shikimate and aromatic amino acids biosynthesis pathways in plants. Mol. Plant. 3, 956–972 (2010).
Shen, J. W. et al. Lycorine: A potential broad-spectrum agent against crop pathogenic fungi. J. Microb. Biotechnol. 24, 354–358 (2014).
Diamond, A. & Desgagné-Penix, I. Metabolic engineering for the production of plant isoquinoline alkaloids. Plant Biotechnol. J. 14, 1319–1328 (2016).
Palacios, C. & Gonzalez, L. Is vitamin D deficiency a major global public health problem?. J. Steroid Biochem. Mol. Biol. 144, 138–145 (2014).
Van Meer, G. & Holthuis, J. C. Sphingolipid transport in eukaryotic cells. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1486, 145–170 (2000).
Strack, D. & Fester, T. Isoprenoid metabolism and plastid reorganization in arbuscular mycorrhizal roots. New Phytol. 172, 22–34 (2006).
Stanley, L. & Yuan, Y. W. Transcriptional regulation of carotenoid biosynthesis in plants: so many regulators, so little consensus. Front. Plant Sci. 10, 1017 (2019).
Felemban, A., Braguy, J., Zurbriggen, M. D. & Al-Babili, S. Apocarotenoids involved in plant development and stress response. Front. Plant Sci. 10, 478231 (2019).
Quan, J. et al. Glutamic Acid and Poly-γ-glutamic Acid Enhanced the Heat Resistance of Chinese Cabbage (Brassica rapa L. ssp. pekinensis) by Improving Carotenoid Biosynthesis, Photosynthesis, and ROS Signaling. Int. J. Mol. Sci. 23, 11671 (2022).
Das, A., Rushton, P. J. & Rohila, J. S. Metabolomic profiling of soybeans (Glycine max L.) reveals the importance of sugar and nitrogen metabolism under drought and heat stress. Plants 6, 21 (2017).
Rincón, M. A., Narvaez-Ortiz, H. Y., López, A. J. & Zimmermann, B. H. Characterization of the pyrimidine catabolic pathway of Oryza sativa. FASEB J. 32, 537–539 (2018).
Lopez, A. J. et al. New Insights into rice pyrimidine catabolic enzymes. Front. Plant Sci. 14, 1079778 (2023).
Garavito, M. F., Narváez-Ortiz, H. Y. & Zimmermann, B. H. Pyrimidine metabolism: Dynamic and versatile pathways in pathogens and cellular development. J. Genet. Genomics 42, 195–205 (2015).
Galili, G. New insights into the regulation and functional significance of lysine metabolism in plants. Annu. Rev. Plant Biol. 53, 27 (2002).
Ali, Q. et al. Role of amino acids in improving abiotic stress tolerance to plants. In Plant tolerance to environmental stress (pp. 175–204) (CRC Press, 2019).
Yang, S. et al. Pepper CaMLO6 negatively regulates Ralstonia solanacearum resistance and positively regulates high temperature and high humidity responses. Plant Cell Physiol. 61, 1223–1238 (2020).
Khare, S. et al. Plant secondary metabolites synthesis and their regulations under biotic and abiotic constraints. J. Plant Biol. 63, 203–2016 (2020).
Gupta, M. & Gupta, S. An overview of selenium uptake, metabolism, and toxicity in plants. Front. Plant Sci. 7, 2074 (2017).
Liu, M. & Lu, S. Plastoquinone and ubiquinone in plants: Biosynthesis, physiological function and metabolic engineering. Front. Plant Sci. 7, 1898 (2016).
Lin, Z. et al. Caffeine synthesis and its mechanism and application by microbial degradation: A review. Foods. 12, 2721 (2023).
Seifert, G. J., Strasser, R. & Van Damme, E. J. Plant glycobiology—a sweet world of glycans, glycoproteins, glycolipids, and carbohydrate-binding proteins. Front. Plant Sci. 12, 751923 (2021).
Amara, A. et al. Networks and graphs discovery in metabolomics data analysis and interpretation. Front. Mol. Biosci. 9, 841373 (2022).
Cardoso, L. L., Freire, F. B. S. & Daloso, D. M. Plant metabolic networks under stress: A multi-species/stress condition meta-analysis. J. Soil Sci. Plant Nutr. 23, 4–21 (2023).
Cohen, S. P. & Leach, J. E. High temperature-induced plant disease susceptibility: more than the sum of its parts. Curr. Opin. Plant biol. 56, 235–241 (2020).
Chen, Y. & Fernando, W. G. D. Induced resistance to blackleg (Leptosphaeria maculans) disease of canola (Brassica napus) caused by a weakly virulent isolate of Leptosphaeria biglobosa. Plant Dis. 90, 1059–1064 (2006).
Koch, E., Song, K., Osborn, T. C. & Williams, P. H. Relationship between pathogenicity and phylogeny based on restriction fragment length polymorphism in Leptosphaeria maculans. Mol. Plant Microbe Interact. 4, 341–349 (1991).
Abràmoff, M. D., Magalhães, P. J. & Ram, S. J. Image processing with ImageJ. Biophoton. Int. 11, 36–42 (2004).
Tortosa, M., Cartea, M. E., Rodríguez, V. M. & Velasco, P. Unraveling the metabolic response of Brassica oleracea exposed to Xanthomonas campestris pv. campestris. J. Sci. Food Agric. 98, 3675–3683 (2018).
Liu, H., Garrett, T. J., Su, Z., Khoo, C. & Gu, L. UHPLC-Q-Orbitrap-HRMS-based global metabolomics reveal metabolome modifications in plasma of young women after cranberry juice consumption. J. Nutr. Biochem. 45, 67–76 (2017).
Farahbakhsh, F. et al. Comparative metabolomics of temperature sensitive resistance to wheat streak mosaic virus (WSMV) in resistant and susceptible wheat cultivars. J. Plant Physiol. 237, 30–42 (2019).
Xia, J. & Wishart, D. Using MetaboAnalyst 3.0 for comprehensive metabolomics data analysis. Curr. Protoc. Bioinformatics. 55, 10 (2016).
Mahdavi, V., Ghanati, F. & Ghassempour, A. Integrated pathway-based and network-based analysis of GC-MS rice metabolomics data under diazinon stress to infer affected biological pathways. Anal. Biochem. 494, 31–36 (2016).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, 587–592 (2023).
Acknowledgements
Z. Amjadi and H. Hamzehzarghani acknowledge the financial support of Shiraz University. YJ. Huang acknowledges financial support from the Department for Environment, Food and Rural Affairs (OREGIN, CH0110). V.M. Rodriguez acknowledges financial support from the Group of Genetics, Breeding, and Biochemistry of Brassicas.
Author information
Authors and Affiliations
Contributions
H. Hamzehzarghani contributed in conceptualization, methodology, and visualization of the investigation, defining the general hypothesis, designing the experiments based on the hypothesis, supervising the research project throughout, analyzing and interpreting the results, administering the chemical and statistical analysis and writing the original draft of the main manuscript and revising, reviewing and/or editing of the text. Z. Amjadi performed the research throughout her Ph.D. project, contributed in analyzing the results, the chemical and statistical analysis, as well as revising, reviewing and editing of the text. V.M. Rodriguez helped us with resources and hyphenated instrumentation by providing UPLC-QTOF/MSm methodology, and formal analysis Y. Huang helped with methodology and providing resources [two canola cultivars (Drakkar—susceptible without any resistance genes and Excel- resistant carrying the resistance gene Rlm7) and one Lm isolate EM24, which possesses three known effectors (AvrLm1, AvrLm6, and AvrLm7)] F. Farahbakhsh helped with chemical and statistical analysis.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Amjadi, Z., Hamzehzarghani, H., Rodriguez, V.M. et al. Studying temperature’s impact on Brassica napus resistance to identify key regulatory mechanisms using comparative metabolomics. Sci Rep 14, 19865 (2024). https://doi.org/10.1038/s41598-024-68345-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68345-3
- Springer Nature Limited