Abstract
Recent trials have shown the efficacy of trastuzumab deruxtecan (T-DXd) in HER2-negative patients, but there is not yet a way to identify which patients will best respond, especially with the inability of current HER2 IHC and FISH assays to accurately determine HER2 expression in the unamplified setting. Here, we present a heavily pre-treated patient with triple-negative breast cancer (HER2 IHC 0 who had a complete response to T-DXd. In this case, we used a CLIA-certified reverse-phase protein array-based proteomic assay (RPPA) to determine that the patient had moderate HER2 protein expression (HER2Total 2+, 42%) and activation (HER2Y1248 1+, 23%). Using these results, we determined that the patient may benefit from T-Dxd despite being traditionally qualified as HER2 IHC 0. These findings highlight the potential for proteomics-based assays that may more accurately quantitate HER2 and (its activation) in the HER2 unamplified/IHC 0 setting to better select patients whose tumors are classically molecularly defined as HER2 IHC 0, but still could respond to HER2-directed therapy, and give patients access to therapies which for which they otherwise would not be eligible.
Similar content being viewed by others
Introduction
Traditional indications for HER2-directed therapy in breast cancer patients have been based on immunohistochemical (IHC) staining on tumor tissue. Per the National Comprehensive Cancer Network (NCCN) guidelines, these therapies are indicated for patients that score 2+ or 3+ on IHC with evidence of HER2 gene amplification on fluorescent in-situ hybridization (FISH)1. While the current standard IHC and FISH selection criteria are proven to have high predictive value for response to monoclonal antibodies and receptor tyrosine kinase inhibitors (TKI), the emergence of targeted HER2 therapies, including small molecule inhibitors or antibody-drug conjugates, have challenged the existing predictive HER2 IHC and FISH thresholds. HER2-directed therapeutics such as trastuzumab deruxtecan (T-DXd) appear to benefit patients whose tumors were traditionally classified as “HER2 negative.” More specifically, many of these patients actually have low levels of HER2 expression (HER2 LOW tumors defined as HER2 IHC 1+/2+ and FISH negative), yet the exact mechanism for response to HER2-directed therapy is still being elucidated2. While T-DXd is not currently approved for roughly 20% of breast cancer patients with HER2 IHC 0 tumors, recent results identified a subset (~30%) of patients with HER2 IHC 0 tumors who responded to T-DXd2,3.
The limitations of relying on HER2 IHC as a selection marker for HER2-directed therapy include the variability of interpretation by pathologists and tumor cellularity4,5. Furthermore, recent studies have shown the analytical and technical limitations of IHC-based quantitation of HER2 protein expression in HER2 unamplified/negative tumors. A recent analysis of HER2-negative breast cancer patients, including patients with HER2 IHC 0 tumors treated with the HER2 TKI neratinib, found that therapeutic response could be predicted by HER2 activation (i.e. phosphorylation) in the absence of HER2 molecular alterations, including mutation or gene amplification6. Consequently, there has been interest in and clinical need for the development and deployment of better analytical approaches to quantify HER2 activation in HER2-negative tumors to identify patients with HER2 LOW or IHC 0 tumors who may benefit from HER2-directed therapies such as T-DXd3,4,5. A CLIA-certified reverse-phase protein array (RPPA)-based assay was recently developed which quantitatively measures both HER2 protein expression and HER2 phosphorylation in routinely collected FFPE tumor tissue with a significantly increased analytical sensitivity over HER2 IHC that has been demonstrated to have a 15-fold quantitative dynamic range in HER2 IHC 0 tumors7. We have recently incorporated this CLIA-certified proteomic assay in our institutional proteomics feasibility protocol where patients are profiled by both commercially available CLIA-certified next-generation sequencing (NGS) and an RPPA-based proteomic/phosphoproteomic assay. In the case presented here, the excellent clinical response to HER2-directed therapy in a patient with triple-negative breast cancer (TNBC; ER/PR negative, HER2 IHC 0) as 6th line of therapy supports the clinical utility of proteomic/phosphoproteomics-based assays as to help identify patients who otherwise would not be selected for HER2-directed therapy.
Results
Case presentation
We present the case of a 57-year-old woman with a BRCA1 mutation and heavily pre-treated metastatic TNBC (HER2 IHC 0) who had an excellent response to T-DXd after the identification of HER2 protein expression and activation/phosphorylation from a next-generation RPPA-based proteomic analysis. Germline next-generation sequencing (NGS) testing was performed many years ago and was inaccessible in her present medical record, but somatic testing revealed a BRCA1 p.L95fs frameshift mutation with a variant allele fraction (VAF) of 55.6% which is presumed to represent the germline variant. This result prompted a prophylactic double mastectomy and bilateral salpingo-oophorectomy in 2014. In 2018, she developed a mass in the left axillary tail of the reconstructed breast which was biopsy proven to be a triple negative invasive ductal carcinoma (HER2 IHC 1+). She underwent tumor excision and axillary lymph node dissection but declined any adjuvant therapy. In 2020, she experienced recurrent disease with masses in the pectoralis major, adnexa, axillary lymph nodes, and a lymph node posterior to the left clavicle. Biopsy of the left axillary mass confirmed a triple negative invasive ductal carcinoma, (ER/PR negative, HER2 IHC 0), and PD-L1 negative via IHC). NGS performed on the recurrent tumor tissue identified the aforementioned BRCA1 variant as well as an activating PIK3CA mutation (p.N345K, missense variant in exon 4).
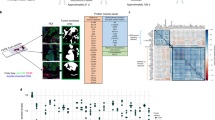
The patient subsequently received several lines of therapy including chemotherapy with dose-dense doxorubicin/cyclophosphamide, olaparib, gemcitabine/carboplatin/pembrolizumab, sacituzumab govitecan and eribulin with progression in various sites (Fig. 1). At the time of progression on her 5th line of therapy (eribulin), she had widespread metastases to the brain, mediastinal lymph nodes, lungs, chest wall, liver, and skin. She received intermittent courses of radiation therapy to the chest wall and regional lymph nodes, and stereotactic radiation therapy to the brain lesion. A skin biopsy obtained from the left flank prior to the start of 5th line therapy confirmed a poorly differentiated adenocarcinoma within the dermis, consistent with metastatic breast carcinoma (ER/PR IHC negative, HER2 IHC 0 and PD-L1 IHC negative). Repeat NGS on this specimen identified clinically relevant mutations in BRCA1, PIK3CA, TP53 and CTCF but no mutation/amplification in ERBB2, HER3, EGFR, nor ERS1. Several additional variants of unknown significance were reported. At the time of the repeat biopsy, a clinical trial at the Inova Schar Cancer Institute (ISCI) evaluating the feasibility of using both a commercially available genomics/NGS-based panel assay and RPPA-based proteomics assay on the same biopsy tissue was available, and the patient was enrolled. Tumor epithelium was harvested via laser microdissection (LMD) for RPPA-based quantification of total HER2 (HER2Total) and phosphorylated (p)HER2Y1248 as part of a 32-protein/phosphoprotein biomarker panel breast cancer assay in a commercial CLIA/CAP-accredited laboratory (Ignite Proteomics) (Table 1)6,7,8. The Ignite Proteomics assay quantitates the total expression levels and activation/phosphorylation levels of several proteins and/or phosphoproteins with known significance in cancers, including breast cancer9. Relative and absolute expression of HER2Total and (p)HER2Y1248 were determined by comparing the RPPA-based quantitative values derived from the patient’s LMD enriched tumor epithelium from the same tissue that was used for NGS against an existing RPPA referent dataset derived from LMD enriched tumor epithelium from 744 breast cancer specimens with centrally determined HER2-positive/amplified (IHC 3+/2+, FISH positive) and HER2-negative/unamplified (HER2 IHC 0 and HER2 IHC 1+/2+, FISH negative (LOW)) status including RPPA-based HER2 and phosphoHER2 reference data from 165 patients with TNBC as a direct comparator with the case study patient (Fig. 2). RPPA-based quantitative analysis and output of the 32 proteins and phosphoprotein analytes on the CLIA assay undergo extensive assay validation and credentialing under design control. Since the CLIA RPPA format is a calibrated assay with external calibrators with known amounts of total and phosphorylated analyte along with high and low controls are printed on every array the individual patient data are interpolated to the external calibrators and then to the calibrated referent population to ultimately provide both a relative and absolute value output. The assay validation data of the RPPA-based total and phosphorylated HER2, which were integral to this case report, include credentialing and correlating the RPPA-based HER2 and phosphorylated HER2 values from the reference population and individual patient value(s) to orthogonal measurements of the analyte such as central IHC and FISH HER2 determinations in both the HER2+ and HR-HER2- setting (Fig. 2). As shown in Fig. 2, the RPPA derived HER2 and phosphorylated HER2 values obtained in the reference population are highly concordant with the HER2 amplified (IHC 3+) cohort, and in keeping with previously described dynamics of quantitative HER2 assay correlations with IHC in the HER2- setting (IHC 0-2+/FISH−)4. The quantitative amount of total HER2 for this patient was ~5000 amol/ug protein loaded and ~40 RAU/ug/ul for phosphorylated HER2, and these patient-specific values are shown in relationship to both the RPPA determined population reference data and the centrally determined HER2 IHC determinations in order to provide a means to contextualize the RPPA quantified patient-specific values (Fig. 2). Moreover, under CLIA RPPA assay validation for total and phosphorylated HER2, the RPPA derived reference population data, the HER2 IHC determinations, and individual patient sample were also compared against a series of 10 breast cancer cell lines with known HER2 amplification status and previously determined HER2 levels4 (Fig. 3). Our quantitative total HER2 values (amol/ug total protein loaded) from our CLIA RPPA platform of the 10 breast cancer cell lines (Supplemental Table 1) compared favorably to previously reported values from an orthogonal total HER2 immunoassay recently described in a subset of these same cell lines4.
Correlation of total HER2 (left) and phosphorylated HER2 Y1248 (Right) with central IHC determined HER2 in HR+/HER2−, HR−/HER2+, HR+/HER2−, HR−/HER2− (a: total breast cancer reference population N = 744 pts) and HR-/HER2- (b: TNBC breast cancer referent population N = 165, bottom). Centrally determined IHC scores are shown: Red = IHC 3+, Yellow = IHC 2+, Green = IHC 1+, and Light Blue = IHC 0). a: pie charts representing the percentage of each HER2 IHC determination at specific RPPA ranges (gray dashed line) are shown for RPPA total HER2 (left) and phosphorylated HER2 Y1248 (right). The value for the case patient is shown as a red line for reference and context.
Comparison of RPPA-based total HER2 levels from 10 breast cancer cell lines (black circles, labeled) with known HER2 amplification status (Amplified: AU656 and SKBR3) with the total breast cancer reference population (open circles, N = 744 pts, left) and the TNBC referent population (open circles, N = 165, right). The case study patient is shown (red circle) for comparison. The specific RPPA quantitative values for the breast cancer cell lines are provided in Supplemental Table 1.
While the tumor was scored as HER2 IHC 0 (Fig. 4), the quantitative RPPA-based proteomics assay identified HER2 protein levels to be moderately expressed (Table 1: HER2Total 2+, 42%; Figs. 2 and 3) and modestly activated (Table 1: HER2Y1248 1+, 23%; Figs. 2 and 3). Evidence for heterodimerization and functional activation of the HER2 receptor was seen by commensurate co-activation/phosphorylation of HER3Y1269 (2+, 67%), EGFRY1068 (1+, 23%), and EGFRT654 (1+, 38%) as well as high levels of downstream AKT-mTOR activation/phosphorylation of mTORC1S2448 (2+, 53%), AKTT308 (3+, 99%), and AKTS473 (3+, 99%) (Table 1).
The RPPA proteomics results were reviewed by the ISCI Molecular Tumor Board (MTB), an expert panel comprised of medical oncologists, genetic counselors, and molecular profiling specialists. Given the high PD-L1 proteomic expression (3+, 72%), pembrolizumab was discussed, but the patient was already known to be refractory to pembrolizumab so this option was not entertained (Table 1). The patient also had an activating PIK3CA mutation and very high levels of AKT activation/phosphorylation. Alpelisib was not recommended, as it is only FDA-approved in combination with endocrine therapy in ER/PR positive patients—which this patient was not—and it was felt to be too poorly tolerated to be given off label. Despite the tumor being HER2 IHC 0, given the moderate levels of HER2 expression and activation seen by the CLIA RPPA quantitative assay along with evidence of HER2-HER3 heterodimerization, the MTB recommended treatment with T-DXd. T-DXd was approved by the patient’s insurance by providing a detailed rationale, the Ignite Proteomics CLIA proteomics report, associated references, and MTB recommendations to the patient’s insurance company. The patient had an excellent response with near-resolution of her hepatic lesions after four cycles of T-DXd. No measurable disease was observed after nine cycles, including no new brain metastases, consistent with a complete response to therapy (Fig. 5).
Discussion
For several decades, HER2-directed therapy was exclusively indicated for patients with HER2-amplified breast cancers (HER2 IHC 3+ or IHC 2+/FISH positive), thus excluding patients whose tumors were classified as HER2 negative by IHC from receiving such therapy. However, recent data has found that approximately 60% of patients who were previously classified as HER2-negative could be considered as HER2 LOW (IHC 1+ or 2+ and FISH negative)10,11. Data from the DESTINY-Breast04 trial found that treatment with T-DXd in previously-treated HER2 LOW breast cancer patients was associated with a significantly longer median PFS (mPFS) and median overall survival (mOS)12. Furthermore, recently published data from the DAISY trial has shown that a cohort of patients with HER2 LOW or HER2 IHC 0 expression had a response to T-DXd with objective response rates (ORR) of 37.5% and 29.7%, respectively2. These data have raised interest in predicting who among patients with HER2 LOW and HER2 IHC 0 expressing tumors could respond to HER2-directed therapy.
Recent quantitative proteomic analysis of HER2 IHC LOW and IHC 0 breast tumors via RPPA-based assay of LMD enriched tumor epithelium revealed that a significant percentage ( ~ 30%) of HER2 IHC 0 TNBC tumors actually expressed modest amounts of HER2Total and phosphorylated/activated pHER27. This frequency approximates a 30% ORR to T-DXd in HER2 IHC 0 patients in the DAISY trial2. These results are further congruent with past evidence that subsets of HER2 IHC 0 breast cancers truly express activated/phosphorylated HER28. In the I-SPY2 trial, RPPA quantification of HER2Total and pHER2 predicted pathologic complete response (pCR) in breast cancer patients with HER2 IHC 0, HER2 LOW, and HER2 HIGH (3+) using HER2-directed therapy and other targeted agents7.
We present a heavily pre-treated metastatic TNBC patient with a HER2 IHC 0 tumor finding who had a complete response to 6th line T-DXd after disease recurrence. The patient was selected for HER2-directed therapy based on HER2 expression and activation identified using a CLIA RPPA-based proteomic assay. This case is notable because both NGS and IHC identified no alterations in HER2, HER3, or EGFR such as mutation or amplification, that would have otherwise predicted the patient as a potential responder to T-DXd.
While the choice to use T-DXd was based solely on the RPPA-based proteomics result, the treatment was not part of our standard MTB protocol. We previously reported a case of proteomics-directed treatment of an inflammatory myofibroblastic tumor, but this is the first patient participating in our protocol that has been selected for HER2-directed therapy based on an RPPA-based proteomics assay13. To our knowledge, this is the first case reported wherein a patient with HER2 IHC 0 breast cancer was selected for T-DXd based on HER2 expression and activation generated from a proteomics-based assay. One consideration regarding the response seen here is that we cannot delineate the contribution of the bystander killing effect of DXd whereby cytotoxicity is extended to neighboring cells regardless of their HER2 expression level14. Prospective trials are needed to evaluate the predictive value of quantitative HER2 and activated/phosphorylated HER2 for T-DXd response in HER2 LOW and IHC 0 tumors, but this case provides a justification for further evaluation in larger prospective trials given the patient’s profound complete response. Moreover, these results could provide a molecular rationale and motivation for prospective validation of the CLIA HER2 expression/activation proteomics assay in clinical samples from retrospective study sets such as DAISY TRIAL (NCT04132960) and DESTINY-Breast06 (in the context of HR+ disease; NCT04494425). T-DXd and other HER2-directed therapies have demonstrated promising results in clinical trials in multiple solid tumor types, including (but not limited to) gastric, colorectal, biliary tract, pancreatic, and endometrial cancers15,16,17. Use of proteomics-based assays to select patients whose tumors are classically defined as HER2 IHC 0 —yet may actually express actionable levels of HER2 protein and protein activation—may result in wider benefit from the growing cadre of HER2-directed therapies and offers new treatment options for patients who otherwise might not be eligible for HER2-targeted therapy.
Methods
Patient specimen
A formalin-fixed paraffin-embedded (FFPE) tissue specimen prepared from a heavily pre-treated (four prior lines of therapy) recurrent skin punch biopsy from a left upper flank skin lesion was obtained from a 57-year-old female patient with metastatic TNBC for multi-omic analysis by NGS and RPPA. Written informed consent was obtained from the patient, which included the provision to analyze and publish information and data regarding the patient’s clinical presentation, and results and data from precision medicine/next-generation genomic sequencing testing, including the RPPA proteomics test results. All experimental protocols involving human data in this study were in accordance with the Declaration of Helsinki.
Laser microdissection
Serial tissue thin sections (8 µm) were sectioned by microtome, placed onto placed onto polyethylene naphthalate (PEN) membrane slides (Leica Microsystems, Wetzlar, Germany), and stained with hematoxylin and eosin (H&E). A micrograph of a representative section on glass (5 µm) was reviwed by a board-certified pathologist in Aperio eSlide Manager and Aperio ImageScope (Leica Microsystems) to annotate and confirm tumor areas of tumor epithelium for LMD enrichment. LMD enrichment of tumor epithelium was performed on the LMD7 (Leica Microsystems). The LMD enriched tumor sample for RPPA analysis was collected into a cylindrical pressure cycling technology (PCT; Pressure Biosciences, Inc., South Easton, MA, USA) microtube. A lysis/extraction buffer comprised of 10% tris(2-carboxyethyl)phosphine (TCEP, 50 mM; ThermoFisher Scientific, Inc., Waltham, MA, USA), 22.5% tris hydrochloride (Tris-HCl, 225 mM; ThermoFisher Scientific, Inc.), 4% sodium dodecyl sulfate (SDS, v/v; ThermoFisher Scientific, Inc.), 10% glycerol (v/v; ThermoFisher Scientific, Inc.), and 37.5% MilliQ type I water was added at a ratio of 2.5 µl buffer per mm18 LMD tissue. The microtube was capped with a PCT microcap and placed into 0.5 ml PCR tubes, briefly centrifuged, and stored at -80°C until sample lysis.
Reverse phase protein microarray
The LMD tumor sample for RPPA analysis was heated at 95 °C for 20 minutes, briefly centrifuged, and heated at 80 °C for 2 hours. After heating, the tube was stored at 4 °C for one minute and then centrifuged for 14,000 rpm for 15 minutes. The lysate supernatant containing the denatured protein was transferred to a fresh low protein binding o-ring screw-top tube (Agilent Technologies, Savage, MD, USA) and stored at -80 °C until shipment on dry ice to Ignite Proteomics (Golden, CO, USA) for RPPA analysis.
RPPA analysis was performed as previously described using a 32-marker, CLIA-based RPPA panel for examination of the total and phosphoprotein abundances of several targets with known relevance in solid tumors9,13,19. Briefly, lysates were printed onto nitrocellulose backed slides in technical triplicates alongside all requisite controls and calibrators. Prior to staining, nitrocellulose slides were pre-treated with ReBlot (MilliporeSigma, Rockville, MD, USA) and a blocking reagent (I-Block; Applied Biosystems, Waltham, MA, USA). Slides were incubated with primary antibodies for 30 min at room temperature, followed by incubation with a secondary antibody. Each staining run included a single negative control slide (only antibody diluent; no primary antibody). Protein detection was amplified via horseradish peroxidase-mediated biotinyl tyramide deposition and visualized using a fluorescent probe (LI-COR Biosciences, Inc., Lincoln, NE, USA). Images of the stained RPPA slides were captured on an InnoScan 710-AL (Innopsys, Inc., Chicago, IL, USA). Individual spots were detected using the InnoScan software program, Mapix, and spots with non-standard morphology and/or staining were flagged and removed from analysis. The average median intensity value was calculated for each sample on the array for each analyte as well as the negative control slide. Background subtracted intensity values for each sample were fit to an analyte-specific calibrator and total protein normalized. Resulting values were compared to a population referent to determine patient sample percentile and quartile score. Antibodies used on the RPPA were validated before use by confirming the presence of a single band at the appropriate molecular weight with a panel of control cell lysates using conventional western blotting as previously described18,20. For the RPPA, the total HER2 antibody (Invitrogen, Inc, Cat# MA5-14509) was used at 1:400 dilution and the phosphoHER2 Y1248 (AbCam, Inc, Catalog # ab13327) was used at 1:20,000 dilution.
Next-generation sequencing
Clinical NGS (specifically, DNA-seq) analysis was performed using the Tempus xT 648-gene panel1,3,21 (Tempus Labs, Inc., Chicago, IL, USA) using the same FFPE tissue specimen as was used for the LMD-RPPA analysis. The sample for DNA-seq was prepared from tissue sections with approximately 70% tumor cellularity after microdissection.
Ethical approval and patient consent
This study was conducted under the Western-Copernicus Group (WRG) IRB-approved protocol under which the patient provided informed consent and was enrolled in Inova Schar Cancer Institute’s Molecular Tumor Board (MTB) study. The written informed consent included the provision to analyze and publish information and data regarding the patient’s clinical presentation, and results and data from precision medicine/next-generation genomic sequencing testing, including the RPPA proteomics test results.
Data availability
The raw data that support the findings of this study are available from Tempus Labs, Inc. but restrictions apply to the availability of these data, which were used under license for the current study, and so are not publicly available. Data are however available from the authors upon reasonable request and with permission of Tempus Labs, Inc. Patient Case level structured NGS data supplied by Tempus Labs, Inc. and structured normalized proteomic data supplied by Ignite Proteomics are available at https://capmm.science.gmu.edu/research/data/.
Code availability
Code sharing not applicable to this article as no codes were generated or analyzed during the current study.
References
Wolff, A. C. et al. Human epidermal growth factor receptor 2 testing in breast cancer: american society of clinical oncology–college of american pathologists guideline update. Arch. Pathol. Lab. Med. 147, 993–1000 (2023).
Mosele, F. et al. Trastuzumab deruxtecan in metastatic breast cancer with variable HER2 expression: the phase 2 DAISY trial. Nat. Med. 29, 2110–2120 (2023).
Viale, G. et al. Retrospective study to estimate the prevalence and describe the clinicopathological characteristics, treatments received, and outcomes of HER2-low breast cancer. ESMO Open 8, 101615 (2023).
Moutafi, M. et al. Quantitative measurement of HER2 expression to subclassify ERBB2 unamplified breast cancer. Lab. Investig. 102, 1101–1108 (2022).
Robbins, C. J. et al. Multi-institutional assessment of pathologist scoring HER2 immunohistochemistry. Mod. Pathol. 36, 100032 (2023).
Wulfkuhle, J. D. et al. Evaluation of the HER/PI3K/AKT family signaling network as a predictive biomarker of pathologic complete response for patients with breast cancer treated with neratinib in the I-SPY 2 TRIAL. JCO Precis. Oncol. https://doi.org/10.1200/po.18.00024 (2018).
Petricoin, E. F. et al. Abstract HER2-17: HER2-17 Novel Quantitative HER2 Assay for determining dynamic HER2 expression in the HER2 IHC 0 “ultra-low” setting: implications for precision therapy in HER2- breast cancer. Cancer Res. 83, HER2-17–HER12-17 (2023).
Wulfkuhle, J. D. et al. Molecular analysis of HER2 signaling in human breast cancer by functional protein pathway activation mapping. Clin. Cancer Res. 18, 6426–6435 (2012).
Wulfkuhle, J. D. et al. Multiplexed cell signaling analysis of human breast cancer applications for personalized therapy. J. Proteome Res. 7, 1508–1517 (2008).
Tarantino, P. et al. HER2-low breast cancer: pathological and clinical landscape. J. Clin. Oncol. 38, 1951–1962 (2020).
Schettini, F. et al. Clinical, pathological, and PAM50 gene expression features of HER2-low breast cancer. NPJ Breast Cancer 7, 1 (2021).
Modi, S. et al. Trastuzumab deruxtecan in previously treated HER2-low advanced breast cancer. N. Engl. J. Med. 387, 9–20 (2022).
Hunt, A. L. et al. Integration of multi-omic data in a molecular tumor board reveals EGFR-associated ALK-inhibitor resistance in a patient with inflammatory myofibroblastic cancer. The Oncologist https://doi.org/10.1093/oncolo/oyad129 (2023).
Ogitani, Y., Hagihara, K., Oitate, M., Naito, H. & Agatsuma, T. Bystander killing effect of DS-8201a, a novel anti-human epidermal growth factor receptor 2 antibody-drug conjugate, in tumors with human epidermal growth factor receptor 2 heterogeneity. Cancer Sci. 107, 1039–1046 (2016).
Li, B. T. et al. 654O Efficacy and safety of trastuzumab deruxtecan (T-DXd) in patients (pts) with solid tumors harboring specific HER2-activating mutations (HER2m): primary results from the international phase II DESTINY-PanTumor01 (DPT-01) study. Ann. Oncol. 34, S459–S460 (2023).
Meric-Bernstam, F. et al. Efficacy and safety of trastuzumab deruxtecan (T-DXd) in patients (pts) with HER2-expressing solid tumors: DESTINY-PanTumor02 (DP-02) interim results. J. Clin. Oncol. 41, LBA3000–LBA3000 (2023).
Harding, J. J. et al. Zanidatamab for HER2-amplified, unresectable, locally advanced or metastatic biliary tract cancer (HERIZON-BTC-01): a multicentre, single-arm, phase 2b study. Lancet Oncol. 24, 772–782 (2023).
Signore, M., and Reeder, K. A. Molecular Profiling Methods in Molecular Biology. pp. 139–155 (eds. Espina, V. & Liotta, L. A.) (Humana Press, 2012).
Baldelli, E. et al. Molecular Profiling: Methods and Protocols (ed Virginia Espina) 149-169 (Springer New York, 2017).
Akbani, R. et al. Realizing the promise of reverse phase protein arrays for clinical, translational, and basic research: a workshop report. Mol. Cell. Proteom. 13, 1625–1643 (2014).
Beaubier, N. et al. Clinical validation of the tempus xT next-generation targeted oncology sequencing assay. Oncotarget 10, 2384–2396 (2019).
Acknowledgements
The authors would like to acknowledge Paulette Mhawech-Fauceglia for histopathology image analysis support, and Aratara Nutcharoen and Brian Corgiat for proteomic sample preparation support.
Author information
Authors and Affiliations
Contributions
Study concept and design: T.L.C., T.P.C., and E.F.P. Data acquisition, analysis, and interpretation: A.L.H., J.R., L.M., J.B.D., E.F.P., T.P.C., T.L.C., and J.H. Writing (original draft): L.E.J., J.R., T.L.C., and J.H. Writing (review and editing): L.E.J., J.R., S.C., M.L., A.L.H., L.M., E.F.P., T.P.C., T.L.C., and J.H. All authors gave final approval of the completed work and are accountable for accuracy and integrity.
Corresponding author
Ethics declarations
Competing interests
T.P.C. is a ThermoFisher Scientific, Inc SAB member and receives research funding from AbbVie. E.F.P. is a SAB member and consultant to Ignite Proteomics, is on the editorial board of npjPO, and receives research funding from Genentech, Pfizer, Mirati, Springworks Therapeutics, Deciphera, AbbVie, and is a co-inventor of the RPPA technology described herein, and related HER2 biomarker patents and receives royalties on the related license agreements and is affiliated J.H. is on the advisory board for Exelixis Pharmaceuticals, Inc.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Johnston, L.E., Randall, J., Chouraichi, S. et al. Proteomics based selection achieves complete response to HER2 therapy in HER2 IHC 0 breast cancer. npj Precis. Onc. 8, 203 (2024). https://doi.org/10.1038/s41698-024-00696-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41698-024-00696-6
- Springer Nature Limited