Abstract
Salix matsudana is a deciduous, rapidly growing willow species commonly cultivated in China, which can tolerate drought, salt, and heavy metal stress conditions. Selection of suitable reference genes for quantitative real-time PCR is important for normalizing the expression of the key genes associated with various stresses. To validate suitable reference genes, we selected 11 candidate reference genes (five traditional housekeeping genes and six novel genes) and analyzed their expression stability in various samples, including different tissues and under different abiotic stress treatments. The expression of these genes was determined using five programs—geNorm, NormFinder, BestKeeper, ΔCt, and RefFinder. The results showed that α-TUB2 (alpha-tubulin 2) and DnaJ (chaperone protein DnaJ 49) were the most stable reference genes across all the tested samples. We measured the expression profiles of the defense response gene SmCAT (catalase) using the two most stable and one least stable reference genes in all samples of S. matsudana. The relative quantification of SmCAT varied greatly according to the different reference genes. We propose that α-TUB2 and DnaJ should be the preferred reference genes for normalization and quantification of transcript levels in future gene expression studies in willow species under various abiotic stress conditions.
Similar content being viewed by others
Introduction
Drought, salt, and heavy metal stresses are major abiotic factors that contribute to the risk of environment and affect forestry productivity worldwide1,2,3,4,5; however, plants need to thrive in adverse circumstances6. Plants with short growth cycles, such as Arabidopsis thaliana7, soybean8, sorghum9, jute10, Sedum alfredii11, rice12, and tobacco13, have been the focus of studies on the effects of various abiotic stresses, and a few studies have been performed on plants with long growth cycles under different stress conditions. Short growth cycle plants are limited by low biomass, while plants (especially woody plants) with high biomass and long growth cycles are more able to deal with severe abiotic stress conditions. Only a small number of reference genes have been reported in trees under drought, salt, and heavy metal stress conditions14,15,16,17,18.
The genus Salix (Salicaceae) contains more than 450 willow species worldwide; 275 of these species grow in China19,20,21,22. Willow species are used for energy production, afforestation, and greening due to their high biomass, rapid growth, and ability to adapt to different stress conditions23,24,25,26,27,28. Salix matsudana is a deciduous, rapidly growing willow species commonly cultivated in China, which can tolerate drought, salt, and heavy metal stresses29,30,31,32,33. Physiological and biochemical properties have been characterized in S. matsudana34,35. Meanwhile, some key genes have been identified to regulate stress response factors in stressed plants at the molecular level36,37,38. Understanding the expression patterns of key stress response genes will help elucidate the mechanisms involved in various stresses of S. matsudana.
Gene expression analysis has been applied to understand different kinds of biological processes39. Quantitative real-time polymerase chain reaction (qRT-PCR) is widely used for gene expression analysis due to its high sensitivity, accuracy, specificity, and reproducibility40,41,42. However, factors such as sample amount, RNA integrity, reverse transcription efficiency, and cDNA quality can significantly influence the reliability of the gene expression results43,44,45. To reduce the influence of these factors, internal reference genes are used to obtain accurate biologically meaningful expression values46; however, unstable reference genes can cause significant biases and misinterpretations of the expression data47,48. Actin (ACT) and β-tubulin (β-TUB) have been used as reference genes for qRT-PCR normalization in gene expression analysis in S. matsudana under salt and copper stresses37,49; however, a systematic study to validate reference genes has not been reported for S. matsudana under abiotic stresses. To obtain accurate expression data, it is necessary to select suitable reference genes for each plant species and to verify their stability under the specific experimental conditions of interest.
In this study, we determined the expression profiles of 11 candidate reference genes from S. matsudana in six different tissues and under three kinds of abiotic stresses. The 11 candidate genes were ACT, alpha-tubulin 1 (α-TUB1), alpha-tubulin 2 (α-TUB2), chaperone protein DnaJ 49 (DnaJ), E3 ubiquitin-protein ligase ARI8 (ARI8), F-box family protein (F-box), histone H2A (H2A), heat shock 70 kDa protein (HSP 70), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), membrane-anchored ubiquitin-fold protein (MUB), and β-TUB. The transcriptome data of S. matsudana were used as the source of the potential reference genes (Unpublished data). The stabilities of the 11 reference genes were analyzed using five statistical algorithms—geNorm43, NormFinder44, BestKeeper50, ΔCt method51, and RefFinder, a web-based software52. The expression levels of the defense response gene SmCAT (catalase) as a target gene were assayed to verify the selected reference genes. The results will provide suitable reference genes for qRT-PCR normalization for accurate gene expression analysis in S. matsudana under different stress conditions.
Materials and Methods
Plant materials and stress treatments
Cuttings (approximately 10 cm long) from annual branches of S. matsudana were grown in hydroponics. Plants were supplemented with water containing 1/4 strength Hoagland53 solution on alternate days under normal conditions (25 °C, 16 h light/8 h dark). After 45 days of culture, groups of S. matsudana seedlings were subjected to different abiotic stresses in solutions containing 1/4 strength Hoagland solution at pH 6.0 as follows: drought (15% PEG 6000), salt (100 mM NaCl), and heavy metal (100 μM CdCl2). Untreated seedlings were used as the control. The roots of the treated plants were sampled at 0 h, 12 h, 24 h, 48 h, and 72 h. Tissues from the root, xylem, bark, stem, leaf, and flower were collected from the untreated plants. All the samples from three biological replicates were carefully harvested, immediately frozen in liquid nitrogen, and stored at −80 °C until total RNA extraction.
Total RNA isolation and cDNA synthesis
Total RNA from each sample was isolated from approximately 0.1 g fresh root using a total RNA kit (NORGEN, Thorold, Canada) and treated with DNase I (TaKaRa, Dalian, China) to remove any genomic DNA contamination. The RNA concentration of each sample was determined using a NanoDrop-2000 spectrophotometer (Thermo, Wilmington, USA). Samples with a 260/280 ratio of 1.9–2.1 and a 260/230 ratio ≥2.0 were chosen to determine the quality and purity of the RNA preparations. The integrity of the purified RNA was checked by 1.0% (p/v) agarose gel electrophoresis. Subsequently, first-strand cDNA was synthesized in a 20-μL reaction mixture in an Invitrogen SuperScript First Strand Synthesis System (Invitrogen, Carlsbad, USA) following the manufacturer’s instructions, and stored at −20 °C until use.
Screening of candidate reference genes and primer design
We identified 11 candidate reference genes and one target gene (Table 1) from the S. matsudana transcriptome data. Primers were designed based on the sequences the 11 genes using Primer3 (http://bioinfo.ut.ee/primer3-0.4.0/primer3/) with the following criteria: GC content 45–65%, optimal Tm 58–61 °C, primer length 18–22 bp, and amplicon length 120–220 bp (Table 1). The specificity of each selected primer pair was observed via standard RT-PCR using Premix Ex Taq (TaKaRa, Dalian, China), and each gene was verified by 2% agarose gel electrophoresis and sequenced to ensure its reliability.
qRT-PCR
qRT-PCR amplification was performed in 96-well plates with a Applied Biosystems 7300 Real-Time PCR System (Applied Biosystems, CA, USA) using SYBR® Premix Ex Taq™ (TaKaRa, Dalian, China). PCR reactions were prepared in 20 μL volumes containing: 2 μL of 50-fold diluted synthesized cDNA, 10 μL 2 × SYBR Premix Ex Taq™, 0.8 μL of each primer, 0.4 μL ROX reference dye (50×), and 6.8 μL ddH2O. The reactions comprised an initial step of 95 °C for 30 s, followed by 40 denaturation cycles at 95 °C for 5 s and primer annealing at 60 °C for 31 s. Next, the melting curves ranging from 60 °C to 95 °C were evaluated in each reaction to check the specificity of the amplicons. Biological triplicates of all the samples were used for the qRT-PCR analysis, and three technical replicates were analyzed for each biological sample. The threshold cycle (Ct) was measured automatically.
Statistical analysis to determine the expression stability of the candidate reference genes
Standard curves were generated in Microsoft Excel 2013 to calculate the gene-specific PCR efficiency and the correlation coefficient from 5-fold series dilution of a mixed cDNA (flower, bark, and stem) template for each primer pair. The amplification plots, melting curves and sequencing peaks were shown in Figure S1a,b,c. The PCR amplification efficiency (E) and the correlation coefficient were calculated using the slope of the standard curve according to the equation E = [5−1/slope − 1] × 100. Stabilities of the 11 selected reference genes were evaluated by four algorithms—geNorm, NormFinder, BestKeeper, and the ΔCt method. Finally, RefFinder (http://www.fulxie.0fees.us), a comprehensive evalution platform integrating the four above algorithms, ranked the overall stabilities of these 11 candidate genes. Pairwise variations based on the geNorm calculation were used to determine the optimal number of candidate reference genes for accurate normalization.
Expression normalization of SmCAT gene based on different reference genes
The defense response gene SmCAT was selected as the target gene to measure the stabilities of the candidate reference genes by quantifying SmCAT expression levels in all the tested samples. SmCAT gene expression levels were normalized with the two most stable candidate reference genes (α-TUB2 and DnaJ), as well as one of the least stable reference genes (β-TUB).
Results
qRT-PCR data for the candidate reference genes
The 11 selected candidate reference genes (Table 1) are orthologs of genes in Salix purpurea, for which the whole genome has been sequenced. The specificity and accuracy of the primers designed for the selected genes were determined by 2% agarose gel electrophoresis (Figure S2a), and further confirmed by a single peak in the melting-curve analysis (Figure S2b). The primer sequences, amplicon length, correlation coefficient, and PCR amplification efficiency are shown in Table 1. Furthermore, the qRT-PCR products were sequenced (File S1) to determine the accuracy of the 11 genes.
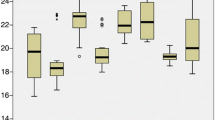
To evaluate the stability of the 11 candidate reference genes at the transcript level under the three abiotic stress conditions, the gene expression levels were determined by the average Ct values, which varied from 17 to 30 (Fig. 1). According to the average Ct values of all the samples, α-TUB1 was the most abundantly expressed gene, followed by DnaJ, α-TUB2, and F-box, while H2A was the least abundantly expressed gene, followed by β-TUB, ACT and MUB.
Analysis of gene expression stability
Expression stabilities of the 11 candidate reference genes were determined using geNorm, NormFinder, ΔCt, and BestKeeper, and their overall stabilities were ranked by RefFinder across all the stress treatments and tissue samples.
geNorm analysis
The stabilities of the 11 candidate reference genes of S. matsudana calculated using geNorm were ranked in the different tissues and abiotic stress treatments according to their M values, as shown in Fig. 2. The lowest M value indicates the most stable reference gene, and the highest M value indicates the least stable one. DnaJ and ARI8 had the highest expression stabilities in the six tissues, and all the genes had M values below the threshold of 1.5 (Fig. 2a). The top two most stable genes were DnaJ and α-TUB2 for drought and heavy metal stresses, and α-TUB2 and MUB for salt stress (Fig. 2b,c,d). When the stabilities from all the samples were combined, DnaJ and α-TUB2 were determined to be the most stable reference genes in all the samples (Fig. 2e), while β-TUB had the less stability.
The pairwise variation (Vn/Vn+1) between two sequential normalization factors NFn and NFn+1 was calculated by the geNorm algorithm to determine the optimal number of reference genes for accurate normalization. A cutoff value of 0.15 is the recommended threshold indicating that an additional reference gene will make no remarkable contribution to the normalization. The V2/3 values in the tissues, salt, and heavy metal were less than 0.15 (Fig. 3), which suggested that the top two reference genes were sufficient for accurate normalization. For the drought stress samples V4/5 was 0.123, indicating that the top four reference genes (DnaJ, α-TUB2, MUB, and ACT) were needed for accurate normalization. For the total samples V3/4 was 0.138, showing that three reference genes (DnaJ, α-TUB2, and MUB) were required.
NormFinder analysis
As shown in Table 2, DnaJ was the most stable gene (lowest stability value) in the salt and drought subsets calculated using NormFinder. For the heavy metal samples, α-TUB2 was the most stable gene, while ARI8 was the most stable gene in the different tissues. When all samples were taken together to determine the stability of reference genes, the three most stable genes were α-TUB2, ARI8, and DnaJ.
ΔCt analysis
The 11 candidate reference genes from the most to least stable expression, as calculated by the ΔCt method, are listed in Table 3. α-TUB2 was the most stable reference gene in the drought, heavy metal, and total samples subsets. MUB and ARI8 were the most stable genes for the salt subset and different tissues, respectively, and were considered the ideal reference genes.
BestKeeper analysis
BestKeeper determined the stabilities of the candidate reference genes based on their standard deviation (SD). Genes with SD > 1 was considered unacceptable reference genes. The genes are listed from most to least stable in Table 4. DnaJ was the most stable gene in the tissue and drought subsets, while GAPDH and α-TUB2 were the most stable genes in the heavy metal and salt subsets.
RefFinder analysis
To acquire reliable results for the expression stabilities of the 11 candidate reference genes of S. matsudana, the rankings of the four algorithms were integrated by RefFinder and the results are shown in Table 5. The 11 genes were ranked from the most to least stable expression by RefFinder (Fig. 4). The expression of α-TUB2 was ranked the most stable under the salt and heavy metal stress treatments, and the expression of DnaJ was ranked the most stable under the drought stress treatment. Overall, the best reference gene for accurate transcript normalization in all of the samples was α-TUB2, which had the lowest Geomean (geometric mean) of the ranking values.
Reference gene validation
To validate the performance of the best ranked candidate reference genes, the expression patterns of SmCAT (catalase) were analyzed (Fig. 5). CAT as abiotic stress inducible genes, are up-regulated by drought54, salt55, and Cd56 treatments. The CAT with low affinity towards H2O2 but with a high processing rate57, can operate through a complex networking machinery to avoid damage caused by ROS58. In this study, we used the most stable reference genes (α-TUB2 and DnaJ) and the least stable gene (β-TUB) as internal controls for normalization of SmCAT according to the RefFinder rankings. The expression profiles of SmCAT were determined in different tissues and under drought, salt, and heavy metal stresses. When the stable reference genes α-TUB2 and DnaJ were used for normalization, SmCAT exhibited similar expression trends. However, when the least stable reference gene β-TUB was used for normalization, the expression patterns of SmCAT were different from that obtained using the two stable reference genes.
Discussion
Abiotic stress conditions including drought, salt, and heavy metals bring great losses to forestry productivity and increase the risk of environment. To guarantee sustainable forestry productivity and decrease the risk of environment, it is imperative to understand the regulation and function of the key genes under different abiotic stresses. To study gene expression variations and determine gene regulation patterns, suitable reference genes are prerequisite to accurately determine the expression levels of target genes. qRT-PCR is a reliable and accurate technique for measuring gene expression levels. Some suitable reference genes under abiotic stresses, such as GAPDH59,60 and DnaJ10, have been detected in plants; however, the number of reference genes evaluated is limited, especially for woody plants.
S. matsudana is an important afforestation and greening material in China that can adapt to harsh environments including drought, salt, and heavy metal. A good understanding of the molecular mechanisms related to abiotic stress responses in woody plants will not only help in improving forestry productivity but also help to decrease the risk of environment. A few studies have explored the ability of S. matsudana to withstand different abiotic stresses; however, the study of reference genes in willows has lagged behind that of other major plant species. To address this problem, we analyzed the expression of 11 candidate reference genes, five traditional reference genes (ACT, α-TUB1, α-TUB2, β-TUB, and GAPDH) and six new genes (DnaJ, ARI8, MUB, HSP70, F-box, and H2A), in various tissues, including the roots of S. matsudana under different abiotic stresses using qRT-PCR methods. The best and worst candidate reference genes were further verified by expression profiling of the defense response gene SmCAT.
We used five different statistical algorithms to determine the stabilities of candidate reference gene(s) under various abiotic stress conditions in S. matsudana. The results listed in Table 5 showed that, for the most parts, geNorm, NormFinder, ΔCt, and RefFinder consistently ranked the same genes as the most stable candidate reference genes. The BestKeeper algorithm is different from the other algorithms, which explains why the BestKeeper results showed the least correlation with the others61. Therefore, we selected the reference gene(s) determined by geNorm, NormFinder, ΔCt, and RefFinder.
α-TUB2 and DnaJ were the two most stable reference genes in all the sample sets according to the four algorithms. α-TUB2 encoding a cytoskeleton structure protein62 and DnaJ encoding a cellular chaperone have the ability to repair heat-induced protein machinery damage63,64. Our results are in agreement with several previous studies, which showed that α-TUB2 and DnaJ were established as the most stable reference genes in plants under abiotic stresses; for example, in Syntrichia caninervis under drought, salt, and heavy metal stresses65, Corchorus capsularis under drought stress10, Buchloe dactyloides under salt stress66, and Platycladus orientalis under salt stress67. Normalization with multiple reference genes is an effective way to avoid erroneous data that may result from using a single reference gene68. In this study, two top ranked reference genes, DnaJ and α-TUB2 under heavy metal stress and α-TUB2 and MUB under salt stress, were appropriate for gene expression normalization, Meanwhile. Four reference genes, DnaJ, α-TUB2, MUB, and ACT under drought stress, were needed for accurate normalization. Two reference genes were found to be sufficient to analyze the expression of target genes in sorghum62, jute10, and moss65.
Significant differences were revealed in the expression patterns of the target gene SmCAT when was normalized with the two most stable genes (α-TUB2 and DnaJ) compared with one of the least stable genes (β-TUB) (Fig. 5), The results emphasize the importance of using stable reference genes for normalization. Our findings indicated that α-TUB2 and DnaJ either singly or in combination are suitable for normalization of gene expression in S. matsudana under different abiotic stresses. Consequently, we recommend α-TUB2 and DnaJ as the most suitable reference genes for normalization of qRT-PCR expression data in S. matsudana under diverse abiotic stress conditions.
To the best of our knowledge, this is the first report on the identification and validation of suitable reference genes for qRT-PCR analysis in S. matsudana under abiotic stresses.
Conclusion
To validate suitable reference genes for gene expression normalization in S. matsudana under drought, salt, and heavy metal stresses, we selected 11 candidate reference genes using four systematic statistical algorithms (geNorm, NormFinder, ΔCt, and BestKeeper). The obtained results were compared and ranked using RefFinder. Based on the gene stability analysis, we identified α-TUB2 and DnaJ as the most stable reference genes for normalization of gene expression under drought, salt, and heavy metal stress conditions. Furthermore, the expression profiles of SmCAT validated α-TUB2 and DnaJ could be used as suitable reference genes. The reference genes identified in this study will facilitate accurate and consistent expression analysis of stress tolerance genes in willows and woody plants under various abiotic stress conditions for functional genomic studies.
Additional Information
How to cite this article: Zhang, Y. et al. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 7, 40290; doi: 10.1038/srep40290 (2017).
Publisher's note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
Farouk, S., Mosa, A. A., Taha, A. A. & Ibrahim, H. M. Protective effect of humic acid and chitosan on radish (Raphanus sativus, L. var. sativus) plants subjected to cadmium stress. Journal of Stress Physiology & Biochemistry 7 (2011).
Agarwal, P. K., Agarwal, P., Reddy, M. K. & Sopory, S. K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Reports 25, 1263–1274 (2006).
Burke, E. J., Brown, S. J. & Christidis, N. Modeling the recent evolution of global drought and projections for the twenty-first century with the Hadley Centre Climate Model. Journal of Hydrometeorology 7, 1113 (2006).
Shao, H.-B., Chu, L.-Y., Jaleel, C. A. & Zhao, C.-X. Water-deficit stress-induced anatomical changes in higher plants. Comptes Rendus Biologies 331, 215–225 (2008).
Zhang, J. et al. The heat shock factor gene family in Salix suchowensis: a genome-wide survey and expression profiling during development and abiotic stresses. Frontiers in Plant Science 6, 748 (2014).
Shivhare, R. & Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Scientific Reports 6 (2016).
Remans, T. et al. Normalisation of real-time RT-PCR gene expression measurements in Arabidopsis thaliana exposed to increased metal concentrations. Planta 227, 1343–9 (2008).
Hu, R., Fan, C., Li, H., Zhang, Q. & Fu, Y. F. Evaluation of putative reference genes for gene expression normalization in soybean by quantitative real-time RT-PCR. Bmc Molecular Biology 10, 93–93 (2009).
Reddy, P. S. et al. Evaluation of sorghum [Sorghum bicolor(L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Frontiers in Plant Science 7 (2016).
Niu, X. et al. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Frontiers in Plant Science 6, 848 (2015).
Sang, J. et al. Selection and validation of reference genes for real-time quantitative PCR in hyperaccumulating ecotype of Sedum alfredii under different heavy metals stresses. Plos One 8, e82927–e82927 (2013).
Jain, M., Nijhawan, A., Tyagi, A. K. & Khurana, J. P. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical & Biophysical Research Communications 345, 646–651 (2006).
Schmidt, G. W. & Delaney, S. K. Stable internal reference genes for normalization of real-time RT-PCR in tobacco (Nicotiana tabacum) during development and abiotic stress. Molecular Genetics & Genomics 283, 233–241 (2010).
Basa, B. Housekeeping gene selection in poplar plants under Cd-stress: comparative study for real-time PCR normalization. Functional Plant Biology 36, 1079–1087 (2009).
Fries, J., Pfeiffer, S., Kuffner, M. & Sessitsch, A. Spirosoma endophyticum sp. nov., isolated from Zn- and Cd-accumulating Salix caprea . International Journal of Systematic & Evolutionary Microbiology 63, 4586–90 (2013).
He, J. et al. A transcriptomic network underlies microstructural and physiological responses to cadmium in Populus x canescens . Plant Physiology 162, 424–39 (2013).
Atkinson, N. J. & Urwin, P. E. The interaction of plant biotic and abiotic stresses: from genes to the field. Journal of Experimental Botany 63, 3523–43 (2012).
Seki, M., Kamei, A., Yamaguchi-Shinozaki, K. & Shinozaki, K. Molecular responses to drought, salinity and frost: common and different paths for plant protection. Current Opinion in Biotechnology 14, 194–199 (2003).
Newsholme, C. Willows: the genus Salix (B.T. Batsford, 1992).
Argus, G. W. Infrageneric classification of Salix (Salicaceae) in the new world. Systematic Botany Monographs 52, 1–121 (1997).
USA, F.o.N.A.E.C. Flora of North America, Volume 7. Magnoliophyta: Salicaceae to Brassicaceae (2010).
Zhang, J. et al. A high-density genetic map of tetraploid Salix matsudana using specific length amplified fragment sequencing (SLAF-seq). Plos One 11 (2016).
Gullberg, U. Towards making willows pilot species for coppicing production. Forestry Chronicle 69, 721–726 (1993).
Åhman, I., Larsson, S. & Dietrichson, J. Genetic improvement of willow (Salix) as a source of bioenergy. Norwegian Journal of Agricultural Sciences (1994).
Lindegaard, K. N. & Barker, J. H. A. Breeding willows for biomass. Aspects of Applied Biology 155–162 (1997).
Greger, M. & Landberg, T. Use of willow in phytoextraction. International Journal of Phytoremediation 1, 115–123 (2006).
Aronsson, P., Dahlin, T. & Dimitriou, I. Treatment of landfill leachate by irrigation of willow coppice – Plant response and treatment efficiency. Environmental Pollution 158, 795–804 (2010).
Mirck, J. & Volk, T. A. Mass balances and allocation of salt ions from Solvay storm water for shrub willow (Salix spp.). Biomass & Bioenergy 39, 427–438 (2012).
Yang, J., Yi, J., Yang, C. & Li, C. Agrobacterium tumefaciens-mediated genetic transformation of Salix matsudana Koidz. using mature seeds. Tree Physiology 33, 628–639 (2013).
Hangs, R. D., Schoenau, J. J., Rees, K. C. J. V. & Steppuhn, H. Examining the salt tolerance of willow (Salix spp.) bioenergy species for use on salt-affected agricultural lands. Canadian Journal of Plant Science 91, 509–517 (2011).
Pulford, I. D. & Watson, C. Phytoremediation of heavy metal-contaminated land by trees—a review. Environment International 29, 529–40 (2003).
Peng, X., Fan, J., Wang, Q. & Warrington, D. Discrepancy of sap flow in Salix matsudana grown under different soil textures in the water-wind erosion crisscross region on the Loess Plateau. Plant & Soil 390, 383–399 (2015).
Yang, W. D., Chen, Y. T. & Ming-Hua, Q. U. Subcellular distribution and chemical forms of cadmium in Salix matsudana . Acta Botanica Boreali-Occidentalia Sinica 29, 1394–1399 (2009).
Hai-Yan, W. U. et al. Cadmium tolerance of and cadmium transportation and accumulation in Salix matsudana. Chinese Journal of Ecology 30, 1222–1228 (2011).
Surhone, L. M., Tennoe, M. T. & Henssonow, S. F. Salix Matsudana (Betascript Publishing, 2013).
Rao, G. et al. De novo transcriptome and small RNA analysis of two Chinese willow cultivars reveals stress response genes in Salix matsudana . Plos One 9, 134–134 (2014).
Yang, J. et al. Overexpression of the Tamarix hispida ThMT 3 gene increases copper tolerance and adventitious root induction in Salix matsudana Koidz. Plant Cell Tissue & Organ Culture 121, 469–479 (2015).
Konlechner, C. et al. Expression of zinc and cadmium responsive genes in leaves of willow (Salix caprea L.) genotypes with different accumulation characteristics. Environmental Pollution 178, 121–127 (2013).
Bustin, S. A., Benes, V., Nolan, T. & Pfaffl, M. W. Quantitative real-time RT-PCR–a perspective. Journal of Molecular Endocrinology 34, 597–601 (2005).
Gachon, C., Mingam, A. & Charrier, B. Real-time PCR: what relevance to plant studies? Journal of Experimental Botany 55, 1445–54 (2004).
Kumar, K., Muthamilarasan, M. & Prasad, M. Reference genes for quantitative real-time PCR analysis in the model plant foxtail millet (Setaria italica L.) subjected to abiotic stress conditions. Plant Cell Tissue & Organ Culture 115, 13–22 (2013).
Bustin, S. A. Absolute quantification of mRNA using real-time reverse transcription polymerase chain reaction assays. Journal of Molecular Endocrinology 25, 169–93 (2000).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biology 3, research0034.1-research0034.11 (2002).
Andersen, C. L., Jensen, J. L. & Ø, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research 64, 5245–50 (2004).
Janská, A. et al. The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mgg Molecular & General Genetics 288, 639–49 (2013).
Gutierrez, L. et al. The lack of a systematic validation of reference genes: a serious pitfall undervalued in reverse transcription-polymerase chain reaction (RT-PCR) analysis in plants. Plant Biotechnology Journal 6, 609–18 (2008).
Ferguson, B. S., Nam, H., Hopkins, R. G. & Morrison, R. F. Impact of reference gene selection for target gene normalization on experimental outcome using real-time qRT-PCR in Adipocytes. Plos One 5, 5525–5556 (2010).
Mafra, V. et al. Reference genes for accurate transcript normalization in citrus genotypes under different experimental conditions. Plos One 7, e31263–e31263 (2012).
Qiao, G. et al. Comparative proteomic analysis of responses to salt stress in Chinese willow (Salix matsudana Koidz). Plant Molecular Biology Reporter 32, 814–827 (2014).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper–Excel-based tool using pair-wise correlations. Biotechnology Letters 26, 509–515 (2004).
Silver, N., Best, S., Jiang, J. & Thein, S. L. Selection of housekeeping genes for gene expression studies in human reticulocytes using real-time PCR. Bmc Molecular Biology 7, 1–9 (2006).
Xie, F., Peng, X., Chen, D., Xu, L. & Zhang, B. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Molecular Biology 80, 75–84 (2012).
Hoagland, D. R. Minerals, Plants, and Men. (Book Reviews: Lectures on the inorganic nutrition of plants). Scientific Monthly 59 (1944).
Yang, G., Zhang, X. Q. & Xie, W. G. Study on the drought tolerance of dactylis glomerata Lines. Hubei Agricultural Sciences (2007).
Fadzillah, N. A. M., Gill, V., Finch, R. P. & Burdon, R. H. Chilling, oxidative stress and antioxidant responses in shoot cultures of rice. Journal of Experimental Botany 199, 552–556 (1996).
Singh, H. P., Batish, D. R., Kaur, G., Arora, K. & Kohli, R. K. Nitric oxide (as sodium nitroprusside) supplementation ameliorates Cd toxicity in hydroponically grown wheat roots. Environmental & Experimental Botany 63, 158–167 (2008).
Scandalios, J. G. Oxidative stress: molecular perception and transduction of signals triggering antioxidant gene defenses. Brazilian Journal of Medical & Biological Research 38, 995–1014 (2005).
Arora, A., Sairam, R. K. & Srivastava, G. C. Oxidative stress and antioxidative system in plants. Soviet Physics Doklady 16, 1227–1238 (2002).
Sinha, P., Saxena, R. K., Singh, V. K., Krishnamurthy, L. & Varshney, R. K. Selection and validation of housekeeping genes as reference for gene expression studies in pigeonpea (Cajanus cajan) under heat and salt stress conditions. Frontiers in Plant Science 6 (2015).
Galli, V. et al. Validation of reference genes for accurate normalization of gene expression for real time-quantitative PCR in strawberry fruits using different cultivars and osmotic stresses. Gene 554, 205–14 (2015).
Guo, J., Hui, L., Wu, Q., Xu, L. & Que, Y. The choice of reference genes for assessing gene expression in sugarcane under salinity and drought stresses. Scientific Reports 4, 7042–7042 (2014).
Palakolanu, S. R. et al. Evaluation of sorghum [Sorghum bicolor(L.)] reference genes in various tissues and under abiotic stress conditions for quantitative real-time PCR data normalization. Frontiers in Plant Science 7 (2016).
Cheetham, M. E. & Caplan, A. J. Structure, function and evolution of DnaJ: conservation and adaptation of chaperone function. Cell Stress & Chaperones 3, 28–36 (1998).
Schröder, H., Langer, T., Hartl, F. U. & Bukau, B. DnaK, DnaJ and GrpE form a cellular chaperone machinery capable of repairing heat-induced protein damage. Embo Journal 12, 4137–44 (1993).
Li, X. et al. Characterization of reference genes for RT-qPCR in the desert moss Syntrichia caninervis in response to abiotic stress and desiccation/rehydration. Frontiers in Plant Science 6, 38 (2015).
Li, W., Qian, Y. Q., Han, L., Liu, J. X. & Sun, Z. Y. Identification of suitable reference genes in buffalo grass for accurate transcript normalization under various abiotic stress conditions. Gene 547, 55–62 (2014).
Chang, E. et al. Selection of reference genes for quantitative gene expression studies in Platycladus orientalis (Cupressaceae) using real-time PCR. Plos One 7, 65–65 (2012).
Le, D. T. et al. Evaluation of candidate reference genes for normalization of quantitative RT-PCR in soybean tissues under various abiotic stress conditions. Plos One 7, 1602–1603 (2012).
Acknowledgements
This work was supported by the National Nonprofit Institute Research Grant of Chinese Academy of Forestry (CAFYBB2017ZY007, No. TGB2013008, No. RISF2014010), the National High Technology Research and Development Program of China (No. 2013AA102701-3), the Science and Technology Department of Zhejiang Province (No. 2016C32027), and a Nonprofit Research Grant of Zhejiang Province (No. 2016C32G3030016).
Author information
Authors and Affiliations
Contributions
Y.Z., X.H. conceived and designed the experiments. Y.Z., X.H., S.C., and L.Z. performed the experiments. Y.Z., X.H., X.H., M.L. and G.Q. analyzed the data, and Y.Z., X.H. wrote the manuscript and coordinated its revision. Y.W. and R.Z. contributed reagents/materials/funds support. All authors read and provided helpful discussions, and approved the final version.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Zhang, Y., Han, X., Chen, S. et al. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci Rep 7, 40290 (2017). https://doi.org/10.1038/srep40290
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/srep40290
- Springer Nature Limited
This article is cited by
-
Selection and validation of reliable reference genes for quantitative real-time PCR in Barnyard millet (Echinochloa spp.) under varied abiotic stress conditions
Scientific Reports (2023)
-
Reference gene selection for quantitative RT-PCR in Miscanthus sacchariflorus under abiotic stress conditions
Molecular Biology Reports (2022)
-
Selection and validation of appropriate reference genes for RT-qPCR analysis of flowering stages and different genotypes of Iris germanica L
Scientific Reports (2021)
-
Selection of reference genes for quantitative real-time PCR normalization in European quail tissues
Molecular Biology Reports (2021)
-
Identification and validation of superior housekeeping gene(s) for qRT-PCR data normalization in Agave sisalana (a CAM-plant) under abiotic stresses
Physiology and Molecular Biology of Plants (2020)