Abstract
Background
The ability to use dissolved organic phosphorus (DOP) is important for survival and competition when phytoplankton are faced with scarcity of dissolved inorganic phosphorus (DIP). However, phosphorus availability to the freshwater dinoflagellate Peridinium bipes has received relatively little attention, the efficiency of glycerol phosphate use by phytoplankton has rarely been investigated, and the regulatory molecular mechanisms remain unclear.
Result
In the present study, cultures of the freshwater dinoflagellate Peridinium bipes were set up in 119 medium (+DIP), DIP-depleted 119 medium (P-free), and β-glycerol phosphate-replacing-DIP medium (+DOP). Gene expression was analyzed using transcriptomic sequencing. The growth rate of cells in DOP treatment group was similar to that in DIP group, but chlorophyll a fluorescence parameters RC/CS0, ABS/CS0, TR0/CS0, ET0/CS0 and RE0/CS0 markedly decreased in the DOP group. Transcriptomic analysis revealed that genes involved in photosynthesis, including psbA, psbB, psbC, psbD, psaA and psaB, were downregulated in the DOP group relative to the DIP group. Glycerol-3-phosphate dehydrogenase and glyceraldehyde-3-phosphate dehydrogenase, rather than alkaline phosphatase, were responsible for β-glycerol phosphate use. Intercellular gluconeogenesis metabolism was markedly changed in the DOP group. In addition, genes involved in ATP synthases, the TCA cycle, oxidative phosphorylation, fatty acid metabolism and amino acid metabolism in P. bipes were significantly upregulated in the DOP group compared with the DIP treatment.
Conclusions
These findings suggested that β-glycerol phosphate could influence the photosynthesis and metabolism of P. bipes, which provided a comprehensive understanding of the phosphorus physiology of P. bipes. The mechanisms underlying the use of β-glycerol phosphate and other DOPs are different in different species of dinoflagellates and other phytoplankton. DIP reduction may be more effective in controlling the bloom of P. bipes than DOP reduction.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
Background
Phosphorus is a vital element involved in many biochemical reactions and metabolic processes, including cell membrane synthesis, signal transduction, photosynthesis, nucleic acid metabolism and energy metabolism [1,2,3,4]. As an essential nutrient for growth, phosphorus availability not only constrains phytoplankton productivity but also plays a very important role in the community structure of phytoplankton [4]. Therefore, phosphorus is regarded as a limiting factor in marine and freshwater ecosystems [5].
In aquatic environments, dissolved inorganic phosphorus (DIP) and dissolved organic phosphorus (DOP) are the two major total dissolved phosphorus (TDP) pools [6]. DIP can be directly used by phytoplankton cells [7]. However, DIP is rapidly consumed and shows very low replenishment from external sources, often resulting in growth-limiting concentrations in aquatic environments [8,9,10]. However, DOP is relatively abundant in aquatic environments. In natural waters, DOP is derived from soil and sediment, excretion by live organisms, and decay of dead organisms [11]. DOP also comes from organic compounds discharged from industrial, agricultural and domestic drainage [12]. According to Benitez-Nelson [8], in coastal marine environments and in the open ocean, the DOP pool can be up to 50% or 75% of the TDP pool, respectively. Other studies have shown that in the North Atlantic Ocean, DOP often accounts for > 80% of the TDP pool [13,14,15]. The DOP pool has even been found to exceed DIP by an order of magnitude in some environments, such as the Sargasso Sea, where DOP:DIP ratios in surface waters can exceed 100 [13, 16]. Bogé et al. [17] observed that the highest DOP concentration in Toulon Bay, France, was 0–0.33 μM, while DIP concentration was approximately 0–0.19 μM.
Because DOP can be only assimilated by phytoplankton cells with the assistance of hydrolases [6, 18], the ability to use DOP is a potential driving factor in both phytoplankton species composition and the initiation and maintenance of harmful algal blooms (HAB) [9]. Glycerol phosphate is an important component of DOP because it is widely used in biology as well as in medicine and other fields of human endeavor [19,20,21]. However, the efficiency of glycerol phosphate use by phytoplankton has rarely been investigated, and the regulatory molecular mechanisms remain unclear.
Dinoflagellates are an important functional component of the phytoplankton community in marine and freshwater ecosystems [22, 23]. Many dinoflagellates, such as Ostreopsis [24], Akashiwo [25,26,27], Alexandrium [28] and Karenia [6, 29], have been reported to cause HABs in marine ecosystems [30, 31]. Phosphorus is believed to affect the abundance, volume, toxin synthesis and motile form of these dinoflagellates [32,33,34] and to be one of the primary factors influencing the frequency of red tides [35]. Numerous studies have indicated that dinoflagellates can assimilate various types of DOP when DIP is insufficient [18, 36,37,38]. However, relative to marine ecosystems, the dinoflagellate species in freshwater ecosystems are little studied.
Recently, species of Peridinium have occurred in dense blooms in freshwater reservoirs and lakes throughout the world. During the bloom of P. cinctum, the organism comprises more than 90% of the phytoplankton biomass in Lake Kinneret, Israel and Lake Torrens, Australia [39, 40]. A bloom of P. bipes was also observed in the mesotrophic Huanglongdai Reservoir (biomass 5138 μg L−1), China, and Juam Reservoirs, South Korea [41, 42]. High-biomass blooms have caused mass mortalities of a variety of aquatic organisms through predation, starvation, shading, or creation of anoxic conditions [43,44,45,46].
Wynne et al. [39] suggested that the rapid decline of Peridinium in Lake Kinneret in June is brought about by a combination of physical and chemical factors, such as pH, temperature, irradiation and limitation of P, N or other micronutrients. Previous studies have been proposed that phosphorus not only affects the abundance, toxin synthesis and motile form of these dinoflagellates [32,33,34], but also regulates the frequency of dinoflagellate species [35]. However, it is regretful that phosphorus availability to the freshwater dinoflagellate P. bipes has received relatively little attention. Thus, the aims of this study were (1) to evaluate the growth and photosynthetic responses of P. bipes to DIP and DOP; and (2) to identify potential genes and pathways involved in the use of DIP and DOP by comparative transcriptomic analysis.
Materials and methods
Algal strain and culture conditions
The strain of Peridinium bipes used in this study was isolated from a tributary of Three Gorges Reservoir, China. This strain was grown in 119 medium (http://algae.ihb.ac.cn/Products/ProductDetail.aspx?product=10) at 25 ± 1 °C under white light with intensity 50 mmol photons m−2s−1 and a 12-h light/12-h dark cycle. A mixture of ampicillin (final concentration 200 µg mL−1), kanamycin (final concentration 100 µg mL−1) and streptomycin (final concentration 100 µg mL−1) was used to inhibit the growth of bacteria to < 1% of the culture biomass, which was confirmed using a Nikon CE-I fluorescence microscope (Nikon, Tokyo, Japan). Before the experiment, all glassware was soaked in 0.1 mol L−1 HCl for 24 h and then rinsed with ultrapure water to prevent external phosphorus contamination. Cells in the logarithmic growth phase (after growth for about 10 days) were collected by centrifugation at 2683×g for 5 min, washed three times with P-free medium (119 medium without phosphorus), and then inoculated in P-free medium for 5 days to remove excess phosphate [6].
When the DIP concentration in the culture was below the detection limit of 0.2 µmol L−1, the cells were inoculated into 500-mL glass flasks containing 250 mL 119 medium. Three different conditions were used: (i) 119 medium without P (P-free medium); (ii) P-free 119 medium supplemented with 52 µmol L−1 K2HPO4 (DIP treatment, as the control); and (iii) P-free 119 medium with 52 µmol L−1 β-glycerol phosphate (DOP treatment). All phosphorus reagents were purchased from Sigma–Aldrich (USA; > 99% purity, analysis- or chromatography analysis-grade). The initial concentration of P. bipes in each treatment was 0.37 × 104 cells L−1.
Cell density and specific growth rate
After inoculation, cell density was determined using a hemocytometer chamber under a Nikon CE-I microscope. Specific growth rate (μ) was calculated as μ = (lnN1 − lnN0)/(t1 − t0), where N0 and N1 are the biomass at times t0 and t1, and t0 and t1 are the times that the exponential growth phase started and ended.
Polyphasic Chl a fluorescence transient and energy pathway model
Samples in logarithmic growth were dark-adapted for 20 min before conducting chlorophyll a fluorescence measurement using a Plant Efficiency Analyzer (Hansatech Instruments Ltd., UK) with an actinic light of 3000 μmol photon m−2s−1. Fluorescence signals were recorded within a time scan of 10 μs to 2 s. Baesd on the theory of energy flux in PSII, several fluorescence parameters were obtained from the polyphasic Chl a fluorescence transient (OJIP) curve [47]. An energy pathway model of photosynthesis in P. bipes was established according to the energy flow model of Appenroth et al. [48] with specific activity parameters of the unit area of photosynthetic apparatus and the unit active reaction centers (RC) of P. bipes.
DIP concentration and alkaline phosphatase activity
DIP concentrations were measured by the phosphorus molybdenum blue method [49]. The supernatant in samples was used to determine the activity of extracellular phosphatases. Alkaline phosphatase activity (APA) was detected with p-nitrophenyl phosphate (Amresco, USA) as the substrate [50].
Transcriptome analysis
After 11-day inoculation, samples from the DIP and DOP treatments were harvested by suction filtration. The supernatant was discarded, and the centrifuge tube was immersed in liquid nitrogen to freeze the cell pellet for later processing. RNA isolation, library preparation and sequencing referred to the methods of Dong et al. [51]. Transcriptome measurement was undertaken by Novogene Company (Beijing, China). The data were submitted to NCBI (Accession No. PRJNA608149).
Clean data (clean reads) were obtained by removing reads containing adapters or poly-N and low-quality reads from the raw data. Q20, Q30, GC-content and sequence duplication level of the clean data were calculated. All downstream analyses were based on high-quality clean data. Gene function was annotated based on the following databases: Nr (NCBI non-redundant protein sequences), Nt (NCBI non-redundant nucleotide sequences), Pfam (Protein family), KOG/COG (Clusters of Orthologous Groups of proteins), Swiss-Prot (a manually annotated and reviewed protein sequence database), KO (Kyoto Encyclopedia of Genes and Genomes Ortholog database), and GO (Gene Ontology). Transcript relative abundance was aligned to annotated gene models using the HTSeq method (HTSeq v0.6.1) and was expressed as the number of reads. Fragments per kilobase of exon per millions of fragments mapped (FPKM) values were normalized to obtain the gene expression levels from RNA-Seq. Then, analysis of differential expression in different treatments was conducted using the DEGseq method in the DESeq R package (1.18.0). The cutoff value for analyzing gene transcriptional activity was determined based on the 95% confidence interval for all FPKM values. Fold expression changes between different samples were calculated using transcript relative abundance log2 ratios. A differentially gene expression was accepted when genes showed at least q value < 0.005 and |log2FoldChange| > 1.
Statistical analysis
All experiments were performed in triplicate. Data obtained in this study were analyzed using a least significant difference test with one-way analysis of variance (ANOVA) in SPSS software version 17.0 (IBM, USA). Statistical significance was determined at p < 0.05. Data were plotted using Origin software version 6.1 (OriginLab Corporation, USA).
Results
Growth
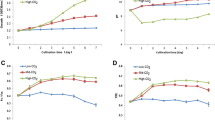
The growth of P. bipes under different treatments after culture for 20 days is shown in Fig. 1a. Compared with the DIP group, a significant decrease was found in the P-free treatment (p < 0.01, ANOVA). However, no significant difference was determined when cells were cultured in DIP and DOP (the β-glycerol phosphate group). After culture for 20 days, the cell density reached 1.95 × 104 cells L−1 (DIP group) and 1.79 × 104 cells L−1 (DOP group). A significant decrease in specific growth rate was observed in the P-free treatment compared with the DIP and DOP treatments (Fig. 1b) (p < 0.01, ANOVA). Specific growth rates (μ) in the P-free, DIP and DOP treatments were 0.06 day−1, 0.12 day−1, and 0.11 day−1, respectively.
DIP concentration and APA
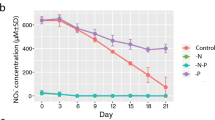
When P. bipes was inoculated into the DIP-containing medium, the concentration of DIP showed a rapid depletion by the 10th day compared with day 1 (p < 0.01), while it was consistently low in the P-free group. In contrast, the DIP concentration in the DOP group increased sharply to reach 0.91 mg L−1 on the 5th day (p < 0.01), and then declined slowly (Fig. 2a). On day 15, there was about 0.41 mg L−1 residual DIP in the DOP culture. APA was barely detectable on the first day in all three groups (Fig. 2b). On day 5 and thereafter, APA increased in the P-free group. In contrast, APA remained low in the other two groups.
Chlorophyll a fluorescence of PSII
OJIP curves for P. bipes treated in P-free, DIP and DOP conditions are shown in Fig. 3. Phase J increased clearly from the normalized OJIP curve in the P-free and DOP groups compared with that for the DIP group. A total of 18 parameters calculated from chlorophyll a fluorescence induction curves are shown in Additional file 1: Table S1. The values of PIabs (performance index based on absorption of light energy), Sm (multiple turnover in the closure of the RCs), N (QA has been reduced in the time span from 0 to tFmax), and \(\psi_{(E_{0})}\) (probability [at t = 0] that a trapped exciton moves an electron into the electron transport chain beyond QA) exhibited a remarkable decrease in the DOP group compared with those in the DIP group (p < 0.05). The values of VJ (relative variable fluorescence in the J-step) and M0 (approximate initial slope of the fluorescence transient) increased in the DOP group compared with that in the DIP group (p < 0.05). Compared with the DIP group, a significant decline in PIabs, Sm, N, \(\psi_{(E_{0})}\), and \(\varphi_{(E_{0})}\) (quantum yield of electron transport), and an increase in VJ and M0 was found in the P-free group (p < 0.05).
A simple energy flow model is shown in Fig. 4. Referring to the RC in the membrane and thus dealing with specific energy flux ABS/RC (absorption flux per RC), TR0/RC (trapped energy flux per RC) and DI0/RC (dissipated energy flux per RC) increased significantly, but ET0/RC (electron transport flux per RC) and RE0/RC (reduction of end acceptors at the PSI electron acceptor side per RC) declined markedly in the P-free group compared with those in the DIP group (p < 0.05). Similarly, a significant difference in TR0/RC, ET0/RC and RE0/RC was also found in the DOP group compared with that in the DIP group (p < 0.05). However, a significant decline in RC/CS0 (density of PSII RCs), ABS/CS0 (absorption flux per cross-section), TR0/CS0 (trapped energy flux per cross-section), ET0/CS0 (electron transport flux per cross-section), DI0/CS0 (dissipated energy flux per cross-section), and RE0/CS0 (reduction of end acceptors at the PSI side per cross-section) in the P-free and DOP treatments was observed in the algal model in contrast to the treatment of DIP. The most striking decease was the number of active RCs in the model.
Energy pipeline of the PSII behavior of P. bipes treated with P-free, DIP and DOP. The membrane model (left) demonstrates the specific energy fluxes (per reaction center, RC) and algae model (right) demonstrates the phenomenological energy fluxes (per excited cross-section, CS). The value of each energy flux is expressed by the appropriate adjustment of the width of the corresponding arrow. The small circles in the algae model show the number of active RC/CS0. *Indicates a p value < 0.05
Transcriptomic analysis
GO analyses revealed 31 downregulated genes when P. bipes was cultured in DOP compared with DIP that were mostly distributed in categories membrane, photosynthesis (photosynthetic electron transport chain, photosynthesis light reaction, and electron transport chain), generation of precursor metabolites and energy, thylakoid, and chlorophyll binding (Fig. 5a; Additional file 2). For example, the downregulated genes involved in photosynthesis included psbA (encoding PSII P680 RC D1 protein), psbD (encoding PSII P680 RC D2 protein), psbC (encoding PSII CP43 chlorophyll apoprotein), psbB (encoding PSII CP47 RC protein), psbW (encoding PSII RC W protein), petD (encoding electron transport protein cytochrome b (C-terminal)/b6), psaA (encoding PSI P700 chlorophyll a apoprotein A1), and psaB (encoding PSI P700 chlorophyll a apoprotein A2), which respectively showed 2.65-, 2.07-, 2.43-, 2.12-, 2.87-, 1.94-, 2.09- and 1.99-fold downregulation in P. bipes cultured in DOP compared with these genes of cells cultured in DIP (p < 0.05) (Fig. 5b).
Down-regulated expression genes in P. bipes between DOP and DIP condition. Results are summarized for the three main GO categories: biological processes, cellular component and molecular function (a). Differential gene expression related to photosynthesis in P. bipes between DOP and DIP conditions (b)
One hundred and forty-eight genes were significantly upregulated in DOP treatment compared with DIP treatment (Additional file 3). Significant shifts in gluconeogenesis were observed, and the genes encoding phosphoenolpyruvate carboxykinase, glycerol-3-phosphate dehydrogenase (GPDH), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), malate dehydrogenase, fructose-1,6-bisphosphatase (FBP), and isocitrate lyase were respectively upregulated 4.70-, 9.64-, 4.52-, 4.17-, 3.73- and 5.13-fold in the DOP culture compared with these genes in DIP culture (p < 0.05). In addition, changes were observed in expression of genes involved in the TCA (tricarboxylic acid) cycle, oxidative phosphorylation, sulfur metabolism, membrane transport, ribosome and transcription (p < 0.05) (Fig. 6).
A schematized network of cellular metabolites’ network derived from transcriptome analyses in P. bipes between DOP and DIP treatments. The biochemical pathways and metabolites in blue are up-regulation. Cit citrate, Aco aconitate, Iso isocitrate, Oxa oxalosuccinate, Ket ketoglutarate, Suc-CoA succinyl-CoA, Suc succinate, Fum fumarate, Mal malate, Oxal oxaloacetate, Glu glutamine, Cyt cytochrome, FBP fructose-1,6-bisphosphatase, ETHE1 sulfur dioxygenase, GAPDH glyceraldehyde-3-phosphate dehydrogenase, PEP phosphoenolpyruvate, PEPCK phosphoenolpyruvate carboxykinase, F6P fructose-6-phosphate, G3P glycerol-3-phosphate, GPDH G3P dehydrogenase, GAP glyceraldehyde-3-phosphate, DHAP dihydroxyacetone phosphate
Discussion
Mechanism of β-glycerol phosphate uptake and use
In the face of DIP limitation, algae have evolved a set of mechanisms to obtain sufficient P [18, 52], including enhancement of the ability to assimilate low abundance inorganic phosphorus via high-affinity phosphate transporters [53]; decreasing the demand for phosphorus by replacing membrane phospholipids with non-phosphorus lipids and accelerating the turnover of phospholipid to provide a short-term phosphorus supply [54,55,56]; bypassing phosphorus-consuming processes in glycolysis reactions and lipid recycling [57,58,59]; and obtaining P from organic phosphorus, which is the major alternative phosphorus supply to DIP in ecosystems [34, 60,61,62,63], via alkaline phosphatase (AP) and other hydrolytic enzymes [6, 18, 64].
Oh et al. [36] found that Alexandrium tamarense grew poorly on glycerophosphate, while Gymnodinium catenatum was able to use glycerophosphate as well as DIP, suggesting that dinoflagellates show different responses to glycerophosphate. DIP can be directly absorbed and used by cells, but the application of DOP requires cells to consume more energy to break bonds to obtain active phosphorus [62, 65]. In the present study, P. bipes grew well using β-glycerol phosphate—only slightly lower growth was found in the DOP group compared with the DIP group (Fig. 1), this indicates that glycerophosphate could be used as the sole phosphorus source to support the growth of P. bipes.
Measurement of PSII activity can provide information about photosynthetic apparatus, the absorption and distribution of energy, and the transportation of electrons in PSII of intact algal cells [66, 67]. Chlorophyll fluorescence is a very useful method to analyze the energy transfer and photochemical events of PSII [47, 68, 69]. In this study, a remarkable decrease in density of reaction centers (RC/CS0), absorption flux per cross-section (ABS/CS0), dissipated energy flux per cross-section (DI0/CS0), trapped energy flux per cross-section (TR0/CS0), electron transport flux per cross-section (ET0/CS0), and reduction of end acceptors at the PSI electron acceptor side per cross-section (RE0/CS0) was found in P-free and DOP conditions compared with the culture grown in DIP (Additional file 1: Table S1; Figs. 3 and 4), indicating that photosynthesis decreased significantly in P-free and DOP conditions [70,71,72].
Moreover, an increase in the closure of active RCs (Vj) and the maximum rate at which QA was reduced (M0), and a decrease in the energy required for QA to be fully reduced (Sm) and for QA to be reduced in the time span from 0 to tFmax (N), was also observed in the P-free and DOP groups (Additional file 1: Table S1; Figs. 3 and 4), suggesting that the probability that an electron residing on Q−A would enter the transport chain was reduced; which in turn reduced the proportion of quanta in the electron transport chain [47, 68, 69]. These findings indicated that photosynthesis was inhibited in the DOP culture of P. bipes due to blockage of the electron transport chain from Q−A to QB.
Transcriptomic analyses revealed that 31 genes, including many encoding photosynthesis-related proteins such as psbA, psbD, psbC, psbB, psbW, petD, psaA and psaB, were downregulated when P. bipes was cultured in DOP compared with that in DIP (Fig. 5a; Additional file 2). The RC of PSII consists of proteins D1 (encoded by psbA) and D2 (encoded by psbD), which provide the binding sites for the electron transfer chain cofactors [73]. Protein D1 is connected with RC P680, the primary electron acceptor pheophytin, and the secondary electron acceptor QB, while the D2 protein provides a binding site for the electron acceptor QA [74, 75]. Due to strongly oxidative chemistry of PSII water splitting, the D1 protein is prone to constant photodamage, whereas most of the other PSII subunits ordinarily remain undamaged [76]. When subjected to environmental stress, the degradation rate of D1 and D2 proteins is greater than their synthesis rate, which drives destruction of the RC and blocks electron transfer [77]. In this study, both psbA and psbD were downregulated in the DOP group (Fig. 5a; Additional file 2), suggesting that the synthesis rate of D1 and D2 proteins might be impeded, resulting in damage to the RCs and blocked electron transfer. CP43 and CP47 are important subunits of PSII, which together form the central light-harvesting pigment complex and are closely connected to the RC [78]. Expression of psbC and psbB was downregulated (Fig. 5b) in the DOP treatment, indicating that the ability to capture light was reduced. These data collectively indicated that PSII activity was inhibited in the DOP-cultured P. bipes.
Previous studies have proposed that some phytoplankton can excrete AP to hydrolyze organic phosphorus when DIP is scarce [79, 80]. It is clear that in our study, β-glycerol phosphate was hydrolyzed because the concentration of DIP in the medium increased (Fig. 2a). Luo et al. [6] proposed that three mechanisms might underlie the use of DOP: (1) extracellular DOP hydrolysis is slow and coupled tightly to uptake, leaving no residual DIP; (2) DOP is absorbed directly without extracellular hydrolysis; and (3) both extracellular hydrolysis and direct uptake of DOP or its hydrolysis products occur. In the present study, however, there was no significant increase in APA when P. bipes was cultured in DOP (Fig. 2b), suggesting that P. bipes did not employ AP for β-glycerol phosphate hydrolysis. Huang et al. [81] showed that APA increased only when low-molecular-weight DOP was exhausted in marine microalgae. Luo et al. [6] found that the dinoflagellate Karenia mikimotoi did not release AP to hydrolyze DOP when it was cultured in ATP, and 5ʹ-nucleotidase enzymatic activity was responsible for the utilization of ATP in K. mikimotoi [6], Prochlorococcus MED4 [82], Thalassiosira pseudonana [63] and Emiliania huxleyi [83]. These results suggested that AP was not the only enzyme to hydrolyze DOP. Therefore, the mechanism of DOP use in P. bipes needs to be further studied in the future.
Modulation of metabolic pathways by β-glycerol phosphate
One hundred and forty-eight significantly upregulated genes were detected in transcriptomic analyses on growth of P. bipes in DOP compared with DIP (Fig. 6; Additional file 3). ATP synthases are membrane-embedded rotary motors that produce or consume ATP and control the pH within cells [84]. The Na+/K+-ATPase is an ion pump belonging to the P-type ATPase family. Using energy derived from ATP hydrolysis, Na+/K+-ATPase generates electrochemical gradients for Na+ and K+ across plasma membranes, as required for electrical excitability, cellular uptake of ions, nutrients and neurotransmitters, and regulation of cell volume and intracellular pH [85]. Kdp is a P-type ATPase of unique structure, a complex of three membrane proteins (KdpFABC); this complex is a high-affinity ATP-dependent potassium uptake system, which plays a fundamental role in the steady supply of potassium to cells [86]. The Ca2+-transporting ATPase is the most abundant in the sarcoplasmic reticulum membrane [87]. It couples the energy derived from hydrolysis of ATP to transport of Ca2+ ions across the membrane against a concentration gradient [87]. In the present study, genes encoding these P-type ATPases were upregulated in the DOP group (Fig. 6; Additional file 3), suggesting that DOP affected the regulation of ion transport and osmotic pressure of cells. F-type ATPases are highly conserved enzymes used primarily for the synthesis of ATP [84]. Upregulation of the genes encoding the F-type synthase (ATPeF1A, ATPeF1B) (Fig. 6; Additional file 3) indicated increased ATP synthesis and energy metabolism in the DOP group.
The metabolism of glycerol-3-phosphate (G3P) is important for environmental stress responses of eukaryotic microalgae. All cells contain polar lipids with a glycerol backbone; the glycerol backbone in the polar lipids of bacteria and eukaryotes is G3P [88]. G3P can be used in various metabolic pathways via glycolysis [88]. GPDH activity is important for glycerol production via the metabolism of G3P to provide osmotic stress tolerance or for lipid synthesis in many organisms [89, 90]. There is a G3P shuttle in mitochondria, consisting essentially of two components: an NAD+-GPDH (EC 1.1.1.8) in the cytosol, and a membrane-bound FAD-GPDH (EC 1.1.99.5) located in the inner mitochondrial membrane. The NAD+-GPDH reduces dihydroxyacetone phosphate (DHAP) to G3P using NADH as a reducing agent, while the FAD-GPDH catalyzes the conversion of G3P to DHAP and forms FADH2 which enters the electron transport chain [91]. In this study, the expression of glpA encoding the FAD-GPDH was 9.64-fold higher in the DOP treatment than in the DIP treatment (Additional file 3), indicating the enhancement of DHAP generation.
The structure of β-glycerol phosphate used as the DOP in the experiments in this study is similar to that of G3P. Thus, GDPH might be an important enzyme for catalysis of conversion of β-glycerol phosphate to DHAP. In the DOP culture group, the genes encoding GAPDH and FBP were significantly upregulated. GAPDH and FBP are key enzymes in gluconeogenesis. GAPDH can reversibly catalyze the oxidation and phosphorylation of 1,3-diphosphoglycerate to glyceraldehyde-3-phosphate to form Pi [92]. FBP (encoded by FBP) and phosphoenolpyruvate carboxykinase (encoded by pckA) regulate the irreversible steps of gluconeogenesis [93, 94]. In the present study, the expression of FBP and pckA in P. bipes was significantly upregulated in the DOP group compared with the DIP group (Fig. 6; Additional file 3), suggesting that the gluconeogenesis pathway was fueled by β-glycerol phosphate. As photosynthesis was downregulated on culture in DOP, intracellular organic synthesis and insufficient supply of glucose would be replenished by gluconeogenesis. Moreover, gluconeogenesis releases Pi, resulting in the observed increase of DIP in the culture medium (Fig. 2). The enhanced Pi could make up for the deficiency in the intracellular phosphorus supply if DOP could not be directly used, or the use was delayed.
The TCA cycle is the final common catabolic pathway for the oxidation of fuel molecules. In this cycle, succinyl-CoA synthetase converts succinyl-CoA to succinate and yields a high-energy phosphate bond [95]. Malate dehydrogenase, encoded by the gene MDH2, is responsible for the regeneration of oxaloacetic acid and a new turn of the TCA cycle, and catalyzes conversion of malate to oxaloacetate and generates NADH. Oxidative decarboxylation is an important stage in the TCA cycle. Complex I (NADH-Q reductase), Complex II (succinate-Q reductase), Complex III (cytochrome c reductase), Complex IV (cytochrome c oxidase) and Complex V (ATP synthase) are involved in oxidative phosphorylation to catalyze the transfer of electrons coupled with protons across the membrane using substrates generated in the TCA cycle [96]. In this study, genes encoding succinyl-CoA synthase (e.g. LSC1, LSC2) and malate dehydrogenase (e.g. MDH2) were significantly upregulated in the DOP group compared with the DIP group (Fig. 6; Additional file 3). Moreover, genes involved in oxidative phosphorylation, such as those encoding cytochrome c reductase (GO: 0004553) and cytochrome c oxidase (e.g. CYC, COX1), also showed significantly higher expression in the DOP group compared with the DIP group (Fig. 6; Additional file 3). These data indicate that P. bipes could enhance the production of energy to support metabolism when it was cultured in DOP.
The gene Amt is associated with ammonium transport and is induced substantially when ammonium is limiting for growth. The assimilation of ammonium is regulated by the combined activities of glutamine synthetase, encoded by glnA [97]. In this study, Amt and glnA were respectively upregulated 6.77- and 3.92-fold in the DOP group compared with the DIP group (Additional file 3), suggesting that the enhancement of ammonium transport is possibly resulted from the decline of the N:P ratio due to increased P concentration in the DOP group. Similar results have been observed in other dinoflagellates, such as K. mikimotoi [6]. In addition, regulation of ammonium transporters is not only a function of substrate availability resulting from external supply, but also the extent of internal metabolic pathways that are ammonium-generating, namely photorespiration and the extent of mixotrophic nutrition [98].
Krüßel et al. [99] demonstrated that sulfur dioxygenase (encoded by ETHE1) plays an important role in plant cysteine catabolism. Sulfur dioxygenase can oxidize persulfide to sulfite and is involved in metabolic homeostasis in mitochondria. It can also bind to nuclear transcription factors [100]. The cysteine desulfurase IscS can catalyze formation of alanine and sulfane sulfur from cysteine, and provides sulfur for iron–sulfur cluster synthesis [101]. The genes ETHT1 and IscS were upregulated 3.66- and 7.23-fold, respectively, in the DOP group in our study compared with the DIP group (Additional file 3), suggesting that DOP improved sulfur metabolism and respiratory electron transport [99].
Conclusions
The present study combined physiological and transcriptome sequencing analyses to determine the response of P. bipes to different phosphorus conditions. The data indicate that the RC of PSII was damaged and the electron transfer in PSII was blocked when P. bipes was cultured in DOP conditions, due to the downregulation of photosynthesis genes. GPDH and GAPDH are most likely responsible for β-glycerol phosphate use, and alkaline phosphatase activity was an indicator of DIP stress, but not an exclusive indicator for DOP. Moreover, gluconeogenesis played a vital role in β-glycerol phosphate use. In addition, ATPases, the TCA cycle, oxidative phosphorylation, and fatty acid, ammonium, sulfur and amino acid metabolism were significantly upregulated in DOP conditions compared with culture in DIP, suggesting their involvement in the use of β-glycerol phosphate or its hydrolysis products in P. bipes. Collectively, these findings provided a comprehensive understanding of the phosphorus physiology of P. bipes. In addition, our results underscore that GPDH and GAPDH should be studied further, and that the mechanisms underlying the use of β-glycerol phosphate and other DOPs are different in different species of dinoflagellates and other phytoplankton, suggesting that the control of dinoflagellate blooms by targeted nutrient reduction is largely dependent upon the dominant species. DIP reduction may be more effective in controlling the bloom of P. bipes than DOP reduction.
Availability of data and materials
The material contained in this manuscript has not been published or submitted to another publisher in part or in whole. All authors have reviewed the final version of the manuscript and approve it for publication.
Abbreviations
- DIP:
-
Dissolved inorganic phosphorus, it also represents K2HPO4 here
- DOP:
-
Dissolved organic phosphorus, it also represents glycerol phosphate here
- TCA:
-
Tricarboxylic acid
- TDP:
-
Total dissolved phosphorus
- OJIP:
-
The polyphasic Chl a fluorescence transient
- RC:
-
Active reaction centers
- APA:
-
Alkaline phosphatase activity
- FPKM:
-
Fragments per kilobase of exon per millions of fragments mapped
- PIabs:
-
Performance index based on absorption of light energy
- S m :
-
Multiple turnover in the closure of the RCs
- N :
-
QA has been reduced in the time span from 0 to tFmax
- \(\psi_{(E_{0})}\) :
-
Probability [at t = 0] that a trapped exciton moves an electron into the electron transport chain beyond QA
- V J :
-
Relative variable fluorescence in the J-step
- M 0 :
-
Approximate initial slope of the fluorescence transient
- \(\varphi_{(E_{0})}\) :
-
Quantum yield of electron transport
- ABS/RC:
-
Absorption flux per RC
- TR0/RC:
-
Trapped energy flux per RC
- DI0/RC:
-
Dissipated energy flux per RC
- ET0/RC:
-
Electron transport flux per RC
- RE0/RC:
-
Reduction of end acceptors at the PSI electron acceptor side per RC
- RC/CS0 :
-
Density of PSII RCs
- ABS/CS0 :
-
Absorption flux per cross-section
- TR0/CS0 :
-
Trapped energy flux per cross-section
- ET0/CS0 :
-
Electron transport flux per cross-section
- DI0/CS0 :
-
Dissipated energy flux per cross-section
- RE0/CS0 :
-
Reduction of end acceptors at the PSI side per cross-section
- GPDH:
-
Glycerol-3-phosphate dehydrogenase
- GAPDH:
-
Glyceraldehyde-3-phosphate dehydrogenase
- FBP:
-
Fructose-1,6-bisphosphatase
- AP:
-
Alkaline phosphatase
- G3P:
-
Glycerol-3-phosphate
- DHAP:
-
Dihydroxyacetone phosphate
References
Takeda E, Taketani Y, Nashiki K, Nomoto M, Shuto E, Sawada N, Yamamoto H, Isshiki M (2006) A novel function of phosphate-mediated intracellular signal transduction pathways. Adv Enzyme Regul 46(1):154–161
Paytan A, Mclaughlin K (2007) The oceanic phosphorus cycle. Chem Rev 107(2):563–576
Wang X, Huang B, Zhang H (2014) Phosphorus deficiency affects multiple macromolecular biosynthesis pathways of Thalassiosira weissflogii. Acta Oceanol Sin 33(4):85–91
Yuan Y, Bi Y, Hu Z (2017) Phytoplankton communities determine the spatio-temporal heterogeneity of alkaline phosphatase activity: evidence from a tributary of the Three Gorges Reservoir. Sci Rep 7(1):16404
Schindler DW (1977) Evolution of phosphorus limitation in lakes. Science 195(4275):260–262
Luo H, Lin X, Li L, Lin L, Zhang C, Lin S (2017) Transcriptomic and physiological analyses of the dinoflagellate Karenia mikimotoi reveal non-alkaline phosphatase-based molecular machinery of ATP utilization. Environ Microbiol 19(11):4506–4518
Currie DJ, Kalff J (1984) The relative importance of bacterioplankton and phytoplankton in phosphorus uptake in freshwater. Limnol Oceanogr 29(2):311–321
Benitez-Nelson CR (2000) The biogeochemical cycling of phosphorus in marine systems. Earth-Sci Rev 51(1):109–135
Dyhrman ST, Ruttenberg KC (2006) Presence and regulation of alkaline phosphatase activity in eukaryotic phytoplankton from the coastal ocean: implications for dissolved organic phosphorus remineralization. Limnol Oceanogr 51(3):1381–1390
Meseck SL, Alix JH, Wikfors GH, Ward JE (2009) Differences in the soluble, residual phosphate concentrations at which coastal phytoplankton species up-regulate alkaline-phosphatase expression, as measured by flow-cytometric detection of ELF-97® fluorescence. Estuar Coast 32(6):1195–1204
Taylor WD, Lean DRS (2018) Observations on the dynamics and fate of dissolved organic phosphorus in lake water and a new model of eplimnetic P cycling. Aquat Sci 80(2):13
Tahara M, Kubota R, Nakazawa H, Tokunaga H, Nishimura T (2005) Use of cholinesterase activity as an indicator for the effects of combinations of organophosphorus pesticides in water from environmental sources. Water Res 39(20):5112–5118
Wu J, Sunda W, Boyle EA, Karl DM (2000) Phosphate depletion in the western North Atlantic Ocean. Science 289(5480):759–762
Cavender-Bares KK, Karl DM, Chisholm SW (2001) Nutrient gradients in the western North Atlantic Ocean: relationship to microbial community structure and comparison to patterns in the Pacific Ocean. Deep-Sea Res Pt I 48(11):2373–2395
Ammerman JW, Hood RR, Case DA, Cotner JB (2003) Phosphorus deficiency in the Atlantic: an emerging paradigm in oceanography. Eos Trans AGU 84(18):165–170
Mclaughlin K, Sohm JA, Cutter GA, Lomas MW, Paytan A (2013) Phosphorus cycling in the Sargasso Sea: investigation using the oxygen isotopic composition of phosphate, enzyme-labeled fluorescence, and turnover times. Glob Biogeochem Cycle 27(2):375–387
Bogé G, Lespilette M, Jamet D, Jamet JL (2013) The relationships between particulate and soluble alkaline phosphatase activities and the concentration of phosphorus dissolved in the seawater of Toulon Bay (NW Mediterranean). Mar Pollut Bull 74(1):413–419
Lin S, Litaker RW, Sunda WG (2016) Phosphorus physiological ecology and molecular mechanisms in marine phytoplankton. J Phycol 52(1):10–36
Duke SA, Rees DA, Forward GC (1979) Increased plaque calcium and phosphorus concentrations after using a calcium carbonate toothpaste containing calcium glycerophosphate and sodium monofluorophosphate. Caries Res 13(1):57–59
Langenbach F, Handschel J (2013) Effects of dexamethasone, ascorbic acid and β-glycerophosphate on the osteogenic differentiation of stem cells in vitro. Stem Cell Res Ther 4(5):117
Deng A, Kang X, Zhang J, Yang Y, Yang S (2017) Enhanced gelation of chitosan/β-sodium glycerophosphate thermosensitive hydrogel with sodium bicarbonate and biocompatibility evaluated. Mat Sci Eng C Mater 78:1147–1154
Taylor FJR, Hoppenrath M, Saldarriaga JF (2008) Dinoflagellate diversity and distribution. Biodivers Conserv 17(2):407–418
Le Bescot N, Mahé F, Audic S, Dimier C, Garet M, Poulain J, Wincker P, Vargas C, Siano R (2016) Global patterns of pelagic dinoflagellate diversity across protist size classes unveiled by metabarcoding. Environ Microbiol 18(2):609–626
Casabianca S, Perini F, Casabianca A, Pugliese L, Giussani V, Chiantore M, Penna A (2014) Monitoring toxic Ostreopsis cf. ovata in recreational waters using a qPCR-based assay. Mar Pollut Bull 88(1–2):102–109
Du X, Peterson W, Mcculloch A, Liu G (2011) An unusual bloom of the dinoflagellate Akashiwo sanguinea off the central Oregon, USA, coast in autumn 2009. Harmful Algae 10:784–793
Yang C, Li Y, Zhou Y, Zheng W, Tian Y, Zheng T (2012) Bacterial community dynamics during a bloom caused by Akashiwo sanguinea in the Xiamen sea area, China. Harmful Algae 20:132–141
Chen T, Liu Y, Song S, Li C, Tang Y, Yu Z (2015) The effects of major environmental factors and nutrient limitation on growth and encystment of planktonic dinoflagellate Akashiwo sanguinea. Harmful Algae 46:62–70
Anderson DM, Alpermann TJ, Cembella AD, Collos Y, Masseret E, Montresor M (2012) The globally distributed genus Alexandrium: multifaceted roles in marine ecosystems and impacts on human health. Harmful Algae 14:10–35
Yamaguchi H, Hirano T, Yoshimatsu T, Tanimoto Y, Matsumoto T, Suzuki S, Hayashi Y, Urabe A, Miyamura K, Sakamoto S, Yamaguchi M, Tomaru Y (2016) Occurrence of Karenia papilionacea (Dinophyceae) and its novel sister phylotype in Japanese coastal waters. Harmful Algae 57(Part A):59–68
Gobler CJ, Berry DL, Anderson OR, Burson A, Koch F, Rodgers BS, Moore LK, Goleski JA, Allam B, Bowser P, Tang Y, Nuzzi R (2008) Characterization, dynamics, and ecological impacts of harmful Cochlodinium polykrikoides blooms on eastern Long Island, NY, USA. Harmful Algae 7(3):293–307
Richlen ML, Morton SL, Jamali EA, Rajan A, Anderson DM (2010) The catastrophic 2008–2009 red tide in the Arabian Gulf region, with observations on the identification and phylogeny of the fish-killing dinoflagellate Cochlodinium polykrikoides. Harmful Algae 9(2):163–172
Vanucci S, Pezzolesi L, Pistocchi R, Ciminiello P, Dell’Aversano C, Iacovo ED, Fattorusso E, Tartaglione L, Guerrini F (2012) Nitrogen and phosphorus limitation effects on cell growth, biovolume, and toxin production in Ostreopsis cf. ovata. Harmful Algae 15:78–90
Abadiea E, Chiantellaa C, Crottiera A, Rhodesb L, Masseretc E, Berteauxa T, Laabir M (2018) What are the main environmental factors driving the development of the neurotoxic dinoflagellate Vulcanodinium rugosum in a Mediterranean ecosystem (Ingril lagoon, France)? Harmful Algae 75:75–86
Ou L, Wang D, Huang B, Hong H, Qi Y, Lu S (2008) Comparative study of phosphorus strategies of three typical harmful algae in Chinese coastal waters. J Plankton Res 30(9):1007–1017
Park J, Jeong HJ, Yoo YD, Yoon EY (2013) Mixotrophic dinoflagellate red tides in Korean waters: distribution and ecophysiology. Harmful Algae 30S:S28–S40
Oh SJ, Yamamoto T, Kataoka Y, Matsuda O, Matsuyama Y, Kotani Y (2002) Utilization of dissolved organic phosphorus by the two toxic dinoflagellates, Alexandrium tamarense and Gymnodinium catenatum (Dinophyceae). Fish Sci 68(2):416–424
Huang B, Ou L, Hong H, Luo H, Wang D (2005) Bioavailability of dissolved organic phosphorus compounds to typical harmful dinoflagellate Prorocentrum donghaiense Lu. Mar Pollut Bull 51(8–12):838–844
Richardson B, Corcoran AA (2015) Use of dissolved inorganic and organic phosphorus by axenic and nonaxenic clones of Karenia brevis and Karenia mikimotoi. Harmful Algae 48:30–36
Wynne D, Patni NJ, Aaronson S, Berman T (1982) The relationship between nutrient status and chemical composition of Peridinium cinctum during the bloom in Lake Kinneret. J Plankton Res 4(1):125–136
Regel RH (2004) Vertical migration, entrainment and photosynthesis of the freshwater dinoflagellate Peridinium cinctum in a shallow urban lake. J Plankton Res 26(2):143–157
Wu JT, Kuo-Huang LL, Lee J (1998) Algicidal effect of Peridinium bipes on Microcystis aeruginosa. Curr Microbiol 37(4):257–261
Sun Y, You J, Lei L, Han B (2008) The feature of particulate fatty acids composition in a dinoflagellates bloom in Huanglongdai Reservoir. South China. Ecol Sci 27(5):394–397 (in Chinese, with English abstract)
Shumway S (1990) A Review of the effects of algal blooms on shellfish and aquaculture. J World Aquacult Soc 21(2):65–104
Gainey LF, Shumway SE (1991) The physiological effect of Aureococcus anophagefferens (“Brown Tide”) on the Lateral Cilia of Bivalve Mollusks. Biol Bull 181(2):298–306
Smayda TJ (1997) Harmful algal blooms: their ecophysiology and general relevance to phytoplankton blooms in the sea. Limnol Oceanogr 42(5, Part 2):1137–1153
Landsberg JH (2002) The effects of harmful algal blooms on aquatic organisms. Rev Fish Sci 10(2):113–390
Strasser RJ, Tsimilli-Michael M, Srivastava A (2004) Analysis of the chlorophyll a fluorescence transient. In: Papageorgiou GC, Govindjee (eds) Chlorophyll a fluorescence. Springer, Dordrecht
Appenroth KJ, Stöckel J, Srivastava A, Strasser RJ (2001) Multiple effects of chromate on the photosynthetic apparatus of Spirodela polyrhiza as probed by OJIP chlorophyll a fluorescence measurement. Environ Pollut 115(1):49–64
Martin JB, Doty DM (1949) Determination of inorganic phosphate. Anal Chem 21(8):965–967
Shen H, Song L (2007) Comparative studies on physiological responses to phosphorus in two phenotypes of bloom-forming Microcystis. Hydrobiologia 592(1):475–486
Dong C, Zhang H, Yang Y, He X, Liu L, Fu J, Shi J, Wu Z (2019) Physiological and transcriptomic analyses to determine the responses to phosphorus utilization in Nostoc sp. Harmful Algae 84:10–18
Dyhrman S, Ammerman J, Van Mooy B (2007) Microbes and the marine phosphorus cycle. Oceanography 20(2):110–116
Orchard ED, Webb EA, Dyhrman ST (2009) Molecular analysis of the phosphorus starvation response in Trichodesmium spp. Environ Microbiol 11(9):2400–2411
Van Mooy BAS, Rocap G, Fredricks HF, Evans CT, Devol AH (2006) Sulfolipids dramatically decrease phosphorus demand by picocyanobacteria in oligotrophic marine environments. Proc Natl Acad Sci USA 103(23):8607–8612
Van Mooy BAS, Fredricks HF, Pedler BE, Dyhrman ST, Karl DM, Koblížek M, Lomas MW, Mincer TJ, Moore LR, Moutin T, Rappé MS, Webb EA (2009) Phytoplankton in the ocean use non-phosphorus lipids in response to phosphorus scarcity. Nature 458(7234):69–72
Martin P, Van Mooy BAS, Heithoff A, Dyhrman ST (2011) Phosphorus supply drives rapid turnover of membrane phospholipids in the diatom Thalassiosira pseudonana. ISME J 5(6):1057–1060
Theodorou ME, Elrifi IR, Turpin DH, Plaxton WC (1991) Effects of phosphorus limitation on respiratory metabolism in the green-alga Selenastrum minutum. Plant Physiol 95(4):1089–1095
Duff SMG, Moorhead GBG, Lefebvre DD, Plaxton WC (1989) Phosphate starvation inducible bypasses of adenylate and phosphate dependent glycolytic enzymes in Brassica nigra suspension cells. Plant Physiol 90:1275–1278
Wurch LL, Bertrand EM, Saito MA, Van Mooy BAS, Dyhrman ST (2011) Proteome changes driven by phosphorus deficiency and recovery in the brown tide-forming alga Aureococcus anophagefferens. PLoS ONE 6(12):e28949
Lin X, Zhang H, Huang B, Lin S (2011) Alkaline phosphatase gene sequence and transcriptional regulation by phosphate limitation in Amphidinium carterae (Dinophyceae). J Phycol 47(5):1110–1120
Lin X, Zhang H, Huang B, Lin S (2012) Alkaline phosphatase gene sequence characteristics and transcriptional regulation by phosphate limitation in Karenia brevis (Dinophyceae). Harmful Algae 17:14–24
Dyhrman ST, Chappell PD, Haley ST, Moffett JW, Orchard ED, Waterbury JB, Webb EA (2006) Phosphonate utilization by the globally important marine diazotroph Trichodesmium. Nature 439(7072):68–71
Dyhrman ST, Jenkins BD, Rynearson TA, Saito MA, Mercier ML, Alexander H, Whitney LP, Drzewianowski A, Bulygin VV, Bertrand EM, Wu Z, Benitez-Nelson C, Heithoff A (2012) The transcriptome and proteome of the diatom Thalassiosira pseudonana reveal a diverse phosphorus stress response. PLoS ONE 7(3):e33768
Bai F, Liu R, Yang Y, Ran X, Shi J, Wu Z (2014) Dissolved organic phosphorus use by the invasive freshwater diazotroph cyanobacterium, Cylindrospermopsis raciborskii. Harmful Algae 39:112–120
Ashkenazi N, Zade SS, Segall Y, Karton Y, Bendikov M (2005) Selective site controlled nucleophilic attacks in 5-membered ring phosphate esters: unusual C–O vs. common P–O bond cleavage. Chem Commun 37(47):5879–5881
Zhou W, Juneau P, Qiu B (2006) Growth and photosynthetic responses of the bloom-forming cyanobacterium Microcystis aeruginosa to elevated levels of cadmium. Chemosphere 65(10):1738–1746
Perales-Vela HV, González-Moreno S, Montes-Horcasitas C, Cañizares-Villanueva RO (2007) Growth, photosynthetic and respiratory responses to sub-lethal copper concentrations in Scenedesmus incrassatulus (Chlorophyceae). Chemosphere 67(11):2274–2281
Strasser RJ, Strasser BJ (1995) Measuring fast fluorescence transients to address environmental questions: the JIP test. In: Mathis P (ed) Photosynthesis: from light to biosphere. Kluwer Academic Publishers, Dordrecht
Strasser RJ, Srivastava A, Tsimilli-Michael M (2000) The fluorescence transient as a tool to characterize and screen photosynthetic samples. In: Yunus M, Pathre U, Mohanty P (eds) Probing photosynthesis: mechanisms, regulation and adaptation. Taylor and Francis Press, London
Krüger GHJ, Tsimilli-Michael M, Strasser RJ (1997) Light stress provokes plastic and elastic modifications in structure and function of photosystem II in camellia leaves. Physiol Plantarum 101(2):265–277
Force L, Critchley C, Van Rensen JJS (2003) New fluorescence parameters for monitoring photosynthesis in plants. Photosynth Res 78(1):17–33
Eullaffroy P, Frankart C, Aziz A, Couderchet M, Blaise C (2009) Energy fluxes and driving forces for photosynthesis in Lemna minor exposed to herbicides. Aquat Bot 90(2):172–178
Hankamer B, Barber J, Boekema EJ (1997) Structure and membrane organization of photosystem II in green plants. Annu Rev Plant Biol 48(1):641–671
Barber J, Nield J, Morris EP, Zheleva D (1997) The structure, function and dynamics of photosystem II. Physiol Plantarum 100(4):817–827
Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S (2004) Architecture of the photosynthetic oxygen-evolving center. Science 303(5665):1831–1838
Chis C, Mahboob A, Vassilieev S, Bica A, Peca L, Brouce D, Aro E, Sicora CI (2013) D1′—a new member of D1 protein family in cyanobacteria. In: Kuang T, Lu C, Zhang L (eds) Photosynthesis research for food, fuel and the future. Springer, Berlin
Andersson B, Aro EM (1997) Proteolytic activities and proteases of plant chloroplasts. Physiol Plantarum 100(4):780–793
Bricker TM (1990) The structure and function of CPa-1 and CPa-2 in photosystem II. Photosynth Res 24(1):1–13
Gillor O, Hadas O, Post AF, Belkin S (2002) Phosphorus bioavailability monitoring by a bioluminescent cyanobacterial sensor strain. J Phycol 38(1):107–115
Wu Z, Zeng B, Li R, Song L (2012) Physiological regulation of Cylindrospermopsis raciborskii (Nostocales, Cyanobacteria) in response to inorganic phosphorus limitation. Harmful Algae 15:53–58
Huang B, Huang S, Weng Y, Hong H (2000) Effect of dissolved phosphorus on alkaline phosphatase activity in marine microalgae. Acta Oceanol Sin 19(2):29–35
Krumhardt KM, Callnan K, Roache-Johnson K, Swett T, Robinson D, Reistetter EN, Saunders JK, Rocap G, Moore LR (2013) Effects of phosphorus starvation versus limitation on the marine cyanobacterium Prochlorococcus MED4 I: uptake physiology. Environ Microbiol 15(7):2114–2128
Dyhrman ST, Palenik B (2003) Characterization of ectoenzyme activity and phosphate-regulated proteins in the coccolithophorid Emiliania huxleyi. J Plankton Res 25(10):1215–1225
Schmidt C, Zhou M, Marriott H, Morgner N, Politis A, Robinson CV (2013) Comparative cross-linking and mass spectrometry of an intact F-type ATPase suggest a role for phosphorylation. Nat Commun 4:1985
Morth JP, Pedersen BP, Toustrup-Jensen MS, Sørensen TLM, Petersen J, Andersen JP, Vilsen B, Nissen P (2007) Crystal structure of the sodium–potassium pump. Nature 450(7172):1043–1049
Irzik K, Pfrötzschner J, Goss T, Ahnert F, Haupt M, Greie JC (2011) The KdpC subunit of the Escherichia coli K+-transporting KdpB P-type ATPase acts as a catalytic chaperone. FEBS J 278(17):3041–3053
Yamamoto H, Kawakita M (1999) Chemical modification of an arginine residue in the ATP-binding site of Ca2+-transporting ATPase of sarcoplasmic reticulum by phenylglyoxal. Mol Cell Biochem 190(1–2):169–177
Shin-Ichi Y, Yoshiki N, Satoshi A, Akihiko Y (2016) Birth of archaeal cells: molecular phylogenetic analyses of G1P dehydrogenase, G3P dehydrogenases, and glycerol kinase suggest derived features of archaeal membranes having G1P polar lipids. Archaea 2016:1–16
Shen W, Li JQ, Dauk M, Huang Y, Periappuram C, Wei Y, Zou J (2010) Metabolic and transcriptional responses of glycerolipid pathways to a perturbation of glycerol 3-phosphate metabolism in Arabidopsis. J Biol Chem 285(30):22957–22965
Driver T, Trivedi DK, Mcintosh OA, Dean AP, Goodacre R, Pittman JK (2017) Two glycerol-3-phosphate dehydrogenases from Chlamydomonas have distinct roles in lipid metabolism. Plant Physiol 174(4):2083–2097
Shen W, Wei Y, Dauk M, Zheng Z, Zou J (2003) Identification of a mitochondrial glycerol-3-phosphate dehydrogenase from Arabidopsis thaliana: evidence for a mitochondrial glycerol-3-phosphate shuttle in plants. FEBS Lett 536(1–3):92–96
Velasco R, Salamini F, Bartels D (1994) Dehydration and ABA increase mRNA levels and enzyme activity of cytosolic GAPDH in the resurrection plant Craterostigma plantagineum. Plant Mol Biol 26(1):541–546
Lee PC, Lee SY, Hong SH, Chang HN (2002) Cloning and characterization of Mannheimia succiniciproducens MBEL55E phosphoenolpyruvate carboxykinase (pckA) gene. Biotechnol Bioproc E 7(2):95–99
Ghanem A, Kitanovic A, Holzwarth J, Wölfl S (2017) Mutational analysis of fructose-1,6-bis-phosphatase FBP1 indicates partially independent functions in gluconeogenesis and sensitivity to genotoxic stress. Microb Cell 4(2):52–63
Cohen GN (2010) Microbial biochemistry, 3rd edn. Springer, Dordrecht
Letts JA, Sazanov LA (2017) Clarifying the supercomplex: the higher-order organization of the mitochondrial electron transport chain. Nat Struct Mol Biol 24(10):800–808
Arnaud J, Gavin T, Anne-Marie M, Reinhard K, Mike M (2005) In vivo functional characterization of the Escherichia coli ammonium channel AmtB: evidence for metabolic coupling of AmtB to glutamine synthetase. Biochem J 390(1):215–222
Glibert PM, Wilkerson FP, Dugdale RC, Raven JA, Dupont CL, Leavitt PR, Parker AE, Burkholder JM, Kana TM (2016) Pluses and minuses of ammonium and nitrate uptake and assimilation by phytoplankton and implications for productivity and community composition, with emphasis on nitrogen-enriched conditions. Limnol Oceanogr 61(1):165–197
Krüßel L, Junemann J, Wirtz M, Brike H, Thornton JD, Browning LW, Poschet G, Hell R, Balk J, Braun HP, Hildebrandt TM (2014) The mitochondrial sulfur dioxygenase ethylmalonic encephalopathy protein1 is required for amino acid catabolism during carbohydrate starvation and embryo development in Arabidopsis. Plant Physiol 165(1):92–104
Hao Y, Huang J, Liu C, Li H, Liu J, Zeng Y, Yang Z, Li R (2016) Differential protein expression in metallothionein protection from depleted uranium-induced nephrotoxicity. Sci Rep 6(1):38942
Kambampati R (2000) Evidence for the transfer of sulfane sulfur from iscs to thii during the in vitro biosynthesis of 4-thiouridine in Escherichia coli tRNA. J Biol Chem 275(15):10727–10730
Acknowledgements
The research was supported by the National Natural Science Foundation of China (41877410), Fundamental Research Funds for the Central Universities (XDJK2019C044 and XDJK2017B010).
Funding
This work was supported by the National Natural Science Foundation of China (41877410), Fundamental Research Funds for the Central Universities (XDJK2019C044 and XDJK2017B010).
Author information
Authors and Affiliations
Contributions
Each author has participated sufficiently in the work to take public responsibility for all the content. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Table S1.
Parameters calculated using data extracted from the O-J-I-P fast fluorescence transient.
Additional file 2.
Information on down-regulated genes under different phosphorus treatments.
Additional file 3.
Information on up-regulated genes under different phosphorus treatments.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yang, Y., Shi, J., Jia, Y. et al. Unveiling the impact of glycerol phosphate (DOP) in the dinoflagellate Peridinium bipes by physiological and transcriptomic analysis. Environ Sci Eur 32, 38 (2020). https://doi.org/10.1186/s12302-020-00317-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12302-020-00317-6