Abstract
L-type amino acid transporter 1 (LAT1) is upregulated in various cancer types and contributes to disease progression. Previous studies have demonstrated or suggested that hypoxia-inducible factors (HIFs), the key transcription factors in hypoxic responses, control the expression of LAT1 gene in several types of cancer cells. However, this regulatory relationship has not been investigated yet in colorectal cancer (CRC), one of the cancer types in which the increased LAT1 expression holds prognostic significance. In this study, we found that neither LAT1 mRNA nor protein is induced under hypoxic condition (1% O2) in CRC HT-29 cells in vitro, regardless of the prominent HIF-1/2α accumulation and HIFs-dependent upregulation of glucose transporter 1. The hypoxic treatment generally did not increase either the mRNA or protein expression of LAT1 in eight CRC cell lines tested, in contrast to the pronounced upregulation by amino acid restriction. Interestingly, knockdown of von Hippel-Lindau ubiquitin ligase to inhibit the proteasomal degradation of HIFs caused an accumulation of HIF-2α and increased the LAT1 expression in certain CRC cell lines. This study contributes to delineating the molecular mechanisms responsible for the pathological expression of LAT1 in CRC cells, emphasizing the ambiguity of HIFs-dependent transcriptional upregulation of LAT1 across cancer cells.
Similar content being viewed by others
Introduction
Due to increased metabolic demand for oxygen and typically disorganized intratumoral vascularization, solid tumors often develop hypoxic subregions, where the oxygen tension falls below physiological levels1,2. Adaptation to poorly oxygenized environments manifests malignant features of cancers that lead to poor clinical outcomes, including metastasis, chemo/radio-resistance, and immune evasion3,4,5. Master regulators of cellular responses to the decreased oxygen availability are hypoxia-inducible factors (HIFs). HIFs function as heterodimeric transcription factors of an α-subunit (HIF-1α, HIF-2α, or HIF-3α) and a common β-subunit (HIF-1β), namely HIF-1, HIF-2, or HIF-36,7,8,9,10. Among the three α-subunits, HIF-1α and -2α play predominant and partly redundant roles in determining transcriptional responses to hypoxia. HIF-3α lacks the transactivation domain and is supposed to mainly function as a competitive negative regulator of HIF-1/2-dependent gene expression11,12. HIFs in cancer cells coordinate the expression of hundreds to thousands of target genes involved in angiogenesis, glucose metabolism, redox homeostasis, cancer stemness, and so on3,4,5.
L-type amino acid transporter 1 (LAT1; SLC7A5)13 that preferentially uptakes extracellular large neutral amino acids, including most of the essential amino acids13,14, is overexpressed in a wide variety of cancers15,16. Amino acids transported by LAT1 are used as substrates for biosynthetic and bioenergetic reactions to support the rapid proliferation and growth of cancer cells. Moreover, one of the LAT1 substrates, leucine, activates the mechanistic target of rapamycin complex 1 (mTORC1), a nutrient-sensing kinase complex that plays pivotal roles in the regulation of cellular metabolism and is often aberrantly activated in cancers17,18. High LAT1 expression correlates with poor prognosis of multiple cancer types, indicating its pathological functions in disease progression and malignancy15,19,20. LAT1 thus has been recognized as an attractive molecular target for cancer therapeutics and diagnostics15. Anti-cancer effects of LAT1 inhibition have been proven well in various studies, especially with a LAT1-selective inhibitor, nanvuranlat (JPH203 or KYT-0353)15,21. The first randomized phase II clinical trial of nanvuranlat monotherapy was recently performed against pretreated, advanced, and refractory biliary tract cancers and resulted in a significantly improved progression-free survival of patients, supporting the rationale of targeting LAT1 for cancer therapy22. However, our understanding of the molecular mechanisms responsible for the pathological upregulation of LAT1 in cancer cells is still limited.
Previous studies have demonstrated or suggested links between HIFs and the LAT1 expression in cancer. Hypoxic treatment increases the amount of LAT1 mRNA in vitro in differentiated murine neuroblastoma Neuro2A cells via a HIF-2α-dependent manner23. In glioblastoma cell lines and primary glioblastoma multiforme cells, HIF-1/2α enhance the LAT1 expression under hypoxia both at mRNA and protein levels24. LAT1 mRNA expression is also reported to elevate under hypoxic conditions in multiple breast cancer cell lines, even though the contributions of HIFs have not been clarified25. Besides hypoxia, other experimental or genetic conditions that cause the accumulation of HIFs also promote the LAT1 expression. Acidosis induces the expression of LAT1 mRNA and protein in cervical carcinoma SiHa cells via a HIF-2α-dependent manner26. Direct transcriptional regulation of the LAT1 gene expression by HIF-2 was reported in von Hippel-Lindau (VHL) ubiquitin ligase-deficient clear cell renal cell carcinoma (ccRCC) 786-O cells, in which HIF-2α accumulates due to the lack of its VHL-dependent proteasomal degradation27. In the same study, authors have demonstrated by chromatin immunoprecipitation that HIF-2α binds to the proximal promoter region of the LAT1 gene that contains two potential HIF-binding motifs27. An immunohistochemical study on malignant pleural mesothelioma showed that the abundance of LAT1 significantly correlates with that of HIF-1α, as well as with multiple proteins encoded by HIFs target genes, including glucose transporters GLUT1 (SLC2A1) and GLUT3 (SLC2A3), hexokinase 1, and vascular endothelial growth factor28. Moreover, HIF-1α and/or -2α also positively regulate the expression of several other amino acid transporters25,26,29,30,31, implying that they play fundamental roles in controlling amino acid utilization of cancer cells in response to hypoxia.
These observations raise a tempting hypothesis that the accumulation of HIFs induced by intratumoral hypoxia generally contributes to the pathological upregulation of LAT1 across various types of cancer cells. Colorectal cancer (CRC) is one of the cancer types, in which the pathological importance of LAT1 expression has been recognized. A previous study reported that the high LAT1 protein expression correlates with tumor cell proliferation, aggressiveness, metastasis, and poor prognosis of patients in CRC32. Prognostic significance of LAT1 protein expression was also shown in sporadic CRC patients33 and CRC patients treated with oxaliplatin-based adjuvant chemotherapy34. Notably, analyses of the Cancer Genome Atlas (TCGA) and the Clinical Proteomic Tumor Analysis Consortium (CPTAC) cohorts show significantly higher expressions of LAT1 mRNA and protein in colon adenocarcinoma tumor tissues than normal tissues, suggesting that dysregulation at the transcriptional level is at least partly involved in the elevated expression of LAT1 (Supplementary Fig. S1). In the present study, we assessed the possibility of the HIF-1/2 -dependent regulation of the LAT1 gene expression in CRC cells under hypoxia.
Results
VHL knockdown induces accumulation of HIF-2α and LAT1 expression in specific CRC cell lines
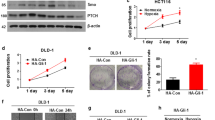
In normoxic conditions, HIF-α subunits accept oxygen-dependent hydroxylation at two proline residues by prolyl hydroxylase domain-containing proteins (PHDs). This hydroxylation recruits VHL ubiquitin ligase for poly-ubiquitination and subsequent proteasomal degradation of HIF-α subunits35,36. In contrast, under hypoxia, non-hydroxylated HIF-α subunits are stabilized, translocated to the nucleus, and assembled with HIF-1β to drive the expression of various target genes. The direct regulation of LAT1 transcription by HIFs has been most extensively demonstrated in ccRCC 786-O cells, in which the mutagenic inactivation of VHL genes causes the HIF-2α accumulation27. As the VHL deficiency is not common in CRC cell lines, we first examined whether the siRNA-mediated knockdown of VHL increases HIF-1α and/or -2α, resulting in the subsequent expression of LAT1. Transfection of siRNA reduced the expression of VHL mRNA by ~ 50–65% in HCT116, SW480, COLO205, and HT-29 cells (Fig. 1a). Analysis by western blotting revealed that the VHL knockdown did not affect the amount of HIF-1α in any of the tested cell lines (Fig. 1b). HIF-1α was detected in two bands between 100 and 150 KDa. The upper stronger and lower weaker bands most likely represent phosphorylated and non-phosphorylated forms of HIF-1α, respectively37. Interestingly, HIF-2α was modestly increased in VHL-reduced HCT116 and HT-29 cells, whereas not in SW480 and COLO205 cells. The expression of LAT1 mRNA was also elevated selectively in HCT116 (2.0-fold) and HT-29 cells (1.6-fold) by VHL knockdown, in contrast to a trend of slight decrease in SW480 (1.1-fold) and COLO205 cells (1.2-fold) (Fig. 1c). The mRNA expression of GLUT1, a well-established HIFs target35,36,37,38, exhibited similar tendencies of an increase in HCT116 (1.4-fold) and HT-29 cells (1.3-fold) and a decrease in SW480 (1.2-fold) and COLO205 cells (1.1-fold) by VHL knockdown. Consistently, the LAT1 and GLUT1 proteins were selectively increased in VHL-reduced HCT116 and HT-29 cells, but not in SW480 and COLO205 cells (Fig. 1b). The smear bands of GLUT1 at ~40–70 KDa probably represent the heterogenous modification by N-glycosylation38.
Effects of VHL knockdown on the accumulation of HIFs and LAT1 expression in CRC cell lines. (a) Cells transfected with siRNA targeting VHL (siVHL) were cultured for 48 h and analyzed for VHL expression by quantitative real-time PCR. Knockdown efficiency was evaluated compared to cells transfected with negative control siRNA (siNC). (b, c) Cells treated as in (a) were analyzed by western blotting for HIF-1α, HIF-2α, LAT1, GLUT1, and β-actin (loading control) (b) or quantitative real-time PCR for LAT1 and GLUT1 (c). Bar graphs in (b) show densitometric quantification of the detected bands. Y-axis in (c) represents the log2FC (siVHL/siNC) values of mRNA expression. Data are shown as mean ± SEM from three independent experiments (n = 3). *p < 0.05, **p < 0.01, unlabeled: not significant (p > 0.05) (one-sample t-test versus hypothetical mean of 1 of each gene in control sample of each cell line treated with siNC). (d) Cells knocked down for VHL alone (siVHL) or with HIF-2α (siVHL/siHIF-2α) were cultured for 48 h and analyzed by quantitative real-time PCR for LAT1 and GLUT1. Data are shown as mean ± SEM from six independent experiments (n = 6). *p < 0.05, ns not significant (p > 0.05) (one-way ANOVA followed by Holm-Sidak’s multiple comparison test).
We then investigated whether the upregulation of LAT1 and GLUT1 under the VHL-reduced condition is dependent on HIF-2, by simultaneously knocking down VHL and HIF-2α in HT-29 cells (Fig. 1d). Under this experimental condition, the VHL knockdown efficiency at mRNA level was lower (reduction by 40.2 ± 11.2%, data not shown) compared to that was observed by single knockdown experiments in Fig. 1a–c (reduction by 65.0 ± 10.3%), presumably because the reduced concentration of VHL-targeted siRNA from 60 to 30 nM for double knockdown. Still, the suppression of VHL caused an increase in the mRNA expression of LAT1 (1.4-fold) and GLUT1 (1.2-fold). Co-transfection of HIF-2α-targeted siRNA reverted the upregulation of LAT1 expression. A similar tendency was also observed with the expression of GLUT1 mRNA, although statistical significance was not reached. These results indicate that the VHL knockdown causes a HIF-2-dependent upregulation of LAT1 in certain CRC cell lines, partially mimicking their regulatory relationships previously reported in VHL-deficient ccRCC cells27.
Accumulation of HIF-1/2α under hypoxia does not induce LAT1 expression in HT-29 cells
We next examined the possibility of HIFs-dependent regulation of LAT1 expression under actual hypoxic culture conditions in HT-29 cells. The median tumor oxygen tensions in many cancer types are reported to be generally 1–2%39. Even though the data on direct oxygen measurement in CRC are limited, a previous study using polarographic needle electrodes showed the median oxygen tension in 15 rectal cancer patients to be 2.5%40. Also, an immunohistochemical study using a hypoxia probe pimonidazole and a PET imaging study have suggested that CRC develops intertumoral hypoxia at a comparable level with other cancer types41,42. We therefore adopted 1% O2 for the hypoxic condition in the present study, which has been commonly used in in vitro studies of cancer hypoxia. As shown in Fig. 2a, treatment of HT-29 cells with hypoxia caused a pronounced accumulation of HIF-1/2α. During 48 h of the hypoxic treatment, HIF-1α became most abundant at 12 h, followed by a considerable decrease between 24 and 48 h. HIF-2α showed a gradual increase to the highest accumulation at 24 h, followed by a moderate reduction at 48 h. The amount of GLUT1 protein was also drastically increased during the 48 h of incubation with 1% O2. In contrast, LAT1 protein was not obviously increased by the hypoxic treatment (Fig. 2a). We also tested the effects of a chemical hypoxia-mimicking agent CoCl2, which suppresses the VHL-dependent proteasomal degradation of HIF-α subunits by inhibiting PHDs43,44. Although less pronounced than actual hypoxia, the treatment with 100 µM CoCl2 also induced a transient increase in the protein amount of HIFs and GLUT1, but not of LAT1, in HT-29 cells (Fig. 2a).
Effects of hypoxic treatment on the accumulation of HIFs and LAT1 expression in HT-29 cells. (a) Cells incubated with normoxic 21% O2 (( −) Hypoxia) or hypoxic 1% O2 (( +) Hypoxia) for 0, 12, 24 and 48 h were analyzed by western blotting for HIF-1α, HIF-2α, LAT1, GLUT1, and ATF4. Cells treated with 100 μM CoCl2 (( +) CoCl2) were analyzed for comparison. (b,c) Cells transfected with siRNA for HIF-1α (siHIF-1α) or HIF-2α (siHIF-2α), or both (siHIF-1α/2α), were treated with hypoxia for 12 h, and analyzed by western blotting for HIF-1α and HIF-2α (b) or quantitative real-time PCR for LAT1 and GLUT1 (c). Data are presented in (c) as mean ± SEM from four independent experiments (n = 4). **p < 0.01, ****p < 0.0001, ns not significant (p > 0.05) (two-way ANOVA followed by Tukey’s multiple comparison test). (d) siRNA-transfected cells treated with hypoxia for 24 h were analyzed by western blotting for LAT1 and GLUT1. Cells transfected with negative control siRNA (siNC) treated with normoxia ( −) or hypoxia ( +) were used as control samples in (c,d). β-actin was detected as a reference for equal protein loading in (a,b,d). Bar graphs in (a,b,d) show densitometric quantification of the detected bands.
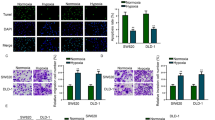
Interestingly, LAT1 protein became even more abundant in untreated control and CoCl2-treated cells than in hypoxia-treated cells at 48 h. Activating transcription factor 4 (ATF4), a stress-induced transcription factor, is known to regulate the expression of several amino acid biosynthetic enzymes and transporters, including LAT1, in response to amino acid deficiency45,46. During a long-time culture under the conditions that allow active proliferation of cells, a substantial amount of amino acids in the medium would be consumed and might promote ATF4-dependent LAT1 expression. As shown in Supplementary Fig. S2a, hypoxia clearly inhibited the proliferation of HT-29 cells compared to the normoxia and CoCl2 treatment. In other CRC cell lines, HCT116, SW480, and COLO205 cells, the proliferation was also suppressed by hypoxia and, unlike HT-29 cells, by CoCl2 treatment as well. Consistently, we detected an increase in the amount of ATF4 and LAT1 protein in normoxia- and CoCl2-treated HT-29 cells (Fig. 2a) as well as in normoxia-treated HCT116, SW480, and COLO205 cells at 48 h compared to at 0 h (Supplementary Fig. S2b). The increase of both LAT1 and ATF4 in the normoxia-treated CRC cells was suppressed by replenishing the culture medium at 24 h, suggesting that the limited amino acid availability causes the induction of LAT1 expression via ATF4 under the experimental conditions (Supplementary Fig. S2b). It is also noteworthy that we made an apparently contradictory observation that the hypoxia even induced a more prompt and pronounced accumulation of ATF4 than normoxia and CoCl2 treatment in HT-29 cells (Fig. 2a). This increase in ATF4, however, is thought not to be attributed to an amino acid deficiency, considering the attenuated proliferation of HT-29 cells under hypoxia (Supplementary Fig. S2a). Consistently, the upregulation of ATF4 in HT-29 cells under hypoxia was not suppressed by replenishing the culture medium at 24 h (Supplementary Fig. S2b). These results suggest a possibility that hypoxia perturbates the ATF4-dependent transcription of LAT1, as discussed later.
We then knocked down HIF-1α and/or -2α to confirm the transcriptional response mediated by their accumulation under hypoxia (Fig. 2b). As shown in Fig. 2c, in the cells transfected with negative control siRNA (siNC), the 12 h of hypoxic treatment resulted in a 5.5-fold increase of GLUT1 mRNA. Individual knockdown of HIF-1α and -2α did not significantly reduce the amount of GLUT1 mRNA in hypoxia-treated cells, although there tends to be a slight decrease by suppressing HIF-1α. However, simultaneous knockdown of HIFs significantly reverted the hypoxia-dependent upregulation of GLUT1 mRNA expression. Consistently, the GLUT1 protein was markedly increased by the 24 h of hypoxic treatment and was suppressed by the double knockdown of HIFs (Fig. 2d). These results indicate the redundant function of HIF-1α and -2α in the regulation of GLUT1 gene expression under hypoxia. Evidently, however, exposure of HT-29 cells to 1% O2 did not alter the expression of LAT1 either at mRNA or protein levels under the tested conditions (Fig. 2c and d), indicating that the accumulation of HIF-1α and -2α under hypoxia does not promote the upregulation of LAT1.
Accumulation of HIF-1/2α under hypoxia does not induce LAT1 expression in multiple CRC cells
We further examined the effects of hypoxia-induced accumulation of HIFs on the expression of LAT1 in seven more CRC cell lines (HCT116, LoVo, DLD-1, SW480, COLO205, WiDr, and SW620 cells). As the accumulation of HIF-1/2α under hypoxia was confirmed to be transient in HT-29 cells and the LAT1 expression exhibited an increase under the normoxic conditions in multiple cells after 48 h of culture (Fig. 2a and Supplementary Fig. S2b), we adopted the hypoxic treatment of 24 h to reveal general trends in multiple cell lines in all the following assays.
The CRC cell lines exhibited a considerable variation in the abundance of HIF-1α and -2α under normoxic conditions (Fig. 3a). Treatment with 1% O2 for 24 h generally increased the amounts of HIFs in all the cell lines. This accumulation of HIFs was accompanied by an apparent increase of GLUT1 mRNA in HCT116 (2.6-fold), LoVo (2.2-fold), DLD-1 (2.2-fold), COLO205 (5.1-fold), HT-29 (5.3-fold), and WiDr cells (7.5-fold), with statistical significance except in LoVo cells (Fig. 3b). GLUT1 mRNA was also slightly increased in SW480 (1.3-fold) and SW620 cells (1.2-fold). Consistently, the hypoxic treatment increased the amount of GLUT1 protein in all the tested cell lines (Fig. 3a). In contrast, LAT1 mRNA was even decreased with statistical significance markedly in SW480 (3.4-fold), COLO205 (5.3-fold), and SW620 cells (3.2-fold), and slightly in HCT116 cells (1.3-fold) (Fig. 3b). The expression of LAT1 mRNA also tended to slightly decrease in LoVo (1.3-fold) and HT-29 cells (1.2-fold). No apparent change in the abundance of LAT1 mRNA was observed in DLD-1 cells. A slight but insignificant increase of LAT1 mRNA (1.5-fold) was detected only in WiDr cells. As shown in Fig. 3a, LAT1 protein expression was not upregulated by the low oxygen tension in any of the tested CRC cell lines. Rather, there was a noticeable decrease of LAT1 protein in hypoxia-treated COLO205 cells. These results indicate that the hypoxia-dependent accumulation of HIF-1α and -2α generally does not induce the LAT1 expression in CRC cells.
Effects of hypoxic treatment on the accumulation of HIFs and LAT1 expression in CRC cell lines. (a,b) Cells incubated with normoxic 21% O2 ( −) or hypoxic 1% O2 ( +) for 24 h were analyzed by western blotting for HIF-1α, HIF-2α, LAT1, GLUT1, and ATF4 (a) or quantitative real-time PCR for LAT1 and GLUT1 (b). β-actin was detected as a reference for equal protein loading in (a). Bar graphs in (a) show densitometric quantification of the detected bands. Y-axis in (b) represents the log2FC (hypoxia/normoxia) values of mRNA expression. Data are shown as mean ± SEM from three independent experiments (n = 3). *p < 0.05, **p < 0.01, ***p < 0.001, unlabeled: not significant (p > 0.05) (one-sample t-test versus hypothetical mean of 1 of each gene in control sample of each cell line treated with normoxia).
As demonstrated in HT-29 cells (Fig. 2a), we found that the amount of ATF4 was also substantially increased by hypoxia in LoVo and WiDr cells, in spite of the mostly unaltered LAT1 expression at both mRNA and protein levels in these cell lines (Fig. 3a and b). Hypoxia did not apparently alter the amount of ATF4 in HCT116 and DLD-1 cells, or even decreased ATF4 in SW480, COLO205, and SW620 cells, demonstrating that the effects of hypoxia on the ATF4 expression are considerably different between cell lines.
Amino acid restriction does not confer hypoxia-dependence to the LAT1 expression in CRC cells
The HIF-2-dependent transcriptional activation of LAT1 previously reported in VHL-deficient ccRCC cells was solely investigated in a low amino acid culture medium (RPMI-1640 medium with 5% of the standard amino acid concentration) with a specific aim to conduct experiments under the conditions where the mTORC1-dependent contributions of LAT1 to the cell proliferation are apparent27. We therefore tested the possibility that the limited amino acid availability confers the hypoxia-dependence to the LAT1 expression in CRC cell lines. Incubation with the low amino acid medium for 24 h under normoxia did not induce the accumulation of HIFs in CRC cells (Fig. 4a). HIF-1α and -2α even tended to decrease in multiple cell lines, such as HIF-1α in HCT116, LoVo, DLD-1, SW480, COLO205, and HT-29 cells and HIF-2α in HCT116, LoVo, DLD-1, SW480, COLO205, HT-29, and SW620 cells. In contrast, LAT1 mRNA was significantly increased in HCT116 (3.9-fold), SW480 (5.4-fold), HT-29 (2.2-fold), WiDr (4.1-fold), and SW620 cells (3.7-fold) (Fig. 4b). A slight and not significant increase was also observed in DLD-1 (1.3-fold) and COLO205 cells (1.4-fold). Only in LoVo cells, no apparent change was detected in the amount of LAT1 mRNA. LAT1 protein was consistently generally increased in the CRC cell lines by the amino acid restriction, except in LoVo cells (Fig. 4a). We confirmed an increase in the ATF4 protein in all cell lines incubated with the low amino acid medium, implying its contribution to upregulating the LAT1 expression (Fig. 4a). The expression of GLUT1 was less responsive to the limited amino acid availability compared to hypoxia. GLUT1 mRNA was significantly increased only in SW480 (2.2-fold) and COLO205 cells (1.4-fold), while tended to slightly increase in HT-29 (1.2-fold), WiDr (1.6-fold), and SW620 cells (1.4-fold) (Fig. 4b). A slight but insignificant decrease of GLUT1 mRNA was detected in HCT116 (1.1-fold), LoVo (1.1-fold), and DLD-1 cells (1.3-fold). At the protein level, GLUT1 was decreased in HCT116, DLD-1, and SW480 cells, whereas increased in LoVo cells (Fig. 4a).
Effects of amino acid restriction on the accumulation of HIFs and LAT1 expression in CRC cell lines. (a,b) Cells incubated with low amino acid RPMI-1640 medium ( +) or normal RPMI-1640 medium ( −) for 24 h were analyzed by western blotting for HIF-1α, HIF-2α, LAT1, GLUT1, and ATF4 (a) or quantitative real-time PCR for LAT1 and GLUT1 (b). β-actin was detected as a reference for equal protein loading in (a). Bar graphs in (a) show densitometric quantification of the detected bands. Y-axis in (b) represents the log2FC (low amino acid medium/normal medium) values of mRNA expression. Data are shown as mean ± SEM from three independent experiments (n = 3). *p < 0.05, **p < 0.01, unlabeled: not significant (p > 0.05) (one-sample t-test versus hypothetical mean of 1 of each gene in control sample of each cell line incubated with normal RPMI-1640 medium).
We then tested the effects of hypoxia under amino acid restriction (Fig. 5). Even in this experimental condition, HIF-1α and -2α showed an accumulation by the treatment with 1% O2 in all cell lines (Fig. 5a). GLUT1 mRNA was generally increased by hypoxia except in SW620 cells, with statistical significance in HCT116 (2.6-fold), DLD-1 (2.1-fold), COLO205 (7.2-fold), and WiDr (5.2-fold) cells. A tendency of increase was also observed in LoVo (1.8-fold), SW480 (1.1-fold), and HT-29 (2.5-fold) cells (Fig. 5b). GLUT1 was also increased at the protein level in all the tested cell lines (Fig. 5a). In contrast, however, the hypoxic treatment in the low amino acid medium even generally reduced LAT1 mRNA. The decrease was significant in LoVo (1.6-fold), DLD-1 (1.4-fold), SW480 (2.8-fold), and SW620 (4.5-fold) cells, while not in HCT116 (2.5-fold), COLO205 (2.5-fold), HT-29 (2.0-fold), and WiDr (1.1-fold) cells (Fig. 5b). A decrease of LAT1 protein was observed in SW480, COLO205, HT-29, and SW620 cells (Fig. 5a). These results indicate that, even under the amino acid restriction, the hypoxia-dependent accumulation of HIF-1α and -2α does not induce the LAT1 expression in CRC cells.
Effects of hypoxic treatment under amino acid restriction on the accumulation of HIFs and LAT1 expression in CRC cell lines. (a,b) Cells cultured in low amino acid RPMI-1640 medium were incubated with normoxic 21% O2 ( −) or hypoxic 1% O2 ( +) for 24 h and analyzed by western blotting for HIF-1α, HIF-2α, LAT1, and GLUT1 (a) or quantitative real-time PCR for LAT1 and GLUT1 (b). β-actin was detected as a reference for equal protein loading in (a). Bar graphs in (a) show densitometric quantification of the detected bands. Y-axis in (b) represents the log2FC (hypoxia/normoxia) values of mRNA expression. Data are shown as mean ± SEM from three independent experiments (n = 3). *p < 0.05, **p < 0.01, unlabeled: not significant (p > 0.05) (one-sample t-test versus hypothetical mean of 1 of each gene in control sample of each cell line treated with normoxia).
Discussion
Elucidating molecular mechanisms responsible for the elevated LAT1 expression in cancer cells will improve our understanding of the disease pathology. It may pave the way to propose more effective diagnostic and therapeutic strategies. Although previous studies have reported the HIF-1/2-dependent transcriptional upregulation of LAT1 in several types of cancer cells23,24,26,27, it has not been investigated yet in CRC cells. The present study explored the relationships between the LAT1 expression and hypoxia-dependent accumulation of HIFs in CRC cells in vitro. As a technical limitation in analyzing multiple CRC cell lines, we collected data from a single time point of hypoxic treatment (24 h) with a single oxygen tension (1% O2). Therefore, we do not exclude the possibility that the results would be different under distinct hypoxic treatments (e.g., longer-term hypoxia or lower oxygen tension). Nevertheless, the LAT1 expression was not significantly upregulated in response to the accumulation of HIF-1/2α under hypoxia in all tested eight CRC cell lines, strongly suggesting that the observed HIFs independence of LAT1 expression represents the general properties of CRC cells. It is also noteworthy that the transcriptional response of LAT1 expression to hypoxia was evidently different from that of a well-established HIFs target, GLUT1.
In contrast to the low oxygen tension, amino acid availability influenced the LAT1 expression in our experimental settings. Amino acid restriction (5% of the normal condition) significantly increased the expression of LAT1 in CRC cells with a concomitant increase of ATF4, a well-known regulator of amino acid metabolism (Fig. 4). Our data suggested that such amino acid dependence of the LAT1 expression accounts for its increase observed in HT-29 cells exposed to normoxia or CoCl2 for 48 h (Fig. 2a and Supplementary Fig. S2). Moreover, we obtained data indicating a complex relationship between hypoxia and ATF4 in the transcriptional regulation of LAT1. Amino acid restriction and hypoxia have been reported to induce ATF4 by the phosphorylation of translation initiation factor eIF2α, albeit mainly via distinct upstream Ser/Thr kinases. Amino acid starvation activates an eIF2α kinase GCN2 (general control nonderepressible 2) by increasing uncharged tRNAs46, whereas hypoxia activates another eIF2a kinase PERK (PKR-like ER kinase) through an ER stress-induced unfolded protein response47. Even though hypoxia significantly induced ATF4 in HT-29, WiDr, and LoVo cells, the LAT1 expression was not upregulated (Fig. 2a and Fig. 3). These results suggest that hypoxia may interfere with the transcriptional function of ATF4 on LAT1, although it is unclear at present whether HIFs are directly involved in this phenomenon or not. It is also of note that hypoxia did not apparently alter the amount of ATF4 in HCT116 and DLD-1 cells, or even decreased ATF4 in SW480, COLO205, and SW620 cells, demonstrating that there are considerable variations between CRC cell lines in the influence of hypoxia on the ATF4 expression.
Pivotal roles of HIFs in the metabolic reprogramming of glycolysis in cancer cells, known as the Warburg effects, have been of great medical and biological interest5. Likewise, although much less is known, an analogous HIFs-mediated reprogramming may also exist for amino acid metabolism. Several studies have reported that HIF-1α and/or -2α regulate the expression of multiple amino acid transporters such as glutamate/aspartate transporters EAAT1 (SLC1A3) and EAAT3 (SLC1A1) in hepatocellular carcinoma cells29, ASCT2 (SLC1A5) in cervix and pancreatic cancer cells26,31, and a cystine/glutamic acid transporter xCT (SLC7A11) in breast cancer cells30. Notably, a study in breast cancer cells showed a hypoxia-dependent upregulation of EAAT3 and neutral amino acid transporter SNAT2 (SLC38A2), in addition to LAT125. The authors have pointed out that amino acids transported by the three upregulated transporters can potentially provide all the amino acid precursors for TCA cycle metabolites, i.e., glutamate and aspartate by EAAT3, gluconeogenic amino acids by SNAT2, and ketogenic amino acids by LAT1. Moreover, HIF-1α and/or HIF-2α control the expression of several amino acid metabolizing enzymes such as a glutaminase GLS1, which promotes glutaminolysis by converting glutamine to glutamate, and a glutamate-cysteine ligase regulatory subunit GCLM, which contributes to oxidative stress resistance through the production of glutathione from glutamate26,30,48.
These previous findings led us to propose the HIFs-mediated upregulation of LAT1 as a part of the generally reprogrammed amino acid utilization in cancer cells under hypoxia. However, our findings in CRC cells were not in line with this possibility, where the expression of LAT1 was not upregulated by hypoxia in a clear contrast to GLUT1. The molecular mechanisms underlying the unresponsiveness of LAT1 to hypoxia in CRC cells are currently unclear. The colon is physiologically highly hypoxic, with the oxygen tensions of ∼2% and ∼0.4% in the lumen of ascending and sigmoid colon of mice, respectively49. Accordingly, immunohistochemistry with hypoxia probes visualized the hypoxia in normal murine colon epithelium50,51. A previous clinicopathological study on colorectal cancer, however, did not detect the LAT1 expression in normal colonic epithelium, suggesting that hypoxia alone does not trigger the LAT1 expression in physiological conditions33. CRC cells derived from adenocarcinoma used in this study might retain such characteristics of the normal colorectal epithelial cells. A possibility worth assessing in future studies is the contribution of methylation because the LAT1 promoter region has been reported to be methylated52,53. We analyzed the methylation of LAT1 promoter region (within 1 kb upstream of transcription start site) in CCLE (Cancer Cell Line Encyclopedia) cell lines and revealed that CRC cell lines (shown as “Bowel” cancer cell lines) exhibit the lowest methylation level among cell lines derived from 23 cancer types and non-cancerous fibroblastic cell lines (Supplementary Fig. S3). “Breast” and “CNS/Brain” cancer cell lines, including those reported to exhibit the hypoxia-dependent upregulation of LAT1 previously24,25, tended to show even higher methylation levels of the LAT1 promoter region compared to the CRC cell lines tested in this study. We therefore currently speculate that the involvement of other mechanisms is more likely, although the methylation status of LAT1 promoter may be distinct under normoxic and hypoxic conditions.
Unresponsiveness of the LAT1 expression to hypoxia in CRC cells we found in this study was somewhat unexpected, especially when considering the direct regulatory relationship between HIF-2α and LAT1 demonstrated in VHL-deficient ccRCC cells27. VHL is a ubiquitin ligase that controls the degradation of various targets besides HIFs, and there possibly could be more unidentified targets54,55. Deficiency of VHL thus may indirectly confer HIF-2-dependence to the LAT1 expression by disrupting the proteostasis of multiple target proteins. We conducted siRNA-mediated knockdown of VHL in selected CRC cell lines (HCT116, SW480, COLO205, and HT-29) to briefly assess this possibility (Fig. 1). As a result, we found an accumulation of HIF-2α, but not HIF-1α, in HCT116 and HT-29 cells. Intriguingly, LAT1 as well as GLUT1 expression was increased at mRNA and protein levels in these two cell lines by knocking down VHL. The increase of LAT1 mRNA in VHL knockdown cells was even more prominent than that of GLUT1. Elucidating the detailed molecular mechanisms, by which the LAT1 expression responds differently to HIF-2α accumulation between hypoxia and VHL deficiency in these specific cell lines, remains as an issue for future research. However, this observation at least raises a possibility that the genetic background of VHL deficiency may account for the HIF-2-dependence of LAT1 expression in specific cancer types such as ccRCC, which is not recapitulated by hypoxia.
Finally, it is also worth noting that few previous studies reported the controversial effects of hypoxia on the expression of LAT1 in cancer cells. Hypoxic treatment of 10% or 1% O2 for 72 h drastically reduced the LAT1 mRNA in glioblastoma T98G and A172 cells56. The same research group demonstrated that hypoxic treatment of 1% O2 for 24 h as well as the treatment with a hypoxia-mimetic deferoxamine significantly reduces the LAT1 mRNA expression in T98G cells, oral squamous cell carcinoma HSC-3 cells, and breast adenocarcinoma MCF-7 cells57. The hypoxia-dependent suppression of LAT1 expression was shown to be reverted by knockdown or inhibition of HIF-1α57. However, as already mentioned above, both mRNA and protein expression of LAT1 were reported to be upregulated HIFs-dependently in distinct glioblastoma U251MG and U87MG cells by hypoxic treatment of 1% O2 for 48 h24. The expression of LAT1 was also shown to be increased in MCF-7 cells by more severe hypoxic treatment of 0.1% O2 for 24 h25. Our findings and these observations in earlier studies emphasize the context-dependent influence of HIFs accumulation on the LAT1 expression in cancer cells. We would like to warn that multiple factors, such as the tissue/organ origin and genetic background of cancer cells, as well as the culture conditions for hypoxic treatments, should be carefully taken into consideration to reveal the relationships between hypoxia and LAT1. Last but not least, the results of our and previous in vitro studies require further verification in vivo to delineate pathologically relevant HIFs-mediated mechanisms underlying the expression of LAT1 in cancer cells.
Methods
Cell culture and hypoxic treatments
Human colorectal cancer cell cells (COLO205 [CCL-222], DLD-1 [CCL-221], HCT116 [CCL-247], HT-29 [HTB-38], LoVo [CCL-229], SW480 [CCL-228], SW620 [CCL-227], and WiDr [CCL-218]; from American Type Culture Collection) were routinely maintained in RPMI-1640 medium (Nacalai tesque) supplemented with 10% FBS (SIGMA) and 100 units/mL penicillin—100 µg/mL streptomycin (Nacalai Tesque), at 37 °C in a humidified CO2 incubator at 21% O2 (95% air supplied with 5% CO2). All experiments were repeated for the indicated times using the same batch of cells.
For all experiments, cells were seeded at either 7.0 × 105 (for LoVo cells) or 3.5 × 105 cells/dish (for other cells) in 60 mm dish containing 4 mL of the culture medium. After two days of culture, the medium was replaced with fresh medium, and cells were treated either with 100 µM CoCl2 or low oxygen tension (1% O2). CoCl2 was dissolved in water to make a stock solution (10 mM) and added to the medium. For the treatment with low oxygen tension, cells were transferred to a multi-gas incubator balanced at 94% N2, 1% O2, and 5% CO2. Control cells were continuously maintained in the CO2 incubator at normoxic culture condition (21% O2). Cells were incubated for the indicated periods and subjected to the analyses by real-time qPCR or western blotting. When described, the amino acid concentrations in the medium were reduced to 5% of the normal condition, by diluting RPMI-1640 medium for 20-fold with amino acid-free RPMI-1640 medium (US Biological) supplemented with 10% dialyzed FBS (Gibco).
Gene knockdown by siRNA
Cells were seeded as described above and immediately transfected with siRNA by using Lipofectamine RNAiMAX (Invitrogen). The following siRNAs were used in this study: HIF-1α (MISSION Predesigned siRNA, SASI_Hs01_00019152), HIF-2α (SASI_Hs02_00332063), VHL (SASI_Hs02_00302782), and MISSION Universal Negative Control #2 (SIC002). siRNA was transfected at the final concentration of 60 nM in all experiments. For double knockdown experiments, co-transfection of two siRNAs was performed at 30 nM each, where the samples knocked down for HIF-1α, HIF-2α, or VHL alone were complemented with 30 nM of the negative control siRNA. Cells knocked down for VHL were cultured for two days and subjected to the analyses by real-time qPCR or western blotting. In other experiments, the transfected cells were cultured for two days and subjected to the treatments with CoCl2 or with low oxygen tension.
Real-time qPCR
Total RNA was extracted from cells by using Isogen II (Nippon Gene, Tokyo, Japan). Quantitative real-time PCR was performed by TB Green Premix Ex Taq (Tli RNase H Plus) (TaKaRa, Shiga, Japan) and QuantStudio 7 Flex real-time PCR system (Thermo Fisher Scientific). Among 32 candidate housekeeping genes on TaqMan Array Human Endogenous Control Plate (Thermo Fisher Scientific), β2-microglobulin (B2M) was used as a control for normalization because its mRNA expression was the least affected by 24 h of hypoxic treatment (1% O2) and/or amino acid restriction (5% of the normal amount in RPMI-1640 medium) in HT-29 cells. Following primer sets were used in this study: LAT1 (forward, 5’-CCGTGAACTGCTACAGCGT-3’; reverse, 5’-CTTCCCGATCTGGACGAAGC-3’), GLUT1 (forward, 5’-GGTTGTGCCATACTCATGACC-3’; reverse, 5’-CAGATAGGACATCCAGGGTAGC-3’), VHL (forward, 5’-GACACACGATGGGCTTCTGGTT-3’; reverse, 5’-ACAACCTGGAGGCATCGCTCTT-3’), and B2M (forward, 5’- CCACTGAAAAAGATGAGTATGCCT -3’; reverse, 5’- CCAATCCAAATGCGGCATCTTCA -3’). All the experiments were performed in quadruplicate for each sample and independently repeated for 3–5 times (n = 3–5).
Western blotting
Cells were lysed on ice by a brief sonication in lysis buffer containing 50 mM Tris–HCl (pH 7.4) 150 mM NaCl, 1 mM EDTA, 1 mM phenylmethylsulfonyl fluoride, a protease inhibitor cocktail (complete EDTA-free; Roche Diagnostics) and 1% NP-40, followed by solubilization for 30 min on ice. After centrifugation at 21,000 × g for 15 min at 4 °C, the supernatants were collected and mixed with Laemmli buffer. Proteins were resolved by SDS-PAGE and transferred to PVDF Blotting Membrane (GE Healthcare). Then, the PVDF membranes were blocked at room temperature for 1 h with 5% skim milk/TBST and incubated with primary antibodies overnight at 4 °C, followed by incubation with horseradish peroxidase-conjugated secondary antibodies for 1 h at room temperature. Signals were developed with ECL Prime Western Blotting Detection System and imaged by Amersham Imager 680 (GE Healthcare). Antibodies used are as follows: anti-LAT1 (KE026, TransGenic), anti-GLUT1 (sc-377228, Santa Cruz Biotechnology), anti-HIF-1α (NB100-479, Novus), anti-HIF-2α (NB100-122, Novus), anti-ATF4 (11,815, Cell Signaling Technology), anti-β-actin (66009-1-Ig, Proteintech), horseradish peroxidase-conjugated goat anti-rabbit IgG (111–035-003, Jackson ImmunoResearch), and horseradish peroxidase-conjugated mouse anti-goat IgG (205–035-108, Jackson ImmunoResearch). All the experiments were independently repeated for 2–3 times to confirm the reproducibility. Densitometric quantification of the detected bands was performed using ImageJ software (National Institutes of Health). Uncropped blot images are presented in Supplementary Figs. S4–S9.
Data availability
All the data analyzed and presented in this study are available from the authors upon reasonable request.
References
Brown, J. M. & Giaccia, A. J. The unique physiology of solid tumors: Opportunities (and problems) for cancer therapy. Cancer Res. 58, 1408–1416 (1998).
Harris, A. L. Hypoxia—A key regulatory factor in tumour growth. Nat. Rev. Cancer 2, 38–47 (2002).
Jing, X. et al. Role of hypoxia in cancer therapy by regulating the tumor microenvironment. Mol. Cancer 18, 1–15 (2019).
Cowman, S. J. & Koh, M. Y. Revisiting the HIF switch in the tumor and its immune microenvironment. Trends Cancer 8, 28–42 (2022).
Wicks, E. E. & Semenza, G. L. Hypoxia-inducible factors: Cancer progression and clinical translation. J. Clin. Investig. 132, e159839. https://doi.org/10.1172/JCI159839 (2022).
Semenza, G. L. & Wang, G. L. A nuclear factor induced by hypoxia via de novo protein synthesis binds to the human erythropoietin gene enhancer at a site required for transcriptional activation. Mol. Cell. Biol. 12, 5447–5454 (1992).
Wang, G. L. & Semenza, G. L. Purification and characterization of hypoxia-inducible factor 1. J. Biol. Chem. 270, 1230–1237 (1995).
Wang, G. L., Jiang, B. H., Rue, E. A. & Semenza, G. L. Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS heterodimer regulated by cellular O2 tension. Proc. Natl. Acad. Sci. U. S. A. 92, 5510–5514 (1995).
Tian, H., McKnight, S. L. & Russell, D. W. Endothelial PAS domain protein 1 (EPAS1), a transcription factor selectively expressed in endothelial cells. Genes Dev. 11, 72–82 (1997).
Hara, S., Hamada, J., Kobayashi, C., Kondo, Y. & Imura, N. Expression and characterization of hypoxia-inducible factor (HIF)-3α in human kidney: Suppression of HIF-mediated gene expression by HIF-3α. Biochem. Biophys. Res. Commun. 287, 808–813 (2001).
Makino, Y., Kanopka, A., Wilson, W. J., Tanaka, H. & Poellinger, L. Inhibitory PAS domain protein (IPAS) is a hypoxia-inducible splicing variant of the hypoxia-inducible factor-3α locus. J. Biol. Chem. 277, 32405–32408 (2002).
Makino, Y. et al. Inhibitory PAS domain protein is a negative regulator of hypoxia-inducible gene expression. Nature 414, 550–554 (2001).
Kanai, Y. et al. Expression cloning and characterization of a transporter for large neutral amino acids activated by the heavy chain of 4F2 antigen (CD98). J. Biol. Chem. 273, 23629–23632 (1998).
Yanagida, O. et al. Human L-type amino acid transporter 1 (LAT1): Characterization of function and expression in tumor cell lines. Biochim. Biophys. Acta 1514, 291–302 (2001).
Kanai, Y. Amino acid transporter LAT1 (SLC7A5) as a molecular target for cancer diagnosis and therapeutics. Pharmacol. Ther. 230, 107964. https://doi.org/10.1016/j.pharmthera.2021.107964 (2022).
Kandasamy, P., Gyimesi, G., Kanai, Y. & Hediger, M. A. Amino acid transporters revisited: New views in health and disease. Trends Biochem. Sci. 43, 752–789 (2018).
Saxton, R. A. & Sabatini, D. M. mTOR signaling in growth, metabolism, and disease. Cell 168, 960–976 (2017).
Liu, G. Y. & Sabatini, D. M. mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020).
Lu, J. J. et al. Prognostic value of LAT-1 status in solid cancer: A systematic review and meta-Analysis. PLoS One 15, 1–16 (2020).
Zhang, J. et al. Review of the correlation of LAT1 with diseases: Mechanism and treatment. Front. Chem. 8, 564809. https://doi.org/10.3389/fchem.2020.564809 (2020).
Oda, K. et al. l-type amino acid transporter 1 inhibitors inhibit tumor cell growth. Cancer Sci. 101, 173–179 (2010).
Furuse, J. et al. Phase II placebo-controlled study of the effect and safety of nanvuranlat in patients with advanced biliary tract cancers previously treated by systemic chemotherapy. Clin. Cancer Res. https://doi.org/10.1158/1078-0432.CCR-24-0461 (2024).
Onishi, Y. et al. Hypoxia affects Slc7a5 expression through HIF-2α in differentiated neuronal cells. FEBS Open Bio. 9, 241–247 (2019).
Zhang, B. et al. Regulation of branched-chain amino acid metabolism by hypoxia-inducible factor in glioblastoma. Cell. Mol. Life Sci. 78, 195–206 (2021).
Morotti, M. et al. Hypoxia-induced switch in SNAT2/SLC38A2 regulation generates endocrine resistance in breast cancer. Proc. Natl. Acad. Sci. U. S. A. 116, 12452–12461 (2019).
Corbet, C. et al. The SIRT1/HIF2α axis drives reductive glutamine metabolism under chronic acidosis and alters tumor response to therapy. Cancer Res. 74, 5507–5519 (2014).
Elorza, A. et al. HIF2α acts as an mTORC1 activator through the amino acid carrier SLC7A5. Mol. Cell. 48, 681–691 (2012).
Kaira, K. et al. L-Type amino acid transporter 1 (LAT1) expression in malignant pleural mesothelioma. Anticancer Res. 31, 4075–4082 (2011).
Hu, H. et al. Hypoxia-inducible factors enhance glutamate signaling in cancer cells. Oncotarget 5, 8853–8868 (2014).
Lu, H. et al. Chemotherapy triggers HIF-1–dependent glutathione synthesis and copper chelation that induces the breast cancer stem cell phenotype. Proc. Natl. Acad. Sci. U. S. A. 112, E4600-4609 (2015).
Yoo, H. C. et al. A variant of SLC1A5 is a mitochondrial glutamine transporter for metabolic reprogramming in cancer cells. Cell Metab. 31, 267-283.e12 (2020).
Ogawa, H. et al. Role of amino acid transporter expression as a prognostic marker in patients with surgically resected colorectal cancer. Anticancer Res. 39, 2535–2543 (2019).
Sakata, T. et al. Positive correlation of expression of L-type amino-acid transporter 1 with colorectal tumor progression and prognosis: Higher expression in sporadic colorectal tumors compared with ulcerative colitis-associated neoplasia. Pathol. Res. Pract. 216, 152972. https://doi.org/10.1016/j.prp.2020.152972 (2020).
Shibasaki, Y. et al. Association of high LAT1 expression with poor prognosis and recurrence in colorectal cancer patients treated with oxaliplatin-based adjuvant chemotherapy. Int. J. Mol. Sci. 24, 2604. https://doi.org/10.3390/ijms24032604 (2023).
Ohh, M. et al. Ubiquitination of hypoxia-inducible factor requires direct binding to the β-domain of the von Hippel—Lindau protein. Nat. Cell Biol. 2, 423–427 (2000).
Jaakkola, P. et al. Targeting of HIF-α to the von Hippel-Lindau ubiquitylation complex by O2-regulated prolyl hydroxylation. Science 292, 468–472 (2001).
Minet, E. et al. ERK activation upon hypoxia: Involvement in HIF-1 activation. FEBS Lett. 468, 53–58 (2000).
Holman, G. D. Structure, function and regulation of mammalian glucose transporters of the SLC2 family. Pflugers Arch. 472, 1155–1175 (2020).
Vaupel, P., Höckel, M. & Mayer, A. Detection and characterization of tumor hypoxia using pO2 histography. Antioxid. Redox Signal. 9, 1221–1236 (2007).
Mattern, J., Kallinowski, F., Herfarth, C. & Volm, M. Association of resistance-related protein expression with poor vascularization and low levels of oxygen in human rectal cancer. Int. J. Cancer 67, 20–23 (1996).
Goethals, L. et al. Hypoxia in human colorectal adenocarcinoma: Comparison between extrinsic and potential intrinsic hypoxia markers. Int. J. Radiat. Oncol. Biol. Phys. 65, 246–254 (2006).
Havelund, B. M. et al. Tumour hypoxia imaging with 18F-fluoroazomycinarabinofuranoside PET/CT in patients with locally advanced rectal cancer. Nucl. Med. Commun. 34, 155–161 (2013).
Ho, V. T. & Bunn, H. F. Effects of transition metals on the expression of the erythropoietin gene: Further evidence that the oxygen sensor is a heme protein. Biochem. Biophys. Res. Commun. 223, 175–180 (1996).
Yuan, Y., Hilliard, G., Ferguson, T. & Millhorn, D. E. Cobalt inhibits the interaction between hypoxia-inducible factor-α and von Hippel-Lindau protein by direct binding to hypoxia-inducible factor-α. J. Biol. Chem. 278, 15911–15916 (2003).
Chen, R. et al. The general amino acid control pathway regulates mTOR and autophagy during serum/glutamine starvation. J. Cell Biol. 206, 173–182 (2014).
Kilberg, M. S., Shan, J. & Su, N. ATF4-dependent transcription mediates signaling of amino acid limitation. Trends Endocrinol. Metab. 20, 436–443 (2009).
Chipurupalli, S., Kannan, E., Tergaonkar, V., D’Andrea, R. & Robinson, N. Hypoxia induced ER stress response as an adaptive mechanism in cancer. Int. J. Mol. Sci. 20, 749. https://doi.org/10.3390/ijms20030749 (2019).
Xiang, L. et al. Glutaminase 1 expression in colorectal cancer cells is induced by hypoxia and required for tumor growth, invasion, and metastatic colonization. Cell Death Dis. 10, 40. https://doi.org/10.1038/s41419-018-1291-5 (2019).
He, G. et al. Noninvasive measurement of anatomic structure and intraluminal oxygenation in the gastrointestinal tract of living mice with spatial and spectral EPR imaging. Proc. Natl. Acad. Sci. U. S. A. 96, 4586–4591 (1999).
Karhausen, J. et al. Epithelial hypoxia-inducible factor-1 is protective in murine experimental colitis. J. Clin. Invest. 114, 1098–1106 (2004).
Kelly, C. J. et al. Fundamental role for HIF-1α in constitutive expression of human β defensin-1. Mucosal Immunol. 6, 1110–1118 (2013).
Vettermann, F. J. et al. L-type amino acid transporter (LAT) 1 expression in 18F-FET-negative gliomas. EJNMMI Res. 11, 124. https://doi.org/10.1186/s13550-021-00865-9 (2021).
Zhang, C. et al. SLC7A5 correlated with malignancies and immunotherapy response in bladder cancer. Cancer Cell Int. 24, 182. https://doi.org/10.1186/s12935-024-03365-7 (2024).
Tarade, D. & Ohh, M. The HIF and other quandaries in VHL disease. Oncogene 37, 139–147 (2018).
Minervini, G., Pennuto, M. & Tosatto, S. C. E. The pVHL neglected functions, a tale of hypoxia-dependent and -independent regulations in cancer. Open Biol. 10, 200109. https://doi.org/10.1098/rsob.200109 (2020).
Wada, Y. et al. Impact of oxygen status on 10B-BPA uptake into human glioblastoma cells, referring to significance in boron neutron capture therapy. J. Radiat. Res. 59, 122–128 (2018).
Harada, T. et al. YC-1 sensitizes the antitumor effects of boron neutron capture therapy in hypoxic tumor cells. J. Radiat. Res. 61, 524–534 (2020).
Acknowledgements
Authors would like to thank Ms. M. Kurauchi, Ms. Y. Hayamizu, and Ms. R. Shioiri for their technical assistance. This study was supported by the Center of Medical Innovation and Translational Research, Graduate School of Medicine, Osaka University.
Funding
This research was financially supported by JSPS Grants-in-Aid for Scientific Research to R.O. [22K06856] and Y.K. [22H02809], the Project for Cancer Research and Therapeutic Evolution from AMED (to Y.K. [JP20cm0106151] and [JP21cm0106151]).
Author information
Authors and Affiliations
Contributions
R.O.: Conceptualization, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—original draft, review, and editing. Y.H.: Investigation, Methodology, Validation. M.X.: Funding acquisition, Writing—review and editing. H.O.: Funding acquisition, Writing—review and editing. Y.K.: Funding acquisition, Project administration, Resources, Supervision, Writing—review and editing.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Ohgaki, R., Hirase, Y., Xu, M. et al. LAT1 expression in colorectal cancer cells is unresponsive to HIF-1/2α accumulation under experimental hypoxia. Sci Rep 14, 19635 (2024). https://doi.org/10.1038/s41598-024-70603-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70603-3
- Springer Nature Limited