Abstract
While circulating metabolites and immune system have been increasingly linked to hypothyroidism risk, the causality underlying these associations remains largely uninterrogated. We used Mendelian randomization to identified putative causal traits for hypothyroidism via integrating omics data. Briefly, we utilized 1180 plasma metabolites and 731 immune cells traits as exposures to identify putatively causal traits for hypothyroidism in the discovery (40,926 cases) and replication cohorts (14,871 cases). By combining MR results from two large-scale cohorts, we ultimately identified 21 putatively causal traits, including five plasma metabolites and 16 immune cell traits. CD3 on CD28+ CD4+ T cell and 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1) demonstrated the most pronounced positive and negative associations with hypothyroidism risk, respectively. The odds ratio and 95% confidence interval were 1.09 (1.07, 1.12) and 0.81 (0.75, 0.87), respectively. No evidence of horizontal pleiotropy, heterogeneity among instrumental variables or reverse causation were found for these 21 significant associations. Our study elucidates key metabolites and immune cell traits associated with hypothyroidism. These findings provide new insights into the etiology and potential therapeutic targets for hypothyroidism.
Similar content being viewed by others
Introduction
Hypothyroidism is a clinical disease of thyroid hormone deficiency1 and leads to a major healthcare concern that impacts approximately 4–10% of the global population2. Levothyroxine administration is the established therapeutic approach for hypothyroidism1, while it often necessitates lifelong treatment3,4. Evidence suggests that systemic metabolic alterations are tied to the hypothyroidism and affect primary bile acid biosynthesis, steroid hormone biosynthesis, lysine degradation, tryptophan metabolism, and purine metabolism5. Immune dysfunction resulting from chronic autoimmune thyroiditis is widely considered as the predominant etiology of primary hypothyroidism1. The infiltration of lymphocytes into thyroid tissue can directly modulate the function of thyroid follicular cells through the actions of IL-1, TNF, and IFNγ6,7. A better understanding of these metabolic and immune features may contribute to uncovering the etiological mechanism of hypothyroidism.
Mendelian randomization (MR) is a statistical method for inferring causality, which is not susceptible to reverse causality and confounding factors8. In recent years, omics-based MR, has gained substantial progress in the identification of disease targets, biomarkers, and potential pathogenic pathways9,10. A previous MR study investigated the causal relationship between 69 metabolites and autoimmune hypothyroidism11. They found that N-(3-furoyl) glycine, pipecolate, phenylalanine, allantoin, indololactate and alanine were associated with autoimmune hypothyroidism. However, the study encompassed a limited number of metabolites and lacked a validation cohort for hypothyroidism, which affected the robustness of the findings to some extent.
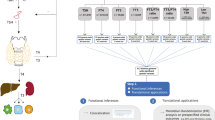
To address these limitations, we meticulously assembled the most comprehensive GWAS data available to date for plasma metabolomics, immune cell traits and two large-scale hypothyroidism cohorts. Leveraging these datasets, we performed a two-sample MR study to systematically investigate the potential causal associations of metabolites and immune system with hypothyroidism. The follow-up analyses comprised five parts. First, colocalization analysis was performed to assess the existence of shared causal variants within the genomic regions. Second, MR based on Bayesian model averaging (MR-BMA) was performed to determine which of a set of related risk factors with common genetic predictors were causal drivers of hypothyroidism risk. Third, the cell type-specific enrichment analysis was employed to evaluate the cellular enrichment features of thyroid tissues in the hypothyroidism datasets. Fourth, the associations between putatively causal traits and four complications of hypothyroidism were investigated. Finally, for identified putatively causal traits, we evaluated their druggability in drug databases.
Results
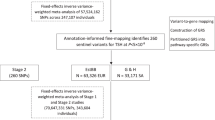
The study design is depicted in Fig. 1. All analyses were based on summary-level data. We used 1180 plasma metabolites and 731 immune cells traits as instrumental variables (IVs) to identify putatively causal traits for hypothyroidism. Details regarding all data sources and sample sizes are summarized in Supplementary Data 1. At the LD clumping criteria of pairwise LD R2 < 0.001 within a 10,000 kb window and the significance threshold of P value < 5 × 10–6, nearly 91% of exposures (1048 exposures for plasma metabolites and 685 exposures for immune cells traits) had at least three SNPs for subsequent analysis (Supplementary Data 2 and 3). The F statistic of the utilized IVs exceeded 10 for all exposure data, which indicates a relatively low risk of weak instrument bias (Supplementary Data 2 and 3). Besides, we performed a series of follow-up analyses, including colocalization analysis, MR-BMA analysis, cell type-specific enrichment analysis, hypothyroidism-related complications association analysis and druggability analysis.
Associations between plasma metabolites and hypothyroidism
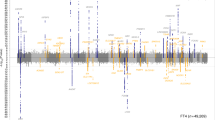
After adjustment for multiple testing by Bonferroni method (P value < 4.77 × 10–5), we identified that 18 plasma metabolites were significantly associated with hypothyroidism risk using inverse-variance weighted method (Fig. 2a and Supplementary Data 4). Genetically predicted levels of five plasma metabolites were positively associated with hypothyroidism, while the other 13 plasma metabolites were inversely associated with hypothyroidism. Among them, the association between kynurenine and hypothyroidism was not passed the sensitivity analysis due to the inconsistent effect direction of the additional method (MR Egger) (Supplementary Data 4). No evidence of horizontal pleiotropy (Egger P for intercept >0.05 and MR-PRESSO global test P > 0.05), heterogeneity among IVs (P for Cochran’s Q > 0.05) or reverse causation (P for MR Steiger test >0.05) were found for other 17 significant associations (Supplementary Data 4).
a Forest plots illustrating the 18 potential causal associations between metabolites and hypothyroidism as revealed in the discovery cohort. b Forest plots illustrating the five potential causal associations between metabolites and hypothyroidism as revealed by MR meta-analysis. OR odds ratio, CI confidence interval.
In the replication stage, two plasma metabolites were successfully validated in the replication cohort with nominal significance (P value < 0.05), including 2’-deoxyuridine and 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1) (Supplementary Data 5). In the meta-analysis of discovery and replication datasets using random effect model, five plasma metabolites displayed nominal significant associations with hypothyroidism (P value < 0.05) (Fig. 2b and Supplementary Data 6). These five metabolites were retained for the following analysis. Among them, genetically predicted sphingomyelin (d18:1/20:0, d16:1/22:0) levels were positively associated with hypothyroidism, while a ratio of alpha-ketoglutarate to kynurenine and levels of cerotoylcarnitine (C26), 2’-deoxyuridine and 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1) were inversely associated with hypothyroidism. Overall, per standard deviation (SD) increase in genetically predicted metabolites levels, the odds ratio (OR) of hypothyroidism ranged from 0.81 (95% confidence interval [CI], 0.75–0.87) for 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1) to 1.05 (95% CI, 1.02–1.07) for sphingomyelin (d18:1/20:0, d16:1/22:0).
Associations between immune cells traits and hypothyroidism
After adjustment for multiple testing by Bonferroni method (P value < 7.30 × 10–5), we identified a total of 27 immune cell traits that were significantly associated with hypothyroidism using inverse-variance weighted method (Fig. 3a and Supplementary Data 7). Genetically predicted 20 immune cell traits were positively associated with hypothyroidism, while the other seven immune cell traits were inversely associated with hypothyroidism. Among them, two associations of TCRgd T cell to lymphocyte ratio and CD28- CD127– CD25++ CD8+ T cell to T cell ratio with hypothyroidism were not passed the sensitivity analysis due to the inconsistent effect direction of the additional method (MR Egger) (Supplementary Data 7).
a Forest plots illustrating the 27 potential causal associations between immune cell traits and hypothyroidism as revealed in the discovery cohort. b Forest plots illustrating the 16 potential causal associations between immune cell traits and hypothyroidism as revealed by MR meta-analysis. OR odds ratio, CI confidence interval.
In the replication stage, nine immune cell traits were successfully validated in the replication cohort with nominal significance (Supplementary Data 8). In the meta-analysis of discovery and replication datasets using random effect model, 16 immune cell traits displayed nominal significant associations with hypothyroidism (Fig. 3b and Supplementary Data 9). These 16 immune cell traits were retained for the following analysis. Among them, genetically predicted Transitional B cell absolute count, CD27 on IgD- CD38+ B cell and CD3 on eight different T cell were positively associated with hypothyroidism, while BAFF-R on two different B cell and three ratios of CD8dim T cell to leukocyte, activated CD4 regulatory T cell to CD4 regulatory T cell, CD11c+ HLA DR++ monocyte to monocyte and CCR2 on CD62L+ myeloid Dendritic Cell, were inversely associated with hypothyroidism. Overall, per standard deviation (SD) increase in genetically predicted immune cell traits, the odds ratio (OR) of hypothyroidism ranged from 0.92 (95% CI, 0.90–0.95) for CD11c+ HLA DR++ monocyte to monocyte ratio to 1.09 (95% CI, 1.07–1.12) for CD3 on CD28+ CD4+ T cell.
Nine metabolites and immune cell traits were supported by colocalization evidence
Colocalization analysis could complement the MR results by addressing the limitations associated with linkage disequilibrium, thereby providing a more nuanced understanding of the shared genetic basis underlying exposure and outcomes12. Among the 21 identified associations (five for metabolites and 16 for immune cell traits), nine (43%) of these 21 putatively causal trait-hypothyroidism pairs showed a high colocalization with hypothyroidism using traditional colocalization method and sum of single effects (SuSiE) method (Fig. 4, Supplementary Data 10 and Supplementary Fig. 1). The result indicated the existence of shared causal variants and further suggested that there may be common biological mechanism between these nine putatively causal traits and hypothyroidism.
MR analysis based on Bayesian model averaging
Interestingly, eight CD3 related immune traits were identified as potential causal traits for hypothyroidism in our study and show similar effect. To determined which of a set of these immune traits with common genetic predictors were causal drivers of hypothyroidism risk, we employed MR-BMA analysis, which is an extension of multivariate MR (MVMR)13. According to the previous study14, we set z to 10,000, the prior probability to 0.1, and prior variance (σ2) to 0.25. Marginal inclusion probability (MIP) and Model-averaged causal effect (MACE) were calculated. MR-BMA result showed CD3 on HLA DR+ CD4+ T cell was the most likely potential causal trait (MIP = 0.424, MACE = 0.018, P value = 7.9 × 10–3, Ranking = 1) (Supplementary Data 11). We also adjusted the prior probability to 0.5 as the sensitivity analysis, and the result is consistent with the previous conclusion (MIP = 0.396, MACE = 0.016, P value = 2.9 × 10–2, Ranking = 1) (Supplementary Data 11).
Cell type-specific enrichment analysis for hypothyroidism
In our MR analysis, the GWAS of immune cell was conducted based on peripheral blood samples. However, there is currently a lack of available GWAS data for immune cells at the tissue level (i.e. thyroid tissue). To understand the enrichment of cell types in thyroid tissues of hypothyroid population, a cell type-specific enrichment analysis (CSEA) was performed. Among the 12 cell types present in thyroid tissue, we identified eight cell types that exhibited nominally significant associations with at least one hypothyroidism dataset (Supplementary Data 12), which were endothelial cell, endothelial cell (APC), smooth muscle cell, T cell, macrophage, dendritic cell, B cell and B cell (Plasmacyte). The result of CSEA could be an important complementary cellular feature of hypothyroidism alongside MR results.
Association between putatively causal traits and complications of hypothyroidism
Previous studies have reported a series of complications associated with hypothyroidism, including diabetes mellitus in pregnancy15, diabetic nephropathy16, heart failure17, chronic kidney disease18. An assessment of the shared metabolic and immune cellular mechanisms underlying hypothyroidism and its complications is crucial for gaining a comprehensive understanding of the underlying pathophysiology and potentially managing disease progression. Therefore, we explored the association between putatively causal traits and the above-mentioned complications.
We found that eight putatively causal traits displayed nominal significant associations with complications of hypothyroidism (Fig. 5 and Supplementary Data 13). Four associations among them were not passed the sensitivity analysis due to the inconsistent effect direction of the additional method (MR Egger), which were 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1)-chronic kidney disease, CCR2 on CD62L+ myeloid Dendritic Cell-Diabetic nephropathy, CD27 on IgD- CD38+ B cell-heart failure and CD3 on CD39+ CD4+ T cell-diabetic nephropathy pairs (Supplementary Data 14).
For other four associations, two putatively causal traits were identified as protective and risk factor for diabetic nephropathy, respectively, which were activated CD4 regulatory T cell to CD4 regulatory T cell ratio (OR 0.77; 95% CI 0.66–0.91) and CD3 on activated CD4 regulatory T cell (OR 1.14; 95% CI 1.02–1.28). Moreover, BAFF-R on IgD+ CD38- unswitched memory B cell (OR 0.98; 95% CI 0.97–0.99) and CD20- B cell (OR 0.94; 95% CI 0.88–1.00) were both identified as protective factor for heart failure (Supplementary Data 14).
Druggability of identified putatively causal traits
In druggability evaluation, we found CD3 and Sphingomyelin (d18:1/20:0, d16:1/22:0) have been targeted for drug development (Supplementary Data 15). A drug targeting CD3 have been developed to treat relapsed or refractory diffuse large B-cell lymphoma (Glofitamab). A drug targeting sphingomyelin (d18:1/20:0, d16:1/22:0) have been developed to treat acid sphingomyelinase deficiency (Olipudase alfa). These results suggest that CD3 and Sphingomyelin (d18:1/20:0, d16:1/22:0), may provide insights into drug repurposing.
Discussion
In the present study, we performed a comprehensive investigation on associations of plasma metabolites and immune system with hypothyroidism risk. We finally identified 21 putatively causal traits associated with hypothyroidism via combining two large-scale hypothyroidism cohorts. The follow-up analysis revealed the roles of four hypothyroidism-associated metabolites and immune cell traits in complications of hypothyroidism. Druggability evaluation prioritized two hypothyroidism-related traits, including CD3 and sphingomyelin that could be modified by drug interventions.
Although the majority of the 21 putatively causal traits were first reported by our study, several of them are consistent with the findings reported in previous research. The positive association between sphingomyelin (d18:1/20:0, d16:1/22:0) and hypothyroidism risk is consistent with a metabolomic analysis involving 27 cases in the Hashimoto’s thyroiditis with subclinical hypothyroidism (HTS) group. They found that the HTS group exhibited elevated levels of sphingomyelin compared to the control group19. The 1-(1-enyl-palmitoyl)-2-oleoyl-GPE (p-16:0/18:1), a derivative of glycerophospholipids (GPL), was found to be associated with decreased hypothyroidism risk in our study. This metabolite in the hypothyroidism group depicted a decreasing trend via UPLC − Q-TOF/MS in a study enrolled discovery and validation sets of 175 and 300 participants, respectively20. All of these showcase the validity of our findings.
Our MR analysis and CSEA together reveal the important role of lymphocytes in the development of hypothyroidism. In fact, lymphocytes are closely related to Hashimoto’s thyroiditis (HT), which one of the most common cause of primary hypothyroidism21,22. The presence of lymphocytic infiltrates is a characteristic pathological feature commonly observed in HT23. To be specific, three thyroidal stromal cell subsets, including ACKR1+ endothelial cells and CCL21+ myofibroblasts and CCL21+ fibroblasts, may potentially facilitate lymphocyte trafficking from the bloodstream to thyroid tissues. Numerous cytokines and chemokines released by infiltrating lymphocytes have been demonstrated to play a pivotal role in the pathogenesis of HT23,24, which may increases the risk of hypothyroidism.
Our study found that activated CD4 regulatory T cell to CD4 regulatory T cell ratio was to be associated with decreased diabetic nephropathy risk. CD4+CD25+FoxP3+ natural regulatory T cells (Tregs) function as a potent immunosuppressive population in inflammatory diseases, capable of counteracting pro-inflammatory cell populations25. Feuerer et al. employed a mouse model to elucidate the significance of Tregs in the pathogenesis of insulin resistance. They found that revealing a substantial reduction in the abundance of Tregs within murine visceral adipose tissue (mVAT) among obese mice compared to lean control animals26. Eller et al. demonstrated that depletion of regulatory T cells (Tregs) in db/db mice resulted in increased manifestations of diabetic nephropathy, including albuminuria and glomerular hyperfiltration. Conversely, adoptive transfer of CD4+FoxP3+Tregs significantly ameliorated insulin sensitivity and diabetic nephropathy27. All of these showcase the validity of our finding.
Our study also provides insights into drug reutilization for hypothyroidism. Both CD3 antigen and Sphingomyelin (d18:1/20:0, d16:1/22:0) were identified as risk factors for hypothyroidism in our study. Glofitamab targeting CD3 have been developed to treat relapsed or refractory diffuse large B-cell lymphoma. Glofitamab is a bi-specific monoclonal antibody that specifically targets the CD3 protein complex on T cells. By binding to CD3, Glofitamab facilitates the formation of immune synapses, which recruit T cells to CD20-expressing B cells and ultimately resulting in the elimination of target B cells28. Therefore, Glofitamab could be a candidate drug candidate for hypothyroidism due to its ability to target lytic B cells. As a recombinant human acid sphingomyelinase, Olipudase alfa exhibits the capability to hydrolyze sphingomyelin29, rendering it a promising therapeutic candidate for the management and prevention of hypothyroidism.
The study has several strengths. First, we utilized a large pool and number of plasma metabolites and immune cells as exposures in the MR studies and two outcome cohorts were employed for validation analysis to ensure the reliability of the results. Second, we systematically removed exposures that had fewer than three IVs, thereby ensuring the conduction of pleiotropy test. Third, the population ancestry of the datasets we used was all from Europe, therefore, population stratification was not a potential bias in our study.
The interpretation of our findings should be approached with caution due to several limitations. First, the major concern in MR analysis lies in the potential pleiotropic effects of SNPs, which may violate the assumptions underlying MR. According to the solution in the previous literature10, we used sensitivity analyses including a variety of analytical methods, such as MR-Egger, to evaluate the consistence of estimates and directions. Second, the scope of our analysis was limited to Europeans, thereby constraining the generalizability of our findings to other populations. Third, the GWAS of hypothyroidism herein did not account for the subtypes of hypothyroidism, encompassing primary, central, and peripheral hypothyroidism. This limitation may have slightly compromised the precision of the targets and markers that we identified. However, given that the extremely low incidence of both central and peripheral hypothyroidism21,30, the population in our study could be broadly categorized as individuals with primary hypothyroidism.
In conclusion, our study identified key plasma metabolites and immune cell traits with a potential causal association with hypothyroidism risk, some of which show potential for drug repurposing. These metabolic and cellular characteristics may provide valuable knowledge for understanding the pathogenesis and prediction of targets related to hypothyroidism, thereby offering insights into future prevention strategies and treatments.
Methods
GWAS data source
The GWAS data for 1180 annotated plasma metabolites were extracted from a metabolomics study in the Canadian Longitudinal Study on Aging (CLSA) cohort31. This study focused on 8299 unrelated European ancestry individuals in CLSA who had circulating plasma metabolites measured by Metabolon HD4 platform. The GWAS data for 731 immune cell traits were obtained from SardiNIA project, and 3757 of volunteers were immune profiled by flow cytometric measurements32.
Data on the associations with hypothyroidism were obtained from FinnGen R9 study33 and UK biobank34. We treated the FinnGen R9 study as the discovery study, which comprised 40,926 cases and 274,069 controls. The other study was considered as the replication study, which comprised 14,871 cases and 406,300 controls.
GWAS summary datasets for above-mentioned complications were downloaded from publicly accessible databases of the IEU OpenGWAS project35 : https://gwas.mrcieu.ac.uk/. Trait names and GWAS IDs are as follows: chronic kidney disease (“finn-b-N14_CHRONKIDNEYDIS”), Diabetes mellitus in pregnancy (“finn-b-O15_PREG_DM”), diabetic nephropathy (“ebi-a-GCST90018832”) and heart failure (“ebi-a-GCST009541”).
Selection of the genetic instrumental variables
For each exposure, we initially filtered SNPs with a threshold of P value < 5 × 10–6 and minor allele frequency (MAF) > 0.05. Subsequently, we performed clumping by the PLINK software using a cut-off of R2 < 0.001 and window size of 10,000 kb, utilizing the 503 European samples data from the 1000 Genomes project as our reference panel36. Only the SNPs with the lowest P value were retained. For metabolites and cell traits, if the screened SNPs were not present in the outcome, we excluded the corresponding SNPs. We also systematically removed exposures that had fewer than three IVs, thereby ensuring the conduction of subsequent pleiotropy test. Moreover, we further introduced RadialMR method, which can identify outlier pleiotropic SNPs37. Before MR analysis, we used this method to exclude outlier pleiotropic SNPs in IVs to reduce the risk of pleiotropy.
Testing instrument strength
F statistic is calculated to test instrument strength through the formula F = β2/SE2, where β denotes the estimated genetic effect on exposure, SE represents standard error of β38. F statistic of ≥10 indicates the absence of significant weak instrumental bias39.
Statistics and reproducibility
Two-sample MR was performed to evaluate the causal associations between exposures and outcomes (R package TwoSampleMR, version 0.5.6). Inverse-variance weighted (IVW) under a multiplicative random-effects model was used for main analysis method40. However, a scenario may arise where the P value of the IVW for random effects is statistically significant, while that of the fixed effects model is not. In such cases, to ensure more conservative results, we employ IVW based on the fixed effects model for analysis.
OR or β value with 95% confidence interval were evaluated to identify causal effects between exposures and outcomes, assuming MR assumptions were met. Adjustment for multiple testing was conducted to avoid the potential influence of false positive findings. Bonferroni method was used across all MR analyses. Meta-analyses of MR result in both the discovery and replication cohorts were calculated using the random effect model (R package meta, version 7.0-0). The I2 statistic was calculated to assess the heterogeneity of each outcome from different data sources, and the I2 values < 25%, 25–75%, and >75% were considered to indicate low, moderate, and high heterogeneity, respectively, as described in the previous literature41.
MR assumptions
Adherence to three fundamental assumptions is crucial for ensuring the validity of causal estimates in MR studies. First, the genetic variants exhibit a robust association with the exposure. Second, genetic variants are not associated with a confounding factor between exposure and outcome. Third, the variants do not affect outcome independently of exposure42,43.
Sensitivity analysis
We performed a series of sensitivity analyses to assess the robustness and reliability of the results. The Cochran’s Q statistic was calculated using a fixed-effect variance weighted analysis, with P value ≤ 0.05 indicating the presence of heterogeneity44. MR-Egger regression with intercept terms to assess the potential presence of horizontal pleiotropy. Deviations from 0 (P value < 0.05) were deemed invalid for all SNPs45. In addition to the MR Egger method, we employed MR-PRESSO as an additional tool for pleiotropy assessment46. We also conducted the MR-RAPS, weighted-median and weighted-mode methods to evaluate the robustness of our calculated results. MR Steiger test was performed to estimate the potential reverse causal impact47.
In our study, a significant association was considered confident if it met a series of stringent criteria: (1) the significance of association passed multiple tests using IVW method as well as the directional consistency of the effect using other extra four MR approaches, (2) there was no significant heterogeneity among IVs (P for Cochran’s Q > 0.05), (3) there was no evidence of horizontal pleiotropy (Egger P for intercept >0.05 and MR-PRESSO global test P > 0.05) and (4) no reverse causal MR effect (P for MR Steiger test >0.05).
Colocalization analysis
To increase power in colocalization analysis, we used the METAL software to meta-analyze the summary statistics of hypothyroidism derived from FinnGen R9 study and UK biobank48. The analysis was conducted using a fixed-effects model and the associations from two data sources were weighted by standard error of GWAS estimates.
Colocalization analysis employed a Bayesian model to evaluate support for five exclusionary hypotheses: (1) no association with either trait; (2) exclusive correlation with trait one; (3) exclusive correlation with trait two; (4) presence of an association between the two traits, but with different causal variants for each trait; and (5) both features exhibit correlation and share the same causal variant 49. The analysis provides posterior probabilities for each hypothesis testing, including H0, H1, H2, H3, and H4. We assigned a prior probability of 1 × 10−4 for the SNP being associated exclusively with trait 1 (p1), and likewise for trait 2 (p2). Additionally, we set the prior probability of the SNP being associated with both traits (p12) at 1 × 10−5. To establish high support evidence of colocalization, we considered two signals to have a posterior probability for shared causal variants (PH4) ≥ 0.8. Given the limitation of traditional colocalization method in detecting exposure and outcome traits with multiple causal genetic variants, we employed the SuSiE method along with a genetic correlation matrix reference panel derived from individuals of European ancestry in Phase III of the 1000 Genome Project to identify multiple causal variants. For each exposure, we selected regions that had 500 kb windows upstream and downstream of each instrumental variable in MR for analysis50, and the instrumental variable with the largest posterior probability for H4 was reported. The analysis was conducted using the coloc package (5.2.3) and susieR package (0.12.40). Locuscomparer package (1.0.0) was used to visualize the results of colocalization.
MR-BMA analysis
In short, taking into account all the exposures specified, MR-BMA iterates over a number of potentially “real” causal models13. For each exposure, a MIP value is calculated, representing the posterior probability that the exposure factor x appears in a true causal model for a given z iteration. The method also estimates the MACE, which represents the exposure’s weighted average direct causal effect on risk across all candidate models. It is worth noting that MACE should not be interpreted in absolute terms. It will be biased towards zero due to the contraction applied in variable selection. But this metric can be used to understand the direction and magnitude of the impact relative to other exposures included in the model.
We combined the IVs from the eight CD3 related immune traits, and further clumped by linkage disequilibrium (R2 < 0.001 within a window of 10,000 kb) to ensure that the IVs were independent. SNP effects and corresponding standard errors were then extracted from the corresponding immune GWAS summary statistics and harmonized with the outcome hypothyroidism GWAS information. We set z to 10,000, the prior probability to 0.1, and prior variance (σ2) to 0.25 in the main analysis. We additionally set prior probability to 0.5 as sensitivity analyses. The analysis was conducted using the mrbma package (0.1.0).
Cell type‑specific enrichment analysis
The Cell type-Specific Enrichment Analysis DataBase (CSEA-DB, https://bioinfo.uth.edu/CSEADB/)51, was used to investigate the enrichment of 12 general cell types in thyroid tissue of hypothyroidism patients. CSEA-DB collected GWAS aggregate statistics from three main sets: the Multi-trait Set (MTC) panel; The British biological bank (UKBB) group; Extended Trait Collection (ETC). After quality control, trait-associated gene (TAG) sets for all collected GWAS studies were calculated by Multi-marker Analysis of GenoMic Annotation method. We used three hypothyroidism datasets for analysis. The trait IDs were B1020 (128 cases), B2212 (17,574 cases) and B322 (118 cases).
Druggability evaluation
The DrugBank database (https://go.drugbank.com/)52, was used to evaluate the druggability of identified putatively causal traits. We searched all drugs that targeted the related metabolite and immune cell traits and recorded their drug name, grade, and indications.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
Our research is based on publicly available summary data and all GWAS are referenced in the article and further organized in Supplementary Data 1. Ethical approval and participant consent were obtained in the original studies. Any additional information required to reanalyze the data reported in this report is available from the corresponding author upon reasonable request.
Code availability
The manuscript used public software those are available online. The names of the software are presented in the Methods and the detailed statistical codes can be found in the related resources.
References
Chaker, L. et al. Hypothyroidism. Nat. Rev. Dis. Prim. 8, 1–17 (2022).
Udovcic, M., Pena, R. H., Patham, B., Tabatabai, L. & Kansara, A. Hypothyroidism and the Heart. Methodist Debakey Cardiovasc J. 13, 55 (2017).
Mehuys, E. et al. Investigating levothyroxine use and its association with thyroid health in patients with hypothyroidism: A community pharmacy study. Thyroid 33, 918–926 (2023).
Biondi, B. & Cooper, D. S. Thyroid hormone therapy for hypothyroidism. Endocrine 66, 18–26 (2019).
Shao, F. et al. Plasma metabolomics reveals systemic metabolic alterations of subclinical and clinical hypothyroidism. J. Clin. Endocrinol. Metab. 108, 13–25 (2022).
Theofilopoulos, A. N., Kono, D. H. & Baccala, R. The multiple pathways to autoimmunity. Nat. Immunol. 18, 716–724 (2017).
Rapoport, B. & McLachlan, S. M. Reflections on thyroid autoimmunity: A personal overview from the past into the future. Horm. Metab. Res. 50, 840–852 (2018).
Emdin, C. A., Khera, A. V. & Kathiresan, S. Mendelian randomization. JAMA 318, 1925–1926 (2017).
Chen, Y. et al. Plasma metabolites and risk of seven cancers: A two-sample Mendelian randomization study among European descendants. BMC Med. 22, 90 (2024).
Han, X. et al. Integrating genetics and metabolomics from multi-ethnic and multi-fluid data reveals putative mechanisms for age-related macular degeneration. Cell Rep Med 4, 101085 (2023).
Liu, X. et al. Investigating causal associations among gut microbiota, metabolites and autoimmune hypothyroidism: A univariable and multivariable Mendelian randomization study. Front Immunol 14, 1213159 (2024).
Zuber, V. et al. Combining evidence from Mendelian randomization and colocalization: Review and comparison of approaches. Am. J. Hum. Genet 109, 767–782 (2022).
Zuber, V., Colijn, J. M., Klaver, C. & Burgess, S. Selecting likely causal risk factors from high-throughput experiments using multivariable Mendelian randomization. Nat. Commun. 11, 29 (2020).
Lord, J. et al. Mendelian randomization identifies blood metabolites previously linked to midlife cognition as causal candidates in Alzheimer’s disease. Proc. Natl Acad. Sci. USA 118, e2009808118 (2021).
Dincgez, B., Ercan, I., Sahin, I. & Erturk, N. K. The risk of developing gestational diabetes mellitus in maternal subclinical hypothyroidism: a systematic review and meta-analysis. Arch. Gynecol. Obstet. 309, 765–774 (2024).
Mansournia, N. et al. Subclinical hypothyroidism and diabetic nephropathy in Iranian patients with type 2 diabetes. J. Endocrinol. Invest 40, 289–295 (2017).
Bielecka-Dabrowa, A., Godoy, B., Suzuki, T., Banach, M. & von Haehling, S. Subclinical hypothyroidism and the development of heart failure: an overview of risk and effects on cardiac function. Clin. Res Cardiol. 108, 225–233 (2019).
You, A. S. et al. Impact of thyroid status on incident kidney dysfunction and chronic kidney disease progression in a nationally representative cohort. Mayo Clin. Proc. 99, 39–56 (2024).
Jiang, X. et al. Serum metabolomic analysis in patients with Hashimoto’s thyroiditis. Front Endocrinol. 13, 1046159 (2022).
Dong, H. et al. Serum lipidomic analysis reveals biomarkers and metabolic pathways of thyroid dysfunction. ACS Omega 8, 10355–10364 (2023).
Chaker, L., Bianco, A. C., Jonklaas, J. & Peeters, R. P. Hypothyroidism. Lancet 390, 1550–1562 (2017).
Ragusa, F. et al. Hashimotos’ thyroiditis: Epidemiology, pathogenesis, clinic and therapy. Best. Pr. Res Clin. Endocrinol. Metab. 33, 101367 (2019).
Zhang, Q.-Y. et al. Lymphocyte infiltration and thyrocyte destruction are driven by stromal and immune cell components in Hashimoto’s thyroiditis. Nat. Commun. 13, 775 (2022).
Ajjan, R. A. & Weetman, A. P. Cytokines in thyroid autoimmunity. Autoimmunity 36, 351–359 (2003).
Siegmund, K. et al. Migration matters: regulatory T-cell compartmentalization determines suppressive activity in vivo. Blood 106, 3097–3104 (2005).
Feuerer, M. et al. Lean, but not obese, fat is enriched for a unique population of regulatory T cells that affect metabolic parameters. Nat. Med 15, 930–939 (2009).
Eller, K. et al. Potential role of regulatory T cells in reversing obesity-linked insulin resistance and diabetic nephropathy. Diabetes 60, 2954–2962 (2011).
Dickinson Michael, J. et al. Glofitamab for relapsed or refractory diffuse large B-cell lymphoma. N. Engl. J. Med. 387, 2220–2231 (2022).
McGovern, M. M., Avetisyan, R., Sanson, B.-J. & Lidove, O. Disease manifestations and burden of illness in patients with acid sphingomyelinase deficiency (ASMD). Orphanet J. Rare Dis. 12, 41 (2017).
Asakura, Y., Tachibana, K., Adachi, M., Suwa, S. & Yamagami, Y. Hypothalamo-pituitary hypothyroidism detected by neonatal screening for congenital hypothyroidism using measurement of thyroid-stimulating hormone and thyroxine. Acta Paediatr. 91, 172–177 (2002).
Chen, Y. et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat. Genet 55, 44–53 (2023).
Orrù, V. et al. Complex genetic signatures in immune cells underlie autoimmunity and inform therapy. Nat. Genet 52, 1036–1045 (2020).
Kurki, M. I. et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023).
Zhou, W. et al. Efficiently controlling for case-control imbalance and sample relatedness in large-scale genetic association studies. Nat. Genet 50, 1335–1341 (2018).
Elsworth, B. et al. The MRC IEU OpenGWAS data infrastructure. Preprint at bioRxiv (2020).
Purcell, S. et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet 81, 559–575 (2007).
Bowden, J. et al. Improving the visualization, interpretation and analysis of two-sample summary data Mendelian randomization via the Radial plot and Radial regression. Int J. Epidemiol. 47, 1264–1278 (2018).
Richardson, T. G., Hemani, G., Gaunt, T. R., Relton, C. L. & Davey Smith, G. A transcriptome-wide Mendelian randomization study to uncover tissue-dependent regulatory mechanisms across the human phenome. Nat. Commun. 11, 185 (2020).
Palmer, T. M. et al. Using multiple genetic variants as instrumental variables for modifiable risk factors. Stat. Methods Med Res. 21, 223–242 (2012).
Bowden, J., Davey Smith, G., Haycock, P. C. & Burgess, S. Consistent estimation in Mendelian randomization with some invalid instruments using a weighted median estimator. Genet Epidemiol. 40, 304–314 (2016).
Chen, J. et al. Gastrointestinal consequences of type 2 diabetes mellitus and impaired glycemic homeostasis: A Mendelian randomization study. Diabetes Care 46, 828–835 (2023).
Angrist, J. D., Imbens, G. W. & Rubin, D. B. Identification of causal effects using instrumental variables: Rejoinder. J. Am. Stat. Assoc. 91, 468–472 (1996).
Didelez, V. & Sheehan, N. Mendelian randomization as an instrumental variable approach to causal inference. Stat. Methods Med. Res. 16, 309–330 (2007).
Bowden, J. et al. A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat. Med. 36, 1783–1802 (2017).
Bowden, J., Davey Smith, G. & Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int J. Epidemiol. 44, 512–525 (2015).
Verbanck, M., Chen, C.-Y., Neale, B. & Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet 50, 693–698 (2018).
Hemani, G., Tilling, K. & Smith, G. D. Orienting the causal relationship between imprecisely measured traits using GWAS summary data. Plos Genet 13, e1007081 (2017).
Willer, C. J., Li, Y. & Abecasis, G. R. METAL: Fast and efficient meta-analysis of genomewide association scans. Bioinformatics 26, 2190–2191 (2010).
Foley, C. N. et al. A fast and efficient colocalization algorithm for identifying shared genetic risk factors across multiple traits. Nat. Commun. 12, 764 (2021).
Liu, D. et al. Association between inflammatory bowel disease and cancer risk: evidence triangulation from genetic correlation, Mendelian randomization, and colocalization analyses across East Asian and European populations. BMC Med. 22, 137 (2024).
Dai, Y. et al. CSEA-DB: An omnibus for human complex trait and cell type associations. Nucleic Acids Res. 49, D862–D870 (2021).
Wishart, D. S. et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res. 46, D1074–D1082 (2018).
Acknowledgements
We acknowledge investigators and participants in the CLSA, the SardiNIA project, the FinnGen study, and cited genome-wide association studies for sharing data. This work was supported by National Natural Science Foundation of China (NSFC) grant 82304149 to L.L. and Natural Science Foundation of Zhejiang Province (LQ23H260003) to Z.L.
Author information
Authors and Affiliations
Contributions
Y.S. and S.T. conceived and supervised research. K.L., J.Y., D.M. and Z.L. participated in the data collection. S.Z. and L.L. conducted research and performed statistical analyses. S.Z. wrote the manuscript and L.L. assisted with proofreading the manuscript. Y.S. and S.T. had primary responsibility for final content. All authors discussed the results and approved the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Consent for publication
All the authors have consented to publication.
Ethics approval and consent to participate
The manuscript does not contain clinical studies or patient data. Our study is based on the large-scale GWAS datasets, and not the individual-level data. All participants gave informed consent in all the corresponding original studies, and no additional ethical approval was applicable.
Peer review
Peer review information
Communications Biology thanks Emmanuel Adewuyi, Chaojie Yang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Primary Handling Editors: Ani Manichaikul and Joao Valente. A peer review file is available.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zheng, S., Liu, L., Liang, K. et al. Multi-omics insight into the metabolic and cellular characteristics in the pathogenesis of hypothyroidism. Commun Biol 7, 990 (2024). https://doi.org/10.1038/s42003-024-06680-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06680-x
- Springer Nature Limited