Abstract
1,4-Naphthoquinone scaffold-derived compounds has shown considerable pharmacological properties against cancer, including acute myeloid leukemia (AML) However, its impact and mechanisms in AML are uncertain. In this study, the mechanisms of 1,4-naphthoquinone scaffold-derived compounds against AML were investigated via network pharmacology, molecular docking and molecular dynamics simulation. ASINEX database was used to collect the 1,4-naphthoquinone scaffold-derived compounds, and compounds were extracted from the software to evaluate their drug similarity and toxicity. The potential targets of compounds were retrieved from the SwissTargetPrediction Database and the Similarity Ensemble Approach Database, while the potential targets of AML were obtained from the GeneCards databases and Gene Expression Omnibus. The STRING database was used to construct a protein–protein interaction (PPI) network, topologically and Cyto Hubb plugin of Cytoscape screen the central targets. After selecting the potential key targets, the gene ontology (GO) function annotation and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis were performed for the intersection targets, and a network map of “compounds-potential targets-pathway-disease” were constructed. Molecular docking of the compounds with the core target was performed, and core target with the strongest binding force and 1,4-naphthoquinone scaffold-derived compounds was selected for further molecular dynamics simulation and further molecular mechanics/Poisson–Boltzmann surface area (MM/PBSA) approach verification. In addition, the Bloodspot database was applied to perform the overall survival of core targets. A total of 19 1,4-naphthoquinone scaffold-derived compounds were chosen out, and then 836 targets of compounds, 96 intersection targets of AML were screened. Core targets include STAT3, TLR4, HSP90AA1, JUN, MMP9, PTPRC, JAK2, PTGS2, KIT and CSF1R. GO functional enrichment analysis revealed that 90 biological processes, 10 cell components and 12 molecular functions were enriched while KEGG pathway enrichment analysis revealed 34 enriched signaling pathways. Analysis of KEGG enrichment hinted that these 10 core genes were located in the pathways in cancer, suggesting that 1,4-naphthoquinone scaffold-derived compounds had potential activity against AML. Molecular docking analysis revealed that the binding energies between 1,4-naphthoquinone scaffold-derived compounds and the core proteins were all higher than − 6 kcal/mol, indicating that the 10 core targets all had strong binding ability with compounds. Moreover, a good binding capacity was inferred from molecular dynamics simulations between compound 7 and MMP9. The total binding free energy calculated using the MM/GBSA approach revealed values of − 6356.865 kcal/mol for the MMP9-7 complex. In addition, Bloodspot database results exhibited that HSP90AA1, MMP9 and PTPRC were associated with overall survival. The findings provide foundations for future studies into the interaction underlying the anti-AML potential of compounds with 1,4-naphthoquinone-based scaffold structures. Compounds with 1,4-naphthoquinone-based scaffold structures exhibits considerable potential in mitigating and treating AML through multiple targets and pathways.
Similar content being viewed by others
Introduction
Acute myeloid leukemia (AML) is a highly heterogeneous hematological malignancies characterized by clonal proliferation and impaired differentiation of myeloid hematopoietic cells1,2. The incidence rate of AML was 4.3 persons/100,000 persons/year, and the mortality rate was 2.8 persons/100,000 persons/year, seriously endangering human health. The 5-year survival rate of patients was only 24%, and the median overall survival time was only 8.5 months1. Since the 1980s, the “3 + 7” standard treatment regimen for AML (3-day anthracyclines + 7-day cytarabine) has remained almost unchanged. In recent years, the emergence of new targeted drugs and the application of immunotherapy have brought new hope for the treatment of AML3. However, compared with young patients, elderly AML patients are characterized by advanced age, more comorbidities, poor drug tolerance, and a higher incidence of mutated genes with poor prognosis. Low-intensity chemotherapy and demethylation therapy are the main treatment methods, and the 5-year survival rate is less than 10%4. Meanwhile, because AML is highly heterogeneous at cytogenetic and molecular levels, different AML patients often carry different gene mutations, and existing targeted drugs can only benefit patients with corresponding gene mutations3. Therefore, the search for new drugs is still a research hotspot.
Natural products are an important source of rich drug diversity, and the mining of active ingredients in natural products has made great contributions to human treatment of various diseases over the past century, and it would continue to provide an indispensable source of bioactive lead compounds for drug research5,6. Natural products and their derivatives are important components of natural medicines and important sources of drugs included in pharmacopoeia. According to rough estimates, half of the medicines sold in the modern market are derived from natural medicines5. 1,4-naphthoquinone derivatives are common compounds in nature7. Besides good anti-tumor activity, they also have many pharmacological activities such as antibacterial, anti-inflammatory, antioxidant and cardiac protection8. A variety of 1, 4-naphthoquinone derivatives, such as plumbagin, shikonin, and juglone, have been shown to have significant efficacy in cancer treatment9,10,11. Moreover, the mechanism of action of naphthoquinone against AML was reviewed, and 1, 4-naphthoquinone showed the anti-AML activity through various apoptotic pathway and cellular signaling pathways12, and thus 1, 4-naphthoquinone scaffold-derived compounds could represent the promising compounds against AML.
However, the molecular interactions between 1,4-naphthoquinone scaffold-based compounds and AML-associated proteins are not fully understood. Network pharmacology is an emerging approach to drug analysis that is based on systems biology, multidirectional pharmacology, and network analysis, enabling the construction of “active component-target-gene-disease” networks to reveal drug-disease interactions from a holistic perspective13,14. Moreover, the method is also widely used to gain insight into the therapeutic mechanisms of cancer-related drugs15,16. Therefore, in the present study, the possible mechanism of 1, 4-naphthoquinone scaffold-based compounds in the treatment of AML was analyzed by means of network pharmacology from the aspects of active targets, signal pathways, molecular docking and molecular dynamics simulation, so as to provided theoretical basis for subsequent experimental and clinical studies. A diagram of the workflow is shown in Fig. 1.
Materials and methods
Collection and screening of 1,4-naphthoquinone scaffold-derived compounds
1,4-Naphthoquinone scaffold-derived compounds were sourced from the ASINEX database (http://www.asinex.com). DataWarrior Chemical Data Analysis (https://openmolecules.org/datawarrior/) and Visualization software V6.0.0 were used to evaluate compounds17. 1,4-Naphthoquinone compounds are plotted in the “Structure” function of the software, looking for compounds with similar 1,4-naphthoquinone base structures. Compounds based on the structural framework of 1,4-naphthoquinone scaffolds were then extracted from the software to evaluate their drug similarity and toxicity.
The prediction of drug‑likeness and toxic parameters
The properties of drug-likeness on the 1,4-naphthoquinone scaffold-derived compounds were performed by SwissADME (http://www.swissadme.ch/index.php)18. The filtering standard was based on Lipinski’s rule: Molecular weight (< 500 g/mol) or Topological Polar Surface Area (TPSA) (< 140 Å2) or Moriguchi octanol–water partition coefficient (MLogP) (≤ 4.15) or Hydrogen Bonding Acceptor (HBA) (< 10) or Hydrogen Bonding Donor (HBD) (≤ 5)19. To accept the rule, the molecules should not be violated more than 2 parameters out of 5 parameters. The toxicity of the 1,4-naphthoquinone scaffold-derived compounds were confirmed by Molecular Properties Prediction (https://www.organic-chemistry.org/prog/peo/), and the mutagenicity, tumorigenicity, irritancy, and reproductive effects of compounds are all “none”, which is the inclusion criterion for non-toxicity of the compound20. Compounds with no violations were chosen for further assessment.
Common targets prediction of 1,4-naphthoquinone scaffold-derived compounds and AML
The potential targets of 1,4-naphthoquinone scaffold-derived compounds were predicted using the SwissTargetPrediction Database (STP) (http://www.swisstargetprediction.ch/) and the Similarity Ensemble Approach Database (SEA) (https://sea.bkslab.org/)21,22. The potential targets of AML were identified using the GeneCards databases (https://www.genecards.org) searching for the keyword “acute myeloid leukemia”23 and Gene Expression Omnibus (GEO) database in GSE37307 (https://www.ncbi.nlm.nih.gov/geo/)24. Then the common targets prediction of 1,4-naphthoquinone scaffold-derived compounds and AML were collected using the Venny Website (https://bioinfogp.cnb.csic.es/tools/venny/index.html), and is represented by a Veen diagram.
Establishment of protein–protein interaction (PPI) network and screening of key targets
The potential targets for 1,4-naphthoquinone scaffold-derived compounds and AML were introduced into STRING platform (https://cn.string-db.org/), and then multi-protein analysis function was then applied and “Homo sapiens” was selected25. Remove hidden 9 disconnected nodes from the network and then downloaded PPI data. Then, the data of PPI was imported into Cytoscape 3.7.1 to draw the PPI network and further obtain the top 10 core targets based on the median value of degree, betweenness centrality, and closeness centrality. PPI network was filtered by molecular complex detection algorithm (MCODE) plugin in Cytoscape.
Establishment of GO enrichment analysis and KEGG pathway analysis
The obtained top 10 key targets were imported into David platform (https://david.ncifcrf.gov/)26. Set the Identifier to “OFFICE_GENE_SYMBOL”, species to “Homo sapiens”, and List Type to “Gene List”, respectively. Then, select “GOTERM _BP_DIRECT”, “GOTERM_CC_DIRECT”, “GOTERM_MF_DIRECT” in Gene_Ontology and “KEGG_PATHWAY” in Pathways. GO enrichment analysis and KEGG pathway analysis were performed, and the results were visualized.
Molecular docking
The AutoDock software was used to perform molecular docking between top 2 key targets (STAT3 and TLR4) and 1,4-naphthoquinone scaffold-derived compounds. The 3D structure of 1,4-naphthoquinone scaffold-derived compounds from ChemBio3D software. The crystal structures of target proteins were obtained from the RCSB database (https://www.rcsb.org/)27. Make AutoDockTools 1.5.6 convert ligand and receptor files to pdbqt format and improve their structure by replacing water molecules with hydrogen atoms. AutoDock Vina 1.1.2 was used for molecular docking to obtain the binding energy value, and the combination with the lowest binding energy was visually analyzed by PyMOL 2.4.0 software.
Molecular dynamics simulation
The complex was simulated with 50 ns MD using Gromacs 2023. The protein uses CHARMM 36 field parameters28, and the ligand topology is constructed by GAFF2 field parameters. The protein–ligand complex is placed in a cube box using periodic boundary conditions. The TIP3P water model was used to fill the box with water molecules29. The particle grid Ewald (PME) and Verlet algorithms are used to deal with electrostatic interactions respectively. The heavy atoms of the protein are confined and the steepest descent method is applied to minimize the energy of 50,000 steps. The simulation system was balanced by 100 ps using a gauge ensemble (NVT) and an isothermal isobaric ensemble (NPT). Both van der Waals and Coulomb interactions are calculated using 1.0 nm cutoff values. Finally, the system performed 100 ns molecular dynamics simulations at constant temperature (300 K) and constant pressure (1 bar) with a time step of 2 fs, saving trajectory data every 10 ps. Binding free energy was calculated using the MM/PBSA.py module.
Survival analysis in relation to core targets
Analysis of the overall survival in relation to 10 core targets was performed using the Booldspot (https://www.bloodspot.eu/) database30. A P value < 0.05 was used as a unified screening criterion.
Results
Screening of 1,4-naphthoquinone scaffold-derived compounds for drug-likeness and toxicity properties
First, we retrieved a comprehensive set of 575,302 compounds from the ASINEX database, and then screened them with DataWarrior chemical data analysis and visualization software V5.5.0 to obtain 26 compounds with 1,4-naphthoquinone scaffold (Table S1), and all of which were accepted by Lipinski’s rules. As a promising chemotherapeutic drug, low toxicity is essential, and thus we further predicted the toxic properties of these compounds. The OSIRIS Property Explore was reported to be able to predict the toxicity properties of the compounds, so in present study, 19 compounds were predicted to have no toxicity properties and screened out for further study through OSIRIS Property Explore (Table 1 and Fig. 2).
Acquisition of common targets of 1,4-naphthoquinone scaffold-derived compounds and AML
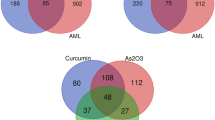
Then, the targets related to the 19 1,4-naphthoquinone scaffold-derived compounds were retrieved from the SwissTargetPrediction database (STP) (535) and Similarity Ensemble Approach Database (460), respectively (Table S2). After sorting out and removing duplicate targets, 836 targets were collected. A total of 15,746 AML-related targets were obtained from GeneCards database, and while 1505 AML-related targets were collected from GSE37307. 96 potential overlapping targets of compounds and AML were obtained by the website of Venny 2.1.0, which was considered to be differentially expressed genes and designated as potential hub genes for the effects of 1,4-naphthoquinone scaffold-derived compounds on AML (Fig. 3).
Protein–protein interaction network analysis
To explore the potential mechanism of 1,4-naphthoquinone scaffold-derived compounds for AML, PPI network was constructed based on the degree centrality (DC), betweenness centrality (BC) and closeness centrality (CC) of targets. It can be seen from Fig. 4A that 21 nodes and 155 edges were obtain with DC ≥ 21 as the criterion. Then 10 nodes and 42 edges were acquired under the condition of DC ≥ 25, BC > 0.015 and CC > 0.540. Finally, 10 top targets with greatest performance were obtained, including STAT3, TLR4, HSP90AA1, JUN, MMP9, PTPRC, JAK2, PTGS2, KIT and CSF1R. Moreover, Table 2 was included gene names, DC, BC and CC of these top 10 targets, and these top 10 targets may play the most important role in the progress of 1,4-naphthoquinone scaffold-derived compounds treatment of AML.
The potential targets identified by PPI analysis. (A) The screening process of PPI network topology. 10 targets were obtained from 87 common targets screened by DC, BC and CC. (B) The PPI network. The node color is proportional to the degree value in the network. (C) The network of core 10 targets. (D) Cluster analysis of PPI network.
Then, the obtained 87 potential common targets of 1,4-naphthoquinone scaffold-derived compounds and AML were visualized in Cytoscape3.7.1 software. As shown in Fig. 4B, the collective targets were used to construct the PPI network composed of 87 nodes with 542 edges. Moreover, the color of node from deep to shallow demonstrated that the node DC was from large to small. The PPI among top 10 targets was illustrated in Fig. 4C, two targets with greater degree were STAT3 and TLR4. Moreover, cluster analysis was carried out through the MCODE plugin of Cytoscape to produce highly connected sub-networks and assign targets to 4 group, including cluster 1 (12 nodes and 50 edges), cluster 2 (14 nodes and 57 edges), cluster 3(8 nodes and 12 edges) and cluster 4 (3 nodes and 3edges) (Fig. 4D).
GO enrichment analysis, KEGG pathway analysis and construction of compound-target-pathway-disease network
To identify the top 10 targets functions, GO enrichment analysis and KEGG pathway analysis were performed through David data. Three categories of GO functional annotated targets were attained, including biological process (BP), molecular function (MF) and cellular component (CC). Specifically, the GO functional enrichment analysis included 90 BP, 10 CC and 12 MF. The top 10 items enriched in BP (Fig. 5A) and MF (Fig. 5B) and CC (Fig. 5C) are presented as bubble plots and histograms (Fig. 5D). In addition, BP was mainly associated with positive regulation of nitric oxide biosynthetic process, positive regulation of cell migration, positive regulation of tumor necrosis factor production, inflammatory response and negative regulation of cell proliferation. CC was associated with plasma membrane, cell surface, receptor complex, external side of plasma membrane and integral component of plasma membrane. MF was associated with protein homodimerization activity, identical protein binding, receptor binding, protein phosphatase binding and protein tyrosine kinase activity.
Furthermore, KEGG pathway analysis-related top 10 targets with 34 signaling pathways were determined, and 10 signaling pathways were selected for mapping and presented them in the form of bubble maps. As shown in Fig. 6A,B, the 10 signaling pathways including pathways in cancer, IL-17 signaling pathway, PI3K-Akt signaling pathway, necroptosis, proteoglycans in cancer, acute myeloid leukemia, TNF signaling pathway, estrogen signaling pathway, NOD-like receptor signaling pathway and MAPK signaling pathway. These data indicate that the protective effects of 1,4-naphthoquinone scaffold-derived compounds against AML are closely linked to these 10 signaling pathways.
Moreover, the annotation of KEGG pathways and the involved potential targets are shown in Table 3. The KEGG pathways of pathways in pathways in cancer was visualized using pathview in Fig. 7A. In addition, the network diagram of the relationship between compounds, key targets, top 10 signaling pathways and AML was also established (Fig. 7B).
Molecular docking
To validate the potential therapeutic effects of network pharmacology, molecular docking was employed to evaluate the binding affinity between small molecule drugs and potential targets. The negative binding energy is used to indicate the affinity between the ligand molecule and the target protein of the receptor. Moreover, the lower and more negative binding energy showed that the active compound could bind better to the target protein. Generally, the binding energy between small molecules and proteins is ≤ − 5.0 kcal/mol, indicating that the two have good binding activity31. The parameters of the docking box are collected (Table 4). Meanwhile, the results of binding energies were illustrated as Fig. 8 and Table 5, and it was found that the binding energies of all compounds and the 10 core genes were all less than − 6 kcal/mol. Furthermore, among the core targets, MMP9 and PTGS2 were have the strongest binding affinity with compound 7 and compound 25, which are − 12.4, − 11.6, − 12.4 and − 11.1 kcal/mol respectively. Furthermore, the magnified image of molecular docking of compound 7 and compound 25 with MMP9 and PTGS2 are shown in Fig. 9.
Molecular dynamics simulation
As per molecular docking findings, MMP9 with the strongest binding force and compound 7 was selected for further molecular dynamics simulation verification. Root mean square deviation (RMSD) is a good indicator of the conformational stability of proteins and ligands, and also a measure of the degree of deviation from the starting position of the atom. The smaller the RMSD, the better the conformational stability. Therefore, RMSD is used to evaluate the balance of the simulation system. As shown in Fig. 10A, the movement of the protein/small molecule complex gradually converges during about 61 ns simulation and maintains relatively stable fluctuations in the middle and late stages of the simulation. Moreover, RMSD was maintained at around 10 Å in the middle and late stages of the simulation, meaning that the binding of small molecules did not lead to sustained and significant changes in protein conformation. Root mean square fluctuation (RMSF) can indicate the flexibility of amino acid residues in a protein. As shown in Fig. 10B,C, the receptor protein is composed of two chains, the protein is relatively flexible as a whole. Radius of Gyration (Rg) can be used to describe the change of the overall structure, and it can be used to characterize the tightness of the protein structure. The greater the change of Rg, the more expanded the system. As can be seen from Fig. 10D, the fluctuation of the binding system is relatively stable after 61 ns, and the Rg value fluctuates around 23A, indicating that the binding of small molecules makes the system denser and more tightly bound.
The binding conformation of complex was used to calculate the binding free energy using the MM/PBSA method. The binding energy of complex was − 26,555.284 kJ/mol. Negative values indicate that the molecule has binding affinity for the target protein, and lower value indicates stronger binding. Additionally, the van der Waals contribution (− 135.952 kJ/mol) and electrostatic contribution (− 26,421.700 kJ/mol) of the small molecule was favorable to the binding of target protein (Fig. 10E). All in all, MMP9 and compound 7 had a good binding effect.
Clinical relevance of core genes
The 10 core targets were selected based on the results in the PPI network and then imported into the Bloodspot database for clinical relevance analysis. The results from the Bloodspot database showed that the expression level of HSP90AA1, MMP9 and PTPRC was significantly correlated with overall survival (P < 0.05), while the expression of the other core targets was not significantly related to the overall survival of AML patients (Fig. 11). This result suggested that 1,4-naphthoquinone scaffold-derived compounds may affect the prognosis of AML patients by regulating targets such as MMP9 and HSP90AA1.
Discussion
AML is a heterogeneous hematological malignancy that occurs in the elderly and has complex cytogenetic and molecular biological features1. Chemotherapy is often the preferred treatment for patients with AML. However, the use of chemotherapy drugs is linked with significant side effects, unpredictable clinical outcomes, ultimately leading to treatment failure3.
Accumulating evidence indicates that natural products have a variety of special structures and novel biological activities, and are an important source of anti-tumor drugs5,6. 1,4-naphthoquinone is a small aromatic ring compound with many biological activities, which has been widely used in the treatment of many diseases, including AML12. Structurally, 1,4-naphthoquinone is a bicarbonyl ring with abundant active modification sites, and it is easy to form hydrogen bonds, hydrophobicity and other forces29. Among them, natural products of 1, 4-naphthoquinone derivatives have been shown to have significant efficacy in cancer treatment10,11,12. The development of 1, 4-naphthoquinone derivatives is one of the important directions of anti-tumor drug research and development. However, the complexes underlying the anti-cancer effects of compounds produced from 1,4-naphthoquinone scaffold with AML remain unclear.
Network pharmacology that uses systems biology theory can determine the course of compounds in disease treatment from the whole level, systematically predict and reveal the mechanism of action of drugs13,14,15,16. Therefore, this may be promising research method in the field of the action of 1,4-naphthoquinone scaffold-derived compounds against AML. Thus, to further clarify the possible targets and pathways underlying the anti-AML effect of 1,4-naphthoquinone scaffold-derived compounds, we used network pharmacology to construct biological action networks, then GO function and KEGG enrichment analyses of core targets were performed, further verified them through molecular docking and molecular dynamics simulation, and tried to explore and analyze the potential targets and mechanism of action of 1,4-naphthoquinone scaffold-derived compounds against AML.
In our study, preliminary results showed that 10 core targets, including STAT3, TLR4, HSP90AA1, JUN, MMP9, PTPRC, JAK2, PTGS2, KIT and CSF1R, that might be the key targets of 1,4-naphthoquinone scaffold-derived compounds against AML were identified. STAT3 is a recognized oncogenic transcription factor, and abnormal activation of STAT3 signaling pathway is associated with a variety of tumors, including AML32,33. Moreover, STAT3 gene is highly expressed in AML patients, and is related to the immune typing and risk of patients34. TLR4 is a pattern recognition receptor that can bind both exogenous and endogenous ligands. It is expressed by acute myeloid leukemia (AML) cells, several bone marrow stromal cells, and non-leukemia cells involved in inflammation35. Moreover, studies showed that TLR4 expression by AML cells is associated with an adverse prognosis, and TLR4 inhibition can have a chemosensitizing and/or direct antileukemic effect36. HSP90AA1, also known as LAP2/HSPC1, encodes a protein-chaperone protein that plays a major role in protein folding and stabilization as a homodimer, and largely determines tumor proliferation, survival, invasion, metastasis, and angiogenesis by regulating multiple cancer-associated proteins37. The study found that AML patients with higher levels of HSP90AA1 had lower remission rates and were associated with poorer prognosis for AML38,39. Moreover, in vivo study found that indirubin could exhibit leukemia cells proliferation via Targeting HSP90AA140. JUN, also called activator protein-1 (AP-1), is an important modulator in several immune disorders and carcinomas41. Higher levels of AP-1 components were found in AML patients, and AP-1 may provide hematologists with important information on diagnostic criteria and can highly predict the survival rate of newly diagnosed AML patients42. MMP9 is a member of the matrix metalloproteinase (MMP) protein family, and it is found to play a critical role in AML by increasing the invasive properties of malignant myeloblasts43. A significantly lower expression level of MMP-9 in AML-patients compared to normal controls44. Moreover, study showed that patients with lower MMP-9 levels tended to have longer survival times44. PTPRC, also known as CD45 (encoded by the PTPRC gene), is a recognized cell surface marker in nucleated hematopoietic cells, and it is thought to contribute to the development of disease and the progression of AML45,46. Guo et al. found that overexpression of PTPRC indicated poor prognosis in pediatric AML patients, and its expression level was correlated with the infiltration level of activated dendritic cells, thus PTPRC could be a promising immunotherapy target for pediatric AML47. JAK2 is a nonreceptor tyrosine kinase which transmits information to the nucleus via the signal transducer and activator of transcriptions (STATs), where it plays various biological roles48. The study found that JAK2 was not only the most commonly mutated gene in patients with myeloproliferative neoplasms (MPN), but was also present in patients with initial AML and AML-MPN49. JAK2 mutation can continuously activate JAK2-STAT signaling pathway and participate in the occurrence and development of acute leukemia49. PTGS2, also known as cyclooxygenase-2 (COX-2), is considered as a component of inflammatory reactions and is directly related to the process of inflammation, pain, angiogenesis, and cancer50. In the AML cell lines, tumor angiogenesis is activated through the induction of COX-2 expression51, moreover methylation of PTGS2 may play a role in the initiation and leukemogenesis phases of AML. Furthermore, PTGS2 can be considered as diagnostic biomarkers for AML52. KIT (CD117) is a transmembrane protein and is detected in a variety of human neoplastic diseases, including AML53. Moreover, KIT is a specific marker for the myeloid lineage, which is expressed early during hematopoietic differentiation and can aid the diagnosis of AML in difficult cases54. However, inhibition of wild-type KIT is not always satisfactory in AML53. CSF1R is a cell surface glycoprotein encoded by the CSF1R gene located on the distal end of the long arm chromosome 5 (5q32)55. More importantly, analysis of CSF1R expression levels in AML patient samples found a correlation between high levels of CSF1R expression and shorter overall survival56. Meanwhile, GO enrichment analysis and KEGG analysis identified many enriched pathways are involved in the effects of 1,4-naphthoquinone scaffold-derived compounds against AML. More importantly, the molecular docking results showed that 1,4-naphthoquinone scaffold-derived compounds could spontaneously interact with these core targets. Among these compounds, compound 7 had a good binding effect with MMP9 through molecular dynamics simulation and MM/GBSA approach. In addition, the results of survival analysis showed that the expression level of HSP90AA1, MMP9 and PTPRC was significantly correlated with overall survival. However, the present study has certain limitations. 1, 4-naphthoquinone scaffold-derived compounds were not commercially available and hard to synthesize by ourselves in our study, and it is challenging to fully elucidate the anti-AML function of the compounds through additional biological experiments, including in vivo and in vitro models. Thus, this is another difficult subject for us to study in the future.
Conclusion
In conclusion, the current study combines network pharmacology, molecular docking and molecular dynamics simulation to firstly elucidate the molecular and pharmacological mechanisms of 1,4-naphthoquinone scaffold-derived compounds against AML. However, the in-depth study of the underlying molecular mechanisms needs to be further explored and verified.
Data availability
All data generated or analysed during this study are included in this published article [and its supplementary information files] and available from the corresponding author on reasonable request.
Abbreviations
- AML:
-
Acute myeloid leukemia
- PPI:
-
Protein–protein interaction
- GEO:
-
Gene Expression Omnibus
- MCODE:
-
Molecular complex detection algorithm
- GO:
-
Gene ontology
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- DC:
-
Degree centrality
- BC:
-
Betweenness centrality
- CC:
-
Closeness centrality
- MM/PBSA:
-
Molecular mechanics/Poisson–Boltzmann surface area
References
Shallis, R. M., Wang, R., Davidoff, A., Ma, X. & Zeidan, A. M. Epidemiology of acute myeloid leukemia: Recent progress and enduring challenges. Blood Rev. 36, 70–87 (2019).
Madanat, Y. F., Kalaycio, M. E. & Nazha, A. Advances in acute myeloid leukemia genomics, where do we stand in 2018?. Acta Med. Acad. 48(1), 35–44 (2019).
Greiner, J., Götz, M. & Wais, V. Increasing role of targeted immunotherapies in the treatment of AML. Int. J. Mol. Sci. 23(6), 3304 (2022).
Martínez-Cuadrón, D. et al. Evolving treatment patterns and outcomes in older patients (≥60 years) with AML: Changing everything to change nothing?. Leukemia 35(6), 1571–1585 (2021).
Yang, G. X. et al. Advanced natural products chemistry research in China between 2015 and 2017. Chin. J. Nat. Med. 16(12), 881–906 (2018).
Mansoori, B. et al. Photodynamic therapy for cancer: Role of natural products. Photodiagnosis Photodyn. Ther. 26, 394–404 (2019).
Klaus, V. et al. 1,4-Naphthoquinones as inducers of oxidative damage and stress signaling in HaCaT human keratinocytes. Arch. Biochem. Biophys. 496(2), 93–100 (2010).
Aminin, D. & Polonik, S. 1,4-Naphthoquinones: Some biological properties and application. Chem. Pharm. Bull. (Tokyo) 68(1), 46–57 (2020).
Liu, Y., Cai, Y., He, C., Chen, M. & Li, H. Anticancer properties and pharmaceutical applications of plumbagin: A review. Am. J. Chin. Med. 45(3), 423–441 (2017).
Lu, J. J. et al. Quinones derived from plant secondary metabolites as anti-cancer agents. Anticancer Agents Med. Chem. 13(3), 456–463 (2013).
Ahmad, T. & Suzuki, Y. J. Juglone in oxidative stress and cell signaling. Antioxidants (Basel) 8(4), 91 (2019).
Lee, M. H., Lapidus, R. G., Ferraris, D. & Emadi, A. Analysis of the mechanisms of action of naphthoquinone-based anti-acute myeloid leukemia chemotherapeutics. Molecules 24(17), 3121 (2019).
Zhao, L. et al. Network pharmacology, a promising approach to reveal the pharmacology mechanism of Chinese medicine formula. J. Ethnopharmacol. 309, 116306 (2023).
Yuan, Z. et al. Progress and prospects of research ideas and methods in the network pharmacology of traditional Chinese medicine. J. Pharm. Pharm. Sci. 25, 218–226 (2022).
Sun, Z., Wang, Y., Pang, X., Wang, X. & Zeng, H. Mechanisms of polydatin against spinal cord ischemia-reperfusion injury based on network pharmacology, molecular docking and molecular dynamics simulation. Bioorg. Chem. 140, 106840 (2023).
Sun, T. et al. Network pharmacology-based strategy combined with molecular docking and in vitro validation study to explore the underlying mechanism of Huo Luo Xiao Ling Dan in treating atherosclerosis. Drug Des. Dev. Ther. 16, 1621–1645 (2022).
López-López, E., Naveja, J. J. & Medina-Franco, J. L. DataWarrior: An evaluation of the open-source drug discovery tool. Expert Opin. Drug Discov. 14(4), 335–341 (2019).
Daina, A., Michielin, O. & Zoete, V. SwissADME: A free web tool to evaluate pharmacokinetics, drug-likeness and medicinal chemistry friendliness of small molecules. Sci. Rep. 7, 42717 (2017).
Nendza, M. & Müller, M. Screening for low aquatic bioaccumulation. 1. Lipinski’s “Rule of 5” and molecular size. SAR QSAR Environ. Res. 21(5–6), 495–512 (2010).
Ismail, N. Z. et al. Molecular docking and molecular dynamic simulations of apoptosis proteins with potential anticancer compounds present in Clinacanthus nutans extract using gas chromatography-mass spectrometry. J. Biomol. Struct. Dyn. 41(13), 6104–6120 (2023).
Daina, A., Michielin, O. & Zoete, V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res. 47(W1), W357–W364 (2019).
Gu, S. & Lai, L. H. Associating 197 Chinese herbal medicine with drug targets and diseases using the similarity ensemble approach. Acta Pharmacol. Sin. 41(3), 432–438 (2020).
Stelzer, G. et al. The GeneCards Suite: From gene data mining to disease genome sequence analyses. Curr. Protoc. Bioinform. 54, 1.30.1-1.30.33 (2016).
Jia, G. et al. Decoding the mechanism of Shen Qi Sha Bai decoction in treating acute myeloid leukemia based on network pharmacology and molecular docking. Front. Cell Dev. Biol. 9, 796757 (2021).
Szklarczyk, D. et al. The STRING database in 2023: Protein–protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res. 51(D1), D638–D646 (2023).
Dennis, G. Jr. et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 4(5), P3 (2003).
Burley, S. K. et al. RCSB Protein Data bank: Tools for visualizing and understanding biological macromolecules in 3D. Protein Sci. 31(12), e4482 (2022).
Jo, S. et al. CHARMM-GUI: A web-based graphical user interface for CHARMM. J. Comput. Chem. 29(11), 1859–1865 (2008).
Mark, P. & Nilsson, L. Structure and dynamics of the TIP3P, SPC, and SPC/E water models at 298 K. J. Phys. Chem. A 105, 9954–9960 (2001).
Bagger, F. O. et al. BloodSpot: A database of gene expression profiles and transcriptional programs for healthy and malignant haematopoiesis. Nucleic Acids Res. 44(D1), D917–D924 (2016).
Vanommeslaeghe, K. et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. J. Comput. Chem. 31(4), 671–690 (2010).
Hirose, R. et al. A method for detecting covalent modification of sensor proteins associated with 1,4-naphthoquinone-induced activation of electrophilic signal transduction pathways. J. Toxicol. Sci. 37(5), 891–898 (2012).
Furtek, S. L. et al. Strategies and approaches of targeting STAT3 for cancer treatment. ACS Chem. Biol. 11(2), 308–318 (2016).
Bruserud, Ø. et al. STAT3 as a possible therapeutic target in human malignancies: Lessons from acute myeloid leukemia. Expert Rev. Hematol. 8(1), 29–41 (2015).
Curran, E., Corrales, L. & Kline, J. Targeting the innate immune system as immunotherapy for acute myeloid leukemia. Front. Oncol. 5, 83 (2015).
Bruserud, Ø., Reikvam, H. & Brenner, A. K. Toll-like receptor 4, osteoblasts and leukemogenesis; The lesson from acute myeloid leukemia. Molecules 27(3), 735 (2022).
Taipale, M., Jarosz, D. F. & Lindquist, S. HSP90 at the hub of protein homeostasis: Emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11(7), 515–528 (2010).
Flandrin, P. et al. Significance of heat-shock protein (HSP) 90 expression in acute myeloid leukemia cells. Cell Stress Chaperones 13(3), 357–364 (2008).
Zhang, Z. et al. Integrated bioinformatic analysis of microarray data reveals shared gene signature between MDS and AML. Oncol. Lett. 16(4), 5147–5159 (2018).
Yao, Y. et al. Indirubin, an active component of Indigo naturalis, exhibits inhibitory effects on leukemia cells via targeting HSP90AA1 and PI3K/Akt pathway. Anticancer Agents Med. Chem. 24(9), 718–727 (2024).
Ye, N. et al. Small molecule inhibitors targeting activator protein 1 (AP-1). J. Med. Chem. 57(16), 6930–6948 (2014).
Trop-Steinberg, S. & Azar, Y. AP-1 expression and its clinical relevance in immune disorders and cancer. Am. J. Med. Sci. 353(5), 474–483 (2017).
Klein, G. et al. The possible role of matrix metalloproteinase (MMP)-2 and MMP-9 in cancer, e.g. acute leukemia. Crit. Rev. Oncol. Hematol. 50(2), 87–100 (2004).
Lin, L. I. et al. Marrow matrix metalloproteinases (MMPs) and tissue inhibitors of MMP in acute leukaemia: Potential role of MMP-9 as a surrogate marker to monitor leukaemic status in patients with acute myelogenous leukaemia. Br. J. Haematol. 117(4), 835–841 (2002).
Qian, D. et al. JAK2 and PTPRC mRNA expression in peripheral blood mononuclear cells from patients with systemic lupus erythematosus. Clin. Rheumatol. 39(2), 443–448 (2020).
Guo, L., Chen, L. & Wang, H. CD45 correlates with adverse risk stratification, decreased treatment response and unfavorable survival profiles in elderly acute myeloid leukemia patients. Cancer Biomark. 23(3), 455–463 (2018).
Guo, G. et al. PTPRC overexpression predicts poor prognosis and correlates with immune cell infiltration in pediatric acute myeloid leukemia. Clin. Lab. 68(7), 210940 (2022).
Staerk, J. & Constantinescu, S. N. The JAK-STAT pathway and hematopoietic stem cells from the JAK2 V617F perspective. JAKSTAT 1(3), 184–190 (2012).
Bader, M. S. & Meyer, S. C. JAK2 in myeloproliferative neoplasms: Still a protagonist. Pharmaceuticals (Basel) 15(2), 160 (2022).
Akhtar, M. et al. Promoter methylation regulates Helicobacter pylori-stimulated cyclooxygenase-2 expression in gastric epithelial cells. Cancer Res. 61(6), 2399–2403 (2001).
Chien, M. H. et al. Vascular endothelial growth factor-C (VEGF-C) promotes angiogenesis by induction of COX-2 in leukemic cells via the VEGF-R3/JNK/AP-1 pathway. Carcinogenesis 30(12), 2005–2013 (2009).
Kiani-Zadeh, M. et al. Studying the potential of upregulated PTGS2 and VEGF-C besides hyper-methylation of PTGS2 promoter as biomarkers of acute myeloid leukemia. Mol. Biol. Rep. 49(8), 7849–7862 (2022).
Szatkowski, D. & Hellmann, A. The overexpression of KIT proto-oncogene in acute leukemic cells is not necessarily caused by the gene mutation. Acta Haematol. 133(1), 116–123 (2015).
Valverde, L. R. et al. C-kit receptor (CD117) expression in acute leukemia. Ann. Hematol. 72(1), 11–15 (1996).
Cannarile, M. A. et al. Colony-stimulating factor 1 receptor (CSF1R) inhibitors in cancer therapy. J. Immunother. Cancer 5(1), 53 (2017).
Edwards, D. K. V. et al. CSF1R inhibitors exhibit antitumor activity in acute myeloid leukemia by blocking paracrine signals from support cells. Blood 133(6), 588–599 (2019).
Acknowledgements
We thank all authors for their contributions and support.
Author information
Authors and Affiliations
Contributions
Conception and design: R C and JY S; Provision of study materials: R C, HF L and JY S; Collection and assembly of data: R C, HF L, WK M; Data analysis and interpretation: R C, HF L, WK M; Manuscript writing: all authors; Final approval of the manuscript: all authors. All authors agree to publish this paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, R., Liu, H., Meng, W. et al. Analysis of action of 1,4-naphthoquinone scaffold-derived compounds against acute myeloid leukemia based on network pharmacology, molecular docking and molecular dynamics simulation. Sci Rep 14, 21043 (2024). https://doi.org/10.1038/s41598-024-70937-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-70937-y
- Springer Nature Limited