Abstract
Capacitation is an essential post-testicular maturation event endowing spermatozoa with fertilizing capacity within the female reproductive tract, significant for fertility, reproductive health, and contraception. By using a human-relevant large animal model, the domestic boar, this study focuses on furthering our understanding of the involvement of the ubiquitin-proteasome system (UPS) in sperm capacitation. The UPS is a universal, evolutionarily conserved, cellular proteome-wide degradation and recycling machinery, that has been shown to play a significant role in reproduction during the past two decades. Herein, we have used a bottom-up proteomic approach to (i) monitor the capacitation-related changes in the sperm protein levels, and (ii) identify the targets of UPS regulation during sperm capacitation. Spermatozoa were capacitated under proteasomal activity-permissive and inhibiting conditions and extracted sperm proteins were subjected to high-resolution mass spectrometry. We report that 401 individual proteins differed at least two-fold in abundance (P < 0.05) after in vitro capacitation (IVC) and 13 proteins were found significantly different (P < 0.05) between capacitated spermatozoa with proteasomal inhibition compared to the vehicle control. These proteins were associated with biological processes including sperm capacitation, sperm motility, metabolism, binding to zona pellucida, and proteasome-mediated catabolism. Changes in RAB2A, CFAP161, and TTR during IVC were phenotyped by immunocytochemistry, image-based flow cytometry, and Western blotting. We conclude that (i) the sperm proteome is subjected to extensive remodeling during sperm capacitation, and (ii) the UPS has a narrow range of distinct protein substrates during capacitation. This knowledge highlights the importance of the UPS in sperm capacitation and offers opportunities to identify novel pharmacological targets to modulate sperm fertilizing ability for the benefit of human reproductive health, assisted reproductive therapy, and contraception, as well as reproductive management in food animal agriculture.
Similar content being viewed by others
Introduction
Sperm capacitation and protein degradation via the ubiquitin-proteasome system (UPS) are two concurrent events to which our group has dedicated a substantial amount of research effort. Sperm capacitation, for the first time observed independently by Austin and Chang in 19511,2 is the terminal sperm maturation process taking place in the oviductal sperm reservoir3. Our current understanding of sperm capacitation is that it entails a sum of biophysical and biochemical changes in the plasma membrane and the reactivation of multiple dormant metabolic and signaling pathways that enable spermatozoa to reach and fertilize the freshly ovulated oocyte(s)4,5,6. These capacitation-related events take place in a compartmentalized and orchestrated manner, together conveying sperm hyperactivated motility, responsiveness to chemoattractants such as progesterone7,8,9,10,11 and chemorepellents such as Zn divalent cation12,13, and the ability to bind to zona pellucida and undergo acrosomal exocytosis14,15,16. Spermatozoa, stored within the female oviductal sperm reservoir, are ready to be released in waves for several hours after an ovulatory signal, thus maximizing the window of opportunity for fertilization17.
Protein ubiquitination is a stable, reversible, posttranslational protein modification where a small (76 AA, 8.5 kDa), ubiquitous, species-conserved chaperone protein ubiquitin is covalently ligated through its terminal G76 residue to the ε-NH2 group of an internal lysine of the substrate18. The same isopeptide bond is formed between the ubiquitin moieties of the polyubiquitin chain, in particular, between G76 and K4819 residues that serves as a proteolysis recognition signal for the downstream 26S proteasome20,21. The cascade reaction of ubiquitin conjugation is facilitated by three enzymes (i) E1—the ubiquitin-activating enzyme, (ii) E2—the ubiquitin-conjugating/carrier enzyme, and (iii) E3—the ubiquitin protein ligase22,23. The 26S proteasome is a large (∼ 1.5 MDa), multicatalytic protease that degrades polyubiquinated proteins into small peptides. The subunit composition of canonical 26S proteasome as well as the components’ functions can be found elsewhere24,25. Functional 26S proteasome participates in cellular proteostasis, i.e., the balance between protein synthesis, activity, and recycling by performing protein degradation and turnover of misfolded proteins or proteins at the end of their lifespan. Regulation of a variety of cellular processes like cell cycle and division, regulation of transcription factors, and assurance of cellular quality control is also performed via the degradation of intracellular proteins by the UPS26,27,28.
In more than twenty years of research effort that our group has dedicated to understanding the role of the UPS in mammalian reproduction, we showed that UPS modulates many aspects of mammalian reproduction; including gametogenesis13,16,25,29,30,31,32,33,34,35,36,37,38,39, epididymal sperm maturation40,41, sperm binding to oviductal epithelium42,43, sperm capacitation12,13,32,44,45,46,47, fertilization16,25,34,37,39, post-fertilization paternal mitophagy33,36,48,49,50,51, and zygotic genome activation52,53. Our understanding of the UPS role during mammalian sperm capacitation is becoming more extensive. Briefly, components of the UPS have been shown to participate in capacitation-associated events such as degradation of protein kinase PRKAR1 and the kinase-binding AKAP3 sperm protein54,55,56; acrosomal remodeling, sperm protein processing, and compartmentalization46,47,57,58; hyperactivation56,59 as well as spermadhesin de-aggregation from the sperm surface necessary for sperm detachment from the oviductal epithelium44. Studies of in vitro capacitation (IVC) during sperm coincubation with oviductal epithelial explants and oviductal glycan-coated beads indicate that the 26S proteasome is physiologically important for the detachment of spermatozoa from oviductal sperm reservoir-mimicking surfaces in vitro and ex vivo42,43. Further, the 26S proteasome may be one of two key regulators of mammalian sperm capacitation pointed out in silico60. Lastly, the proteolytic activity of the 26S proteasome seems to be important during the early stages of sperm capacitation45,60 and is regulated by PRKA in a feedback loop61,62.
Sperm proteome remodeling during sperm capacitation is necessary for spermatozoa to perform their function during gamete binding and fusion. Studies referenced below show that the UPS is involved in the regulation of multiple sperm proteins during sperm capacitation. We hypothesize that the UPS is involved in the modulation of a multitude of sperm proteins during sperm capacitation and the previously reported UPS substrates are just a fraction of the complete UPS target protein list. The objective of this study was therefore to (i) isolate the porcine sperm proteome from non-capacitated and capacitated spermatozoa with or without proteasomal inhibition, (ii) identify targets of UPS regulation by using high-resolution mass spectrometry, and (iii) screen three significantly different UPS target proteins during sperm capacitation by using our established sperm protein phenotyping pipeline.
Results
Proteins identified in non-capacitated versus in vitro capacitated boar spermatozoa
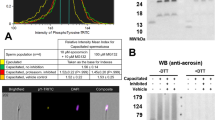
A fresh, sperm-rich fraction of ejaculates (n = 6) from two fertile, non-transgenic white large boars (Sus scrofa) was used to conduct the experiment. The sperm-rich fraction was split into quarters; three quarters were in vitro capacitated (IVC) under (i) proteasomal activity-permissive conditions, (ii) 100 µM MG132 proteasome-inhibiting conditions including 0.2% (v/v) EtOH, and 0.2% EtOH vehicle control, while the last quarter was used as non-capacitated (NC) control. Spermatozoa were successfully capacitated as demonstrated by an increase in protein tyrosine phosphorylation (Supplementary Fig. 1).
Sperm proteins from all three IVC groups and NC spermatozoa were extracted and processed for high-resolution mass spectrometry. Six biological replicates of each treatment group were performed, and proteomic analysis was performed in all working replicates that were used for statistical and bioinformatic analyses. Altogether, we have identified 999 UniProtKB annotated accessions (Supplementary Data File 1), which translates into 896 gene-encoded proteins.
The identified whole sperm protein dataset was assessed by cluster analyses—including heat map with dendrogram clustering and principal component analysis; and GO enrichment analysis including biological process, molecular function, and cellular component analyses. The heat map analysis (Fig. 1a) shows the clustering of non-capacitated control samples, while the proteome differences in all capacitated treatment groups were not sufficient to cluster them in distinct groups. This observation was confirmed with principal component analysis (Fig. 1b), where non-capacitated samples formed a distinct cluster. Two distinct clusters were formed of capacitated samples, however, both capacitated sperm clusters contained representatives of all three treatment groups. The GO biological process enrichment analysis (Supplementary Fig. 2, and Supplementary Data File 2) identified 27 categories (Padj < 0.05) with some of the most enriched ones being cilium movement involved in cell motility (fold enrichment = 15.84, Padj = 1.37 × 10−05), mitochondrial ATP synthesis coupled proton transport (fold enrichment = 14.52, Padj = 3.92 × 10−06), and binding of spermatozoa to zona pellucida (fold enrichment = 14.12, Padj = 2.16 × 10−22). The GO cellular component enrichment analysis (Supplementary Fig. 3, and Supplementary Data File 2) identified 37 different categories (Padj < 0.05) with some of the most enriched categories being proteasome regulatory particle (fold enrichment = 21.35, Padj = 8.30 × 10−08), zona pellucida receptor complex (fold enrichment = 21.35, Padj = 1.59 × 10−06), and sperm fibrous sheath (fold enrichment = 21.36, Padj = 5.53 × 10−04). Furthermore, GO molecular function enrichment analysis (Supplementary Fig. 4, and Supplementary Data file 2) identified 21 different categories (Padj < 0.05) with some of the most enriched categories being pyruvate dehydrogenase (NAD +) activity (fold enrichment = 22.14, Padj = 1.54 × 10−02), proteasome-activating ATPase activity (fold enrichment = 18.97, Padj = 2.80 × 10−03), and proton-transporting ATP synthase activity (fold enrichment = 14.76, Padj = 1.87 × 10−04).
Comparison of pig sperm proteomes before and after in vitro capacitation including proteasomal inhibition and vehicle control. (a) Heat map analysis with non-constrained differences, and (b) principal component analysis of non-capacitated, capacitated, 100 µM MG132 IVC, and vehicle IVC pig sperm proteomes. In vitro capacitated cluster 1—IVC-CL1, in vitro capacitated cluster 2—IVC-CL2, and non-capacitated cluster—NC-CL.
Changes in sperm proteome after in vitro capacitation (non-capacitated vs. capacitated sperm proteomes comparison)
Changes in sperm protein abundances of IVC spermatozoa were investigated by using volcano plot analysis63 and are presented in Supplementary Fig. 5a. The fold change threshold was set to 2, and the level of significance was P < 0.05. After sperm IVC, 262 UniProtKB accessions translating to 228 unique proteins were more abundant while 187 UniProtKB accessions translating to 173 unique proteins were less abundant in IVC spermatozoa when compared to NC spermatozoa. These significantly different proteins were subjected to GO enrichment and KEGG pathway64,65,66 analyses (Figs. 2, 3, and 4, Supplementary Fig. 6, and Supplementary Data File 2). Some of the most enriched biological processes (Padj < 0.05) in capacitated spermatozoa were regulation of cilium beat frequency (fold enrichment = 44.24, Padj = 3.23 × 10−02), cilium-dependent cell motility (fold enrichment = 38.71, Padj = 2.43 × 10−03), and cilium movement involved in cell motility (fold enrichment = 35.19, Padj = 3.78 × 10−03). Meanwhile, positive regulation of establishment of protein localization to telomere (fold enrichment = 58.90, Padj = 1.84 × 10−05), proteasomal ubiquitin-independent protein catabolic process (fold enrichment = 27.18, Padj = 9.87 × 10−04), and binding of spermatozoa to zona pellucida (fold enrichment = 23.88, Padj = 1.98 × 10−07) were some of the most enriched biological processes (Padj < 0.05) in non-capacitated spermatozoa. Some of the most enriched cellular compartments (Padj < 0.05) of the identified sperm proteomes in IVC spermatozoa were inner dynein arm (fold enrichment = 82.52, Padj = 9.40 × 10−04), pyruvate dehydrogenase complex (fold enrichment = 66.01, Padj = 2.33 × 10−03), and outer dynein arm (fold enrichment = 51.57, Padj = 1.90 × 10−04); while some of the most enriched cellular components (Padj < 0.05) in non-capacitated spermatozoa were chaperonin-containing T-complex (fold enrichment = 70.68, Padj = 8.58 × 10−10), proteasome core complex, alpha-subunit complex (fold enrichment = 70.68, Padj = 1.67 × 10−06), and zona pellucida receptor complex (fold enrichment = 66.26, Padj = 1.01 × 10−04). Molecular functions that were among the most significantly enriched (Padj < 0.05) in IVC spermatozoa were pyruvate dehydrogenase (NAD +) activity (fold enrichment = 75.55, Padj = 3.02 × 10−03), ubiquitin protein ligase binding (fold enrichment = 51.94, Padj = 5.63 × 10−13), and dynein intermediate chain binding (fold enrichment = 24.84, Padj = 1.08 × 10−02). Molecular functions that were among the most significantly enriched (Padj = 0.05) in non-capacitated spermatozoa were proteasome-activating ATPase activity (fold enrichment = 55.27, Padj = 1.18 × 10−02), protein binding involved in protein folding (fold enrichment = 26.38, Padj = 3.83 × 10−07), and endopeptidase activity (fold enrichment = 21.49, Padj = 2.17 × 10−07). Lastly, among the most significantly enriched pathways (Padj < 0.05) in IVC spermatozoa were citrate cycle (fold enrichment = 38.09, Padj = 5.20 × 10−13), 2-oxocarboxylic acid metabolism (fold enrichment = 23.76, Padj = 4.06 × 10−07), and carbon metabolism (fold enrichment = 13.15, Padj = 9.15 × 10−11). The most significantly enriched pathways (Padj < 0.05) in non-capacitated spermatozoa were proteasome (fold enrichment = 26.98, Padj = 6.28 × 10−11), glycolysis/gluconeogenesis (fold enrichment = 13.35, Padj = 3.55 × 10−04), and carbon metabolism (fold enrichment = 10.16, Padj = 1.86 × 10−05).
Targets of the ubiquitin-proteasome system during sperm in vitro capacitation
Similarly, as in the previous section, changes in sperm protein abundances between IVC-proteasomally-inhibited and IVC-vehicle control spermatozoa were compared using volcano plot analysis (Supplementary Fig. 5b). No significantly different proteins were found between these two groups by using this technique. Therefore, the student’s T-test was used to identify additional differentially abundant sperm proteins, with a level of significance P < 0.05. In total, 13 proteins were found significantly different between proteasomally-inhibited and vehicle groups. Nine proteins were significantly more abundant in the proteasomally-inhibited spermatozoa including ACAD10, ALB, BTBD16, CFAP161, CLYBL, CST3, FAM205C, H2AC1, and RAB2A. The significantly more abundant proteins in the vehicle control spermatozoa were EEF1A1, KLK8, MT-ND2, and TTR (Table 1).
A literature search of these 13 proteins was performed by searching each protein individually in the PubMed.gov database for their reported/known function and localization specifically in spermatozoa (Table 1). Proteins ACAD10, ALB, CST3, H2AC1, MT-ND2, and RAB2A were characterized in spermatozoa previously (see Table 1 for references), with known biological functions. Localization-wise; ALB, H2AC1, and RAB2A are localized in the head; ACAD10, CLYBL, and MT-ND2 are localized in midpiece/mitochondrial sheath; flagellar localization was reported for CFAP161 and EEF1A1; and finally, CST3 was reported in both the head and flagellum. Three significantly different sperm proteins (RAB2A, CFAP161, and TTR) between capacitated, proteasomally inhibited, and vehicle control groups were phenotyped in spermatozoa within all treatment groups.
Localization and dynamics of selected UPS-regulated sperm proteins during sperm in vitro capacitation
Candidates for UPS modulation during sperm IVC—RAB2A, member RAS onco family, cilia and flagella associated protein 161 (CFAP161), and transthyretin (TTR) were subjected to phenotype studies employing immunocytochemistry (ICC), image-based flow cytometry (IBFC), Western blotting (WB), and immunoprecipitation (IP; only TTR) (Figs. 5, 6 and 7).
Phenotyping of RAB2A protein in the non-capacitated (NC) and in vitro capacitated (IVC) spermatozoa with and without proteasomal inhibition, including vehicle control. The ICC detection of RAB2A (red) in NC (a1) and IVC, non-inhibited (a2) spermatozoa, including non-immune serum control (a3). Spermatozoa were co-stained for acrosomal presence with peanut agglutinin (PNA, green), and nuclear stain DAPI (blue). All fluorescence channels are superimposed with the differential interference contrast (DIC) brightfield channel. Scale bars represent 20 µm. The IBFC of formaldehyde fixed and Triton X-100 permeabilized NC and IVC spermatozoa with or without proteasomal inhibition, including a vehicle and negative, normal rabbit serum control (b1). Immunofluorescence images of NC (b2) and IVC spermatozoa (b3), including normal mouse serum (b4) obtained by IBFC to confirm labeling. The IBFC was performed in 4 replicates with consistent results. Detection of RAB2A by WB (c1) in the extracts obtained from NC and IVC spermatozoa under proteasome permissive and inhibiting conditions including vehicle control. The red arrows point to the 24 and 27 kDa RAB2A band doublet reported previously80, and the blue arrow points to the 31 kDa form observed here for the first time. The membrane was stripped and reprobed with anti-TUBB antibody (c2) and stained with CBB R-250 (c3) for protein load normalization purposes. Western blotting was performed in 6 replicates. Statistical evaluation of the IBFC results (b5) and WB results (c4) was performed by using ANOVA with Sidak’s post hoc test (α = 0.05). Full, uncropped membranes of all six replicates of TUBB and RAB2A WB detections are presented in Fig. S7 A and B, respectively. RAB2A blocking peptide assay and Western blotting of recombinant human RAB2A are presented in Fig. S8 and S9, respectively.
Phenotyping of CFAP161 protein in the non-capacitated (NC) and in vitro capacitated (IVC) spermatozoa with and without proteasomal inhibition, including vehicle control. The ICC detection of CFAP161 (red) in NC (a1) and IVC, non-inhibited (a2) spermatozoa, including non-immune serum control (a3). Spermatozoa were co-stained for acrosomal presence with peanut agglutinin (PNA, green), and nuclear stain DAPI (blue). All fluorescence channels are superimposed with the differential interference contrast (DIC) brightfield channel. Scale bars represent 20 µm. The IBFC of formaldehyde fixed and Triton X-100 permeabilized NC and IVC spermatozoa with or without proteasomal inhibition, including a vehicle and negative, normal rabbit serum control of the total sperm fluorescence intensity (b1) and the sperm flagellum fluorescence intensity (c1). Immunofluorescence images of NC (b2) and IVC spermatozoa (b3), including normal rabbit serum (b4) obtained by IBFC to confirm labeling. The IBFC was performed in 4 replicates with consistent results. Statistical evaluation of the total sperm fluorescence intensity (b5), and the sperm flagellum fluorescence intensity (c2) was performed by using ANOVA with Sidak’s post hoc test (α = 0.05).
Phenotyping of TTR in the non-capacitated (NC) and in vitro capacitated (IVC) spermatozoa, with and without proteasomal inhibition including vehicle control. The ICC detection of TTR (red) in NC (a1) and IVC, non-inhibited (a2) spermatozoa, including non-immune serum control (a3); the insets show TTR channel only. Spermatozoa were co-stained for acrosomal presence with peanut agglutinin (PNA, green), and a nuclear stain DAPI (blue). All fluorescence channels are superimposed with the DIC brightfield channel. Scale bars represent 20 µm. The IBFC of formaldehyde fixed and Triton X-100 permeabilized NC and IVC spermatozoa with or without proteasomal inhibition, including vehicle control and negative immunolabeling control, normal rabbit serum control (b1). Immunofluorescence images of NC (b2) and IVC spermatozoa (b3), including normal rabbit serum (b4) obtained by IBFC to confirm labeling. The IBFC was performed in 4 replicates with consistent results. Statistical evaluation of the IBFC results (b5) was performed by using ANOVA with Sidak’s post hoc test (α = 0.05). The relative amounts of TTR median fluorescence intensity are related to the non-capacitated sperm control with an assigned value of 100%. The statistical significance P < 0.05 is denoted with small letters.
Starting with RAB2A, the protein localized to the acrosomal region in both non-capacitated (NC) and in vitro capacitated spermatozoa (Fig. 5a1,a2, resp.). A decrease in fluorescence intensity median after IVC was observed by IBFC, resulting in two distinct sperm cohorts (Fig. 5b1). The decrease in the RAB2A signal intensity was observed in the acrosomal region (Fig. 5b2,b3), corroborating ICC images. The RAB2A sperm-cohort redistribution was significant between NC and IVC groups (P < 0.0001), but no significance was observed between spermatozoa that were capacitated under proteasome-inhibiting conditions (100 µM MG132) when compared to the vehicle control group (0.2% EtOH; P = 0.29, Fig. 5b4). By using WB, we detected RAB2A in all sperm treatment groups as a protein band triplet at ~ 24, 27, and 31 kDa (Fig. 5b1–b3). The RAB2A abundance/extractability increased significantly after IVC (P = 0.013, Fig. 5c4), while WB detected no significant abundance change between the proteasomally-inhibited IVC versus the vehicle control group (P = 0.97, Fig. 5c4).
The second studied protein, CFAP161, localized to the acrosomal region of the sperm head and the flagellum in NC spermatozoa (Fig. 6a1), and the acrosomal region only in IVC spermatozoa (Fig. 6a2). The median signal intensity decreased after IVC, resulting in two distinct sperm cohorts (Fig. 6b1). The higher median signal intensity in NC spermatozoa was attributed to the stronger fluorescence intensity in the acrosomal region as well as to the presence of flagellar labeling (Fig. 6b2,b3), as also observed by ICC. The sperm cohort redistribution was significant (P = 0.022) after IVC when compared to NC control, and no significance was observed in the proteasomally-inhibited IVC treatment group compared to its vehicle IVC control (P = 0.131, Fig. 6b4). Further, we observed a drop in sperm cohort with tail labeling after IVC (Fig. 6c1), however, without statistical significance between IVC vs NC control spermatozoa (P = 0.129, Fig. 6c2) as well as between the proteasomally-inhibited IVC treatment group compared to its vehicle IVC control (P = 0.653, Fig. 6c2).
The last studied protein, TTR, localized in both the acrosomal region and postacrosomal sheath of the sperm head as well as in the flagellum of NC and IVC spermatozoa (Fig. 7a1,a2). The fluorescence signal intensity decreased significantly after IVC when compared to NC control (P = 0.04, Fig. 7b1,b5), however, no significance was observed between the proteasomally-inhibited IVC treatment group compared to its vehicle IVC control (P = 0.845, Fig. 7b1,b5). The acrosomal, postacrosomal, and flagellar labeling of TTR was corroborated by IBFC in both NC and IVC spermatozoa (Fig. 7b2,b3, resp.).
Discussion
This study is a continuation of ongoing research efforts to understand the regulation mechanism of sperm capacitation by the ubiquitin-proteasome system (UPS). In the present study, we monitored how UPS modulates the whole sperm proteome remodeling during sperm capacitation. Adding to the previous studies, where a handful of UPS targets were identified during sperm capacitation including MFGE8, ADAM5, ACRBP, CSEL1, and PFDN4 to name a few44,46,47,81, we are adding 13 more potential UPS targets in the present study. The relatively narrow range of UPS targets in the whole pig sperm proteome during sperm capacitation confirms our previous study46, where we reported 14 candidate UPS targets on the boar sperm surface during capacitation. In both our proteomic studies, we reason that a relatively small number of UPS targets during IVC might be due to (i) a possible compensatory mechanism of PRKA that might drive capacitation forward in proteasomally-inhibited spermatozoa60, (ii) the residual caspase-like and trypsin-like activities of 20S core that can compensate for the inhibited chymotrypsin-like activity (MG132 is a potent inhibitor of the chymotrypsin-like activity of the 20S core, which also inhibits caspase-like and trypsin-like activities, albeit with lesser efficiency82), and (iii) later stages of sperm capacitation might be less dependent on the proteasomal activity as the proteasome abundance diminishes after 4 h of IVC45. The mechanism that leads to diminished proteasomal abundance after 4 h may likely be due to the shedding of proteasomes from the sperm surface, loss of proteasome-rich acrosomes by premature/spontaneous exocytosis, or other mechanisms. The UPS targets, identified in the current study, deserve further attention as the majority was not phenotyped or characterized.
We phenotyped three targets of the UPS during capacitation in the present study, namely RAB2A, CFAP161, and TTR. We have previously identified RAB2A as a sperm surface protein with zona pellucida binding activity80. Other groups implicated RAB2A in acrosomal biogenesis in mice77 and bulls78. Recently, RAB2A was found to be present in perinuclear theca fraction79 implicating a role in post-fertilization events. In the present study, we monitored capacitation-related changes in RAB2A and made two observations, (i) the median fluorescence intensity of RAB2A decreases, while (ii) RAB2A extractability increases after IVC. Presumably, the decrease in median fluorescence intensity of RAB2A is caused by the decrease in accessibility to the antibody. We hypothesize that during the protein remodeling that occurs during capacitation, RAB2A may assemble into multimeric high molecular weight (HMW) ZP-binding complexes on the sperm anterior head plasma membrane (APM)83,84, and the epitope recognized by our antibody may be masked, therefore becoming less accessible. Indeed, RAB2A has previously been found among protein aggregates in lipid rafts in human and mouse spermatozoa85,86. Kongmanas et al.83 did not find RAB2A within the pig’s HMW complexes, however, the protein was detected in the extracts from APM vesicles from both non-capacitated and capacitated pig spermatozoa. The increase in RAB2A abundance/extractability after IVC could be explained by the fact that RAB2A is a component of detergent-resistant membrane (DRM) domains, insoluble membrane fractions also known as lipid rafts, as shown in aforementioned proteomic studies in human and mouse spermatozoa85,86. Capacitation induces changes in the membrane distribution of proteins in these lipid rafts87, and these may become more susceptible to extraction. A proteomic comparison of DRM-associated proteins from capacitated and non-capacitated mouse spermatozoa had previously shown a decrease in DRM-associated proteins in the capacitated spermatozoa88. This supports our hypothesis that extraction resistance is higher in non-capacitated spermatozoa and that extractability/solubility increases after capacitation, although other studies have observed an increase in the presence of lipid rafts in capacitated spermatozoa85,89,90,91. Nixon & Aitken90 suggested that the conflicting results may be due to the different methods used to isolate lipid rafts. Another possible explanation for the increased RAB2A abundance after IVC is better extractability from the perinuclear theca. We have shown a similar pattern previously for perinuclear theca protein CCIN92. Our WB results are consistent with our previous study in boar spermatozoa80, but not with other studies reporting a decrease in RAB2A after capacitation in mouse spermatozoa93, and RAB2 (the authors do not specify RAB2A/B) in boar spermatozoa94. Our antibody (5C5) recognizes RAB2A protein in pigs, uniquely identified by MS after immunoprecipitation of the protein from sperm extract and known to be present in the male reproductive tract and gametes80. We therefore assume that it is a specific form of RAB2A, which may also differ between mammalian species. The authors in the latter study94 even proposed that RAB2/RAB2A is involved in structural modification of the acrosome to induce acrosomal exocytosis after capacitation, as has been documented for RAB3A in human spermatozoa95,96,97. We can speculate that the contradictory findings of these studies may be caused by the use of different antibodies which might recognize different epitopes of the RAB2A protein. One finding supporting this hypothesis could be that RAB2A was observed in both the head and tail of boar spermatozoa98, in contrast to our observation of RAB2A localization being restricted to the acrosomal region of the sperm head. These studies used a commercially available polyclonal antibody raised against a recombinant fragment corresponding to the C-terminal amino acids of human RAB2 expressed in E. coli99. In addition, Kwon et al.98 used a commercially available polyclonal anti-RAB2A antibody to detected only one RAB2 (presumably RAB2A) isoform of 24 kDa. Bae et al.93 also detected only one RAB2 isoform of 25 kDa in mouse spermatozoa using a rabbit polyclonal antibody. We have previously reported two RAB2A isoforms with molecular masses of 24 and 27 kDa80 in boar spermatozoa, and in the current study, we detected isoforms of 24, 27, and 31 kDa by using the same mouse monoclonal antibody 5C5. The reason for this difference could be more sensitive detection or it may be due to boar breed or individual variability or even a different strength of the detergent used.
Little is known about the second phenotyped protein, cilia and flagella associated protein 161 (CFAP161), in spermatozoa. At the time of this paper's publication, only one functional study done in Cfap161-/- mice was available69. The authors detected CFAP161 in the flagellum of wild-type (WT) mouse spermatozoa. Mutant Cfap161-/- mice bred normally, the disruption of the Cfap161-/- gene did not lead to motile cilia-related phenotypes, no abnormalities were found in sperm count and motility when compared to WT, and no obvious structural abnormalities were observed in the axonemes of the mutant mice revealing no evidence for essential CFAP161 function in mice sperm flagellum. However, the authors noticed that genes related to microtubules and cilia, microtubule motor activity, and inner dyneins were dysregulated, suggesting compensation for Cfap161-/- knockout. We localized CFAP161 in the pig sperm flagellum of NC boar spermatozoa that vanished after sperm IVC capacitation (P = 0.13), implying a role in sperm hyperactivation. The UPS does not seem to be involved in CFAP161 shedding from the sperm flagellum (P = 0.65). Furthermore, we observed acrosomal localization of CFAP161 in both NC and IVC spermatozoa with lower signal intensity after IVC (P = 0.022). Proteasomal inhibition did not reduce the signal intensity in a significant manner (P = 0.131). The abundance of CFAP161 in spermatozoa detected by WB seems to be consistent as no significant change was detected among all treatment groups (P > 0.05).
The last phenotyped protein, transthyretin, was identified in sperm proteome previously100. Besides the carrier role of TTR for thyroxin and retinol throughout the human body including the brain101, it also seems to play a role in human receptivity and normal pregnancy102 and has been identified in the bovine sperm proteome of epididymal and ejaculated bull spermatozoa100. We are reporting TTR in NC and IVC pig spermatozoa for the first time. We observed a significant decrease in TTR labeling after IVC (P = 0.04) by using IBFC, however, we were not able to corroborate our proteomic data that suggested TTR as the target of proteasomal inhibition by our phenotyping pipeline, most likely due to a very low abundance of TTR in spermatozoa.
Apart from identifying new UPS targets/substrates during sperm capacitation, our study adds new knowledge to the pig sperm proteome remodeling during sperm capacitation, reported previously in comparative proteomic studies46,83,94,103,104,105. Sperm capacitation is a multifaceted event as demonstrated by GO and KEGG pathway enrichment analyses of 401 proteins that changed significantly in their abundance after IVC, identified in this study, and its complete understanding is far from complete.
Even though this study did not aim to reveal the underlying mechanisms, we reason that the increase in protein abundance after IVC might be due to altered characteristics of sperm protein solubility. Spermatozoa carry highly condensed accessory structures such as the hypercondensed, protaminated DNA, the perinuclear theca, the outer dense fibers, and the fibrous sheath. The condensed chromatin and protein structures have unique solubilization properties and varied resistance to detergent extraction, likely affected by IVC. We have shown previously that this was the case for the perinuclear theca protein CCIN, as we observed better extractability of this protein after IVC92; this is also likely the reason for the higher abundance of RAB2A protein after IVC, reported in this study. On the other hand, proteins whose abundance decreased after IVC might have been shed from the sperm surface and released into the capacitation medium (as is the case of sperm surface spermadhesins44), while the intracellular proteins were processed by intrinsic proteases (such as acrosin, matrix metalloproteases, or proteasomes). Nevertheless, the actual mechanism of these sperm protein content changes warrants further investigation.
A deeper understanding of the sperm capacitation process will benefit assisted reproductive technologies and therapies (ART) in animals and humans, respectively. Boar semen extenders could be improved by the addition of proteasomal inhibitors, effectively preventing the UPS-target proteins from capacitation-related modifications, which might ward off undesirable premature sperm capacitation. Conversely, including small molecule activators of the proteasomal activity in the in vitro fertilization (IVF) media may boost sperm fertility during human ART, in particular intrauterine insemination and conventional IVF. Intracytoplasmic sperm injection (ICSI) could benefit from controlled, proteasome-optimized sperm capacitation treatment before individual spermatozoa are selected for the procedure, to boost post-ICSI disassembly of accessory sperm structures, the release of sperm-borne oocyte activating factors and sperm nuclear decondensation. Lastly, the unique cellular surface exposure of sperm proteasomes and their sensitivity to non-permeant proteasomal inhibitors give the possibility for the development of pharmacological contraceptives for humans.
Methods
Antibodies and reagents
Proteasomal inhibitor MG132 (cat # BML-PI102) was purchased from ENZO Life Sciences (Farmingdale, NY, USA). Reagents for indirect immunofluorescence, image-based flow cytometry, and Western blotting, were sourced as follows: rabbit polyclonal anti-CFAP161 antibody (cat # NBP2-14,378, Novus Biochemicals, Centennial, CO, USA); mouse monoclonal anti-TTR antibody (cat # MAB7505, R&D Systems, Minneapolis, MN, USA); mouse monoclonal anti-phosphotyrosine antibody, clone 4G10 (cat # 05-321) was bought from Millipore Sigma, Burlington, MA, USA; mouse IgG isotype control (cat # 31903), and normal rabbit serum (cat # 31883) were both purchased from Invitrogen, Carlsbad, CA, USA. Mouse monoclonal anti-RAB2A, clone 5C5 was made in-house80 and the antibody specificity was confirmed by blocking peptide assay using recombinant human RAB2A (cat # PR0373, Canvax Reagents, Boecillo, Valladolid, Spain) as well as WB detection of recombinant human RAB2A (NPB1-72344, Novus Biologicals, Centennial, CO, USA) by using 5C5 antibody. Mouse monoclonal anti-β-tubulin antibody, clone E7 (Antibody Registry ID: AB_2315513) was bought from Developmental Studies Hybridoma Bank of the University of Iowa, Iowa City, IA, USA. Goat anti-mouse IgG, AF488 (cat # A11001), goat anti-mouse IgG, HRP (cat # 31430), goat anti-rabbit IgG, AF488, goat anti-rabbit IgG, AF647 (cat # A21244), goat anti-rabbit IgG, HRP (cat # 31460), lectin PNA, AF568 (cat # L32458), and DAPI (cat # D1306) were purchased from Invitrogen, Carlsbad, CA, USA. For Western blotting, the following reagents were used: Halt™ Protease and Phosphatase inhibitor cocktail, EDTA free (cat # 78443) was purchased from ThermoFisher Scientific, Rockford, IL, USA; NuPAGE™ 4–12% Bis-Tris gel (cat# NP0329BOX) and Novex® Sharp Pre-stained Protein Standard purchased from Invitrogen, Carlsbad, CA, USA; bicinchoninic acid assay (BCA, cat #23227) was purchased from ThermoFisher Scientific, Rockford, IL, USA; PVDF Immobilon Transfer Membrane and Luminata Crescendo Western HRP Substrate were bought from Millipore Sigma, Burlington, MA, USA; and sequencing grade modified trypsin (cat # V5111a) was purchased from Promega Corporation, Madison, WI, USA. All other chemicals used in this study were purchased from Sigma-Aldrich, St. Louis, MO, USA.
Compliance with ethical standards
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All studies involving vertebrate animals were completed under the strict guidance of an Animal Care and Use protocol, approved by the Animal Care and Use Committee (ACUC) of the University of Missouri. This article does not contain any studies with human participants performed by any of the authors.
The study is reported in accordance with ARRIVE guidelines (https://arriveguidelines.org).
Semen processing and in vitro sperm capacitation (IVC)
Fresh boar spermatozoa were collected weekly from healthy, non-transgenic fertile large white boars (n = 2) at the National Swine Resource and Research Center, University of Missouri, used for routine in vitro fertilization trials with high blastocyst yield. The sperm-rich fraction (n = 6) was used for the study purposes. The concentration and motility of ejaculates were evaluated by conventional semen analysis methods under a light microscope. Sperm concentration was measured by hemocytometer (ThermoFisher Scientific) and ranged from 300 to 500 million/mL; only ejaculates with > 80% motile spermatozoa and < 20% morphological abnormalities were used for the study. Collections contaminated with urine were discarded. The collected semen fractions were free of contaminants other than the expected minimal content of sperm cytoplasmic droplets, thus not necessitating gradient purification. Spermatozoa were separated from seminal plasma by centrifugation (2000 RPM ~ 400 × g, 10 min; IEC Centra CL2, ThermoFisher Scientific), then washed with pre-warmed HEPES buffered Tyrode lactate medium supplemented with polyvinyl alcohol (TL-HEPES-PVA), containing 10 mM sodium lactate, 0.2 mM sodium pyruvate, 2 mM NaHCO3, 2 mM CaCl2, 0.5 mM MgCl2 and 0.01% (w/v) polyvinyl alcohol (PVA); pH = 7.4, t = 37 °C. After the final wash, the sperm pellet was split into quarters; the first quarter was not allowed to capacitate, processed immediately as described below, and used as a non-capacitated (NC) control sample. The other three quarters were in vitro capacitated for 4 h, 37 °C, 5% (v/v) CO2 in TL-HEPES-PVA described above, supplemented with 5 mM sodium pyruvate, 11 mM glucose, and 2% (w/w) BSA as described previously45 either in the absence of the proteasomal inhibitor (IVC control), in the presence of 100 µM MG132 and 0.2% (v/v) ethanol (IVC, proteasomally-inhibited group), or the presence of 0.2% (v/v) ethanol alone (IVC vehicle control). We have demonstrated that our IVC protocol successfully capacitated spermatozoa under the conditions used in this study (Supplementary Fig. 1) as well as that the concentration of 100 µM MG132 represents strong proteasome-inhibiting conditions12,44,45,46,47,59. Spermatozoa after IVC were washed from BSA. Non-capacitated and IVC-capacitated groups were processed for (i) mass spectrometry of sperm proteins, (ii) immunocytochemistry and image-based flow cytometry, and (iii) Western blotting and immunoprecipitation.
Proteomics
The whole sperm proteomes from all four treatment groups, i.e., non-capacitated, capacitated without proteasomal inhibition, capacitated with 100 µM MG132, and IVC vehicle control were isolated lysing approximately 100 million spermatozoa by using 1% (w/v) 3-(4-Heptyl)phenyl-3-hydroxypropyl)dimethylammoniopropanesulfonate (C7BzO, cat # C0856, Millipore Sigma), 7 M urea, 2 M thiourea, 40 mM Tris∙HCl, pH = 7.4 supplemented with Halt™ Protease and Phosphatase inhibitor cocktail for 1 h at 4 °C with end-over-end rotation. Samples were centrifuged at 16,000 × g, 4 °C, for 20 min and the supernatants were transferred to Protein LoBind tubes (cat # 022431102, Eppendorf, Hamburg, DEU). Samples were lyophilized and shipped to the University of Newcastle, NSW, Australia.
To generate the spectral library, peptides were analyzed by LC-MS in DDA mode on a Sciex 6600 triple ToF mass spectrometer. Mobile phase A was 2% Acetonitrile, 0.1% Formic Acid, and Mobile Phase B was 80% Acetonitrile, 0.1% Formic acid. Tryptic digests of samples were loaded onto a Sciex trapping column (200 µM × 0.5 mm ChromXP C18-CL, 3 µM, 120 Å), washed for 5 min at 2 mL/min with 2% mobile phase B, then eluted directly onto the separation column (75 μm × 15 cm ChromXP, C18, 3 μm, 120 Å). The gradient used to separate the peptides was 5–9% B in 2 min, 9–35% B in 52 min, and 35–50% B over 7 min, all as linear increases at a flow rate of 5 µl/min. For peptides entering the spectrometer, the scan range was 400–1250 m/z, with the top 20 peaks selected for fragmentation with a 30 s exclusion time, and the collision energy spread was set at 5 eV.
The files were searched against the Sus scrofa UniProtKB database (all isoforms, downloaded 13/06/19). To obtain peptide quantitative data, the samples were again run through liquid chromatography (LC), coupled to a 6600 Triple ToF mass spectrometer, running a SWATH acquisition method. Briefly, the LC methods were identical to the DDA acquisition, but fragments were acquired by sequentially scanning the total mass range for fragmentation in set m/z ranges (windows). Window size was calculated using Sciex’s provided method and based on the DDA results. In order to quantify peptides, the spectral library was processed using PeakView (2.2), and the raw data were then exported into MarkerView (version 1.3.0.1). From there, the data were again exported as a raw text file, which allowed for analysis using the freeware Perseus software (version 2.0.3.1) (www.perseus-framework.org). In Perseus, the data were log2-transformed and then normalized to the median of each individual sample. Sample groups were compared by using a paired Welch’s t-test and FDR analysis. Proteins with a fold change greater than 2.0 and an adjusted p-value > 0.05 were considered significant. The experiment was replicated six times, and results are presented in the Excel spreadsheet as Supplementary Data 1.
Immunofluorescence
Non-capacitated and IVC-capacitated spermatozoa with and without proteasomal inhibition were fixed in 2.0% (v/v) formaldehyde in PBS for 10 min at room temperature, and permeabilized in 0.1% (v/v) TrX-100 in PBS (PBST) or fixed and permeabilized in 50% (v/v) ice-cold acetone in PBS without prior formaldehyde fixation. Approximately 20 million spermatozoa per sample were blocked with PBST supplemented with 5% (v/v) normal goat serum (NGS). All the antibodies used for immunofluorescence studies were previously characterized. Primary antibodies used were as follows: anti-CFAP161 (1:100 dilution, NBP2-14,378, Novus Biochemicals), anti-TTR (1:100 dilution, MAB7505, R&D Systems), and anti-RAB2A (1:200 dilution80) all diluted in PBST supplemented with 5% (v/v) NGS. Primary antibodies were added to sperm sample tubes and incubated overnight at 4 °C. For the primary control, non-immune rabbit serum of comparable protein concentration or mouse IgG isotype of comparable immunoglobulin concentration was used instead of primary antibodies and processed in the same fashion. The following morning, spermatozoa were washed twice with PBST with 1% (v/v) NGS. Next, an appropriate species-specific secondary antibody, such as the goat anti-mouse conjugated to AF488, or goat anti-rabbit conjugated to AF647 (1:200 dilution) in PBST with 1% (v/v) NGS was added and allowed to incubate for 40 min at room temperature. For acrosomal integrity assessment, peanut agglutinin conjugated to Alexa Fluor 568 (PNA-AF568, 1:1500 dilution; Invitrogen) was used, and 4’,6-Diamidino-2-Phenylindole Dilactate (DAPI), a DNA stain (1:1000 dilution) was used as a reference and nuclear contrast stain. Both PNA-AF568 and DAPI were mixed and coincubated with secondary antibodies. After incubation with secondary antibodies, spermatozoa were washed twice with PBST with 1% (v/v) NGS. For the secondary antibody control, secondary antibodies were omitted as well as DAPI and PNA-AF647 and processed in the same fashion. All steps were accomplished in suspensions.
Epifluorescence microscopy
The fluorescently labeled samples were mounted on microscope slides with VectaShield (Vector Laboratories, Inc., Burlingame, CA, #H-1000) and imaged by using a Nikon Eclipse 800 microscope (Nikon Instruments, Inc., Melville, NY, USA) with Retiga QI-R6 camera (Teledyne QImaging, Surrey, BC, Canada) operated by MetaMorph software (ver. 7.10.2.240, Molecular Devices, San Jose, CA, USA). Images were adjusted for contrast and brightness in Adobe Photoshop 2020 (Adobe Systems, Mountain View, CA, USA), to match the fluorescence intensities viewed through the microscope eyepieces.
Image-based flow cytometry (IBFC)
The fluorescently labeled samples were measured with an Amnis FlowSight Imaging Flow Cytometer (AMNIS Cytek Biosciences, Fremont, CA, USA) as described previously106. The instrument was fitted with a 20 × microscope objective (numerical aperture of 0.9) with an imaging rate of up to 2000 events per sec. The sheath fluid was PBS (without Ca2+ or Mg2+). The flow-core diameter and speed were 10 µm and 66 mm per second, respectively. The raw image data were acquired by using INSPIRE® software (AMNIS Cytek Biosciences, Fremont, CA, USA). To maximize resolution, the camera setting was at 1.0 µm per pixel of the charge-coupled device. Samples were excited by using four lasers concomitantly: a 405-nm line (20 mW), a 488/642-nm line (20 mW), a 561-nm line (20 mW), a 785-nm line (70 mW, side scatter), and two LEDs (32.57 mW and 19.30 mW respectively). Signals were recorded in the following channels: channels 1 and 9—brightfield, channel 2—green fluorescence (AF488, 505–560 nm), channel 4—orange fluorescence (AF568, 595–642 nm), channel 6 (SSC), channel 7—blue fluorescence (DAPI, 435–505 nm), and channel 11—infrared fluorescence (AF647, 642–745 nm). A total of 10,000 events were collected per sample, and data were analyzed by using IDEAS® software (Version 6.2.189.0; AMNIS Cytek Biosciences, Fremont, CA, USA). A focused, single-cell population gate was used for the histogram display of mean pixel intensities by frequency for collected channels. Intensity histograms of individual channels were then used for drawing regions of subpopulations with varying intensity levels and visual confirmation. The intensity of DAPI was used for histogram normalization among NC and IVC spermatozoa. Relevant masks were applied for analysis of specific sperm compartments as well as to exclude autofluorescent debris.
Sperm protein isolation
The processing of an ejaculate to obtain spermatozoa as well as sperm IVC is described in previous sections. A total of 100 million TBS-washed spermatozoa per treatment (NC, IVC spermatozoa with or without proteasomal inhibition, vehicle control) were resuspended in 100 µL of lithium dodecyl sulfate (LDS) loading buffer (106 mM Tris HCl, 141 mM Tris base, 2% (w/v) LDS, 10% (w/v) glycerol, 0.51 mM (0.75%) EDTA, 0.22 mM (0.075%) Coomassie Brilliant Blue G250, 0.175 mM (0.025%) Phenol Red, pH = 8.5) supplemented with Halt™ protease and phosphatase inhibitor cocktail, and left to incubate overnight at 4 °C with end-over-end rotation. The following day, the suspensions were spun at 13,000 × g and 4 °C, for 15 min and the supernatants were transferred to new Eppendorf tubes. Before the PAGE loading, 2-mercaptoethanol was added to the final concentration of 2.5% (v/v), and incubated at 70 °C for 10 min.
Immunoprecipitation
The TTR protein was immunoprecipitated from NC and IVC spermatozoa by using the Thermo Scientific Pierce crosslink magnetic IP/Co-IP kit (cat # 88805) according to manufacturer ‘s protocol. Briefly, 5 µg of mouse monoclonal anti-TTR antibody (MAB7505, R&D Systems) was bound to the Protein A/G magnetic beads. Sperm proteins were extracted from 100 million NC or IVC spermatozoa by the addition of 500 µL of lysis buffer supplemented with Halt™ Protease and Phosphatase inhibitor cocktail and left to incubate for 30 min at 4 °C with end-over-end rotation. Protein concentration was determined with a BCA assay (cat #23227, ThermoFisher Scientific). One milligram of the total protein was added to the magnetic beads with the bound antibody and incubated for 60 min at room temperature with end-over-end rotation. The antigen was eluted with a low pH and neutralized. Resultant immunoprecipitates were concentrated by using MilliporeSigma Amicon centrifugal filters (cat # UFC500396). A 4 × LDS sample buffer and 2-mercaptoethanol were added to the immunoprecipitates, to prepare a 1 × LDS sample buffer and 2.5% (v/v) 2-mercaptoethanol, respectively. Samples were incubated at 70 °C for 10 min before WB detection.
Western blotting (WB)
For polyacrylamide gel electrophoresis (PAGE), a NuPAGE® electrophoresis system was used (Invitrogen, Carlsbad, CA). A protein equivalent of 10 million spermatozoa (~ 20 µg of the total protein) was loaded per single lane. The PAGE was carried on an Owl™ Dual-Gel Vertical Electrophoresis System (cat # P8DS, ThermoFischer Scientific) with NuPAGE™ 4–12% Bis–Tris gel (cat # NP0329BOX, Invitrogen) using TRIS-MOPS SDS Running Buffer [50 mM Tris Base, 50 mM 3-(N-morpholino)propanesulfonic acid (MOPS), 0.1% (w/v) SDS, 1 mM EDTA, pH = 7.7]. Anode buffer (the upper one) was supplemented with 5 mM NaHSO3 to prevent disulfide bond reoxidation. The molecular masses of separated proteins were estimated using Novex® Sharp Pre-stained Protein Standard (cat # LC5800, Invitrogen, Carlsbad, CA) run in parallel. The PAGE was carried out for 5 min at 80 V to let the samples delve into the gel and then for another 60–70 min at 160 V. The power was limited to 20 W. After PAGE, proteins were electrotransferred onto a PVDF Immobilon Transfer Membrane (Millipore Sigma) by using an Owl™ Mini Tank Electroblotting System (cat # VEP-2, ThermoFisher Scientific) at 300 mA for 90 min for immunodetection, by using Bis-Tris-Bicine transfer buffer (25 mM Bis–Tris base, 25 mM Bicine, 1 mM EDTA, pH = 7.2) supplemented with 10% (v/v) methanol, and 2.5 mM NaHSO3.
The PVDF membrane with the transferred proteins was blocked with 10% (w/v) non-fat milk (NFM) in TBS with 0.05% (v/v) Tween 20 (TBST) and incubated with the primary antibody overnight. Primary antibodies were diluted as follows: anti-CFAP161 (1:2000 dilution; NBP2-14,378, Novus Biochemicals), anti-TTR (1:1000 dilution, MAB7505, R&D Systems), and anti-RAB2A (1:200 dilution80) all diluted in TBST and supplemented with 5% (w/v) NFM. The next day, the membrane was incubated for 40 min with an appropriate species-specific secondary antibody, such as the HRP-conjugated goat anti-rabbit antibody (GAR-IgG-HRP, 1:10,000 dilution; cat # 31460 Invitrogen) or goat anti-mouse antibody (GAM-IgG-HRP, 1:10,000 dilution; cat # 31430 Invitrogen). The membrane was reacted with a chemiluminescent substrate (Luminata Crescendo Western HRP Substrate; Millipore Sigma) and the blot was screened with ChemiDoc Touch Imaging System (Bio-Rad, Hercules, CA, USA), to record the protein bands. Membranes were stripped with Restore WB stripping buffer (cat # 21059) and reprobed with anti-TUBB antibody (clone E7, AB_2315513) for protein load normalization purposes. The images were analyzed by Image Lab Touch Software (ver. 5.2.1, Bio-Rad, Hercules, CA, USA). Unless specified otherwise, procedures were carried out at room temperature. Membranes were stained with CBB R-250 after chemiluminescence detection for additional protein load control.
Statistics and reproducibility
Six biological and technical replicates were conducted for high-resolution mass spectrometry and used for MS data analysis. The MS data were log2-transformed and then normalized to the median of each individual sample. Sample groups were compared by using a paired Welch’s t-test and FDR analysis. Proteins with a fold change greater than 2.0 and an adjusted p-value > 0.05 were considered significant. Four IBFC replicates and six WB replicates were performed to ensure scientific rigor. The numbers of replicates are also denoted in the figure legends. Each data point is presented as a mean ± SD. Datasets were tested for normal distribution by using the Shapiro–Wilk normality test and processed by using one-way analysis of variance (ANOVA) in GraphPad Prism (ver. 9.5.1, GraphPad Software, Inc., La Jolla, CA, USA) in a completely randomized design. Sidak’s multiple comparison test was used to compare the mean values of individual treatments with a 95% confidence interval. A value of P < 0.05 was considered statistically significant. No data was excluded from the statistical evaluation.
Data availability
The MS proteomics data in this paper have been deposited in the ProteomeXchange Consortium (http://proteomecentral.proteomexchange.org) via the MassIVE partner repository with the data set identifier MSV000093882. All source data are included as supplementary data. All other data are available upon reasonable request. Correspondence and requests for materials should be addressed to MZ.
References
Austin, C. R. Observations on the penetration of the sperm in the mammalian egg. Aust. J. Sci. Res. Ser. B Biol. Sci. 4, 581–596 (1951).
Chang, M. C. Fertilizing capacity of spermatozoa deposited into the fallopian tubes. Nature 168, 697–698 (1951).
Suarez, S. S. The oviductal sperm reservoir in mammals: Mechanisms of formation. Biol. Reprod. 58, 1105–1107. https://doi.org/10.1095/biolreprod58.5.1105 (1998).
Florman, H. M. & Ducibella, T. Fertilization in Mammals. In Knobil and Neill’s Physiology of Reproduction Vol. 1 (ed. Neill, J. D.) 55–112 (Elsevier Academic Press, Inc., 2006).
Florman, H. M. & Fissore, R. A. Fertilization in Mammals. In Knobil and Neill’s Physiology of Reproduction (eds Plant, T. N. & Zeleznik, A. J.) 149–196 (Elsevier, 2015).
Yanagimachi, R. in The Physiology of Reproduction Vol. 1 (eds E. Knobil & J.D. Neill) Ch. 2, 189–317 (Raven Press, 1994).
Guidobaldi, H. A., Teves, M. E., Uñates, D. R., Anastasía, A. & Giojalas, L. C. Progesterone from the cumulus cells is the sperm chemoattractant secreted by the rabbit oocyte cumulus complex. PloS One 3, e3040. https://doi.org/10.1371/journal.pone.0003040 (2008).
Oren-Benaroya, R., Orvieto, R., Gakamsky, A., Pinchasov, M. & Eisenbach, M. The sperm chemoattractant secreted from human cumulus cells is progesterone. Hum. Reproduct. 23, 2339–2345. https://doi.org/10.1093/humrep/den265 (2008).
Pérez-Cerezales, S., López-Cardona, A. P. & Gutiérrez-Adán, A. Progesterone effects on mouse sperm kinetics in conditions of viscosity. Reproduction 151, 501–507. https://doi.org/10.1530/rep-15-0582 (2016).
Teves, M. E. et al. Progesterone at the picomolar range is a chemoattractant for mammalian spermatozoa. Fertil. Steril. 86, 745–749. https://doi.org/10.1016/j.fertnstert.2006.02.080 (2006).
Villanueva-Díaz, C., Arias-Martínez, J., Bermejo-Martínez, L. & Vadillo-Ortega, F. Progesterone induces human sperm chemotaxis. Fertil. Steril. 64, 1183–1188. https://doi.org/10.1016/s0015-0282(16)57982-5 (1995).
Kerns, K., Zigo, M., Drobnis, E. Z., Sutovsky, M. & Sutovsky, P. Zinc ion flux during mammalian sperm capacitation. Nat. Commun. 9, 2061. https://doi.org/10.1038/s41467-018-04523-y (2018).
Kerns, K., Zigo, M. & Sutovsky, P. Zinc: A necessary ion for mammalian sperm fertilization competency. Int. J. Mol. Sci. 19, 4097. https://doi.org/10.3390/ijms19124097 (2018).
Bhakta, H. H., Refai, F. H. & Avella, M. A. The molecular mechanisms mediating mammalian fertilization. Development https://doi.org/10.1242/dev.176966 (2019).
Siu, K. K., Serrão, V. H. B., Ziyyat, A. & Lee, J. E. The cell biology of fertilization: Gamete attachment and fusion. J. Cell Biol. https://doi.org/10.1083/jcb.202102146 (2021).
Tumova, L., Zigo, M., Sutovsky, P., Sedmikova, M. & Postlerova, P. Ligands and receptors involved in the sperm-zona pellucida interactions in mammals. Cells 10, 133. https://doi.org/10.3390/cells10010133 (2021).
Bailey, J. L. Factors regulating sperm capacitation. Syst. Biol. Reproduct. Med. 56, 334–348. https://doi.org/10.3109/19396368.2010.512377 (2010).
Hunt, L. T. & Dayhoff, M. O. Amino-terminal sequence identity of ubiquitin and the nonhistone component of nuclear protein A24. Biochem. Biophys. Res. Commun. 74, 650–655. https://doi.org/10.1016/0006-291x(77)90352-7 (1977).
Hershko, A. & Heller, H. Occurrence of a polyubiquitin structure in ubiquitin-protein conjugates. Biochem. Biophys. Res. Commun. 128, 1079–1086. https://doi.org/10.1016/0006-291x(85)91050-2 (1985).
Hough, R., Pratt, G. & Rechsteiner, M. Ubiquitin-lysozyme conjugates. Identification and characterization of an ATP-dependent protease from rabbit reticulocyte lysates. J. Biol. Chem. 261, 2400–2408 (1986).
Waxman, L., Fagan, J. M. & Goldberg, A. L. Demonstration of two distinct high molecular weight proteases in rabbit reticulocytes, one of which degrades ubiquitin conjugates. J. Biol. Chem. 262, 2451–2457 (1987).
Ciechanover, A., Elias, S., Heller, H. & Hershko, A. “Covalent affinity” purification of ubiquitin-activating enzyme. J. Biol. Chem. 257, 2537–2542 (1982).
Hershko, A., Heller, H., Elias, S. & Ciechanover, A. Components of ubiquitin-protein ligase system. Resolution, affinity purification, and role in protein breakdown. J. Biol. Chem. 258, 8206–8214 (1983).
Ciechanover, A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Best Pract. Res. Clin. Haematol. 30, 341–355. https://doi.org/10.1016/j.beha.2017.09.001 (2017).
Sutovsky, P. Sperm proteasome and fertilization. Reproduction 142, 1–14. https://doi.org/10.1530/rep-11-0041 (2011).
Glickman, M. H. & Ciechanover, A. The ubiquitin-proteasome proteolytic pathway: Destruction for the sake of construction. Physiol. Rev. 82, 373–428. https://doi.org/10.1152/physrev.00027.2001 (2002).
Pickart, C. M. & Cohen, R. E. Proteasomes and their kin: Proteases in the machine age. Nat. Rev. Mol. Cell Biol. 5, 177–187. https://doi.org/10.1038/nrm1336 (2004).
Ciechanover, A. Proteolysis: From the lysosome to ubiquitin and the proteasome. Nat. Rev. Mol. Cell Biol. 6, 79–87. https://doi.org/10.1038/nrm1552 (2005).
Manandhar, G., Schatten, H. & Sutovsky, P. Centrosome reduction during gametogenesis and its significance. Biol. Reproduct. 72, 2–13. https://doi.org/10.1095/biolreprod.104.031245 (2005).
Mtango, N. R., Latham, K. E. & Sutovsky, P. Deubiquitinating enzymes in oocyte maturation, fertilization and preimplantation embryo development. Adv. Exp. Med. Biol. 759, 89–110. https://doi.org/10.1007/978-1-4939-0817-2_5 (2014).
Sutovsky, P. Ubiquitin-dependent proteolysis in mammalian spermatogenesis, fertilization, and sperm quality control: Killing three birds with one stone. Microscopy Res. Tech. 61, 88–102. https://doi.org/10.1002/jemt.10319 (2003).
Kerns, K., Morales, P. & Sutovsky, P. Regulation of sperm capacitation by the 26S proteasome: An emerging new paradigm in spermatology. Biol. Reproduct. 94, 117. https://doi.org/10.1095/biolreprod.115.136622 (2016).
Song, W. H., Ballard, J. W., Yi, Y. J. & Sutovsky, P. Regulation of mitochondrial genome inheritance by autophagy and ubiquitin-proteasome system: implications for health, fitness, and fertility. BioMed Res. Int. 2014, 981867. https://doi.org/10.1155/2014/981867 (2014).
Sutovsky, P. Sperm-egg adhesion and fusion in mammals. Expert Rev. Mol. Med. 11, e11. https://doi.org/10.1017/s1462399409001045 (2009).
Sutovsky, P. & Song, W. H. Post-fertilisation sperm mitophagy: the tale of Mitochondrial Eve and Steve. Reproduct. Fertil. Dev. 30, 56–63. https://doi.org/10.1071/rd17364 (2017).
Sutovsky, P., Van Leyen, K., McCauley, T., Day, B. N. & Sutovsky, M. Degradation of paternal mitochondria after fertilization: Implications for heteroplasmy, assisted reproductive technologies and mtDNA inheritance. Reproduct. Biomed. Online 8, 24–33 (2004).
Yi, Y. J., Manandhar, G., Oko, R. J., Breed, W. G. & Sutovsky, P. Mechanism of sperm-zona pellucida penetration during mammalian fertilization: 26S proteasome as a candidate egg coat lysin. Soc. Reproduct. Fertil. Suppl. 63, 385–408 (2007).
Zigo, M. et al. Porcine model for the study of sperm capacitation, fertilization and male fertility. Cell Tissue Res. 380, 237–262. https://doi.org/10.1007/s00441-020-03181-1 (2020).
Zimmerman, S. & Sutovsky, P. The sperm proteasome during sperm capacitation and fertilization. J. Reproduct. Immunol. 83, 19–25. https://doi.org/10.1016/j.jri.2009.07.006 (2009).
Sutovsky, P. et al. A putative, ubiquitin-dependent mechanism for the recognition and elimination of defective spermatozoa in the mammalian epididymis. J. Cell Sci. 114, 1665–1675 (2001).
Baska, K. M. et al. Mechanism of extracellular ubiquitination in the mammalian epididymis. J. Cell. Physiol. 215, 684–696. https://doi.org/10.1002/jcp.21349 (2008).
Sharif, M. et al. Hyperactivation is sufficient to release porcine sperm from immobilized oviduct glycans. Sci. Rep. 12, 6446. https://doi.org/10.1038/s41598-022-10390-x (2022).
Sharif, M., Kerns, K., Sutovsky, P., Bovin, N. & Miller, D. J. Progesterone induces porcine sperm release from oviduct glycans in a proteasome-dependent manner. Reproduction 161, 449–457. https://doi.org/10.1530/rep-20-0474 (2021).
Zigo, M., Jonakova, V., Manaskova-Postlerova, P., Kerns, K. & Sutovsky, P. Ubiquitin-proteasome system participates in the de-aggregation of spermadhesin and DQH protein during boar sperm capacitation. Reproduction 157, 283–295. https://doi.org/10.1530/rep-18-0413 (2019).
Zigo, M., Kerns, K., Sutovsky, M. & Sutovsky, P. Modifications of the 26S proteasome during boar sperm capacitation. Cell Tissue Res. 372, 591–601. https://doi.org/10.1007/s00441-017-2786-6 (2018).
Zigo, M., Kerns, K. & Sutovsky, P. The ubiquitin-proteasome system participates in sperm surface subproteome remodeling during boar sperm capacitation. Biomolecules 13, 996. https://doi.org/10.3390/biom13060996 (2023).
Zigo, M., Manaskova-Postlerova, P., Jonakova, V., Kerns, K. & Sutovsky, P. Compartmentalization of the proteasome-interacting proteins during sperm capacitation. Sci. Rep. 9, 12583. https://doi.org/10.1038/s41598-019-49024-0 (2019).
Song, W. H. & Sutovsky, P. Porcine cell-free system to study mammalian sperm mitophagy. Methods Mol. Biol. 1854, 197–207. https://doi.org/10.1007/7651_2018_158 (2019).
Song, W. H., Yi, Y. J., Sutovsky, M., Meyers, S. & Sutovsky, P. The ART and science of sperm mitophagy. Autophagy 12, 2510–2511. https://doi.org/10.1080/15548627.2016.1239004 (2016).
Song, W. H., Yi, Y. J., Sutovsky, M., Meyers, S. & Sutovsky, P. Autophagy and ubiquitin-proteasome system contribute to sperm mitophagy after mammalian fertilization. Proc. Natl. Acad. Sci. USA 113, E5261-5270. https://doi.org/10.1073/pnas.1605844113 (2016).
Zuidema, D., Jones, A., Song, W. H., Zigo, M. & Sutovsky, P. Identification of candidate mitochondrial inheritance determinants using the mammalian cell-free system. Elife https://doi.org/10.7554/eLife.85596 (2023).
Mao, J. et al. Ubiquitin A-52 residue ribosomal protein fusion product 1 (Uba52) is essential for preimplantation embryo development. Biol. Open https://doi.org/10.1242/bio.035717 (2018).
Benesova, V., Kinterova, V., Kanka, J. & Toralova, T. Potential involvement of SCF-complex in zygotic genome activation during early bovine embryo development. Methods Mol. Biol. 1605, 245–257. https://doi.org/10.1007/978-1-4939-6988-3_17 (2017).
Hillman, P., Ickowicz, D., Vizel, R. & Breitbart, H. Dissociation between AKAP3 and PKARII promotes AKAP3 degradation in sperm capacitation. PloS One 8, e68873. https://doi.org/10.1371/journal.pone.0068873 (2013).
Zapata-Carmona, H., Barón, L., Kong, M. & Morales, P. Protein kinase a (PRKA) activity is regulated by the proteasome at the onset of human sperm capacitation. Cells 10, 3501. https://doi.org/10.3390/cells10123501 (2021).
Qu, X. et al. Inhibition of 26S proteasome enhances AKAP3-mediated cAMP-PKA signaling during boar sperm capacitation. Anim Reprod. Sci. 247, 107079. https://doi.org/10.1016/j.anireprosci.2022.107079 (2022).
Sanchez, R. et al. Participation of the sperm proteasome during in vitro fertilisation and the acrosome reaction in cattle. Andrologia 43, 114–120. https://doi.org/10.1111/j.1439-0272.2009.01031.x (2011).
Yi, Y. J., Sutovsky, M., Kennedy, C. & Sutovsky, P. Identification of the inorganic pyrophosphate metabolizing, ATP substituting pathway in mammalian spermatozoa. PloS One 7, e34524. https://doi.org/10.1371/journal.pone.0034524 (2012).
Hackerova, L. et al. Modulatory effect of MG-132 proteasomal inhibition on boar sperm motility during in vitro capacitation. Front. Vet. Sci. https://doi.org/10.3389/fvets.2023.1116891 (2023).
Taraschi, A. et al. Two-player game in a complex landscape: 26S Proteasome, PKA, and intracellular calcium concentration modulate mammalian sperm capacitation by creating an integrated dialogue-a computational analysis. Int. J. Mol. Sci. 21, 6256. https://doi.org/10.3390/ijms21176256 (2020).
Kong, M., Diaz, E. S. & Morales, P. Participation of the human sperm proteasome in the capacitation process and its regulation by protein kinase A and tyrosine kinase. Biol. Reproduct. 80, 1026–1035. https://doi.org/10.1095/biolreprod.108.073924 (2009).
Zapata-Carmona, H. et al. The activation of the chymotrypsin-like activity of the proteasome is regulated by soluble adenyl cyclase/cAMP/protein kinase a pathway and required for human sperm capacitation. Mol. Hum. Reproduct. 25, 587–600. https://doi.org/10.1093/molehr/gaz037 (2019).
Blighe, K., Rana, S. & Lewis, M. EnhancedVolcano: Publication-ready volcano plots with enhanced colouring and labeling. R package version 1.8.0, 2020).
Kanehisa, M. & Goto, S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 28, 27–30. https://doi.org/10.1093/nar/28.1.27 (2000).
Kanehisa, M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 28, 1947–1951. https://doi.org/10.1002/pro.3715 (2019).
Kanehisa, M., Furumichi, M., Sato, Y., Kawashima, M. & Ishiguro-Watanabe, M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 51, D587-d592. https://doi.org/10.1093/nar/gkac963 (2023).
He, M. et al. Identification and characterization of new long chain acyl-CoA dehydrogenases. Mol. Genet. Metab. 102, 418–429. https://doi.org/10.1016/j.ymgme.2010.12.005 (2011).
Arroteia, K. F. et al. Albumin is synthesized in epididymis and aggregates in a high molecular mass glycoprotein complex involved in sperm-egg fertilization. PloS One 9, e103566. https://doi.org/10.1371/journal.pone.0103566 (2014).
Beckers, A. et al. The highly conserved FOXJ1 target CFAP161 is dispensable for motile ciliary function in mouse and Xenopus. Sci. Rep. 11, 13333. https://doi.org/10.1038/s41598-021-92495-3 (2021).
Strittmatter, L. et al. CLYBL is a polymorphic human enzyme with malate synthase and β-methylmalate synthase activity. Hum. Mol. Genet. 23, 2313–2323. https://doi.org/10.1093/hmg/ddt624 (2014).
Lee, R. K. et al. Expression of cystatin C in the female reproductive tract and its effect on human sperm capacitation. Reproduct. Biol. Endocrinol. RB&E 16, 8. https://doi.org/10.1186/s12958-018-0327-0 (2018).
Eckhardt, K. et al. Male germ cell expression of the PAS domain kinase PASKIN and its novel target eukaryotic translation elongation factor eEF1A1. Cell Physiol. Biochem. 20, 227–240. https://doi.org/10.1159/000104169 (2007).
Tovich, P. R. & Oko, R. J. Somatic histones are components of the perinuclear theca in bovine spermatozoa. J. Biol. Chem. 278, 32431–32438. https://doi.org/10.1074/jbc.M303786200 (2003).
Hamilton, L. E. et al. Core histones are constituents of the perinuclear theca of murid spermatozoa: an assessment of their synthesis and assembly during spermiogenesis and function after gametic fusion. Int. J. Mol. Sci. 22, 8119. https://doi.org/10.3390/ijms22158119 (2021).
Torres-Flores, U. & Hernández-Hernández, A. The interplay between replacement and retention of histones in the sperm genome. Front. Genet. 11, 780. https://doi.org/10.3389/fgene.2020.00780 (2020).
Hirst, J. Mitochondrial complex I. Ann. Rev. Biochem. 82, 551–575. https://doi.org/10.1146/annurev-biochem-070511-103700 (2013).
Morohoshi, A. et al. FAM71F1 binds to RAB2A and RAB2B and is essential for acrosome formation and male fertility in mice. Development https://doi.org/10.1242/dev.199644 (2021).
Mountjoy, J. R., Xu, W., McLeod, D., Hyndman, D. & Oko, R. RAB2A: A major subacrosomal protein of bovine spermatozoa implicated in acrosomal biogenesis. Biol. Reproduct. 79, 223–232. https://doi.org/10.1095/biolreprod.107.065060 (2008).
Zhang, M. et al. High resolution proteomic analysis of subcellular fractionated boar spermatozoa provides comprehensive insights into perinuclear theca-residing proteins. Front. Cell Dev. Biol. 10, 836208. https://doi.org/10.3389/fcell.2022.836208 (2022).
Zigo, M. et al. Panel of monoclonal antibodies to sperm surface proteins as a tool for monitoring localization and identification of sperm-zona pellucida receptors. Cell Tissue Res. 359, 895–908. https://doi.org/10.1007/s00441-014-2072-9 (2015).
Miles, E. L. et al. Transgenic pig carrying green fluorescent proteasomes. Proc. Natl. Acad. Sci. USA 110, 6334–6339. https://doi.org/10.1073/pnas.1220910110 (2013).
Kisselev, A. F. & Goldberg, A. L. Proteasome inhibitors: From research tools to drug candidates. Chem. Biol. 8, 739–758. https://doi.org/10.1016/s1074-5521(01)00056-4 (2001).
Kongmanas, K. et al. Proteomic characterization of pig sperm anterior head plasma membrane reveals roles of acrosomal proteins in ZP3 binding. J. Cell. Physiol. 230, 449–463. https://doi.org/10.1002/jcp.24728 (2015).
Tanphaichitr, N. et al. Remodeling of the plasma membrane in preparation for sperm-egg recognition: Roles of acrosomal proteins. Asian J. Androl. 17, 574–582. https://doi.org/10.4103/1008-682x.152817 (2015).
Nixon, B. et al. Composition and significance of detergent resistant membranes in mouse spermatozoa. J. Cell. Physiol. 218, 122–134. https://doi.org/10.1002/jcp.21575 (2009).
Asano, A., Nelson, J. L., Zhang, S. & Travis, A. J. Characterization of the proteomes associating with three distinct membrane raft sub-types in murine sperm. Proteomics 10, 3494–3505. https://doi.org/10.1002/pmic.201000002 (2010).
van Gestel, R. A. et al. Capacitation-dependent concentration of lipid rafts in the apical ridge head area of porcine sperm cells. Mol. Hum. Reproduct. 11, 583–590. https://doi.org/10.1093/molehr/gah200 (2005).
Sleight, S. B. et al. Isolation and proteomic analysis of mouse sperm detergent-resistant membrane fractions: Evidence for dissociation of lipid rafts during capacitation. Biol. Reproduct. 73, 721–729. https://doi.org/10.1095/biolreprod.105.041533 (2005).
Bou Khalil, M. et al. Sperm capacitation induces an increase in lipid rafts having zona pellucida binding ability and containing sulfogalactosylglycerolipid. Dev. Biol. 290, 220–235. https://doi.org/10.1016/j.ydbio.2005.11.030 (2006).
Nixon, B. & Aitken, R. J. The biological significance of detergent-resistant membranes in spermatozoa. J. Reproduct. Immunol. 83, 8–13. https://doi.org/10.1016/j.jri.2009.06.258 (2009).
Nixon, B. et al. Proteomic and functional analysis of human sperm detergent resistant membranes. J. Cell. Physiol. 226, 2651–2665. https://doi.org/10.1002/jcp.22615 (2011).
Zigo, M. et al. Zinc is a master-regulator of sperm function associated with binding, motility, and metabolic modulation during porcine sperm capacitation. Commun. Biol. 5, 538. https://doi.org/10.1038/s42003-022-03485-8 (2022).
Bae, J. W. et al. Ras-related proteins (Rab) are key proteins related to male fertility following a unique activation mechanism. Reproduct. Biol. 19, 356–362. https://doi.org/10.1016/j.repbio.2019.10.001 (2019).
Kwon, W. S. et al. A comprehensive proteomic approach to identifying capacitation related proteins in boar spermatozoa. BMC Genom. 15, 897. https://doi.org/10.1186/1471-2164-15-897 (2014).
Yunes, R., Michaut, M., Tomes, C. & Mayorga, L. S. Rab3A triggers the acrosome reaction in permeabilized human spermatozoa. Biol. Reproduct. 62, 1084–1089. https://doi.org/10.1095/biolreprod62.4.1084 (2000).
Belmonte, S. A. et al. Cholesterol content regulates acrosomal exocytosis by enhancing Rab3A plasma membrane association. Dev. Biol. 285, 393–408. https://doi.org/10.1016/j.ydbio.2005.07.001 (2005).
Lopez, C. I., Belmonte, S. A., De Blas, G. A. & Mayorga, L. S. Membrane-permeant Rab3A triggers acrosomal exocytosis in living human sperm. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 21, 4121–4130. https://doi.org/10.1096/fj.06-7716com (2007).
Kwon, W. S., Rahman, M. S., Ryu, D. Y., Park, Y. J. & Pang, M. G. Increased male fertility using fertility-related biomarkers. Sci. Rep. 5, 15654. https://doi.org/10.1038/srep15654 (2015).
Kwon, W. S., Rahman, M. S., Ryu, D. Y., Khatun, A. & Pang, M. G. Comparison of markers predicting litter size in different pig breeds. Andrology 5, 568–577. https://doi.org/10.1111/andr.12332 (2017).
Zoca, S. M., Northrop-Albrecht, E. J., Walker, J. A., Cushman, R. A. & Perry, G. A. Proteomic analyses identify differences between bovine epididymal and ejaculated spermatozoa that contribute to longevity. Theriogenology 184, 51–60. https://doi.org/10.1016/j.theriogenology.2022.02.021 (2022).
Magalhães, J., Eira, J. & Liz, M. A. The role of transthyretin in cell biology: Impact on human pathophysiology. Cell. Mol. Life Sci. CMLS 78, 6105–6117. https://doi.org/10.1007/s00018-021-03899-3 (2021).
Wang, Q., Liu, C. & Zhang, Z. Transthyretin and normal human pregnancy: Mini review. Crit. Rev. Eukaryot. Gene. Expr. 26, 273–277. https://doi.org/10.1615/CritRevEukaryotGeneExpr.2016017323 (2016).
Choi, Y. J. et al. Cytochrome c upregulation during capacitation and spontaneous acrosome reaction determines the fate of pig sperm cells: linking proteome analysis. J. Reproduct. Dev. 54, 68–83. https://doi.org/10.1262/jrd.19116 (2008).
Zigo, M., Jonakova, V. & Manaskova-Postlerova, P. Electrophoretic and zymographic characterization of proteins isolated by various extraction methods from ejaculated and capacitated boar sperms. Electrophoresis 32, 1309–1318. https://doi.org/10.1002/elps.201000558 (2011).
Zigo, M., Jonakova, V., Sulc, M. & Manaskova-Postlerova, P. Characterization of sperm surface protein patterns of ejaculated and capacitated boar sperm, with the detection of ZP binding candidates. Int. J. Biol. Macromol. 61, 322–328. https://doi.org/10.1016/j.ijbiomac.2013.07.014 (2013).
Kennedy, C. E. et al. Protein expression pattern of PAWP in bull spermatozoa is associated with sperm quality and fertility following artificial insemination. Mol. Reproduct. Dev. 81, 436–449. https://doi.org/10.1002/mrd.22309 (2014).
Acknowledgements
We thank the National Swine Resource and Research Center (NSRRC) staff for boar semen collection, and Ms. Miriam Sutovsky for technical and managerial assistance. This research was funded by the USDA National Institute of Food and Agriculture, Agriculture and Food Research Initiative Competitive Grants no. 2021-67015-33404 (PS) and 2019-67012-29714 (KK); The National Health and Medical Research Council (NHMRC) Grant no. 2023952 (MB); the University of Missouri CAFNR Joy of Discovery Seed Grant award (PS, MZ); seed funding University from the College of Agriculture, Food and Natural Resources (CAFNR) University of Missouri (PS); the Grant Agency of the Czech Republic No. GA22-31156S (PP); the project BIOCEV CZ.1.05/1.1.00/02.0109 (PP) from the ERDF, and the support of the Institute of Biotechnology RVO: 86652036 (PP); and the Internal Grant Agency of Czech University of Life Sciences in Prague awards no. SV23-22-21230 (NZ).
Author information
Authors and Affiliations
Contributions
Conceptualization, M.Z. and P.S.; methodology, M.Z., J.N. and V.K.; validation, M.Z., J.N., and K.K.; formal analysis, M.Z., and J.N; resources, P.S., and M.B.; writing—original draft preparation, M.Z., and N.Z.; writing—review and editing, J.N, K.K., P.P., M.B., and P.S.; visualization, M.Z., and J.N.; supervision, P.S., and M.B.; project administration, P.S.; funding acquisition, K.K., P.P., M.B., and P.S. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests. It is disclosed that PS and KK are the founders and owners of AndroLabb LLC, Columbia, MO, a biotech startup involved with sperm processing and sperm quality diagnostics, which did not play any role in the investigations described herein.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zigo, M., Netherton, J., Zelenková, N. et al. Bottom-up approach to deciphering the targets of the ubiquitin-proteasome system in porcine sperm capacitation. Sci Rep 14, 20159 (2024). https://doi.org/10.1038/s41598-024-71056-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-024-71056-4
- Springer Nature Limited