Abstract
Fish sex is largely influenced by steroid hormones, especially sex hormones. Here, we established a steroid hormone-free genetic model by mutation of cyp11a1 in Nile tilapia, which was confirmed by EIA assay. Gonadal phenotype and transcriptome analyses showed that the XX mutants displayed sex reversal from female to male but with defective spermatogenesis. Despite the sex reversal, the aromatase encoding gene cyp19a1a was continuously expressed in the gonads of the XX mutants, which might be caused by androgen deficiency. Whole-mount fluorescence in situ hybridization and transcriptome analysis showed that the gonads of the XX mutants firstly developed towards ovary but shifted to testis between 10 to 15 days after hatching. Detailed expression analysis of key sex differentiation pathway genes foxl3 and dmrt1 combined with apoptosis analysis revealed transdifferentiation of germ cells from female to male during sex reversal. Rescue experiments showed that both P5 and E2 treatment rescued the sex reversal of cyp11a1 mutant XX fish. Overall, our results revealed a transient ovary-like stage and transdifferentiation of germ cells from female to male in the early gonads of the steroid hormone-deprived cyp11a1 mutant XX fish.
Similar content being viewed by others
Introduction
Steroid hormones, including glucocorticoids, mineralocorticoids, estrogens, androgens, and progestins, are important endocrine regulators in vertebrates1. cyp11a1 encodes cholesterol side-chain cleavage enzyme, the only enzyme currently known to catalyze the conversion of cholesterol to pregnenolone (P5), the precursor of all steroid hormones2. Case reports in humans show that mutation in CYP11A1 is lethal due to severe adrenal insufficiency3,4,5,6. The lethality is also observed in rabbits and mouse after Cyp11a1 mutation7,8. These studies in mammals have provided solid evidence for the critical role of Cyp11a1 in steroid hormone production and homeostasis maintenance. Different from the lethality of Cyp11a1 mutation reported in mammals, recent studies have shown that zebrafish cyp11a2 (the counterpart of cyp11a1 in vertebrates) mutants survive to adult9,10. However, unlike other fish species and tetrapods, zebrafish has two cyp11a genes (cyp11a1 and cyp11a2), and both of them are expressed in steroidogenic tissues11. Compensation of cyp11a1 may exist when cyp11a2 is mutated. Therefore, it remains unknown whether cyp11a1 mutants are viable in other fish species.
Fish sex is largely influenced by steroid hormones, especially sex hormones. The essential role of estrogen in fish ovary development has been widely accepted. Blocking estrogen synthesis has been shown to result in the transition of undifferentiated ovary to testis in many fish species12,13,14,15,16,17,18,19,20,21,22, even differentiated ovary to testis in medaka and tilapia23,24. However, the dependence of early ovary differentiation on estrogen seems to vary among different fish species. In medaka, the expression of the aromatase encoding gene cyp19a1a is first detected in the ovary from 4 to 10 days after hatching (dah), after initiation of oogenesis25,26. Blocking estrogen synthesis through aromatase inhibitor (AI) treatment or cyp19a1a mutation does not affect the early oogenesis and folliculogenesis in female medaka21,26. Zebrafish is considered a juvenile hermaphroditic fish, as the gonads develop as juvenile ovary first in both females and males27. Mutation of cyp19a1a, cyp17a1, or cyp11a2 leads to estrogen deficiency and all-male development but does not affect the formation of juvenile ovary and oocyte-like germ cells at early developmental stages in zebrafish10,18,22. These results indicate that estrogen is not necessary for early ovary differentiation in medaka and zebrafish. In Nile tilapia, cyp19a1a expression is detected in the gonads of XX fish at 5 dah, the critical time for tilapia molecular sex differentiation28,29. Blocking estrogen synthesis in XX fish by cyp19a1a or cyp17a1 mutation has been reported to result in female-to-male sex reversal20,30, whether sex reversal caused by estrogen deficiency in tilapia experiences an ovary stage like medaka and zebrafish or experiences an ovary-like stage in gene expression remains to be investigated.
The cyp11a1-free genetic model lacks all steroid hormones. When combining this model with hormone treatment, it is possible to study the physiological effects of a certain hormone. To date, this genetic model has not been established in fish with only one cyp11a gene. Nile tilapia (Oreochromis niloticus) is a gonochoristic fish with an XX/XY sex-determining system. The availability of sex-linked genetic marker31, well-established gene editing technique32, and medium body size which facilitates blood collection and hormone analysis, make tilapia a good model for gene function analysis and endocrine research. In this study, we established a viable steroid hormone-free genetic model in Nile tilapia by mutation of cyp11a1, investigated the role of sex steroid hormone in fish gonad differentiation by analyzing the gonad development of the mutants, combined with rescue experiments and luciferase assay.
Results
Establishment of steroid hormone-free genetic model by mutation of cyp11a1 in tilapia
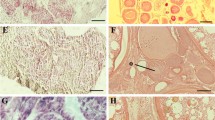
In tilapia, cyp11a1 was mainly expressed in the liver, ovary, testis, and head kidney as revealed by qPCR (Supplementary Fig. 1A). Gonadal transcriptome analysis showed that cyp11a1 displayed sexual dimorphic expression during development, with higher expression in XX gonads at 5, 7, 20, 30, and 40 dah and higher expression in XY gonads at 90 and 180 dah (Supplementary Fig. 1B). In situ hybridization results showed that cyp11a1 was expressed in ovarian theca cells, testicular Leydig cells and head kidney interrenal cells (Supplementary Fig. 1C–K). To get a better understanding of the physiological role of steroid hormones, we established a steroid hormones-free genetic model in tilapia by mutation of cyp11a1 using CRISPR/Cas9 (Fig. 1A, B). Homozygous mutant (cyp11a1−/−) fish with 22 bp deletions were successfully identified by Sau96I digestion and PAGE (Fig. 1C). The deletion was further verified by Sanger sequencing (Fig. 1D). This deletion resulted in a truncated protein lacking adrenodoxin binding domain and heme binding domain due to the premature termination of cyp11a1 translation (Supplementary Fig. 2A). EIA results showed that the cyp11a1−/− fish displayed background levels of serum P5, cortisol, E2, DHP, T and 11-KT compared with wild-type (WT) fish (Fig. 1E–J). Even though the cyp11a1−/− fish displayed decreased survivability under hypoxic conditions (Supplementary Fig. 2B, C), the fish could survive under normal water conditions with enough oxygen. The survival of cyp11a1−/− fish makes it a feasible steroid hormone-free genetic model.
A Simplified diagram displaying the position of cyp11a1 in the steroidogenic pathway. B Target site in cyp11a1. A target site with a Sau96I restriction enzyme site (underlined) was designated in the fourth exon of cyp11a1. The PAM sequence is marked in red. C Mutation identification by Sau96I digestion and PAGE. “+/+”, “+/−”, “−/−” indicate wild-type fish, heterozygous mutants, and homozygous mutants, respectively. D Verification of mutation by Sanger sequencing. A 22 bp deletion was detected in cyp11a1−/− fish. E–J Serum P5, cortisol, E2, DHP, T, and 11-KT level of the WT and cyp11a1−/− fish. Values are presented as mean ± SD (n ≥ 3/genotype). “***” above the error bar in (E, F) indicates statistically significant differences at P < 0.001 as determined by a two-tailed independent Student’s t-test. Different letters above the error bar indicate statistically significant differences at P < 0.05 as determined by one-way ANOVA followed by the Tukey test for multiple comparisons.
Mutation of cyp11a1 results in female-to-male sex reversal of XX fish and defects in spermatogenesis
Histological analyses showed that the gonads of the WT XX fish and WT XY fish at 90 dah developed into ovaries and testes, respectively, while the gonads of both cyp11a1−/− XX and cyp11a1−/− XY fish developed into testes, indicating female to male sex reversal of XX fish after cyp11a1 mutation (Fig. 2A–H). IF results showed that Cyp19a1a was expressed in the gonads of WT XX, but not in the gonads of WT XY and cyp11a1−/− XY fish. Unexpectedly, it was still expressed in the gonads of cyp11a1−/− XX fish. Cyp11c1 was expressed in the gonads of WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish, but not in the gonads of WT XX fish (Fig. 2I–P). To get a comprehensive understanding of gene expression in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish, transcriptome sequencing was performed. Pearson correlation analysis revealed that the global gene expression pattern in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish was highly similar to that in the gonads of WT XY fish but not WT XX fish (Fig. 2Q). Accordingly, more differentially expressed genes were detected in the gonads when comparing cyp11a1−/− XX or cyp11a1−/− XY fish to WT XX fish than comparing to WT XY fish (|log2FC|≥1, P < 0.01) (Fig. 2R). Detailed gene expression analysis revealed downregulation of female pathway gene foxl2, and upregulation of male pathway gene dmrt1 in the gonads of the cyp11a1−/− XX fish compared with WT XX fish (Fig. 2S). Consistent with the IF results, both cyp19a1a and cyp11c1 were expressed in the gonads of the cyp11a1−/− XX fish (Fig. 2S). Further analysis revealed upregulation of steroidogenic factor sf1, steroidogenic acute regulatory protein coding gene star1, star2, steroidogenic enzyme coding genes cyp17a1, hsd3b1, estrogen receptor gene esr2a, androgen receptor genes ar1, ar2, glucocorticoids receptor genes gr1, gr2, progestin receptor gene pgr in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish compared with WT XX and WT XY fish (Fig. 2S). The expression of foxl2, dmrt1, cyp19a1a, and cyp11c1 were further confirmed by qPCR (Fig. 2T). Intriguingly, we found that androgen (11-KT) treatment eliminated the expression of Cyp19a1a in the gonads of the cyp11a1−/− XX fish as revealed by IF (Fig. 3A–E). In luciferase assay, 11-KT treatment inhibited the sf1-activated cyp19a1a transcription via AR1 and AR2. Also, synergistic inhibitory effects were observed for tilapia AR2 and Dmrt1 on cyp19a1a transcription (Fig. 3F).
A–H H&E staining of gonads from the WT XX, WT XY, cyp11a1−/− XX, cyp11a1−/− XY fish at 90 dah. E–H is the higher magnification of the boxed area (black boxes) in (A–D). Black dotted lines outline oogonia (OG), oocytes (OC), spermatogonia (SG), spermatocytes (SC), and spermatids (ST) in the gonads. Scale bar: A–D 50 μm; E–H 20 μm. I–P Immunofluorescence for Cyp19a1a and Cyp11c1 in the gonads of the WT XX, WT XY, cyp11a1−/− XX, cyp11a1−/− XY fish at 90 dah. The positive signal corresponds to green fluorescence. The nuclei were stained with DAPI (blue fluorescence). Scale bar, 50 μm. Q Pearson correlation analysis of gene expression in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. The number inside the square is the Pearson correlation coefficient between two samples. The closer the correlation coefficient gets to 1, the higher the similarity of global gene expression patterns between samples. R The number of DEGs in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. Values on the right side of the column represent the number of up- or down-regulated genes. S The expression of genes related to sex differentiation (foxl2 and dmrt1), steroid hormone synthesis (cyp19a1a, hsd20b2, cyp11c1, sf1, star1, star2, cyp17a1, cyp17a2, and hsd3b1) and response (esr1, esr2a, esr2b, ar1, ar2, gr1, gr2, and pgr) in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. The number inside the square is the RPKM value of genes in each sample. The color from blue to red represents the expression of genes from low to high in each sample. T Relative mRNA expression level of foxl2, dmrt1, cyp19a1a, and cyp11c1 in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah by qPCR (n = 6/genotype). Values are presented as mean ± SD. Different letters above the error bar indicate statistically significant differences at P < 0.05 as determined by one-way ANOVA followed by the Tukey test for multiple comparisons. dah days after hatching.
A–D Immunofluorescence for Cyp19a1a in the gonads of WT XX, WT XY, cyp11a1−/− XX, and 11-KT treated cyp11a1−/− XX fish at 90 dah. The positive signal corresponds to green fluorescence. The nuclei were stained with DAPI (blue fluorescence). Scale bar, 50 μm. E Quantification of average fluorescence intensity of Cyp19a1a positive signal in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and 11-KT treated cyp11a1−/− XX fish at 90 dah (n = 4/genotype). F Effects of AR1, AR2, Sf1, Dmrt1 on the promoter activity of tilapia cyp19a1a (n = 3, technical replicates). Values are presented as mean ± SD. Different letters above the error bar indicate statistically significant differences at P < 0.05 as determined by one-way ANOVA followed by the Tukey test for multiple comparisons. dah days after hatching.
The cyp11a1−/− fish show androgen deficiency raised the question that if the mutants could process spermatogenesis. IF results revealed significantly reduced Vasa-positive germ cells in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish compared to those in the gonads of WT XY fish at 90 dah (Fig. 4A–F). Consistently, gonad transcriptome analysis revealed downregulation of germ cell marker genes vasa, dazl, spermatogonia marker genes nanos2, hells, primary spermatocytes marker genes sycp3, dmc1, secondary spermatocytes marker genes ccna1, aruka, spermatids marker genes tex36, spata18 and mitosis-related genes cenpf, ccnb3, plk1, mis18bp1, kif20a, mastl in the gonads of the cyp11a1−/− XX and cyp11a1−/− XY fish compared with WT XY fish at 90 dah (Fig. 4G). Histological analysis showed that only a few spermatocytes and spermatids exist in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish at 180 dah, indicating severe defects in spermatogenesis (Fig. 4H–M). Notably, both cyp11a1−/− XX and cyp11a1−/− XY fish displayed undersized testes at 90 and 180 dah, as reflected by the significantly decreased cross-sectional area of testes and GSI compared with WT XY fish (Supplementary Fig. 3A–C). To check whether fertile sperm was produced in the mutants, Pap staining, mobility test, and IVF assay were performed. Pap staining and mobility tests showed that no tailed and motile sperm was produced in the gonads of cyp11a1−/− XX and cyp11a1−/− XY fish at 180 dah (Fig. 4N–S). IVF results showed that both cyp11a1−/− XX and cyp11a1−/− XY fish were infertile (Fig. 4T). These results showed that the gonads of steroid hormone-free tilapia developed into testes in both XX and XY fish with defective spermatogenesis.
A–F Immunofluorescence for Vasa in the gonads of the WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. D–F is the higher magnification of the boxed area (white boxes) in (A–C). Red, yellow, and brown dotted lines outline spermatogonia (SG), spermatocytes (SC), and spermatids (ST), respectively. The positive signal corresponds to green fluorescence. The nuclei were stained with DAPI (blue fluorescence). Scale bar: A–C 50 μm; D–F 20 μm. G The expression of germ cell marker genes vasa, dazl, spermatogonia marker genes nanos2, hells, primary spermatocyte marker genes sycp3, dmc1, secondary spermatocyte marker genes ccna1, aruka, spermatid marker genes tex36, spata18, and mitosis-related genes cenpf, ccnb3, plk1, mis18bp1, kif20a, mastl in the gonads of the WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. The number inside the square is the RPKM value of genes in each sample. The color from blue to red represents the expression of genes from low to high in each sample. H–M H&E staining of gonads from the WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 180 dah. K–M is the higher magnification of the boxed area (black boxes) in (H–J). Red, yellow, and brown dotted lines outline spermatogonia (SG), spermatocytes (SC), and spermatids (ST), respectively. Scale bar: H–J 100 μm; K–M 20 μm. N–P Papanicolaou (Pap) staining of testis mash in WT XY, cyp11a1−/− XX, cyp11a1−/− XY fish at 180 dah. Black arrowheads indicate spermatozoa. Scale bar, 10 μm. Q–S Detection of motile sperm in testis homogenate of WT XY, cyp11a1−/− XX, cyp11a1−/− XY fish at 180 dah. The red, green, and blue curves are the trajectories of progressive (PR), non-progressive (NR), and inmotile (IM) cells, respectively. The progressive cells in the testis were usually motile sperm. Scale bar, 50 μm. T Fertility assessment of WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 180 dah. IVF in vitro fertilization. Values are presented as mean ± SD. Different letters above the error bar indicate statistically significant differences at P < 0.05 as determined by one-way ANOVA followed by the Tukey test for multiple comparisons. dah days after hatching.
Transient ovary-like stage during sex reversal of cyp11a1 −/− XX fish
To make the sex reversal process clear, we tracked the expression of the male pathway gene gsdf33,34 in the gonads of the WT XX, WT XY, cyp11a1−/− XX, cyp11a1−/− XY fish at 5, 10, 15, and 25 dah by FISH. The results showed that gsdf was highly expressed in the gonads of the cyp11a1−/− XY and WT XY fish but not in the gonads of the WT XX fish at 5, 10, 15, and 25 dah. In the gonads of the cyp11a1−/− XX fish, gsdf expression was not detected at 5 and 10 dah but gradually upregulated at 15 and 25 dah (Fig. 5A–P, Q). IF results showed that Cyp19a1a was continuously expressed in the gonads of the cyp11a1−/− XX fish at 5, 10, 15, and 25 dah (Fig. 5A–P, R). These results implied that the gonad of the cyp11a1−/− XX fish may first develop toward the ovary but shifted its direction toward the testis during 10 to 15 dah. To investigate whether female germ cells exist in the gonads of the cyp11a1−/− XX fish, we checked the expression of the female germ cell marker gene foxl335 at 15 dah by FISH combined with Vasa antibody staining. The results showed that even though the number of foxl3-positive germ cells in the gonads of the cyp11a1−/− XX was greatly decreased compared with that in the gonads of the WT XX fish, there were still some foxl3-positive germ cells exist, indicating the existence of female germ cells in the gonads of the cyp11a1−/− XX at 15 dah (Fig. 6A–G).
A–P Combined whole-mount fluorescence in situ hybridization for gsdf (red fluorescence) and immunofluorescence for Cyp19a1a (green fluorescence) in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 5, 10, 15, 25 dah. The nuclei were stained with DAPI (blue fluorescence). Scale bar, 50 μm. White dash lines outline the gonads. Q, R Quantification of relative gsdf (Q) and Cyp19a1a (R) positive signal area in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 5, 10, 15, and 25 dah (n = 5/genotype). Values are presented as mean ± SD. Different letters above the error bar indicate statistically significant differences at P < 0.05, as determined by one-way ANOVA followed by the Tukey test for multiple comparisons. dah days after hatching.
A–F Combined whole-mount fluorescence in situ hybridization for foxl3 (red fluorescence) and immunofluorescence for Vasa (green fluorescence) in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah. The nuclei were stained with DAPI (blue fluorescence). Arrowheads indicate the foxl3-positive germ cell. Scale bar, 50 μm. White dash lines outline the boundary of the gonads. G Quantification of foxl3-positive germ cell in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah (n = 6/genotype). H Hierarchical clustering of gene expression in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah. The color from blue to red indicates the expression of genes from low to high. I Schematic diagram displaying two possible fates of the female germ cells (FGC) in the early gonads of cyp11a1−/− XX fish: undergo apoptosis or transdifferentiate into male germ cells (MGC). J Volcano plot of DEGs in the gonads of the WT XX and cyp11a1−/− XX fish. K GO enrichment analysis of the DEGs in the gonads of the WT XX and cyp11a1−/− XX fish. L The expression of genes related to mitosis (pcna, ccnf, ccnb3, plk1, mis18bp1, kif20a, and mastl) and meiosis (rec8a, dmc1, and meioc) in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish. The number inside the square is the RPKM value of genes in each sample. The color from blue to red represents the expression of genes from low to high in each sample. M–T TUNEL staining in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah. The positive signal corresponds to red fluorescence. Antibody against Vasa was used to stain the germ cell (green fluorescence), and the nuclei were stained with DAPI (blue fluorescence). White dash lines outline the gonads. Scale bar, 50 μm. U Quantification of TUNEL-positive signal area in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish (n = 4/genotype). Values are presented as mean ± SD. Differences were determined by one-way ANOVA followed by Tukey test for multiple comparisons. ns not significant. V–A’ Colocalization of foxl3 and Dmrt1 in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah. The nuclei were stained with DAPI (blue fluorescence). White dash lines outline the boundary of the gonads. FGC female germ cells express foxl3 (red fluorescence), MGC male germ cells express Dmrt1 (green fluorescence), TGC germ cells undergoing transdifferentiation expression both foxl3 and Dmrt1. Scale bar: V–X 50 μm; Y–A’ 12.5 μm. dah days after hatching.
Transcriptome sequencing was performed to investigate the global gene expression profile in the gonads of the cyp11a1−/− XX fish at 15 dah. Totally, we detected 1385 DEGs (|log2FC|≥1, P < 0.05) between the gonads of the WT XX and WT XY fish, of which 871 and 514 genes were highly expressed in XX and XY gonads, respectively (Supplementary Fig. 4A). Clustering gene expression analysis revealed similar expression pattern of these DEGs in the gonads of the WT XX and cyp11a1−/− XX fish (Fig. 6H). The delayed gsdf upregulation, retained foxl3-positive germ cells, and ovary-like global gene expression pattern demonstrated that the gonads of the cyp11a1−/− XX fish experienced an ovary-like stage.
Transdifferentiation of germ cells from female to male during sex reversal in cyp11a1 −/− XX fish
Two possible fates of female germ cells exist in the early gonads of cyp11a1−/− XX fish during sex reversal: either undergo apoptosis or transdifferentiate into male germ cells (Fig. 6I). To determine the exact germ cell fate, we first analyzed the gonadal transcriptome of the cyp11a1−/− XX and WT XX fish at 15 dah. We detected 403 upregulated and 629 downregulated genes in the gonads of the cyp11a1−/− XX fish and WT XX fish (Fig. 6J). Go enrichment analysis showed that the upregulated genes were mainly involved in chemokine activity, chemokine receptor binding, myofibril, contractile fiber, and cytokine activity (Fig. 6K). These genes are mainly related to immune and inflammatory responses. Notably, no apoptosis-related gene was significantly upregulated and enriched (Supplementary Fig. 4B and Supplementary Table 1). The downregulated genes are mainly involved in phosphatidic acid biosynthesis and metabolism, meiosis, thyroid hormone response, and hepoxilin metabolic process (Fig. 6K and Supplementary Table 2). Detailed gene expression analysis revealed decreased expression of genes related to germ cell proliferation (pcna, ccnf, ccnb3, plk1, mis18bp1, kif20a, and mastl) and meiosis (rec8a, dmc1, and meioc) (Fig. 6L). TUNEL staining was also performed to evaluate the apoptotic feature in the gonads of cyp11a1−/− XX fish. Consistent with the transcriptome data, we did not detect any increase of apoptosis in the gonads of cyp11a1−/− XX fish at 15 dah (Fig. 6M–U). In tilapia, foxl3 has been proven to determine germ cell fate by antagonizing with dmrt135. Therefore, we checked the colocalization of foxl3 and Dmrt1 in the gonads of the cyp11a1−/− XX fish by FISH combined with IF. The results showed that both foxl3 and Dmrt1 were expressed in the gonads of the cyp11a1−/− XX fish at 15 dah, and they were colocalized in some germ cells (Fig. 6V–A’). These results demonstrate that the female germ cells in the gonads of the cyp11a1−/− XX fish did not undergo apoptosis, but directly transdifferentiated into male germ cells during sex reversal.
P5 and E2 treatment rescued the sex reversal of cyp11a1 −/− XX fish
The direct product of Cyp11a1 (P5) and two main female hormones (E2, DHP) were used to rescue the sex reversal of cyp11a1−/− XX fish. FISH results from fish at 25 dah showed that both P5 and E2 treatment rescued the sex reversal of cyp11a1−/− XX fish as indicated by the disappearance of gsdf expression in the gonads (Fig. 7A–E), while DHP treatment failed to rescue the sex reversal of cyp11a1−/− XX fish (Fig. 7F). Histological analyses showed that a large number of oocytes were present in the gonads of WT XX fish, as well as in P5- and E2-treated cyp11a1−/− XX fish at 90 dah, while no oocytes was found in the gonads of cyp11a1−/− XX fish without treatment, further confirming the rescue effects of P5 and E2 (Fig. 7G–J).
A–F Whole-mount fluorescence in situ hybridization of gsdf in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and P5, E2, or DHP-treated cyp11a1−/− XX fish at 25 dah. The positive signal corresponds to red fluorescence. The nuclei were stained with DAPI (blue fluorescence). White dash lines outline the boundary of the gonads. Scale bar, 50 μm. G–J H&E staining of gonads from WT XX, cyp11a1−/− XX, and P5 or E2-treated cyp11a1−/− XX fish at 90 dah. OC oocytes. Scale bar: G, I, J 40 μm; H 20 μm. dah days after hatching.
Discussion
cyp11a1 is the only gene currently known to catalyze the production of P5, the precursor of all steroid hormones. In tilapia, only one cyp11a1 gene exists, as reported in a previous study11. In this study, we established a steroid hormone-free genetic model by mutation of cyp11a1 in tilapia. Even though basal levels of steroid hormones were detected in cyp11a1 mutant fish, it was more likely due to the non-specificity of the EIA kit. The mutant fish displayed decreased survivability under hypoxic conditions, but they survived under normal water conditions with enough oxygen. Histological analysis, transcriptome analysis, IF, and qPCR analyses revealed female-to-male sex reversal of XX mutants. Detailed FISH and transcriptome analyses captured a transient ovary-like stage at the molecular level and transdifferentiation of germ cells from female to male in the early gonads of the XX mutants.
Fish sex is largely influenced by steroid hormones, especially sex hormones36,37,38,39,40,41,42,43. Regarding the way that sex hormones determine sex in fish, there are two theories, which we call the balance theory36,37 and the absence theory40,41. The former holds that fish sex is determined by the ratio of estrogen to androgen in the gonads during sexual differentiation, while the latter believes that fish sex is determined by the presence or absence of estrogen during sex differentiation. The discrepancy between the two lies in whether androgen is involved in sex determination. With the deepening of research in fish sex determination, the absence theory has been widely supported. In hermaphroditic fish, sex change from male-to female (protandrous) or from female to male (protogynous) is associated with the upregulation or downregulation of estrogen levels, respectively44,45,46,47,48,49. In gonochoristic fish, it has been suggested that cyp19a1a is expressed in the gonads of genetic female fish at the critical window of sex determination28,29,39. Blocking estrogen synthesis through AI treatment12,13,14,15,16,17,50,51,52 or gene mutation16,18,19,20,22,30,53 leads to female-to-male sex reversal, and E2 treatment41,42,54 leads to male-to-female sex reversal in many fish species. In this study, we established a steroid hormone-free genetic model by mutation of cyp11a1 in tilapia. We found that the estrogen- and androgen-deprived cyp11a1 mutant XX and XY fish developed as males, consistent with the phenotype of all-male development in zebrafish after cyp11a2 mutation9,10. Rescue experiments showed that E2 treatment rescued the sex reversal of the cyp11a1 mutant XX fish, indicating that it is the presence or absence of estrogen determines the sex. The facts that the testicular fate of the gonads of cyp11a1 mutant XY fish was not affected and the gonads of cyp11a1 mutant XX fish could still transform into testes even in the absence of androgens supports the point that androgen is not involved in fish sex determination. Most importantly, no steroidogenic/androgenic enzyme genes were found to be expressed in the WT XY gonads of tilapia by IHC, IF, qPCR, and transcriptomic analyses28,29, that explains why the testicular fate of cyp11a1 mutant XY fish was not affected in the present study. These results support the absence theory. Even though androgen treatment has been reported to result in masculinization or female-to-male sex reversal in some fish species, such as Japanese flounder, tilapia, rainbow trout, medaka, zebrafish, and orange-spotted grouper17,50,55,56,57,58, no sex reversal has been reported when androgen synthesis or androgen receptor is disrupted59,60,61,62,63,64,65,66. The androgen-induced sex reversal in fish is thought to be achieved by inhibition of female pathway genes67 or induction of male pathway genes56. In this study, we observed a continuous expression of cyp19a1a in the gonads of the cyp11a1 mutant XX fish. The expression of cyp19a1a was eliminated by 11-KT treatment. Luciferase assay revealed direct inhibition of androgen to the sf1-activated cyp19a1a transcription via AR1 and AR2 and synergistic inhibitory effects of androgen and dmrt1 to cyp19a1a transcription. Taken together, our results confirm that it is estrogen, not androgen, that determines the sex of fish. The androgen-induced sex reversal in fish can be mediated by the inhibition of Sf1-activated cyp19a1a transcription.
Even though the role of estrogen in ovary development has been widely accepted, the dependence of early ovary differentiation on estrogen seems to vary among different fish species. In medaka, cyp19a1a expression is detected after the initiation of oogenesis25,26. Blocking estrogen synthesis through AI treatment leads to female-to-male sex reversal but does not affect early oogenesis26. The gonad of cyp19a1a mutant female medaka first developed into an ovary but gradually transformed into a testis with the degeneration of ovarian tissue21. In zebrafish, blocking estrogen synthesis by mutation of cyp19a1a, cyp17a1, or cyp11a2 leads to all-male development but the juvenile ovary and oocyte-like germ cells formed at early developmental stage10,18,22. These results demonstrate that sex reversal caused by estrogen deficiency in medaka and zebrafish experience an ovary stage. Gonad differentiation is a continuous process involving expression changes of a large number of genes. In tilapia, the molecular differentiation of the gonads initiates around 5 dah, and the first sign of morphological differentiation of the gonads occurs around 23 to 26 dah28. Previously, the morphological features of gonads or the expression of few marker genes were used to characterize the sex reversal process of cyp19a1a, cyp17a1 mutant fish in tilapia20,30, which ignored the global gene expression profiles. In this study, we characterized the sex reversal process of the cyp11a1 mutant XX fish comprehensively at a molecular level. Transcriptome analysis showed that at 15 dah, the global gene expression pattern in the gonads of the mutant XX fish was relatively more similar to those of the WT XX fish, but at 90 dah, it was relatively more similar to those of the WT XY fish. Consistently, FISH analyses revealed that the expression of gsdf was detected in the gonads of WT XY fish at 5 dah, but it was not detected in the gonads of the mutant XX fish until 10–15 dah. Even at 15 dah, foxl3 was still expressed in the gonads of the mutant XX fish. These results demonstrate that the steroid hormone-deprived sex reversal of cyp11a1 mutant XX fish in tilapia experiences an ovary-like stage at a molecular level. The fact that E2 treatment rescued the sex reversal of the cyp11a1 mutant XX fish in this study indicates that the cause of the sex reversal of the cyp11a1 mutant XX fish is estrogen deficiency. Whether the sex reversal caused by estrogen deficiency experience an ovary-like stage at molecular level in other genetic model in tilapia and other fish species is worthy of investigation.
Transdifferentiation describes a direct transition of one differentiated cell type to another differentiated cell type during development68, and it has been observed in adult gonads in both mammals and fishes during sex reversal. In mouse, Foxl2 ablation in adults leads to transdifferentiation of ovarian granulosa and theca cells into testicular Sertoli-like and Leydig-like cells69, and Dmrt1 ablation leads to transdifferentiation of Sertoli cells into granulosa cells70. In tilapia, AI treatment of XX fish from 90 to 180 dah induces the successful transition of the differentiated ovary into the testis, during which transdifferentiation is observed in somatic cells24. Whether germ cells undergo transdifferentiation in the early gonads during sex reversal is worth investigating. In this study, we captured a transient ovary-like stage and witnessed the existence of foxl3-positive female germ cells in the early gonads of the steroid hormone-deprived cyp11a1 mutant XX fish. In theory, these foxl3-positive female germ cells have two different fates during sex reversal: either undergo apoptosis or transdifferentiate into male germ cells. Detailed gene expression analysis of apoptosis-related genes and TUNEL staining excluded the possibility of apoptosis in these cells. Recently, we have proved that in tilapia, foxl3 is expressed in female germ cells but not in male germ cells. dmrt1 is expressed both in male germ cells and somatic cells but not in female germ cells and somatic cells. The sexual fate of tilapia germ cells is determined by the antagonistic interaction of dmrt1 and foxl335. In this study, we detected colocalization of foxl3 and dmrt1 in germ cells in the early gonads of the mutant XX fish, which has never been detected in the WT XX and WT XY gonads. The germ cells expressing both foxl3 and dmrt1 may be undergoing transdifferentiation. Even though we cannot rule out the possibility that male germ cells in the early gonads of cyp11a1 mutant XX fish can be directly differentiated from primordial germ cells, we demonstrate that germ cells can undergo transdifferentiation at the early stage of steroid hormone-deprived sex reversal in this study.
In summary, we established a steroid hormone-free genetic model in tilapia by mutation of cyp11a1. We showed that the gonads of steroid hormone-free tilapia developed into testis in both XX and XY fish with defective spermatogenesis. The gonads of the mutant XX fish first developed toward the ovary, but shifted to the testis during 10–15 dah. Due to the simultaneous loss of androgen and estrogen, cyp19a1a was continuously expressed in the testis of the mutant XX fish. Both P5 and E2 treatment rescued the sex reversal of the mutant XX fish (Fig. 8). This study revealed a transient ovary-like stage and transdifferentiation of germ cells from female to male in the early gonads of tilapia after steroid hormone deprivation.
A steroid hormone-free tilapia model was established by mutation of cyp11a1. Both XX and XY cyp11a1 mutants developed as infertile males due to defects in spermatogenesis. The gonads of the cyp11a1 mutant XX fish first developed toward the ovary but shifted to the testis during 10 to 15 dah. Both P5 and E2 treatment rescued the ovary development of the cyp11a1 mutant XX fish. Due to the simultaneous loss of androgen, cyp19a1a was continuously expressed in the gonads of the cyp11a1 mutant XX fish. PGC primordial germ cells, OG oogonia, SG spermatogonia, OC oocytes, SC spermatocytes, ST spermatids, SZ spermatozoa, Gc granulosa cells, PSc pre-supporting cells, FSc female supporting cells, MSc male supporting cells, PSgc pre-steroidogenic cells, Ic interstitial cells, Lc Leydig cells.
Methods
Animals
Nile tilapia (O. niloticus) used in this study was first introduced by Prof. Nagahama (Laboratory of Reproductive Biology, National Institute for Basic Biology, Okazaki, Japan) and kept in aerated recirculating freshwater tanks at 26 °C under a natural photoperiod in our lab. Animal experiments were conducted in accordance with the regulations of the Guide for Care and Use of Laboratory Animals and were approved by the Committee of Laboratory Animal Experimentation at Southwest University.
Establishment of cyp11a1 mutant line
The sequence of tilapia cyp11a1 (Gene ID: 100692956) was obtained from NCBI. The target site with a Sau96I restriction site (underlined) was designed in the 4th exon of cyp11a1 using the online software ZiFit (http://zifit.partners.org/ZiFiT/). The guide RNA and Cas9 mRNA were prepared as previously reported32. Embryos at the one-cell stage were co-injected with gRNA and Cas9 mRNA with a final concentration of 500 and 1000 ng/µl, respectively. After mutation screening by polyacrylamide gel electrophoresis (PAGE) and Sanger sequencing, F0 chimeric adult XY males were crossed with wild-type (WT) XX females to produce F1 progeny. Siblings in F1 carrying a 22 bp deletion at the same locus were incrossed to produce F2 progeny. Sau96I digestion and PAGE was used for mutation screening in the F2 population. The genetic sex of each fish was identified by a sex-linked marker as described previously31. Primers used for gRNA synthesis, mutation screening, and genetic sex identification are listed in Supplementary Table 3. After mutation screening, the proportion of cyp11a1 homozygous mutant (cyp11a1−/−) fish in the F2 population at 5, 15, 25, 35, and 90 dah was calculated (n = 4 populations at each time point, more than 48 fish were randomly selected in each population).
Measurement of suffocation point
After genotyping, the 90-dah-old WT XY and cyp11a1−/− XY fish (n = 6/genotype) with similar body size (body length: WT XY 6.63 ± 0.34 cm, cyp11a1−/− XY 6.60 ± 0.30 cm; body weight: WT XY 10.37 ± 0.90 g; cyp11a1−/− XY 10.35 ± 0.78 g) were kept in a tank containing 20 L of water at 26 °C. Oxygen in the water was deprived by continuous filling of nitrogen. The suffocation point, which refers to the minimal oxygen concentration at which the fish loses balance, was determined by using the average value of six fish after measurement of the dissolved oxygen concentration when each fish loses balance. The dissolved oxygen concentration was measured by HQ30D portable dissolved oxygen meter (HACH, Colorado, USA).
Measurement of steroid hormones
Enzyme immunoassay (EIA) kit was used to measure the serum P5 (Spbio, Wuhan, China) and cortisol level of WT XY and cyp11a1−/− XY fish, and serum estradiol (E2), 17,20 β-dihydroxy-4-pregnen-3-one (DHP), testosterone (T) and 11-ketotestosterone (11-KT) (Cayman, Michigan, USA) level of WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah. After anesthesia with MS-222 (250 mg/L, Sigma-Aldrich, Missouri, USA), blood was collected from the caudal vasculature of fish from each genotype (serum from three fish as one sample, n ≥ 3/genotype) and placed at 4 °C overnight to allow clot. The serum was separated by blood centrifugation (1000×g, 10 min, 4 °C) and stored at −80 °C. EIA was performed according to the manufacturer’s instructions. All samples were run in triplicate on one 96-well plate to ensure the comparability of each assay.
Quantitative real-time PCR
Twelve tissues (brain, pituitary, gill, heart, spleen, liver, intestine, ovary, testis, kidney, head kidney, and muscle) of WT XX and WT XY fish at 180 dah (n = 4/sex) were collected for expression analysis of cyp11a1 in different tissues. Gonads of WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah were collected for gene expression analysis of foxl2, dmrt1, cyp19a1a, and cyp11c1 (n = 6/genotype). Total RNA was isolated using RNAiso Plus (Takara, Tokyo, Japan). RNA quality and concentration were determined using NanoDrop 2000, and ≥500 ng total RNA was used for cDNA synthesis with PrimeScript RT Master Mix Perfect Real Time kit (Takara, Tokyo, Japan). Quantitative real-time PCR (qPCR) was performed using a TB Green Premix Ex Taq II kit (Takara, Tokyo, Japan). All experiments were performed according to the manufacturer’s instructions. The value was detected on the StepOne Plus Real-time PCR system (Thermo Fisher Scientific, Massachusetts, USA). Gene expression was normalized to β-actin using the 2−ΔΔCt method71. Primers used for qPCR are listed in Supplementary Table 3.
In situ hybridization
In situ hybridization was performed to investigate the cellular location of cyp11a1 in gonads and head kidneys. Ovaries, testes, and head kidneys of WT fish at 240 dah were dissected and fixed in 4% PFA (4 °C, overnight). After fixation, the gonads were embedded in paraffin and sliced into 5 µm sections (cross-section) for use. The digoxigenin (DIG)-labeled sense and antisense probes of cyp11a1 were prepared as follows: a fragment (604 bp) of cyp11a1 were amplified from cDNA by PCR and recovered. After subjected to TA clone, the plasmid containing T7 promoter and insertion or inverted insertion of cyp11a1 fragment were extracted and amplified by PCR. The sense and antisense probes were synthesized by in vitro transcription using the recovered PCR products as templates and a kit containing T7 RNA polymerase and DIG-labeled rNTP mix according to the manufacturer’s instructions (Roche, Basel, Switzerland). The in situ hybridization were performed as follows: Briefly, after deparaffinization in xylene, the slides were placed in 100, 90, 80, and 70% ethanol and 1× PBS for rehydration. Then, the slides were fixed in 4% PFA again, and rinsed in 2 mg/mL glycine, 0.1 M TEA, 0.25% acetic anhydride/0.1 M TEA, successively. Next, the slides were placed in pre-hybridization buffer (66% formamide; 10% 20× SSC) at 65 °C for 2 h and in hybridization buffer (60% formamide; 7.5% dextran sulfate; 0.3 M NaCl, 0.02 M Tris-HCL PH 8.0; 0.0025 M EDTA; 1X Denhardt solution) containing tRNA (20 μg/mL) and antisense probe (500 ng/mL) for hybridization (65 °C, 16 h). After hybridization, the slides were washed stringently with 50% formamide/ 2× SSC, 1× SSC, 0.2× SSC, DIG buffer (0.1 M Maleic acid; 0.15 M NaCl; PH 7.4) and blocked in DIG buffer containing 5% BSA at 37 °C for 1 h. After these steps, the slides were incubated with anti-DIG-AP (Roche, Basel, Switzerland) at 4 °C overnight. Finally, the slides were rinsed in a DIG buffer five times and hatched in a detection buffer (1 M Tris-HCL PH 9.5; 5 M NaCl; 1 M MgCL2) containing NBT/BCIP (Roche, Basel, Switzerland) for signal detection. Images were captured under a BX53 microscope (Olympus, Tokyo, Japan). Primers used for probe preparation are listed in Supplementary Table 3.
Hematoxylin and eosin staining
Gonads from WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah and WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 180 dah were dissected and fixed in Bouin’s solution (24 h, room temperature). After fixation, gonads were embedded in paraffin and sliced into 5 µm sections (middle cross-section) for use. For hematoxylin and eosin (H&E) staining, tissue sections were firstly deparaffinized in xylene and rehydrated in decreasing concentration of ethanol. Then, tissue sections were stained with hematoxylin and eosin successively. Finally, sections were dehydrated in increasing concentration of ethanol and mounted. Images were captured under a BX51 optical microscope (Olympus, Tokyo, Japan). The cross-sectional area of testes of WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 and 180 dah was quantified by Image J Pro 1.51 software using default parameters (n = 6 fish/genotype). The gonadosomatic index (GSI) of WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 180 dah was calculated as (gonad weight/body weight) × 100% (n = 6 fish/genotype).
Papanicolaou staining, mobility test, and fertilization assay
Since no sperm could be squeezed out from the genital pore of cyp11a1 mutants at 180 dah, the testes of WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish were dissected and minced with scissors for further Papanicolaou (Pap) staining, mobility test and in vitro fertilization (IVF) assay to check whether fertile sperm was produced in the mutants (n = 6/genotype). For Pap staining, the homogenates were firstly fixed in 4% PFA for 10 min then stained with Papanicolaou solution EA50 (Solarbio, Beijing, China) for 3 min. Images were captured under a BX51 optical microscope (Olympus, Tokyo, Japan). For the mobility test, the homogenates were mixed with 20 times the volume of water and analyzed under a Sperm Quality Analyzer (Zoneking Software, Beijing, China). For the IVF assay, the homogenates of WT and mutant fish was used to inseminate eggs from WT XX fish. The embryos were checked at 16 h post-fertilization under Leica M205 FA Stereomicroscope (Leica, Wetzlar, Germany) to assess whether fertilization is successful. The fertilization rate was calculated as (the number of eggs with normal blastoderm at 16 hpf/the total number of eggs) × 100%.
Immunofluorescence
Immunofluorescence (IF) was performed to evaluate the expression of Cyp19a1a, Cyp11c1, and Vasa in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah (n = 4/genotype). Gonads were dissected and fixed in Bouin’s solution (24 h, room temperature). After fixation, gonads were embedded in paraffin and sliced into 5 µm sections (cross-section) for use. After deparaffinization and hydration, sections were subjected to antigen retrieval and blocked in donkey serum (37 °C, 1 h). Sections were then incubated with primary antibody (37 °C, 1 h). The rabbit polyclonal antibodies against Cyp19a1a (2 mg/mL, 1:2000) and Cyp11c1 (2 mg/mL, 1:500) were produced by our lab, and their specificity has been verified previously24,65. Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody (1:500, Thermo Fisher Scientific, Massachusetts, USA) was used to detect the primary antibodies (37 °C, 40 min). The nuclei were stained with DAPI (1:1000, Sigma-Aldrich, Missouri, USA). Images were captured under the FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan).
Whole-mount fluorescence in situ hybridization and immunofluorescence
Whole-mount fluorescence in situ hybridization (FISH) and IF were combined to track the expression of gsdf and Cyp19a1a in the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 5, 10, 15, 25 dah (n = 5/genotype) and the expression of foxl3 and Dmrt1 in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah (n = 6/genotype). The DIG-labeled antisense probes of foxl3 and gsdf were prepared as described in the in situ hybridization section of this study. Primers used are listed in Supplementary Table 3. The rabbit antibodies against tilapia Cyp19a1a (2 mg/mL, 1:2000), Dmrt1 (2 mg/mL, 1:500), and Vasa (2 mg/mL, 1:1000) were produced by our lab, and their specificity has been verified previously24,35. After removing the viscera, fish with gonads were fixed in 4% PFA at 4 °C overnight, and placed in 100% methanol at −20 °C for at least 2 h. Then, the fish were placed in 75, 50, 25% methanol, and 1× PBS successively for rehydration and in a 3% H2O2/0.5% KOH medium under light to remove pigment (room temperature, 10 min). After permeabilization in precooled acetone (−20 °C, 30 min), the fish were rinsed four times in 1× PBS and were placed in pre-hybridization buffer (50% formamide; 25% 20× SSC; 0.01 M citric acid; 0.1% Tween-20) at 65 °C for 2 h. Subsequently, the fish were placed in a pre-hybridization buffer containing tRNA (500 μg/mL), heparin (50 μg/mL), and an antisense probe (50–500 ng/mL) for hybridization (65 °C, 16 h). After hybridization, the fish were washed stringently with pre-hybridization buffer, 1× SSC, 0.2× SSC, TN buffer (0.1 M tris-HCL; 0.15 M NaCl; PH 7.5) successively and blocked in TN buffer containing blocking regent (Lot 46925300, Roche, Basel, Switzerland) and 5% donkey serum (37 °C, 1 h). Next, the fish were incubated with anti-DIG-POD (Roche, Basel, Switzerland) at 4 °C overnight. For signal detection, tyramide signal amplification (TSATMR) was performed according to the manufacturer’s instructions (Akoya Biosciences, Marlborough, USA). After TSA amplification, the fish were rinsed four times in 1× PBS and incubated with primary antibody (37 °C, 1 h). Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody (1:500, Thermo Fisher Scientific, Massachusetts, USA) was used to detect the primary antibody (37 °C, 40 min). The nuclei were stained with DAPI (1:1000, Sigma-Aldrich, Missouri, USA). Images were captured under the FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan). The positive signal of gsdf and Cyp19a1a was quantified by Image J Pro 1.51 software using default parameters. The zones selected for quantification were in the middle of the gonads. The number of foxl3-positive germ cells was quantified manually.
TUNEL staining
Terminal deoxynucleotidyl transferase (TdT) dUTP Nick-End Labeling (TUNEL) kit (Roche, Basel, Switzerland) was used to assess the apoptosis in the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah (n = 4/genotype). Rabbit antibody against tilapia Vasa (2 mg/mL, 1:500) was used to mark the germ cells. The TUNEL staining was conducted as follows: after removing the viscera, fish with gonads were fixed in 4% PFA at 4 °C overnight, and placed in 100% methanol at −20 °C for at least 2 h. Then, the fish were placed in 75, 50, 25% methanol, and 1× PBS successively for rehydration. After permeabilization in 0.1% triton X-100 (37 °C, 10 min), fish were rinsed four times in 1× PBS and were incubated with TUNEL reaction mixture containing 5 μL enzyme solution and 45 μL label solution (37 °C, 1 h). The positive control was treated with 3 U/mL DNase I (37 °C, 10 min) before reaction mixture incubation. The negative control was incubated only with 50 μL label solution. After TUNEL incubation, the fish were rinsed four times in 1× PBS and blocked in 5% donkey serum (37 °C, 1 h). Fish were next incubated with Vasa antibody (37 °C, 1 h). Alexa Fluor 488-conjugated donkey anti-rabbit secondary antibody (1:500, Thermo Fisher Scientific, Massachusetts, USA) was used to detect the primary antibody (37 °C, 40 min). The nuclei were stained with DAPI (1:1000, Sigma-Aldrich, Missouri, USA). Images were captured under the FV3000 confocal laser scanning microscope (Olympus, Tokyo, Japan). The positive signal of TUNEL in the gonads was quantified using Image J Pro 1.51 software.
Transcriptome analyses
The expression of cyp11a1 in the gonads of tilapia at 5, 7, 20, 30, 40, 90, and 180 dah was determined by analyzing the gonadal transcriptome in our lab29,72. To evaluate the global gene expression in the gonads of the cyp11a1 mutants, the gonads of the WT XX, WT XY, and cyp11a1−/− XX fish at 15 dah (pooled sample, 30 fish/genotype) and the gonads of the WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah (pooled sample, 4 fish/genotype) were collected. After total RNA was extracted, mRNA was enriched by Oligo(dT) beads. Then the enriched mRNA was reversely transcribed into cDNA and disrupted into short fragments for cDNA library construction. The resulting cDNA library was sequenced on Illumina Novaseq 6000 by Gene Denovo Biotechnology Co. (Guangzhou, China). Raw reads were filtered by fastp to remove adapters and low-quality reads. Reads were submitted to the NCBI SRA database (Accession number: PRJNA1074672). Clean reads from each library were aligned to the reference genome (https://www.ncbi.nlm.nih.gov/datasets /genome/ GCF_001858045.2/) using HISAT2 with default parameters73. The reads per kb per million mapped reads (RPKM) method was used to quantify the gene expression level. Pearson correlation analysis of gene expression in the gonads of WT XX, WT XY, cyp11a1−/− XX, and cyp11a1−/− XY fish at 90 dah was performed to evaluate the sample relationship. It was calculated by sample relationship analysis tools on the Omicshare web server (https://www.omicshare.com) based on the global gene expression of each sample. Differential expression analysis was performed by edgeR software74. The p values was calculated according to the suggested BCV (square-root-dispersion) value (BCF = 0.1). The log2 (FC, fold change) ≥2, P < 0.01 (90 dah), or P < 0.05 (15 dah) were set as the threshold for significantly differential expression. GO enrichment analysis and clustering gene expression analysis were also finished on the Omicshare web server (https://www.omicshare.com).
Rescue experiment and androgen treatment
The direct product of Cyp11a1 (P5) and two main female hormones (E2, DHP) were used to rescue the sex reversal of cyp11a1−/− XX fish. After randomized grouping, the fish with mixed genotypes were firstly raised in aerated water containing 100 µg/L P5, E2, or DHP or the same volume of ethanol (ctrl group) from 5 to 25 dah. The rescue effect of P5, E2, and DHP was first evaluated by FISH at 25 dah using gsdf antisense probe (n = 5/genotype). After evaluation, the successfully rescued groups were subjected to treatment with the same hormone until 90 dah. The treatment of fish from 25 to 90 dah was conducted with a diet containing P5 or E2 at a dosage of 50 µg/g. The gonad phenotype of rescued cyp11a1−/− XX fish was evaluated by histological analysis (n = 5/genotype). Androgen treatment of cyp11a1−/− XX fish was conducted from 75 to 90 dah with a diet containing 11-KT at a dosage of 50 μg/g. The expression of Cyp19a1a was analyzed by IF after treatment (n = 4/genotype). The positive signal of Cyp19a1a was quantified by Image J Pro 1.51 software using default parameters. The zones selected for quantification were the whole gonads.
Dual-luciferase reporter assay
The promoter of cyp19a1a and the open reading frame of sf1, dmrt1 were cloned into pGL3 and pcDNA3.1 vector, respectively, in our previous study75,76. The open reading frame of ar1 and ar2 was inserted into Hind III and Xho I restriction sites of the pcDNA3.1 vector in this study. The Luciferase assay was conducted as follows: HEK293 cells were plated on a 24-well plate at a density of 1 × 106 cells per well. After growing to 70–80% confluence, cells were transfected with pGL3-cyp19a1a (250 ng/well), pcDNA3.1-sf1, -dmrt1, -ar1, -ar2 (100 ng/well), and pRL-TK (internal control, 50 ng/well) constructs using Lipofectamine 2000 (Invitrogen, California, USA). At 12 h post-transfection, 200 ng 11-KT was added to each well to activate the ARs. At 48 h post-transfection, the cells were harvested and lysed. The firefly luciferase enzyme activity (pGL3-) was measured and normalized to the Renilla luciferase enzyme activity (pRL-TK) according to the manufacturer’s instruction (Promega, Wisconsin, USA). Primers used for plasmid construction are listed in Supplementary Table 3.
Statistics and reproducibility
Values are presented as mean ± SD. A two-tailed independent Student’s t-test was used to determine the differences between the two groups. One-way ANOVA, followed by Tukey multiple comparison, was used to determine the significance of differences in more than two groups. All analyses were performed using SPSS 22.0 (IBM, New York, USA). P < 0.05 was used as a threshold for statistically significant differences. Experiments except transcriptome sequencing in this study were performed twice to ensure the reproducibility.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Data availability
RNA-seq data reported in this article could be accessed at the NCBI SRA database (Accession number: PRJNA1074672). The newly generated plasmids, along with maps and sequences, could be obtained in Addgene (ID: 225219, 225220). Supplementary information file contains Supplementary Figs. Supplementary Data 1 contains Supplementary Tables and the source data for all graphs in this study.
References
Miller, W. L. Molecular biology of steroid hormone synthesis. Endocr. Rev. 9, 295–318 (1988).
Hu, M. C., Hsu, H. J., Guo, I. C. & Chung, B. C. Function of Cyp11a1 in animal models. Mol. Cell Endocrinol. 215, 95–100 (2004).
Hiort, O. et al. Homozygous disruption of P450 side-chain cleavage (CYP11A1) is associated with prematurity, complete 46,XY sex reversal, and severe adrenal failure. J. Clin. Endocrinol. Metab. 90, 538–541 (2005).
al Kandari, H., Katsumata, N., Alexander, S. & Rasoul, M. A. Homozygous mutation of P450 side-chain cleavage enzyme gene (CYP11A1) in 46, XY patient with adrenal insufficiency, complete sex reversal, and agenesis of corpus callosum. J. Clin. Endocrinol. Metab. 91, 2821–2826 (2006).
Papadimitriou, D. T., Bothou, C., Zarganis, D., Karantza, M. & Papadimitriou, A. Heterozygous mutations in the cholesterol side-chain cleavage enzyme gene (CYP11A1) can cause transient adrenal insufficiency and life-threatening failure to thrive. Hormones 17, 419–421 (2018).
Kara, O., Gorukmez, O., Ekici, A. & Celik, F. A novel homozygous mutation in CYP11A1 gene is associated with severe adrenal insufficiency in 46, XX patient. Fetal Pediatr. Pathol. 40, 518–522 (2021).
Pang, S. et al. Inherited congenital adrenal hyperplasia in the rabbit: absent cholesterol side-chain cleavage cytochrome P450 gene expression. Endocrinology 131, 181–186 (1992).
Hu, M. C. et al. Steroid deficiency syndromes in mice with targeted disruption of Cyp11a1. Mol. Endocrinol. 16, 1943–1950 (2002).
Li, N., Oakes, J. A., Storbeck, K. H., Cunliffe, V. T. & Krone, N. P. The P450 side-chain cleavage enzyme Cyp11a2 facilitates steroidogenesis in zebrafish. J. Endocrinol. 244, 309–321 (2020).
Wang, Y. et al. Cyp11a2 is essential for oocyte development and spermatogonial stem cell differentiation in zebrafish. Endocrinology 163, bqab258 (2022).
Parajes, S. et al. Redefining the initiation and maintenance of zebrafish interrenal steroidogenesis by characterizing the key enzyme cyp11a2. Endocrinology 154, 2702–2711 (2013).
Piferrer, F. et al. Brief treatment with an aromatase inhibitor during sex differentiation causes chromosomally female salmon to develop as normal, functional males. J. Exp. Zool. 270, 255–262 (1994).
Guiguen, Y. et al. Involvement of estrogens in the process of sex differentiation in two fish species: the rainbow trout (Oncorhynchus mykiss) and a tilapia (Oreochromis niloticus). Mol. Reprod. Dev. 54, 154–162 (1999).
Afonso, L. O., Wassermann, G. J. & Terezinha de Oliveira, R. Sex reversal in Nile tilapia (Oreochromis niloticus) using a nonsteroidal aromatase inhibitor. J. Exp. Zool. 290, 177–181 (2001).
Komatsu, T., Nakamura, S. & Nakamura, M. Masculinization of female golden rabbitfish Siganus guttatus using an aromatase inhibitor treatment during sex differentiation. Comp. Biochem. Physiol. C. Toxicol. Pharmacol. 143, 402–409 (2006).
Sato, T., Suzuki, A., Shibata, N., Sakaizumi, M. & Hamaguchi, S. The novel mutant scl of the medaka fish, Oryzias latipes, shows no secondary sex characters. Zool. Sci. 25, 299–306 (2008).
Vizziano, D. et al. Rainbow trout gonadal masculinization induced by inhibition of estrogen synthesis is more physiological than masculinization induced by androgen supplementation. Biol. Reprod. 78, 939–946 (2008).
Lau, E. S., Zhang, Z., Qin, M. & Ge, W. Knockout of zebrafish ovarian aromatase gene (cyp19a1a) by TALEN and CRISPR/Cas9 leads to all-male offspring due to failed ovarian differentiation. Sci. Rep. 6, 37357 (2016).
Yin, Y. et al. Targeted disruption of aromatase reveals dual functions of cyp19a1a during sex differentiation in zebrafish. Endocrinology 158, 3030–3041 (2017).
Zhang, X. et al. Mutation of foxl2 or cyp19a1a results in female to male sex reversal in XX Nile tilapia. Endocrinology 158, 2634–2647 (2017).
Nakamoto, M. et al. Ovarian aromatase loss-of-function mutant medaka undergo ovary degeneration and partial female-to-male sex reversal after puberty. Mol. Cell Endocrinol. 460, 104–122 (2018).
Zhai, G. et al. Characterization of sexual trait development in cyp17a1-deficient zebrafish. Endocrinology 159, 3549–3562 (2018).
Paul-Prasanth, B. et al. Estrogen oversees the maintenance of the female genetic program in terminally differentiated gonochorists. Sci. Rep. 3, 2862 (2013).
Sun, L. N. et al. Transdifferentiation of differentiated ovary into functional testis by long-term treatment of aromatase inhibitor in Nile tilapia. Endocrinology 155, 1476–1488 (2014).
Yoshitaka, N. & Nobuo, E. Development of the tissue architecture in the gonads of the medaka Oryzias latipes. Zool. Sci. 2, 695–706 (1985).
Suzuki, A., Tanaka, M., Shibata, N. & Nagahama, Y. Expression of aromatase mRNA and effects of aromatase inhibitor during ovarian development in the medaka, Oryzias latipes. J. Exp. Zool. A Comp. Exp. Biol. 301, 266–273 (2004).
Takahashi, H. Juvenile hermaphroditism in the zebrafish, Brachydanio rerio. Bull. Fac. Fish. Hokkaido Univ. 28, 57–65 (1977).
Ijiri, S. et al. Sexual dimorphic expression of genes in gonads during early differentiation of a teleost fish, the Nile tilapia Oreochromis niloticus. Biol. Reprod. 78, 333–341 (2008).
Tao, W. J. et al. Characterization of gonadal transcriptomes from Nile tilapia (Oreochromis niloticus) reveals differentially expressed genes. PLoS ONE 8, e63604 (2013).
Yang, L. Y. et al. Cyp17a1 is required for female sex determination and male fertility by regulating sex steroid biosynthesis in fish. Endocrinology 162, bqab205 (2021).
Sun, Y. L. et al. Screening and characterization of sex-linked DNA markers and marker-assisted selection in the Nile tilapia (Oreochromis niloticus). Aquaculture 433, 19–27 (2014).
Li, M. H. et al. Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics 197, 591–599 (2014).
Shibata, Y. et al. Expression of gonadal soma derived factor (GSDF) is spatially and temporally correlated with early testicular differentiation in medaka. Gene Expr. Patterns 10, 283–289 (2010).
Jiang, D. N. et al. gsdf is a downstream gene of dmrt1 that functions in the male sex determination pathway of the Nile tilapia. Mol. Reprod. Dev. 83, 497–508 (2016).
Dai, S. F. et al. Germline sexual fate is determined by the antagonistic action of dmrt1 and foxl3/foxl2 in tilapia. Development 148, dev199380 (2021).
Yamamoto, T. In Fish physiology (eds Hoar, W. S. & Randall, D. J.) Ch. 3, 117–175 (Academic press, 1969).
Bogart, M. H. Sex determination: a hypothesis based on steroid ratios. J. Theor. Biol. 128, 349–357 (1987).
Nakamura, M., Kobayashi, T., Chang, X. T. & Nagahama, Y. Gonadal sex differentiation in teleost fish. J. Exp. Zool. 281, 362–372 (1998).
Baroiller, J. F., Guiguen, Y. & Fostier, A. Endocrine and environmental aspects of sex differentiation in fish. Cell Mol. Life Sci. 55, 910–931 (1999).
Nagahama, Y. Molecular mechanisms of sex determination and gonadal sex differentiation in fish. Fish. Physiol. Biochem. 31, 105–109 (2005).
Guiguen, Y., Fostier, A., Piferrer, F. & Chang, C. F. Ovarian aromatase and estrogens: a pivotal role for gonadal sex differentiation and sex change in fish. Gen. Comp. Endocrinol. 165, 352–366 (2010).
Li, M. H., Sun, L. N. & Wang, D. S. Roles of estrogens in fish sexual plasticity and sex differentiation. Gen. Comp. Endocrinol. 277, 9–16 (2010).
Zhou, L. Y., Li, M. H. & Wang, D. S. Role of sex steroids in fish sex determination and differentiation as revealed by gene editing. Gen. Comp. Endocrinol. 313, 113893 (2021).
Yeung, W. S. & Chan, S. T. The plasma sex steroid profiles in the freshwater, sex-reversing teleost fish, Monopterus albus (Zuiew). Gen. Comp. Endocrinol. 65, 233–242 (1987).
Guiguen, Y., Jalabert, B., Thouard, E. & Fostier, A. Changes in plasma and gonadal steroid hormones in relation to the reproductive cycle and the sex inversion process in the protandrous seabass, Lates calcarifer. Gen. Comp. Endocrinol. 92, 327–338 (1993).
Chang, C., Lee, M. & Chen, G. Estradiol‐17β associated with the sex reversal in protandrous black porgy, Acanthopagrus schlegeli. J. Exp. Zool. 268, 53–58 (1994).
Bhandari, R. K., Komuro, H., Nakamura, S., Higa, M. & Nakamura, M. Gonadal restructuring and correlative steroid hormone profiles during natural sex change in protogynous honeycomb grouper (Epinephelus merra). Zool. Sci. 20, 1399–1404 (2003).
Li, G. L., Liu, X. C. & Lin, H. R. Seasonal changes of serum sex steroids concentration and aromatase activity of gonad and brain in red-spotted grouper (Epinephelus akaara). Anim. Reprod. Sci. 99, 156–166 (2007).
Peng, C. et al. Natural sex change in mature protogynous orange-spotted grouper (Epinephelus coioides): gonadal restructuring, sex hormone shifts and gene profiles. J. Fish. Biol. 97, 785–793 (2020).
Kitano, T., Takamune, K., Nagahama, Y. & Abe, S. I. Aromatase inhibitor and 17alpha-methyltestosterone cause sex-reversal from genetical females to phenotypic males and suppression of P450 aromatase gene expression in Japanese flounder (Paralichthys olivaceus). Mol. Reprod. Dev. 56, 1–5 (2000).
Andersen, L., Holbech, H., Gessbo, A., Norrgren, L. & Petersen, G. I. Effects of exposure to 17 alpha-ethinylestradiol during early development on sexual differentiation and induction of vitellogenin in zebrafish (Danio rerio). Comp. Biochem Physiol. C. Toxicol. Pharm. 134, 365–374 (2003).
Kobayashi, H. & Iwamatsu, T. Sex reversal in the medaka Oryzias latipes by brief exposure tip of early embryos to estradiol-17. Zool. Sci. 22, 1163–1167 (2005).
Li, L. et al. The role of StAR2 gene in testicular differentiation and spermatogenesis in Nile tilapia (Oreochromis niloticus). J. Steroid Biochem. Mol. Biol. 214, 105974 (2021).
Piferrer, F. Endocrine sex control strategies for the feminization of teleost fish. Aquaculture 197, 229–281 (2001).
Bhandari, R. K., Nakamura, M., Kobayashi, T. & Nagahama, Y. Suppression of steroidogenic enzyme expression during androgen-induced sex reversal in Nile tilapia (Oreochromis niloticus). Gen. Comp. Endocrinol. 145, 20–24 (2006).
Horie, Y. et al. Androgen induces gonadal soma-derived factor, Gsdf, in XX gonads correlated to sex-reversal but not Dmrt1 directly, in the teleost fish, northern medaka (Oryzias sakaizumii). Mol. Cell Endocrinol. 436, 141–149 (2016).
Lee, S. L. J. et al. Histological and transcriptomic effects of 17α-methyltestosterone on zebrafish gonad development. BMC Genomics 18, 557 (2017).
Huang, M. et al. The co-administration of estradiol/17α-methyltestosterone leads to male fate in the protogynous orange-spotted grouper, Epinephelus coioides. Biol. Reprod. 100, 745–756 (2019).
Crowder, C. M., Lassiter, C. S. & Gorelick, D. A. Nuclear androgen receptor regulates testes organization and oocyte maturation in zebrafish. Endocrinology 159, 980–993 (2018).
Tang, H. et al. Fertility impairment with defective spermatogenesis and steroidogenesis in male zebrafish lacking androgen receptor. Biol. Reprod. 98, 227–238 (2018).
Yu, G. et al. Zebrafish androgen receptor is required for spermatogenesis and maintenance of ovarian function. Oncotarget 9, 24320–24334 (2018).
Alward, B. A. et al. Modular genetic control of social status in a cichlid fish. Proc. Natl Acad. Sci. USA 117, 28167–28174 (2020).
Oakes, J. A., Barnard, L., Storbeck, K. H., Cunliffe, V. T. & Krone, N. P. 11β-Hydroxylase loss disrupts steroidogenesis and reproductive function in zebrafish. J. Endocrinol. 247, 197–212 (2020).
Zhang, Q. et al. Zebrafish cyp11c1 knockout reveals the roles of 11-ketotestosterone and cortisol in sexual development and reproduction. Endocrinology 161, 1–20 (2020).
Zheng, Q. Y. et al. Loss of Cyp11c1 causes delayed spermatogenesis due to the absence of 11-ketotestosterone. J. Endocrinol. 244, 487–499 (2020).
Ogino, Y. et al. Evolutionary differentiation of androgen receptor is responsible for sexual characteristic development in a teleost fish. Nat. Commun. 14, 1428 (2023).
Baron, D., Houlgatte, R., Fostier, A. & Guiguen, Y. Expression profiling of candidate genes during ovary-to-testis trans-differentiation in rainbow trout masculinized by androgens. Gen. Comp. Endocrinol. 156, 369–378 (2008).
Knapp, D. & Tanaka, E. M. Regeneration and reprogramming. Curr. Opin. Genet. Dev. 22, 485–493 (2012).
Uhlenhaut, N. H. et al. Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell 139, 1130–1142 (2009).
Matson, C. K. et al. DMRT1 prevents female reprogramming in the postnatal mammalian testis. Nature 476, 101–104 (2011).
Livak, K. J. & Schmittgen, T. D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 25, 402–408 (2001).
Tao, W. J. et al. Transcriptome display during tilapia sex determination and differentiation as revealed by RNA-Seq analysis. BMC Genomics 19, 363 (2018).
Kim, D., Langmead, B. & Salzberg, S. L. HISAT: a fast spliced aligner with low memory requirements. Nat. Methods 12, 357–360 (2015).
Robinson, M. D., McCarthy, D. J. & Smyth, G. K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010).
Wang, D. S. et al. Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol. 21, 712–725 (2007).
Wang, D. S. et al. Doublesex- and Mab-3-related transcription factor-1 repression of aromatase transcription, a possible mechanism favoring the male pathway in tilapia. Endocrinology 151, 1331–1340 (2010).
Acknowledgements
This work was supported by grants 31861123001 and 32172953 from National Natural Science Foundation of China, grant 2022YFD1201600 from National Key Research and Development Program of China, grant CQFTIU2024-08 from Chongqing Fishery Technology Innovation Union, grant 20232005276101 from Science Foundation of School of Life Sciences SWU.
Author information
Authors and Affiliations
Contributions
D.W. and H.X. conceived and designed the experiments; H.X., L.W., S.Y., H.M., Z.X., F.W., and J.W. performed the experiments; H.X. and W.T. analyzed the data, interpreted the results, and drafted the manuscript; D.W. critically edited the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Peer review
Peer review information
Communications Biology thanks the anonymous reviewers for their contribution to the peer review of this work. Primary Handling Editors: Simona Chera and Mengtan Xing. [A peer review file is available].
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Xiao, H., Wang, L., Yan, S. et al. Steroid hormone-deprived sex reversal in cyp11a1 mutant XX tilapia experiences an ovary-like stage at molecular level. Commun Biol 7, 1154 (2024). https://doi.org/10.1038/s42003-024-06853-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s42003-024-06853-8
- Springer Nature Limited