Abstract
Although previous studies have reported that pre-mRNA splicing factors (SFs) are involved in the repair of DNA double-strand breaks (DSBs) via homologous recombination (HR), their exact role in promoting HR remains poorly understood. Here, we showed that SART1, an SF upregulated in several types of cancer, promotes DSB end resection, an essential first step of HR. The resection-promoting function of SART1 requires phosphorylation at threonine 430 and 695 by ATM/ATR. SART1 is recruited to DSB sites in a manner dependent on transcription and its RS domain. SART1 is epistatic with BRCA1, a major HR factor, in the promotion of resection, especially transcription-associated resection in the G2 phase. SART1 and BRCA1 accumulate at DSB sites in an interdependent manner, and epistatically counteract the resection blockade posed by 53BP1 and RIF1. Furthermore, chromosome analysis demonstrated that SART1 and BRCA1 epistatically suppressed genomic alterations caused by DSB misrepair in the G2 phase. Collectively, these results indicate that SART1 and BRCA1 cooperatively facilitate resection of DSBs arising in transcriptionally active genomic regions in the G2 phase, thereby promoting faithful repair by HR, and suppressing genome instability.
Similar content being viewed by others
Introduction
Dysregulated RNA splicing has recently been linked to both promotion and suppression of tumorigenesis, as mutations and altered expression (both upregulation and downregulation) of splicing factors (SFs) have been found in a variety of human malignancies1. In addition to their central role in shaping the transcriptome, recent studies have revealed the involvement of SFs in suppressing genomic instability, which is a hallmark of cancer2. For example, a previous study demonstrated that small interfering RNA (siRNA)-mediated knockdown of SFs leads to elevated levels of histone H2AX phosphorylation at serine 139, which represents DNA damage, including DNA double-strand breaks (DSBs)3. Moreover, two genome-wide screening studies have identified many SFs as factors that promote homologous recombination (HR), which is a major DSB repair pathway4,5. Previous studies have shown that both the splicing-dependent and splicing-independent functions of SFs contribute to HR promotion6,7,8,9,10,11. However, the splicing-independent function (i.e., the direct function) of SFs in HR is poorly understood.
In addition to HR, there is another major DSB repair pathway, non-homologous end-joining (NHEJ). NHEJ is active throughout the cell cycle, whereas HR operates mainly in the S/G2 phase, when sister chromatids are available12. HR is believed to be accurate in somatic cells because conservative synthesis-dependent strand annealing, an HR sub-pathway, is predominant in this cell type13. Thus, in the S/G2 phase, both NHEJ and HR can operate, and the mechanism of choice between these two pathways has been extensively studied. Accumulating evidence indicates that DSB end resection, which generates the 3’-overhang required to initiate HR, is a critical determinant of pathway choice between NHEJ and HR13,14,15. It is known that BRCA1 directs the repair pathway toward HR by promoting resection16. BRCA1 antagonizes 53BP1, which inhibits resection17. Upon DSBs, 53BP1 is phosphorylated by ATM kinase and phosphorylated 53BP1 recruits RIF1, a downstream effector of 53BP1, to DSBs, thereby blocking resection17. BRCA1 facilitates resection by promoting protein phosphatase 4C-dependent 53BP1 dephosphorylation and RIF1 release from DSBs18. However, the involvement of SFs in the antagonism between BRCA1 and 53BP1 over the choice of repair pathway remains largely unknown.
Among the SFs implicated in HR in the previous studies, we focused on SART1 in this study because of its upregulated expression in several types of human cancers and its correlation with tumor resistance to DNA-damaging agents4,19,20,21. In this study, we revealed that SART1 is recruited to DSB sites and promotes resection of DSBs, particularly those arising in transcriptionally active genomic regions in the G2 phase. We obtained evidence that ATM/ATR-dependent phosphorylation at threonine 430 and 695 is critical for SART1 to promote resection. This resection-promoting function of SART1 appears to be closely interlinked with that of BRCA1, where SART1 operates epistatically with BRCA1 to counteract 53BP1/RIF1-mediated resection blockade. Moreover, our results demonstrate that recruitment of SART1 and BRCA1 to DSB sites is interdependent. Consistent with the epistatic relationship between SART1 and BRCA1 during resection, these factors cooperatively suppress genomic alterations caused by DSB misrepair in the G2 phase. These results indicate that when DSBs occur in transcriptionally active genomic regions, SART1 is rapidly recruited to DSB sites and recruits BRCA1, which facilitates resection by antagonizing the 53BP1/RIF1 resection barrier, thereby promoting faithful DSB repair by HR.
Results
SART1 promotes HR by facilitating DSB end resection
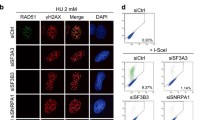
This study aimed to investigate the mechanism by which SART1 promotes HR. First, we reassessed the involvement of SART1 in HR using a well-established HR reporter assay (direct repeat (DR)-green fluorescent protein (GFP) assay)22. This assay uses a human osteosarcoma U2OS cell line with a chromosomally integrated recognition site for the I-Sce I endonuclease, and the cells express GFP when the I-Sce I-induced DSB is repaired by HR. As shown in Fig. 1a–b, siRNA-mediated knockdown of SART1 significantly decreased the percentage of HR( +) cells although S/G2 phase cells, where HR occurs, were increased in SART1 knockdown cells (knockdown of SART1 protein was confirmed by western blotting (WB) in Supplementary Fig. S1a–b). We also confirmed that mRNA level of transfected I-Sce I was not changed by SART1 knockdown (Supplementary Fig. S1c–d). Thus, in consistent with the previous finding, SART1 is involved in HR4. Next, RNA sequencing (RNA-seq) was performed to determine the effect of SART1 knockdown on the transcriptome of human telomerase reverse transcriptase-immortalized retinal pigment epithelial (RPE-hTERT) cells. Analysis of RNA-seq data demonstrated that SART1 knockdown did not affect the mRNA expression levels of most genes, including known HR factors (Supplementary Fig. S2a–b). Furthermore, SART1 knockdown did not affect the protein levels of major HR factors, such as MRE11, CtIP, BRCA1, and RAD51, in RPE-hTERT cells (Supplementary Fig. S3). To further confirm the involvement of SART1 in HR, we examined nuclear foci of RAD51, another established indicator of HR5. RAD51 foci in the G2 phase cells after exposure to ionizing radiation (IR) were analyzed because in this study, we mostly focused on HR of two-ended DSBs induced by IR in the G2 phase. To identify G2 phase cells, CENPF protein (S/G2 phase marker) and 5-ethynyl-2′-deoxyuridine (EdU, S phase marker) were concomitantly visualized and EdU( −)/CENPF( +) cells were regarded as G2 phase cells (Fig. 1c). SART1 knockdown by two distinct siRNAs significantly decreased the number of IR-induced RAD51 foci (Fig. 1c–e). Next, we examined the role of SART1 in DSB end resection, an essential first step in HR23. To visualize the sites of DSBs that underwent resection, the nuclear foci of replication protein A (RPA), a widely used indicator of resected DSBs, was employed (Fig. 1f)18,24. SART1 knockdown significantly decreased the number of RPA foci in G2-irradiated cells (Fig. 1f–h). Collectively, these results indicate that SART1 promotes HR by facilitating DSB end resection.
SART1 promotes homologous recombination (HR) by facilitating DSB end resection. (a) Effect of SART1 knockdown on HR efficiency. U2OS cells harboring the HR reporter were transfected with the indicated siRNA and subjected to the DR-GFP assay. The data are presented as median with interquartile range (N = 4). (b) Effect of SART1 knockdown on the cell cycle. U2OS cells harboring the HR reporter were transfected with the indicated siRNA. Cells were then fixed, and the nuclei were stained with propidium iodide (PI). The cell cycle status was identified by flow cytometry. (c) Immunofluorescence images of RAD51 foci in SART1 knockdown cells. Wild-type RPE-hTERT cells (RPE-hTERT WT) were transfected with indicated siRNAs. Two days after siRNA transfection, cells were irradiated with 2 Gy γ-rays and fixed 2 h later. The cells were treated with EdU 30 min before irradiation until fixation to label S-phase cells. Fixed cells were subjected to RAD51/CENPF immunofluorescence and EdU detection. RAD51 foci in CENPF( +)/EdU( −) G2 phase cells are shown. (d) Number of RAD51 foci in siSART1#1-transfected cells. Immunofluorescence samples were prepared as described in (c). (e) Number of RAD51 foci in siSART1#2-transfected cells. Immunofluorescence samples were prepared as described in (c). (f) Immunofluorescence images of RPA foci in SART1 knockdown cells. siRNA transfection, EdU treatment, irradiation, and fixation were performed, as described in (c). Fixed cells were subjected to RPA/CENPF immunofluorescence and EdU detection. RPA foci in CENPF( +)/EdU( −) G2 phase cells are shown. (g) Number of RPA foci in siSART1#1-transfected cells. Immunofluorescence samples were prepared as described in (f). (h) Number of RPA foci in siSART1#2-transfected cells. Immunofluorescence samples were prepared as described in (f). In (d), (e), (g) and (h), the number of foci in 100 G2 cells from two independent experiments (50 G2 cells/experiment/sample) is shown. Symbols and red bars in (d), (e), (g) and (h) represent the number of foci per cell and the median number of foci in each sample, respectively. Scale bars in (c) and (f), 5 µm.
ATM/ATR-dependent phosphorylation is important for the resection-promoting function of SART1
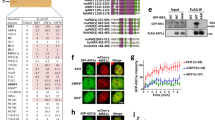
To identify a posttranslational modification or a domain required for the resection-promoting function of SART1, we prepared vectors expressing SART1 mutants. SART1 has two threonine residues (T430 and T695) that are phosphorylated by ATM/ATR kinases25. Thus, we prepared two SART1 mutants that lacked (T430A/T695A) or mimicked (T430E/T695E) phosphorylation (Fig. 2a, these mutants are hereafter referred to as 2A and 2E, respectively). Furthermore, SART1 has a leucine zipper motif that is involved in dimerization and DNA binding26. SART1 also possesses an arginine/serine-rich domain at its amino-terminus known as the RS domain, which is shared by the SR protein family of SFs27,28. Therefore, we constructed SART1 mutants that lacked the leucine zipper motif (ΔLZ) or the RS domain (ΔRS) (Fig. 2a). Silent mutations were introduced into the wild-type (WT) and mutant SART1 cDNAs to make them resistant to SART1 siRNA (siSART1#1). Using these WT or mutant SART1 cDNAs, we made lentiviral vectors that express FLAG-tagged WT or mutant SART1 protein in a doxycycline (Dox)-dependent manner. Then, FLAG-SART1-inducible RPE-hTERT cell lines were established after viral infection and puromycin selection. Dox-inducible expression of FLAG-tagged WT or mutant SART1 proteins in the presence of siSART1#1 was confirmed by WB (Fig. 2b and Supplementary Fig. S4). We examined resection-promoting activity of WT or mutant SART1 by quantifying IR-induced RPA foci in these cells that had been treated with siSART1#1 and Dox. We found that the 2A mutant did not rescue RPA foci number in SART1 knockdown cells while other mutants, including the phospho-mimic 2E mutant, rescued them comparably to WT (Fig. 2c), indicating that ATM/ATR-dependent phosphorylation is important for the resection-promoting activity of SART1.
ATM/ATR-dependent phosphorylation is important for the resection-promoting function of SART1. (a) Diagram of wild-type and mutant SART1 used for the add-back experiments. The domains and ATM/ATR-dependent phosphorylation sites in SART1 are shown. (b) Doxycycline (Dox)-dependent expression of FLAG-tagged WT and mutant SART1 proteins. FLAG-SART1-inducible RPE-hTERT cells were transfected with siSART1#1 and were lysed two days later. Dox was added 16–24 h before cell lysis to induce FLAG-SART1 protein. Then, the cell lysates were subjected to western blotting. (c) The resection-promoting activity of WT and mutant SART1. FLAG-SART1-inducible RPE-hTERT cells were treated with siSART1#1 and Dox for 2 days and 16–24 h, respectively. Then, the cells were irradiated with 2 Gy γ-rays and fixed 2 h later. The cells were treated with EdU from 30 min before irradiation until fixation to label S phase cells. The fixed cells were subjected to RPA/CENPF immunofluorescence and EdU detection. The number of RPA foci in the CENPF( +)/EdU( −) G2 phase cells was quantified. The number of foci in 100 G2 cells from two independent experiments (50 G2 cells/experiment/sample) is shown. Symbols and red bars represent the number of foci per cell and the median number of foci in each sample, respectively. ns, not significant.
SART1 is recruited to DSB sites in a manner dependent on transcription and its RS domain
If SART1 plays a direct role in HR, it should be recruited to DSBs. Therefore, we performed live-cell imaging to examine SART1 recruitment to DSBs using a near-infrared (730 nm) two-photon microbeam that induces DSBs in the presence of a photosensitizer24,29. For this purpose, the WT SART1 cDNA were inserted into the GFP vector to visualize SART1 behavior in live cells. When U2OS cells expressing GFP-SART1 and mCherry-Geminin (a marker of S/G2 phases) were irradiated with the laser, GFP-SART1 accumulated along the laser track in S/G2 phase cells (Fig. 3a). Accumulation began 10 s after irradiation, peaked at 30–40 s, and persisted for at least 110 s (Fig. 3b). The treatment of a transcription inhibitor (DRB, 5,6-dichloro-1-b-D-ribofuranosylbenzimidazole) significantly reduced the recruitment of GFP-SART1 to the laser-irradiated regions without affecting expression level of GFP-SART1 protein (Fig. 3c–d and Supplementary Fig. 5a–b). We next examined the recruitment of the WT and mutant SART1 to DSBs. U2OS cells expressing mCherry-Geminin were transfected with SART1 siRNA to deplete endogenous SART1 and then transfected with the WT or the mutant GFP-SART1 vector. GFP( +)/mCherry( +) cells were subjected to laser irradiation, and GFP-SART1 accumulation along the laser track was assessed. We found that the ΔRS mutant failed to accumulate along the laser track, whereas the 2A, 2E, and ΔLZ mutants showed accumulation (Figs. 3e–f). Consistent with the results of the phosphorylation mutants, treatment with the ATM inhibitor did not influence the recruitment of the WT GFP-SART1 to the laser track (Supplementary Fig. S5c).
SART1 is recruited to DSB sites in a manner dependent on transcription and its RS domain (a) SART1 is recruited to laser-induced DSBs. U2OS cells expressing mCherry-Geminin (an S/G2 phase marker) were transfected with GFP-SART1 vector. One day later, GFP( +)/mCherry( +) cells were irradiated with the 730 nm laser along the line flanked by two red arrowheads. A photosensitizer (Hoechst33342, 10 µg/mL) was added 30 min before irradiation. (b) Kinetics of SART1 recruitment to the laser track. Vector transfection and laser irradiation were performed as described in (a). The intensity of GFP-SART1 in the laser-irradiated regions of S/G2-phase U2OS cells was recorded every 10 s until 110 s after irradiation. N = 6. (c) Effect of transcription inhibition on SART1 recruitment to the laser track. Vector transfection and laser irradiation were done as described in (a). The cells were treated with DRB (100 µM) or DMSO (vehicle control) from 30 min before irradiation. (d) Transcription inhibition suppresses SART1 recruitment to the laser track. Vector transfection, chemical treatment, and laser irradiation were performed as described in (c). The intensity of GFP-SART1 was recorded as descried in (b). N = 6. (e) Live cell images of the wild-type and mutant SART1 proteins after laser irradiation. U2OS cells expressing mCherry-Geminin (an S/G2 phase marker) were transfected with wild-type or mutant GFP-SART1 vectors. One day later, GFP( +)/mCherry( +) cells were irradiated as described in (a). (f) Kinetics of recruitment of wild-type and mutant SART1 to the laser track. Vector transfection and laser irradiation were performed as described in (e). The intensity of GFP-SART1 was recorded as descried in (b). N = 8. Scale bars in (a), (c), and (e), 5 µm. Error bars in (b), (d), and (f) represent standard deviation.
SART1 promotes HR and resection in an epistatic manner with BRCA1 in the G2 phase
It is well-established that BRCA1 promotes HR by facilitating resection16. Therefore, we next examined the role of SART1 in BRCA1-dependent HR and resection. We first compared HR efficiency in BRCA1 and/or SART1 knockdown cells by DR-GFP assay with the cell cycle distribution being concomitantly monitored (Supplementary Fig. S6a–b). Although the knockdown of either BRCA1 or SART1 reproducibly reduced HR( +) cells, the percentage of HR( +) cells in BRCA1 knockdown cells was constantly lower than that in SART1 knockdown cells (Supplementary Fig. S6c–d). The double knockdown of BRCA1 and SART1 reduced HR( +) cells similarly to the single knockdown of BRCA1 (Supplementary Fig. S6c–d). These results indicate that BRCA1 makes a greater contribution than SART1 to overall HR events. Next we focused on HR in the G2 phase by quantifying IR-induced RAD51 foci. Single knockdown of BRCA1 and SART1 similarly reduced the number of RAD51 foci in the G2 phase (Fig. 4a). The double knockdown of BRCA1 and SART1 did not additively decrease the number of RAD51 foci when compared to the single knockdown of BRCA1 or SART1, indicating that these two proteins epistatically promote HR in the G2 phase (Fig. 4a, the knockdown of BRCA1 and SART1 was confirmed by WB or immunofluorescence (IF) in Supplementary Fig. S7a–b and S8a–b). Moreover, the number of IR-induced RPA foci in the G2 phase were similarly reduced by the single knockdown of BRCA1 and SART1, and the double knockdown of these proteins did not show an additive effect, indicating epistasis between BRCA1 and SART1 in resection in the G2 phase (Fig. 4b).
Epistatic relationship between SART1 and BRCA1 in HR and resection in the G2 phase. (a) Number of RAD51 foci in BRCA1 and/or SART1 knockdown cells. RPE-hTERT WT cells were transfected with indicated siRNA(s). Two days after siRNA transfection, the cells were irradiated with 2 Gy γ-rays and fixed 2 h later. The cells were treated with EdU from 30 min before irradiation until fixation to label S phase cells. The fixed cells were subjected to RAD51/CENPF immunofluorescence and EdU detection. The number of RAD51 foci in the CENPF( +)/EdU( −) G2 phase cells was quantified. (b) Number of RPA foci in BRCA1 and/or SART1 knockdown cells. siRNA transfection, EdU treatment, irradiation, and fixation were performed as described in (a). The fixed cells were subjected to RPA/CENPF immunofluorescence and EdU detection. The number of RPA foci in the CENPF( +)/EdU(−) G2 phase cells was quantified. (c) Immunofluorescence images of BRCA1 foci in Rap80-knockout cells. Rap80-knockout RPE-hTERT cells (RPE-hTERT ΔRap80) were transfected with indicated siRNAs. EdU treatment, irradiation, and fixation were performed as described in (a). Fixed cells were subjected to BRCA1/CENPF immunofluorescence and EdU detection. BRCA1 foci in CENPF( +)/EdU( −) G2 phase cells are shown. (d) Effect of SART1 knockdown on the number of BRCA1 foci in Rap80-knockout cells. The immunofluorescence samples were prepared as described in (c). The number of BRCA1 foci in the CENPF( +)/EdU( −) G2 phase cells was quantified. (e) SART1 recruitment to DSB sites is BRCA1-dependent. The laser irradiation was performed as described in Methods. The intensity of GFP-SART1 in the laser-irradiated regions was recorded every 10 s until 110 s after irradiation. (f) SART1 promotes transcription-associated resection in an epistatic manner with BRCA1. RPA immunofluorescence samples were prepared as described in (b). Cells were treated with DRB (100 µM) or DMSO from 30 min before irradiation until fixation. In (a), (b), (d) and (f), the number of foci in 100 G2 cells from two independent experiments (50 G2 cells/experiment/sample) is shown. Symbols and red bars in (a), (b), (d) and (f) represent the number of foci per cell and the median number of foci in each sample, respectively. Scale bars in (c), 5 µm.
SART1 and BRCA1 are recruited to DSB sites in an interdependent manner
Upon DSBs, BRCA1 is recruited to DSB sites and forms discrete nuclear foci30. To gain more insight into the role of SART1 in BRCA1-dependent resection, we examined the impact of SART1 knockdown on the recruitment of BRCA1 to DSBs by assessing BRCA1 foci formation. We found that SART1 knockdown significantly decreased the number of IR-induced BRCA1 foci in WT RPE-hTERT cells (Supplementary Figs. S7c–d). Previous studies identified several distinct complexes of BRCA1 at DSB sites; among them, the BRCA1-Rap80 complex suppresses HR, whereas other complexes promote HR31. To visualize recruitment of only HR-promoting BRCA1, we next examined the number of BRCA1 foci in cells lacking Rap80 (RPE-hTERT ΔRap80 cells). Although the number of BRCA1 foci in ΔRap80 cells was much lower than that in WT cells, SART1 knockdown significantly reduced the number of BRCA1 foci in G2-irradiated ΔRap80 cells (Fig. 4c–d), indicating that SART1 promotes the recruitment of the HR-promoting portion of BRCA1 to DSBs. We next examined if SART1 recruitment to DSBs is affected by BRCA1 knockdown. The laser irradiation experiments revealed that BRCA1 knockdown significantly decreased the recruitment of SART1 along the laser track (Fig. 4e). Collectively, these results indicate that the recruitment of BRCA1 and SART1 to DSB sites is interdependent.
SART1 promotes transcription-associated resection in an epistatic manner with BRCA1
We previously discovered an HR sub-pathway that repairs DSBs arising in transcriptionally active genomic regions in the G2 phase and found that BRCA1 is involved in this transcription-associated HR24. Considering that SFs, such as SART1, should be located at transcription sites, we hypothesized that SART1 also participates in the transcription-associated HR. Indeed, SART1 was recruited to DSB sites in a transcription-dependent manner (Fig. 3c–d). We previously demonstrated that transcriptional inhibition by DRB reduced the number of RPA foci in G2-irradiated cells, indicating that active transcription is required for the resection of a subset of DSBs24. Therefore, we examined the role of SART1 in the transcription-associated resection. Consistent with our previous results, transcriptional inhibition by DRB reduced the number of RPA foci in siControl-transfected cells (Fig. 4f, cf. siControl DMSO and siControl DRB). However, the single knockdown of BRCA1 or SART1 or the double knockdown of BRCA1 and SART1 did not further decrease the number of RPA foci in DRB-treated cells (Fig. 4f, cf. siControl, siBRCA1, siSART1#1, and siBRCA1 + siSART1#1 in the DRB-treated group). Moreover, DRB treatment did not additively decrease RPA foci in BRCA1, SART1, or BRCA1+SART1 knockdown cells (Fig. 4f, cf. DMSO vs DRB in siBRCA1-, siSART1#1-, and siBRCA1 + siSART1#1-treated samples). These results indicate that SART1 promotes transcription-associated resection in an epistatic manner with BRCA1.
SART1 and BRCA1 epistatically counteract 53BP1/RIF1-mediated resection blockade
Current evidence indicates that BRCA1 promotes resection by antagonizing the resection barrier posed by 53BP1 and its downstream effectors such as RIF113,15,16. Thus, we next examined the role of SART1 in this BRCA1’s function. A previous study reported that loss of 53BP1 restores impaired resection and RAD51 foci in BRCA1-deficient cells32. Consistent with this, 53BP1 knockdown restored the number of RPA foci in BRCA1 knockdown cells comparably to that in siControl-treated cells (Fig. 5a). 53BP1 knockdown also restored the number of RPA foci in SART1 knockdown cells to control level (Fig. 5a, the knockdown of BRCA1, SART1, and 53BP1 was confirmed by IF or WB in Supplementary Fig. S8–9). Consistently, 53BP1 knockdown also restored the number of RAD51 foci in BRCA1 and SART1 knockdown cells (Fig. 5b). A previous study demonstrated that BRCA1 promotes 53BP1 dephosphorylation and RIF1 release from DSBs18. Thus, we next examined the role of SART1 in BRCA1-dependent 53BP1 dephosphorylation and RIF1 release from DSBs. Nuclear foci of threonine 543-phosphorylated 53BP1 (53BP1 pT543) were used to monitor 53BP1 phosphorylation at DSB sites. At both early (30 min) and late (4 h) time points after IR, the number of 53BP1 pT543 foci was significantly higher in BRCA1- or SART1 single knockdown cells or BRCA1+SART1 double knockdown cells than in control cells (Fig. 5c–d). The number of 53BP1 pT543 foci was comparable between BRCA1 knockdown and SART1 knockdown cells, and the BRCA1+SART1 double knockdown did not additively increase the number of 53BP1 pT543 foci compared with the single knockdown of BRCA1 or SART1, at both 30 min and 4 h after IR (Fig. 5c). Next, we examined the impact of BRCA1 and/or SART1 knockdown on RIF1 foci, an indicator of RIF1 localization at DSBs18,33. Like the number of 53BP1 pT543 foci, the number of RIF1 foci in BRCA1 or SART1 single knockdown cells was significantly higher than that in control cells at both 30 min and 4 h after IR (Fig. 5e–f). BRCA1+SART1 double knockdown did not further increase the number of RIF1 foci compared to BRCA1 or SART1 single knockdown at either time point (Fig. 5e). These results indicate that SART1 and BRCA1 epistatically counteract the 53BP1/RIF1-mediated resection blockade by facilitating 53BP1 dephosphorylation and RIF1 release from DSBs.
SART1 and BRCA1 epistatically counteract the 53BP1/RIF1-mediated resection blockade. (a) Effect of 53BP1 knockdown on the number of RPA foci in BRCA1 or SART1 knockdown cells. RPE-hTERT WT cells were transfected with the indicated siRNA(s). Two days after siRNA transfection, the cells were irradiated and fixed as indicated. The cells were treated with EdU from 30 m before irradiation until fixation to label S phase cells. The fixed cells were subjected to RPA/CENPF immunofluorescence and EdU detection. Foci number in CENPF(+)/EdU(−) G2 phase was quantified. (b) Effect of 53BP1 knockdown on the number of RAD51 foci in BRCA1 or SART1 knockdown cells. The experiment and foci number quantification were performed as described in (a) except for immunofluorescence of RAD51. (c) Effect of BRCA1 and/or SART1 knockdown on 53BP1 phosphorylation at DSBs. RPE-hTERT WT cells were transfected with the indicated siRNA(s). The experiment and foci number quantification were performed as described in (a) except for immunofluorescence of phosphorylated 53BP1 at threonine 543 (53BP1 pT543). (d) Immunofluorescence images of 53BP1 pT543 foci 4 h after irradiation. Samples were prepared as described in (c). 53BP1 pT543 foci in G2 cells are shown. (e) Effect of BRCA1 and/or SART1 knockdown on RIF1 foci formation at DSBs. The experiment and foci number quantification were performed as described in (a) except for immunofluorescence of RIF1. (f) Immunofluorescence images of RIF1 foci 4 h after irradiation. The samples were prepared as described in (e). RIF1 foci in G2 cells are shown. In (a–c) and (e), the foci number in 100 G2 cells from two independent experiments (50 G2 cells/experiment/sample) is shown. Symbols and red bars in (a–c) and (e) represent the number of foci per cell and the median number of foci in each sample, respectively. Scale bars in (d) and (f), 5 µm.
SART1 and BRCA1 epistatically suppress genomic alterations in the G2 phase
Finally, we examined the genomic consequences of HR defects caused by SART1 knockdown. For this purpose, mitotic chromosomes in G2-phase-irradiated cells were analyzed. To strictly analyze G2 phase-derived chromosomal aberrations, cells were treated with aphidicolin, an inhibitor of DNA synthesis, from 30 min before IR until harvest of mitotic cells to prevent S phase-irradiated cells from progressing to the G2 phase (Fig. 6a). In this analysis, two types of chromosomal aberrations were observed: chromatid break and interchromatid fusion (Fig. 6b). Chromatid break and interchromatid fusion indicate an unrepaired DSB on a chromatid and misrepair between DSBs on two or more distinct chromatids, respectively. Although the frequency of chromatid break was comparable among siControl-, siBRCA1-, siSART1#1-, and siBRCA1+siSART1#1-transfected cells (Fig. 6c, e), single knockdown of BRCA1 and SART1, or double knockdown of these factors significantly increased the frequency of interchromatid fusion (Fig. 6d, f). The frequency of interchromatid fusion was similar between BRCA1 and SART1 knockdown cells. Consistent with the epistatic relationship between BRCA1 and SART1 in HR, the double knockdown of these factors did not further increase the frequency of interchromatid fusion compared to the single knockdown of BRCA1 or SART1 (Fig. 6f). These results indicated that SART1 and BRCA1 epistatically suppressed genomic alterations when DSBs occur in the G2 phase.
SART1 and BRCA1 epistatically suppress genomic alterations in the G2 phase. (a) Scheme of chromosome experiments in (c–f). (b) Representative images of the detected chromosomal aberrations. Yellow arrowheads indicate aberrations. The chromosomes were stained with DAPI. Scale bars, 2 µm. (c) Frequency of chromatid break in BRCA1 and/or SART1 knockdown cells. RPE-hTERT WT cells were transfected with the indicated siRNA(s). Two days after siRNA transfection, the cells were treated, and chromosome samples were prepared as shown in (a). In total, one-hundred metaphase cells from two independent experiments (50 metaphases/experiment/sample) were analyzed for each sample. The numbers of cells with each number of chromatid break(s) are shown. (d) Frequency of interchromatid fusion in BRCA1 and/or SART1 knockdown cells. Chromosome samples were prepared and analyzed as described in (c). The numbers of cells with each number of interchromatid fusion(s) are shown. (e) BRCA1 and/or SART1 knockdown does not affect the frequency of chromatid break. Chromosome samples were prepared and analyzed as described in (c). The numbers of cells with each number of chromatid break(s) are shown. (f) SART1 and BRCA1 epistatically suppress interchromatid fusion. Chromosome samples were prepared and analyzed as described in (c). The numbers of cells with each number of interchromatid fusion(s) are shown.
Discussion
In this study, we demonstrated that SART1 promotes DSB end resection in the G2 phase. The add-back experiments suggested that the resection-promoting function of SART1 requires phosphorylation at T430 and T695 by ATM/ATR. SART1 was found to be recruited to DSB sites in a manner dependent on transcription and its RS domain. Our results indicate that SART1 was epistatic with BRCA1 in the promotion of resection, especially, transcription-associated resection. SART1 and BRCA1 are recruited to DSB sites in an interdependent manner and epistatically alleviate resection barrier posed by 53BP1 and RIF1. Moreover, chromosome analysis demonstrated that SART1 and BRCA1 epistatically suppressed genomic alterations resulting from DSBs in the G2 phase.
Previous studies have reported that many SFs are involved in HR4,5,6,7,8,9,10,11,34,35. While many SFs promote HR by supporting the proper expression of HR factors, some SFs seem to have a direct role in HR, in addition to their canonical function in splicing7,10. We consider that SART1 may promote HR directly rather than indirectly by maintaining proper expression of HR factors, because RNA-seq results showed that SART1 knockdown did not affect the expression of known HR-related factors (Supplementary Figs. S2–3). The direct role of SART1 in HR is also indicated by the rapid recruitment of this SF to laser-induced DSBs in the S/G2 phase cells (Figs. 3a–b). Previous studies have reported that several SFs, such as PRP19 and PPM1G, are also recruited to laser-induced DSBs36,37. However, this phenotype is not common to all SFs because some SFs do not accumulate in laser-irradiated regions (e.g., PRP3) or are excluded from (e.g., THRAP3) laser tracks36,37. Therefore, it is likely that recruitment to DSB sites is limited to SFs that are directly involved in DSB repair or response.
The add-back experiments that complement WT or mutant SART1 into SART1 knockdown cells demonstrated that the phosphorylation-deficient mutant (2A) could not restore RPA foci number while the phosphorylation-mimic mutant (2E) fully restored it (Fig. 2c). These results suggest that ATM/ATR-dependent phosphorylation at T430 and T695 is required for the resection-promoting function of SART1. This is reminiscent of the phenotype of phosphorylation-deficient mutant (S289A) of SF3B2, which cannot fully rescue the defects of resection and HR in SF3B2 knockdown cells10. It was shown that the resection-promoting function of SF3B2 is attributable to its role in CtIP recruitment to DSBs, which cannot be played by the S289A mutant10. The SART1 phosphorylation does not seem to have a measurable impact on its localization at DSBs because both mutation at T430/T695 and ATM inhibition did not significantly alter the SART1 recruitment to the laser track (Fig. 3e–f and Supplementary Fig. S5c). Thus, in the future study, it should be examined whether SART1 recruits some resection-promoting factor, such as BRCA1, to DSBs in a phosphorylation-dependent manner. Regarding the ΔRS mutant, we obtained contradicting results: the ΔRS mutant was not recruited to laser track but could rescue resection defect (Figs. 2c, 3f). Although it is possible that subdetectable level of SART1 recruitment might be enough to promote resection, further study is needed to clarify this point.
Previous studies have investigated the role of BRCA1 in resection and established that BRCA1 promotes resection by counteracting 53BP1/RIF1-mediated resection blockade13,15,16. Our data demonstrated that SART1 is also involved in this process because (1) the reduced number of RPA foci in SART1 knockdown cells was restored by 53BP1 knockdown and (2) the foci numbers of phosphorylated 53BP1 and RIF1 were increased by SART1 knockdown, which were both similarly observed in BRCA1 knockdown cells (Fig. 5a, c–f). SART1 is likely to contribute to BRCA1 antagonism against 53BP1/RIF1 by recruiting the HR-promoting portion of BRCA1 to DSBs because SART1 knockdown impaired BRCA1 foci formation in Rap80-knockout cells, where only HR-promoting BRCA1 complexes are present (Fig. 4c–d)31. Moreover, the laser microirradiation experiment demonstrated that SART1 requires BRCA1 for efficient recruitment to laser-induced DSBs (Fig. 4e). The interdependence between SART1 and BRCA1 for DSB recruitment support the epistatic relationship between these factors in HR. Thus, our results indicate that upon DSB induction, SART1 is rapidly recruited to DSBs in a BRCA1-dependent manner, and then, SART1 recruits HR-promoting BRCA1 complexes to DSBs, thereby relieving the resection barrier posed by 53BP1 and RIF1.
We previously demonstrated that BRCA1 is involved in transcription-associated HR, in which a subset of DSBs requires ongoing transcription for end resection24. Our data indicate that SART1 participates in the resection step of the transcription-associated HR in an epistatic manner with BRCA1 (Fig. 4f). The involvement of SART1 in the transcription-associated resection was also supported by the transcription-dependent accumulation of SART1 at laser-induced DSBs (Fig. 3c–d). Thus, it is likely that when DSBs occur in transcriptionally active genomic regions, SART1 is recruited to such DSBs and recruits BRCA1, which facilitates resection by promoting 53BP1 dephosphorylation and RIF1 release from DSBs.
Previous studies have reported that DNA breaks in BRCA1-deficient cells tend to form radial chromosomes, which is a type of interchromatid fusion32,38. Consistently, our chromosome analysis demonstrated that BRCA1 knockdown increased the frequency of interchromatid fusion after IR in the G2 phase (Fig. 6d, f). Moreover, our results indicated that BRCA1 and SART1 epistatically suppressed this type of genomic alteration because the frequency of interchromatid fusion was similarly increased by the single knockdown of BRCA1 and SART1, and it was not further increased when both factors were knocked down (Fig. 6f). Given the epistatic relationship between SART1 and BRCA1 in resection, it is likely that these factors collaboratively direct the repair pathway toward HR by promoting resection; otherwise, a more error-prone, non-HR pathway operates, which increases genome instability.
In summary, this study has uncovered an epistatic relationship between SART1 and BRCA1 in HR, which, to the best of our knowledge, has not been reported previously. Since SF genes are mutated or aberrantly expressed in various types of cancer and hematopoietic malignancies, cancer therapies targeting SFs have recently emerged. Current strategies include direct inhibition of SFs (e.g., SF3B1) or indirect inhibition through the modulation of post-translational modifications of SFs (e.g., methylation, phosphorylation, and ubiquitylation) by upstream modifiers1. Because many SFs are involved in DNA repair, a combination of SF inhibitors and DNA-damaging cancer therapies, such as radiotherapy and platinum-based therapies, is expected to improve the efficacy of cancer therapies. Thus, further studies on the roles of SFs in DNA repair will contribute to the development of SF inhibition-based cancer therapies.
Methods
Cell culture and irradiation
Human telomerase reverse transcriptase-immortalized retinal pigment epithelial cells (RPE-hTERT) were cultured in Dulbecco’s modified Eagle’s medium (DMEM)/Ham’s F12 medium supplemented with 10% fetal bovine serum (FBS). U2OS cells were cultured in α-MEM supplemented with 10% FBS. Cells were subjected to gamma-ray irradiation using a 137Cs source.
siRNA transfection
For RPE-hTERT cells, siRNA transfection was performed using DharmaFECT 4 (Horizon Discovery, Cambridge, UK), following the manufacturer’s instructions. Briefly, 1 × 105 cells were seeded in a 35-mm dish and cultured for 1 day. Next, siRNA and DharmaFECT 4 were separately diluted in Opti-MEM, mixed, and incubated for 20 min at room temperature (RT). Cells were incubated with a mixture of siRNA and DharmaFECT 4 diluted in culture medium containing 20% FBS for 1 day. The medium was then replaced with fresh medium containing 10% FBS, and the cells were cultured for another day until the experiments, such as irradiation, fixation, and immunofluorescence. For U2OS cells, reverse siRNA transfection was performed using Lipofectamine RNAiMAX (Thermo Fisher Scientific, MA, USA). First, siRNA and Lipofectamine RNAiMAX were separately diluted in Opti-MEM, mixed, and incubated for 20 min at RT. During the incubation, the cells were harvested by trypsinization and suspended in the medium at a concentration of 2 × 105 cells/mL. The cells were then mixed with diluted siRNA/lipofectamine RNAiMAX, plated in a 35-mm dish, and cultured for 1 day. The medium was then replaced with fresh medium containing 10% FBS, and the cells were cultured for another day until the experiment. Two days after siRNA transfection, the cells were subjected to the experiments. The siRNA sequences used in this study are listed in Supplementary Table S1.
DR-GFP assay
U2OS cells harboring the DR-GFP HR reporter were used to examine the efficiency of HR. The cells were transfected with the indicated siRNAs and cultured overnight. Next, the cells were transfected with the pCBASceI vector (expression vector of I-Sce I endonuclease, Addgene #26477) using Lipofectamine 3000 (Thermo Fisher Scientific, MA, USA). Cells transfected with pEGFP-N1 vector (Clontech, CA, USA) were concomitantly prepared to monitor transfection efficiency. Cells were harvested and suspended in PBS containing 1 mM EDTA 48 h after vector transfection. The percentage of GFP-positive cells was determined using a FACS Verse and Lyric (Beckton Dickinson, NJ, USA). The percentage of GFP( +) pCBA-SceI-transfected cells normalized by the percentage of GFP( +) pEGFP-N1-transfected cells was shown as the percentage of HR( +) cells.
Immunofluorescence analysis, foci counting, and image acquisition
In this study, nuclear foci of DSB repair factors were quantified in the G2 phase. To identify G2 phase cells, fixed cells were routinely stained with EdU (S phase marker) and CENPF (S/G2 phase marker), and foci in EdU(-)/CENPF( +) cells (= G2 phase cells) were counted. For EdU labeling, cells were treated with 10 μM EdU from 30 m before irradiation until fixation. For the detection of RPA foci, cells were pre-extracted with 0.2% Triton X-100 in phosphate-buffered saline (PBS) for 1 min (Fig. 1f–h, 2c, 4b) or pre-extracted twice for 3 min with CSK buffer (10 mM HEPES, 100 mM NaCl, 300 mM sucrose, 3 mM MgCl2, and 0.7% Triton X-100) containing 0.3 mg/mL RNase A (Fig. 4f), followed by fixation with 3% paraformaldehyde (PFA)/2% sucrose in PBS for 10 min (Figs. 1f–h, 2c, 4b) or 2% PFA in CSK buffer for 10 min (Fig. 4f) at RT. For the detection of RAD51 foci, the cells were pre-extracted with 0.2% Triton X-100 in PBS for 1 min, followed by fixation with 3% PFA/2% sucrose in PBS for 10 min at RT. For the detection of BRCA1 foci, cells were pre-extracted with CSK buffer for 5 min, followed by fixation with 4% PFA in PBS for 10 min at RT. For the detection of 53BP1 pT543 and RIF1 foci, the cells were fixed with 3% PFA/2% sucrose in PBS for 10 min at RT, followed by permeabilization with 0.2% Triton X-100 in PBS for 2.5 min. Cells were incubated with primary antibodies against the foci-forming factors and CENPF for 30 min at 37 °C, followed by incubation with Alexa Fluor 488 (for foci) and Alexa 555 (for CENPF)-conjugated secondary antibodies for 30 min at 37 °C. The primary antibodies used for the immunofluorescence are listed in Table S2. All antibodies were diluted in 2% bovine serum albumin in PBS. Next, the samples were incubated with an EdU detection solution (Alexa Fluor 647 azide in 100 mM Tris–HCl, 100 mM L-ascorbic acid, and 4 mM CuSO4) for 30 min at RT in the dark. Coverslips were mounted onto glass slides with Vectashield mounting medium containing 4′,6-diamidino-2-phenylindole (DAPI) (Vector Laboratories, CA, USA). The foci of RPA, RAD51, BRCA1, 53BP1 pT543, and RIF1 in G2 phase cells were counted using a fluorescence microscope (BX53, Olympus, Tokyo, Japan or BZ-X700, Keyence, Tokyo, Japan). Images of the foci were acquired using a confocal fluorescence microscope (LSM800; Zeiss, Oberkochen, Germany) with a 100 × objective. Z-stack images were captured at intervals of 0.2–0.25 µm. Maximal intensity projection images were obtained using ZEN 2.3 image analysis software (Zeiss).
Cloning of SART1 cDNA and generation of SART1 mutants
Human SART1 mRNA (NM_005146) was reverse-transcribed into complementary DNA (cDNA), which was tagged with 3 × FLAG and cloned into pIRES vector (Clontech, CA, USA). For the laser irradiation experiments, the SART1 cDNA was cloned into the pEGFP-C3 vector (Clontech, CA, USA) using the In-Fusion HD cloning kit (Takara Bio, Shiga, Japan) .SART1 mutants were generated using the KOD-Plus-Mutagenesis kit (Toyobo, Osaka, Japan) with some modifications. Silent mutations were introduced into the wild-type and the mutant SART1 cDNAs to make them resistant to SART1 siRNA (siSART1#1).
Creation of wild-type or mutant SART1-inducible cell lines
To create FLAG-SART1-inducible RPE-hTERT cell lines, we employed the pCW57.1 vector, a doxycycline-inducible lentiviral vector with a puromycin-resistant gene (Addgene #41393). The FLAG-tagged wild-type or mutant SART1 cDNA (siSART1#1-resistant) was cloned into the pCW57.1 vector to make pCW-FLAG-SART1 vectors using the In-Fusion HD cloning kit. Then, RPE-hTERT cells were infected with lentiviral particles containing pCW-FLAG-SART1 vectors and were subjected to puromycin selection (10 µg/mL) for 10–14 days. The puromycin-resistant cells were used for the experiments in Fig. 2b–c.
Creation of Rap80 knockout cell line
Rap80 knockout RPE-hTERT (RPE-hTERT ΔRap80) cells were established as previously described24. Briefly, RPE-hTERT cells were transfected with the All-in-One CRISPR-Cas9D10A vector (Addgene #74119) comprising a pair of guide RNAs targeting the Rap80 gene and GFP-labeled Cas9D10A nickase. Transfected cells were enriched using FACS Aria II (Becton Dickinson, NJ, USA) and seeded in 96-well plates. Rap80 knockout clones were screened using polymerase chain reaction (PCR) and further confirmed by western blotting.
Laser track analysis
One day before analysis, U2OS cells expressing mCherry-Geminin seeded on a 35-mm glass-bottom dish (Matsunami, Osaka, Japan) were transfected with the GFP-SART1 vector using Lipofectamine 2000 (Thermo Fisher Scientific, MA, USA), following the manufacturer’s instructions. Next, the cells were incubated with 10 µg/mL Hoechst33342 (FUJIFILM Wako, Osaka, Japan) for 10 min and incubated at 37 °C in a temperature-controlled chamber (Zeiss) during the analysis. GFP( +)/mCherry( +) cells were irradiated with a Mai Tai laser (Spectra-Physics, Santa Clara, CA, USA) at a wavelength of 730 nm and a nominal power of approximately 10 mW along a track with a width of approximately 500 nm. The cells were observed under an LSM510 microscope (Zeiss, Germany) equipped with a 63 × objective. For each condition, three to seven cells from two biological replicates were imaged at intervals of 10 s. For the laser track analysis of mutant GFP-SART1, U2OS cells expressing mCherry-Geminin were transfected with SART1 siRNA (siSART1#1) and cultured for 1 day to deplete endogenous SART1. The cells were then transfected with siSART1#1-resistant GFP-SART1 vector (wild-type or mutant), cultured for another day, and subjected to laser track analysis.
Preparation of the chromosome samples
Chromosomal samples were prepared as previously described, with some modifications39,40. Briefly, siRNA-treated RPE-hTERT cells were plated in two T75 flasks per sample. To analyze chromosomes only in G2-irradiated cells, cells were treated with aphidicolin (3 µM), a DNA synthesis inhibitor, from 30 min before irradiation until fixation, so that the irradiated S phase cells did not progress to the G2 phase. Thirty minutes after the addition of aphidicolin, cells were irradiated with 2 Gy γ-rays and 4 h later, Chk1/2 inhibitor (SB218078, 2.5 µM) and colcemid (0.025 µg/mL) were added to the culture medium. Chk1/2 inhibitor and colcemid were used to avoid bias of the G2 checkpoint and to enrich mitotic cells, respectively. Five hours later (9 h after irradiation), mitotic cells were shaken off from the flasks, suspended in medium, and subjected to hypotonic treatment for 20 min at 37 °C in 0.075 M potassium chloride, followed by fixation with ice-cold Carnoy’s fixative (methanol: acetic acid = 3:1). After centrifugation (1,200 rpm, 2 min, 4 °C), the cells were suspended in Carnoy’s fixative, dropped onto glass slides, and dried for 1–3 day. Chromosomes were stained with DAPI in the Vectashield mounting medium.
Statistical analysis
All statistical analyses, except for the analysis of chromosomal aberration data (Fig. 6e–f), were performed using Prism 8 software (GraphPad Software, San Diego, CA, USA). The statistical difference between two groups were analyzed using unpaired t-test, Welch’s t-test, or two-tailed Mann–Whitney test, whereas that between more than two groups were analyzed using Dunn’s multiple comparisons test. For the analysis of the chromosomal aberration data in Fig. 6e–f, a zero-inflated Poisson (ZIP) regression model was applied using the pscl package in R, because the data contained excessive zeros. Differences were considered statistically significant at P < 0.05.
Data availability
The RNA-seq data have been deposited at DNA Data Bank of Japan (DDBJ) under accession number: DRA017000.
References
Bradley, R. K. & Anczuków, O. RNA splicing dysregulation and the hallmarks of cancer. Nat. Rev. Cancer 23, 135–155 (2023).
Hanahan, D. Hallmarks of cancer: new dimensions. Cancer Discov. 12, 31–46 (2022).
Paulsen, R. D. et al. A genome-wide siRNA screen reveals diverse cellular processes and pathways that mediate genome stability. Mol. Cell 35, 228–239 (2009).
Adamson, B., Smogorzewska, A., Sigoillot, F. D., King, R. W. & Elledge, S. J. A genome-wide homologous recombination screen identifies the RNA-binding protein RBMX as a component of the DNA-damage response. Nat. Cell Biol. 14, 318–328 (2012).
Herr, P. et al. A genome-wide IR-induced RAD51 foci RNAi screen identifies CDC73 involved in chromatin remodeling for DNA repair. Cell Discov. 1, (2015).
Anantha, R. W. et al. Requirement of heterogeneous nuclear ribonucleoprotein C for BRCA gene expression and homologous recombination. PLoS One 8, e61368 (2013).
Awwad, S. W., Darawshe, M. M., Machour, F. E., Arman, I. & Ayoub, N. Recruitment of RBM6 to DNA double-strand breaks fosters homologous recombination repair. Mol. Cell Biol. 43, 130–142 (2023).
MacHour, F. E. et al. RBM6 splicing factor promotes homologous recombination repair of double-strand breaks and modulates sensitivity to chemotherapeutic drugs. Nucl. Acids Res. 49, 11708–11727 (2021).
Onyango, D. O., Lee, G. & Stark, J. M. PRPF8 is important for BRCA1-mediated homologous recombination. Oncotarget 8, 93319–93337 (2017).
Prados-Carvajal, R., López-Saavedra, A., Cepeda-García, C., Jimeno, S. & Huertas, P. Multiple roles of the splicing complex SF3B in DNA end resection and homologous recombination. DNA Repair (Amst) 66–67, 11–23 (2018).
Tanikawa, M., Sanjiv, K., Helleday, T., Herr, P. & Mortusewicz, O. The spliceosome U2 snRNP factors promote genome stability through distinct mechanisms; transcription of repair factors and R-loop processing. Oncogenesis 5, e280 (2016).
Hustedt, N. & Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 19, 1–9 (2017).
Scully, R., Panday, A., Elango, R. & Willis, N. A. DNA double-strand break repair-pathway choice in somatic mammalian cells. Nat. Rev. Mol. Cell Biol. 20, 698–714 (2019).
Ceccaldi, R., Rondinelli, B. & D’Andrea, A. D. Repair pathway choices and consequences at the double-strand break. Trends Cell Biol. 26, 52–64 (2016).
Shibata, A. Regulation of repair pathway choice at two-ended DNA double-strand breaks. Mutat. Res. 803–805, 51–55 (2017).
Tarsounas, M. & Sung, P. The antitumorigenic roles of BRCA1–BARD1 in DNA repair and replication. Nat. Rev. Mol. Cell Biol. 21, 284–299 (2020).
Panier, S. & Boulton, S. J. Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 (2014).
Isono, M. et al. BRCA1 directs the repair pathway to homologous recombination by promoting 53BP1 dephosphorylation. Cell Rep. 18, 520–532 (2017).
Sasatomi, T. et al. Expression of the SART1 tumor-rejection antigens in colorectal cancers. Dis. Colon Rectum 43, 1754–1758 (2000).
Shintaku, I. et al. Expression of the SART1 tumor rejection antigen in renal cell carcinoma. Urol. Res. 28, 178–184 (2000).
Allen, W. L. et al. A systems biology approach identifies SART1 as a novel determinant of both 5-fluorouracil and SN38 drug resistance in colorectal cancer. Mol. Cancer Ther. 11, 119–131 (2012).
Pierce, A. J., Johnson, R. D., Thompson, L. H. & Jasin, M. XRCC3 promotes homology-directed repair of DNA damage in mammalian cells. Genes Dev. 13, 2633–2638 (1999).
Cejka, P. & Symington, L. S. DNA end resection: mechanism and control. Annu. Rev. Genet. 55, 285–307 (2021).
Yasuhara, T. et al. Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558-570.e11 (2018).
Matsuoka, S. et al. ATM and ATR substrate analysis reveals extensive protein networks responsive to DNA damage. Science 316, 1160–1166 (2007).
Shichijo, S. et al. A gene encoding antigenic peptides of human squamous cell carcinoma recognized by cytotoxic T lymphocytes. J. Exp. Med. 187, 277–288 (1998).
Hertel, K. J. & Graveley, B. R. RS domains contact the pre-mRNA throughout spliceosome assembly. Trends Biochem. Sci. 30, 115–118 (2005).
Makarova, O. V., Makarov, E. M. & Lührmann, R. The 65 and 110 kDa SR-related proteins of the U4/U6.U5 tri-snRNP are essential for the assembly of mature spliceosomes. EMBO J. 20, 2553–2563 (2001).
Reynolds, P., Botchway, S. W., Parker, A. W. & O’Neill, P. Spatiotemporal dynamics of DNA repair proteins following laser microbeam induced DNA damage—when is a DSB not a DSB?. Mutat. Res. 756, 14–20 (2013).
Hu, Y. et al. RAP80-directed tuning of BRCA1 homologous recombination function at ionizing radiation-induced nuclear foci. Genes Dev. 25, 685–700 (2011).
Li, M. L. & Greenberg, R. A. Links between genome integrity and BRCA1 tumor suppression. Trends Biochem. Sci. 37, 418–424 (2012).
Bunting, S. F. et al. 53BP1 inhibits homologous recombination in Brca1-deficient cells by blocking resection of DNA breaks. Cell 141, 243–254 (2010).
Escribano-Díaz, C. et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol. Cell 49, 872–883 (2013).
Marchesini, M. et al. ILF2 Is a regulator of RNA splicing and DNA damage response in 1q21-amplified multiple myeloma. Cancer Cell 32, 88-100.e6 (2017).
Savage, K. I. et al. Identification of a BRCA1-mRNA splicing complex required for efficient DNA repair and maintenance of genomic stability. Mol. Cell 54, 445–459 (2014).
Maréchal, A. et al. PRP19 transforms into a sensor of RPA-ssDNA after DNA damage and drives ATR activation via a ubiquitin-mediated circuitry. Mol. Cell 53, 235–246 (2014).
Beli, P. et al. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 46, 212–225 (2012).
Venkitaraman, A. R. Tracing the network connecting BRCA and Fanconi anaemia proteins. Nat. Rev. Cancer 4, 266–276 (2004).
Yamauchi, M. et al. Mode of ATM-dependent suppression of chromosome translocation. Biochem. Biophys. Res. Commun. 416, 1111–1118 (2011).
Yamauchi, M. et al. Regulation of pairing between broken DNA-containing chromatin regions by Ku80, DNA-PKcs, ATM, and 53BP1. Sci. Rep. 7, 41812 (2017).
Acknowledgements
We thank Dr. Yasuyoshi Oka for providing information on the anti-SART1 antibody. We thank the Research Promotion Unit of the Medical Institute of Bioregulation at Kyushu University for technical assistance. We appreciate the technical assistance from the Research Support Center, Research Center for Human Disease Modeling, Kyushu University Graduate School of Medical Sciences, which is partially supported by the Mitsuaki Shiraishi Fund for Basic Medical Research. This work was supported by Grants-in-Aid for Scientific Research (KAKENHI) from the Japan Society for the Promotion of Science (JP18K11645 and JP22K12376 to M.Y., JP17H04713 to A.S., and JP18K18191 to T.Y.), Takeda Science Foundation, and Network-type Joint Usage/Research Center for Radiation Disaster Medical Science.
Author information
Authors and Affiliations
Contributions
M.Y. conceived the study and designed the experiments with K.O., T.Y., A.S., K.M., K.S., and N.M.. K.O. and M.Y. created FLAG-SART1-inducible RPE-hTERT cells. R.K. and T.Y. created RPE-hTERT ΔRap80 cells and performed laser track analysis. K.O., M.Y., Y.U., and A.S. performed the DR-GFP assay. Y.U., R. K-I, and A.S. performed RNA-seq and bioinformatic analyses. K.O., M.Y., M.H., and A.S. constructed vectors. K.O., M.Y., M.H., A. Shakayeva, and P.K. performed immunofluorescence, microscopic work, and western blotting. K.O. and H.S. performed γ-irradiation. M.Y. and Y.A performed the chromosome experiments. M.Y. summarized all data for the figures and prepared the manuscript with support from K.O., T.Y., A.S., K.M., K.S., and N.M. M.Y. supervised this study.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ozaki, K., Kato, R., Yasuhara, T. et al. Involvement of the splicing factor SART1 in the BRCA1-dependent homologous recombination repair of DNA double-strand breaks. Sci Rep 14, 18455 (2024). https://doi.org/10.1038/s41598-024-68926-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-68926-2
- Springer Nature Limited