Abstract
Background
Copy number variants (CNVs) have become increasingly instrumental in understanding the etiology of all diseases and phenotypes, including Neurocognitive Disorders (NDs). Among the well-established regions associated with ND are small parts of chromosome 16 deletions (16p11.2) and chromosome 15 duplications (15q3). Various methods have been developed to identify associations between CNVs and diseases of interest. The majority of methods are based on statistical inference techniques. However, due to the multi-dimensional nature of the features of the CNVs, these methods are still immature. The other aspect is that regions discovered by different methods are large, while the causative regions may be much smaller.
Results
In this study, we propose a regularized deep learning model to select causal regions for the target disease. With the help of the proximal [20] gradient descent algorithm, the model utilizes the group LASSO concept and embraces a deep learning model in a sparsity framework. We perform the CNV analysis for 74,811 individuals with three types of brain disorders, autism spectrum disorder (ASD), schizophrenia (SCZ), and developmental delay (DD), and also perform cumulative analysis to discover the regions that are common among the NDs. The brain expression of genes associated with diseases has increased by an average of 20 percent, and genes with homologs in mice that cause nervous system phenotypes have increased by 18 percent (on average). The DECIPHER data source also seeks other phenotypes connected to the detected regions alongside gene ontology analysis. The target diseases are correlated with some unexplored regions, such as deletions on 1q21.1 and 1q21.2 (for ASD), deletions on 20q12 (for SCZ), and duplications on 8p23.3 (for DD). Furthermore, our method is compared with other machine learning algorithms.
Conclusions
Our model effectively identifies regions associated with phenotypic traits using regularized deep learning. Rather than attempting to analyze the whole genome, CNVDeep allows us to focus only on the causative regions of disease.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Background
A copy number variant is an alteration of some base pairs in the human genome, which can be either deletion or duplication. Previously, due to the imprecision of CNV detection methods, CNVs were characterized as variations more significant than one kbps in size. From another point of view, CNVs can be inherited or de novo. Around 4.8–9.5% of the genome is affected by CNVs [1], a more significant portion compared to single nucleotide variants [2]. Together with single nucleotide polymorphisms and other types of structural variants, they are more likely to be associated to the etiology of genetic diseases. In addition, they are classified by their frequency of occurrence as rare variants or polymorphisms.
CNVs are associated with some disorders and phenotypic traits. For example, 22q11. 2 deletions are widely known to be associated with schizophrenia [3]. Moreover, a 20-kb deletion in the IRGM gene is associated with Crohn's disease, a 45-kb deletion of NEGR1 with body mass index, a 32-kb deletion with psoriasis, a 117-kb deletion of UGT2B17 with osteoporosis is reported in [4] and in Huntington disease the tandem repeat expansion occurs in HTT gene. Several studies have found CNV associations with diseases such as idiopathic learning disabilities, systemic lupus erythematosus, and inflammatory autoimmune disorders [5]. Some of the regions are reported to be associated with the three brain related disorders; for example, according to [6] (which gathered the regions from other papers), schizophrenia is associated with deletions at 1q21.1, 3q29, 15q11.2, 15q13.3, 16p12.1, and 22q11.2, as well as duplications at 1q21.1, 7q11.23, 15q11-q13, 16p13.11, and 16p11.2. Autism spectrum disorder is also associated with deletions in 1q21.1, 2p16.3, 15q11.2, 15q13.3, 16p11.2, and 22q11.2 distal, and duplications in 1q21.1, 7q11.23, 15q11q13, 16p11.2, 22q11.21, and 22q13.33 [7]. Besides, for the schizophrenia, deletions in 1q21.1, 1p36, 15q13.3, 15q24 and 16p11.2 and 17q21.31 and duplications in 16p11.2 and 22q11.2 are reported [8].
According to Fisher's exact test and/or permutation tests, the associative analysis relies primarily on statistical inference techniques. Several papers discuss various p-value problems [9]. The primary problem with CNV association with significance tests is how to construct regions to search for associations. Some papers manipulate single basepairs one at a time. [10] After identifying significant DNA segments, the main challenge is merging significant basepairs. The other idea is to evaluate the regions using the CNVs of cases and/or controls. [11] However, this approach is biased towards long- or short-case CNVs. In addition, there might be a subregion of the CNV that is causative for the disease, but the algorithm may find a larger superregion (instead of the subregion). Another idea is to determine the regions based on the positions of the genes. [12] The processing of this data will require a considerable number of genes. The next idea is to use a constant window size for the CNVs. [13] The problem of subregions (discussed above) is also associated with this idea. This approach may present another challenge in determining the window size and whether the windows overlap.
Several works have discussed the drawbacks of using p-values to measure significance. Another challenge is determining the significance threshold. SNATCNV [10] for autism, Coe et al. [11] and Cooper et al. [12]'s work on developmental delay, and PLINK [13] are several highly cited and state-of-the-art works on statistical significance.
Moreover, these methods cannot handle all the heterogeneous characteristics of CNVs effectively. CNV heterogeneous features consist of the type (a categorical variable), the start and end (numerical variables), and the individual ID (An IDentifier that identifies who the CNVs belong to). For example, when the type of the CNV is ignored, two CNVs with the same starts and ends with different types are considered the same, which in turn affects the analysis results; or if the ID of the person is ignored, each CNV is considered for an independent person, which has its shortcomings.
From another perspective, some methods involve calling and associative analysis, whereas others involve only associative analysis. CNV signals are studied in the first group, whereas the outputs of calling algorithms are examined in the second group. Our work belongs to the second group.
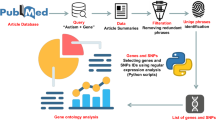
An overview of the pipeline proposed in this study is presented in Fig. 1. According to Fig. 1, we have a stage for model building and choosing the regions; the next step is evaluating the results. To build our model, we need a set of case–control CNVs. We use these CNVs to create a group of regions where changes in their copy number might cause disease, and the model evaluates their associations with the target.
Graphical Abstract: This is the summary of the model building and evaluation of the results. On the left, the model building, which is a collection of CNVs of healthy/sick individuals, is explained. The data is fed into a deep learning model. The next step, on the right, is evaluating the results. The evaluation consists of genes associated with mouse models, brain-enriched data, DECIPHER genotype–phenotype associations, gene ontology analysis, etc.
The proposed model is a multi-layer perceptron (MLP), with group LASSO regularization at the first layer. The group LASSO regularization, an extension of LASSO (Least Absolute Shrinkage and Selection Operator) [14], helps to determine the significance of each region. Each group of features is the weight originating from an input node. According to group LASSO, the selected groups correspond to regions implicated in the cause of the target disease. Training the model includes two steps. The first step is pretraining, in which all of the data for three brain disorders are used, and the model has no regularization. For fine-tuning, the network is regularized with the data for the target disease; after the second step, we have three networks specialized for three disorders. The proximal gradient descent algorithm optimizes the network in the second step.
Using the brain disorders CNVs, we compare our results against state-of-the-art tools. Our tool overlaps a higher percentage of genes overrepresented in the brain (on average 20 percent), and besides, our results have a higher overlap percentage (almost 18 percent) with mouse mutant genes that cause nervous system phenotypes.
In addition, we performed gene ontology (GO) analyses for genes that overlap with the CNVs. GO analyses support the natural association with the target disease. Several terms, such as obsessive–compulsive behavior and axon development, were detected as related to the genes. Further, by utilizing DECIPHER [15], the renowned genotype–phenotype source of information, we analyzed the associated phenotypes with each causative region and examined their relationship to the target disorder. Some phenotypes, such as delayed speech and language development, seizures, microcephaly, and macrocephaly, were detected to be correlated with the causative regions that were found to be associated with the brain disorders. The other analyses involved investigating common genes in three brain disorders and examining more prevalent genes with one disease in one gender. For example, for ASD, duplication in a subregion on 16p11.2 is associated with males, and duplication in a subregion on 21q22.13 is correlated with females.
Results
Associations of the regions with the target disorders
Our model was trained using ~ 195,500 CNVs from patients and healthy individuals (nearly 60 percent from patients and 40 percent from healthy). We use the start and end points of the cases and controls to build the smallest possible regions for investigating possible associations with disease (for each chromosome and type separately). This will create a list of regions with the help of CNV boundaries for each chromosome. The regions are depicted in Fig. 2. As a result, many of the problems discussed in the Background section will be resolved. Then, we compute the amount of overlap of the CNVs of an individual (healthy or patient) with the regions. Each individual has a label of one if he is a patient or zero if he is not ill. This step will convert the case–control study into a format suitable for feeding into our model. In the next step, we have a multi-layer perceptron to train. For training each target disease, we first use the CNVs for all brain disorders in the pretraining. In the fine-tuning phase, we only use the CNV data for the target disease (with labels of the target disease). In the second phase, the training involves adding a regularization term, Group LASSO, to the first layer of the MLP. Using this term, we can identify possible disease-causing regions. The details are discussed in the Method section.
We build the regions with the help of the starts and ends of the CNVs in cases and controls. To create the regions, we sort the starts and ends of the case/control CNVs in chromosomes and create the regions with these main points. In the figure, the blue line represents case CNVs, and the green represents control CNVs. Three regions are formed with (start_CNV_case, start_CNV_control), (start_CNV_control, end_CNV_case), (end_CNV_case, end_CNV_control)
Comparison with machine learning methods
We selected some of the machine learning methods and some evaluation benchmarks to evaluate the algorithm's performance from the machine learning viewpoint. The three chosen methods for comparison are described below.
The permutation feature importance algorithm [16] utilizes the shrinkage in a model performance once a feature value is randomly scrambled. The random forest algorithm [17] employs bagging and feature randomness with multiple decision trees. In Gradient Boosting [18], each classifier advances its predecessor by reducing the miscalculations. It fits a more accurate classifier to the residual errors of the last precursor. The results for ROCAUC and accuracy are reported in Table 1 and Fig. 3. The procedure is as follows: we fed the data of each disorder to every method (we assign label one to cases and label zero to controls), and after that, we evaluate the accuracy of the results (and also ROC AUC). CNVDeep achieves better results than other methods (for every disease, Table 11 lists the top regions discovered by CNVDeep).
Overrepresentation of brain-enriched genes in the candidate regions
Brain disorders are the target diseases for which we seek CNV associations; a deficiency in brain development characterizes this group. As a result, genes that overlap with candidate regions may be overrepresented in the brain [19]. We used the set of brain-enriched genes provided in [10] to measure the percentage of brain-enriched genes that overlap with the candidate regions. Some brain-enriched examples are GABRG3 and GABRA5 duplications for ASD, FAM178B, ANKRD39 deletions for SCZ and SNHG14, and DIP2C duplications for DD. We compare the percentages of coding and noncoding genes for each disease to those found in previous studies. We compared our results to the most extensive study on developmental delay [11], the state-of-the-art results on ASD, and the most commonly used CNV tool (PLINK). They all covered lower percentages of brain-enriched genes than our list. Table 2 lists the results.
Among the chromosomes, the 22nd chromosome possesses the most significant number of brain-enriched genes for brain disorders. Some regions we identified overlap with many brain-enriched genes (coding or noncoding). They are listed in Table 3.
The analysis of the homolog of the genes in mouse associated with nervous system phenotypes
The study of animal models helps us understand disease mechanisms in similar creatures. Mutant mouse models with phenotypic defects in the nervous system are among the models available for exploring neurocognitive disorders.
Our proposed method achieves better results than the other significant methods on these datasets; the details of the results are presented in Table 4. In our method, the overlap of coding genes with the candidate regions is associated with a higher percentage of gene homologs with nervous system traits.
Some regions overlap with numerous mouse mutant genes, such as the ones listed in Table 5. Notably, some genes overlap much with the candidate regions; examples are GABRA5 and DSCAM for ASD. Within the chromosomes, the 22nd chromosome contains most of the genes with such characteristics for ASD, SCZ, and DD.
Phenotypes associated with the candidate regions
To analyze phenotypes associated with the candidate regions of each disease, we can use the DECIPHER [15] data source, which contains genotype–phenotype information for ~ 12,600 patients and ~ 16,600 CNVs with ~ 2,600 phenotypes. Specifically, for each region-phenotype pair, we compute the fraction of patients (with that phenotype) whose CNVs overlap the target region and compare it with the natural expectation. For ASD disease, 1,748 patients with 1,031 phenotypes overlapped with significant regions. The number of overlapped patients for DD was 2,434, with 1,283 phenotypes. For SCZ, these numbers were 976 patients with 688 phenotypes. A heatmap shows the relationship between phenotypes and candidate regions for each target disease. Figures 4, 5, and 6 show the results for ASD, DD, and SCZ, respectively. The detected regions are in the rows, and DECIPHER phenotypes are in the columns. The bold points are regions with overrepresented phenotypes.
The heatmap for DD. The top labels represent DECIPHER phenotypes, and the left labels are candidate regions for developmental delay. The bolder the dots, the stronger the relationship between region and phenotype. Some associated phenotypes are seizures, abnormal facial shape, and specific learning disabilities
As shown in the heatmaps, among the phenotypes in the DECIPHER data source, some examples of ASD disease include 'intellectual disability,' 'global developmental delay,' 'delayed speech and language development,' 'autism,' 'seizures,' 'microcephaly,' 'obesity,' 'muscular hypotonia,' 'short stature,' 'behavioral abnormality,' 'cognitive impairment,' and 'autistic behavior'; for developmental delay (DD), 'intellectual disability,' 'delayed speech, and language development,' 'autism,' 'seizures,' 'microcephaly,' 'behavioral abnormality,' 'short stature,' and 'obesity,' and for SCZ, 'intellectual disability,' 'global developmental delay,' 'delayed speech and language development,' 'microcephaly,' 'autism,' 'seizures,' 'short stature,'' 'behavioral abnormality,' and 'cognitive impairment,' were highlighted as associated phenotypes.
Besides, some regions have the most associations with phenotypes. For ASD, deletion in a region in 16p11.2Footnote 1; For DD, deletion in a subregion in 15q11.2Footnote 2; and for SCZ, deletion in a subregion in 15q11.2.Footnote 3
Genes common to all three disorders and those overrepresented in only one gender
Next, we conduct a cumulative analysis to identify the regions shared by all target disorders and the associated genes. According to our investigation, considering the type of variation (deletion or duplication), some of the genes common in the three disorders are deletions in PRKAB2, CRKL, GJA5, and SLC7A4 and duplications in FAM57B and BCL7B. Some genes common in ASD and DD are deletions in GTF2IRD1, SNAP29, AC083884, and duplication in ACP6; common in ASD and SCZ are duplications in BCL7B, GDPD3, TMEM219, and PRKAB2, and deletion in TANGO2, and common between SCZ and DD are deletions in CDC45, FBXO45, LINC00624, and duplication in WBSCR22.
We performed another analysis for each target disorder using the datasets where their gender was available. We compared the percentage of males and females who were patients and had variation in that region. Accordingly, for ASD, the region duplication in 16p11.2, in subregion from 30,194,353 to 30,199,805, is dominated by males, and females dominate duplication in 21q22.13 in the exact subregion from 38,735,314 to 38,909,325. Finally, for the DD, the following list can be proposed for males and females:
-
Male: Deletion in 3q29, in the exact region, starts from 197,072,247 to 197,300,214.
-
Female: Duplication in 1q21.1 in the exact region starts from 146,852,473 to 146,989,699.
-
Female: Deletion in 15q11.2, the subregion starts from 22,833,499 to 22,873,941.
Gene ontology analyses of the candidate regions
To conduct gene ontology analyses on the overlapped genes, we used WebGestelat [20].
Several analyses were performed, including gene ontology, human phenotype ontology, and disease terms (DisGeNet and GLAD4U), and several brain codes were used as background genes. The other parameters were the ones present on the website.Footnote 4 Tables 6, 7, 8 report the results for each target disease. In these tables, FDR stands for False Discovery Rate. For ASD, some of the results, such as autistic behavior and autism, were trivial. Other nontrivial results were obsessive–compulsive behavior, axon development, cognition, regulation of membrane, abnormal social behavior, and hyperactivity, some of which were also mentioned in [21].
Results of the DD analysis include obsessive–compulsive behavior, cognition, neuron projection organization, regulation of membrane potential, regulation of neuron projection development, regulation of synapse structure or activity, positive regulation of signaling receptor activity, and axon development, as exhibited in [22].
Statistical analysis
We also conducted an independent analysis of the regions of different chromosomes. We used Fisher's exact test (Table 9) to evaluate each region's relative amount of case and control overlaps. The threshold was determined using 100,000 random permutations of case and control labels to ensure the results were not produced randomly. The sample diagrams for the three chromosomes are shown in Fig. 7.
Analysis with synthetic data
The three datasets of available disorders were used to design a new dataset. A random sample of 25,000 patients from cases and 20,000 healthy individuals from controls was selected.
Let src_cnv be (src_ch, src_type, src_strt, src_end) for one of the three data sources. Each patient and healthy individual was subjected to a random perturbation to produce new_cnv = (new_ch, new_type, new_strt, new_end), where:
In this case, p is a random variable with a discrete uniform distribution. The new CNV is constructed in such a manner that the chromosome number will match the source CNV, the type of variation will be random deletion or duplication, and 10 k basepairs will be randomly perturbed at the start and end of the CNV in comparison with the source CNV. To produce these new CNVs, the CNVs for an individual should not overlap.
Table 10 shows the results of evaluating our dataset using machine learning criteria and measuring the percentage of brain-enriched and mouse-mutant genes.
Discussion
The current study presents a novel approach for identifying associations between CNVs based on deep learning. The proposed method detects regions accurately and effectively based on the CNVs of cases and controls. Our training uses all cases and controls of brain disorders in the first step, followed by using CNVs of the target disorder to fine-tune the network. We have used the data of 195,496 CNVs from 132,388 people, 76,528 CNVs for 54,956 healthy, and 118,968 CNVs for 77,432 patients. Since we are looking for associations in brain disorders, we measure the percentage of genes that overlap with our regions that are brain-enriched. Our results were, on average, 20 percent higher than those of other works with similar findings. Furthermore, we study genes whose homologs cause mouse nervous system defects. From this perspective, the genes that overlap with our regions have, on average, 18 percent higher performance compared to previous works. Some regions have many overlaps with brain-enriched genes and genes active in the mouse nervous system; for example, 16p11.2 and 22q11.21 for the NDDs are highlighted regions. Similarly, in SCZ, a duplication in a subregion of 16p11.2 overlaps with brain-enriched and mouse genes. Another aspect of the analysis is that we have some genes that are both brain-enriched and active in the mouse nervous system. Some genes such as SEZ6L2, KCTD13, DOC2A, PRRT2, TBX6, and MAPK3 in 16p11.2 are both brain-enriched and overrepresented in mice, and have more than 600 overlaps with cases of ASD; others, like OTUD7A and CHRNA7 in 15q13.3, have the same features and have more than 150 overlaps with cases for SCZ; and OTUD7A, CHRNA7, MAPK3, TBX6, DOC2A, KCTD13, SEZ6L2, and PRRT2 from 16p11.2 and 15q13.3 have a lot of overlap with DD cases. Interestingly, some genes, such as OTUD7A and CHRNA7, were the top genes associated with all disorders (Table 11).
We further explore the DECIPHER data source to examine which phenotypes correlate most with the discovered regions. It has been measured that intellectual disability (hp:0001249), global developmental delay (hp:0001263), delayed speech and language development (hp:0000750), microcephaly (hp:0000252), seizures (hp:0001250), muscular hypotonia (hp:0001252), autism (hp:0000717), hypertelorism (hp:0000316), low-set ears (hp:0000369), and short stature (hp:0004322) are top phenotypes associated with ASD.
Similarly, for SCZ, some phenotypes such as intellectual disability (hp:0001249), global developmental delay (hp:0001263), delayed speech and language development (hp:0000750), seizures (hp:0001250), microcephaly (hp:0000252), muscular hypotonia (hp:0001252), autism (hp:0000717), hypertelorism (hp:0000316), low-set ears (hp:0000369), strabismus (hp:0000486), short stature (hp:0004322), micrognathia (hp:0000347), and abnormal facial shape (hp:0001999) were identified.
For DD, intellectual disability (hp:0001249), global developmental delay (hp:0001263), delayed speech and language development (hp:0000750), seizures (hp:0001250), microcephaly (hp:0000252), hypertelorism (hp:0000316), muscular hypotonia (hp:0001252), autism (hp:0000717), low-set ears (hp:0000369), strabismus (hp:0000486), abnormal facial shape (hp:0001999) and micrognathia (hp:0000347) were recognized as top phenotypes.
Cumulatively, some phenotypes, such as delayed speech and language development, seizures, and muscular hypotonia, were common among the three disorders. In light of these discoveries, clinicians might doubt the presence of comorbidities if a patient exhibits a variation. We can draw valuable conclusions about their differences and similarities based on our analysis of the three brain disorders separately and jointly.
Conclusions
To explore the effect of variations on neurocognitive disorders, we developed a tool based on deep learning for analyzing CNVs responsible for a target disease. We trained our model with all the CNVs from the three brain related disorders. We made the most effective use of data in the pretraining phase and used CNVs of the target disease in the next stage for fine-tuning. We compared the results with some of the related works for each of the target diseases. Our discovered regions include more coding and lncRNA, which are enriched in the brain, and our results have more homologs in the mouse with nervous system phenotypes. Besides, we used the DECIPHER data source to identify the phenotypes related to the genes of the target disease. Integration with the phenotypic database revealed more attractive characteristics of the detected genes.
In future work, we can model CNV relationships with graph-based classification models. An alternative future path is to use additional evidence, such as protein networks, to analyze the association of CNVs with diseases. Additionally, as a multi-phenotype data source with CNVs for each patient, DECIPHER data can provide a basis for analyzing the relation of the genetic etiology of the disease with the observed phenotypes in the patient and the possible co-occurrence of some phenotypes. Additionally, we can investigate topologically associating domains and their destruction by CNVs as the etiology of the disease. Since our method uses CNV data, it can identify variations associated with a target disease in the context of a case–control study.
Materials and methods
Materials
The primary data we used in our study is from the three brain disorders: autism spectrum disorder, schizophrenia, and developmental delay. The statistics for the three disorders and their references are listed in Table 12.
Some supplementary data were used to analyze the results. The first is FANTOM 5 [23], which lists ~ 21,000 coding and ~ 28,000 noncoding genes. Figure 8 provides the distribution of the genes in different chromosomes. The next is DECIPHER [15], which contains genotype–phenotype information for ~ 12,600 patients and ~ 16,600 CNVs with ~ 2,600 phenotypes. Figure 9 provides the distribution of the genes in different chromosomes. The next is the list of brain-enriched genes [10], which contains 7,339 coding and 7,167 lnc_RNA genes. The distribution of genes across different chromosomes is provided in Fig. 10. The last data source is the genes whose ortholog causes nervous system phenotypes in mice [10].
We gathered the genes that their homologs associate with nervous system phenotypes from the [10]; this resource collects information, Nervous (MP:0003631),Footnote 5 Abnormal morphology (MP:0003632),Footnote 6 Abnormal physiology (MP:0003633),Footnote 7 and Ortholog mappingsFootnote 8 from the MGI repository. The web links for different resources used throughout the research are gathered in Table 13.
The next part is about preprocessing and data cleansing. The first step in all CNV association methods is to filter out regions with less than one kbps (kilobase pairs). Furthermore, in the DECIPHER, those patients without phenotypes were removed. We made sure that all data were in the form of HG19. If not, we convert it with the UCSC Lift Genome Annotation [24]. If a chromosome in a dataset lacks data (for example, X or Y chromosome), it is removed from the analysis. Besides, regions with more overlaps with controls than cases were not the results of the analyses, so they are removed; the last step is the standardization of variables (this step is necessary for our model). The standardization step in machine learning is essential for proximal gradient descent algorithms; it involves centering the variable at 0 (zero mean) and standardizing the variance to 1 (unit variance). As a result, we standardize variables based on the sample mean and standard deviation. In this way, the solution will be independent of the measurement scale.
Method
We use a deep learning model to evaluate the association between CNVs and the target disease. It can be said that a region does not influence the occurrence of disease when all weights emanating from it are zero. The neural network uses regularization to identify the regions that cause the disorder. Consequently, regions are defined as input variables, and the neural network selects causative regions based on the regularization term. The model consists of a multi-layer perceptron (MLP); some terms were added. Our model training includes two phases: pretraining and fine-tuning. Pretraining uses all the data for the three brain disorders. Fine-tuning involves the regularized MLP with the data for the target disease. The regularization we used in this model is Group LASSO (also called L2,1 norm):
where the groups (wg) are weights from a single neuron in the input layer (the blue ovals in Fig. 11), and G is the number of groups. The outer sum is on all the neurons of the input layer. The group LASSO penalty will choose a sparse set of groups. In other words, outgoing weights correspond to one group. We can remove the corresponding region if all the weights are zero.
If the formulation removes a group, all the weights outgoing from the neuron will be zero. The loss function used is the binary cross entropy (since the main problem is binary classification). The activation function in the last layer is sigmoid.
The popular solution is proximal gradient descent [25, 26]. This operator is sometimes called block soft thresholding (for group LASSO). It acts as a soft thresholding operator (Sλ(wg)) for each group. For the group \(w_{g}\)[26], we have:
where \(\lambda\) is the regularization parameter that balances loss and regularization terms. A large λ value delivers results where regularization is more important; thus, there are more zeros among the coefficients [26].
The optimization problem is as follows:
where \(\hat{y}\) is the predicted label, for the actual label y, \(\overset{\lower0.5em\hbox{$\smash{\scriptscriptstyle\smile}$}}{W}_{1}\) is the vector of weights for the first layer, which is the solution to the unconstrained problem:
The proximal operator solves the optimization:
such that:
where \(\ddot{\theta } = \theta^{(i)} - \delta_{i} \nabla_{\theta } L(\theta^{(i)} )\).
The solution of (7) is:
The complete algorithm is shown in Fig. 12. We have two hidden layers for the MLP, and the size of each one is the square root of the last layer. The optimization algorithm is Adam [27].
Complete algorithm used in two phases for training the network. The output of the second phase is the set of nodes whose outgoing weights are nonzero [26]
Availability of data and materials
The source codes used in this Study for clustering and analysis of subtypes are provided in http://git.dml.ir/z_rahaie/DCNV (Table
14).
Notes
(29,720,526, 29,862,986).
(22,833,499, 22,873,941).
(23,034,585, 23,037,636).
References
Zarrei M, MacDonald JR, Merico D, Scherer SW. A copy number variation map of the human genome. Nat Rev Genet. 2015;16(3):172–83. https://doi.org/10.1038/nrg3871.
Redon R, Ishikawa S, Fitch KR, Feuk L, Perry GH, Andrews TD, Fiegler H, Shapero MH, Carson AR, Chen W, Cho EK. Global variation in copy number in the human genome. Nature. 2006;444(7118):444–54. https://doi.org/10.1038/nature05329.
St CD. Copy number variation and schizophrenia. Schizophr Bull. 2009;35(1):9–12. https://doi.org/10.1093/schbul/sbn147.
Forer L, Schönherr S, Weissensteiner H, Haider F, Kluckner T, Gieger C, Wichmann HE, Specht G, Kronenberg F, Kloss-Brandstätter A. CONAN: copy number variation analysis software for genome-wide association studies. BMC Bioinformatics. 2010;11(1):1–9. https://doi.org/10.1186/1471-2105-11-318.
Xu Y, Peng B, Fu Y, Amos CI. Genome-wide algorithm for detecting CNV associations with diseases. BMC Bioinformatics. 2011;12(1):1. https://doi.org/10.1186/1471-2105-12-331.
Warland A, Kendall KM, Rees E, Kirov G, Caseras X. Schizophrenia-associated genomic copy number variants and subcortical brain volumes in the UK Biobank. Mol Psychiatry. 2020;25(4):854–62. https://doi.org/10.1038/s41380-019-0355-y.
Vicari S, Napoli E, Cordeddu V, Menghini D, Alesi V, Loddo S, Novelli A, Tartaglia M. Copy number variants in autism spectrum disorders. Prog Neuropsychopharmacol Biol Psychiatry. 2019;8(92):421–7. https://doi.org/10.1016/j.pnpbp.2019.02.012.
Flore LA, Milunsky JM. Updates in the genetic evaluation of the child with global developmental delay or intellectual disability. In seminars in pediatric neurology. 2012;19(4):173–80. https://doi.org/10.1016/j.spen.2012.09.004.
Goodman SN. Toward evidence-based medical statistics. 1: The P value fallacy. Ann Intern Med. 1999;130(12):995–1004. https://doi.org/10.7326/0003-4819-130-12-199906150-00008.
Alinejad-Rokny H, Heng JI, Forrest AR. Brain-enriched coding and long non-coding RNA genes are overrepresented in recurrent brain disorder CNVs. Cell Rep. 2020;33(4): 108307. https://doi.org/10.1016/j.celrep.2020.108307.
Coe BP, Witherspoon K, Rosenfeld JA, Van Bon BW, Vulto-van Silfhout AT, Bosco P, Friend KL, Baker C, Buono S, Vissers LE, Schuurs-Hoeijmakers JH. Refining analyses of copy number variation identifies specific genes associated with developmental delay. Nat Genet. 2014;46(10):1063–71. https://doi.org/10.1038/ng.3092.
Cooper GM, Coe BP, Girirajan S, Rosenfeld JA, Vu TH, Baker C, Williams C, Stalker H, Hamid R, Hannig V, Abdel-Hamid H. A copy number variation morbidity map of developmental delay. Nat Genet. 2011;43(9):838–46. https://doi.org/10.1038/ng.909.
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, De Bakker PI, Daly MJ, Sham PC. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Human gen. 2007;81(3):559–75. https://doi.org/10.1086/519795.
Tibshirani R. Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Stat Methodol. 1996;58(1):267–88.
Firth HV, Richards SM, Bevan AP, Clayton S, Corpas M, Rajan D, Van Vooren S, Moreau Y, Pettett RM, Carter NP. DECIPHER: database of chromosomal imbalance and phenotype in humans using Ensembl resources. Am J Human Gen. 2009;84(4):524–33. https://doi.org/10.1016/j.ajhg.2009.03.010.
Altmann A, Toloşi L, Sander O, Lengauer T. Permutation importance: a corrected feature importance measure. Bioinformatics. 2010;26(10):1340–7. https://doi.org/10.1093/bioinformatics/btq134.
Qi Y. Random forest for bioinformatics. Ensemble machine learning Methods and applications. 2012. https://doi.org/10.1007/978-1-4419-9326-7_11.
Natekin A, Knoll A. Gradient boosting machines, a tutorial. Front Neurorobot. 2013;4(7):21.
Pinto D, Delaby E, Merico D, Barbosa M, Merikangas A, Klei L, Thiruvahindrapuram B, Xu X, Ziman R, Wang Z, Vorstman JA. Convergence of genes and cellular pathways dysregulated in autism spectrum disorders. Am J Human Gen. 2014;94(5):677–94. https://doi.org/10.1016/j.ajhg.2014.03.018.
Liao Y, Wang J, Jaehnig EJ, Shi Z, Zhang B. WebGestalt 2019: gene set analysis toolkit with revamped UIs and APIs. Nucleic Acids Res. 2019;47(W1):W199-205. https://doi.org/10.1093/nar/gkz401.
Jacob S, Landeros-Weisenberger A, Leckman JF. Autism spectrum and obsessive–compulsive disorders: OC behaviors, phenotypes, and genetics. Autism Res. 2009;2(6):293–311. https://doi.org/10.1002/aur.108.
Riou EM, Ghosh S, Francoeur E, Shevell MI. Global developmental delay and its relationship to cognitive skills. Dev Med Child Neurol. 2009;51(8):600–6. https://doi.org/10.1111/j.1469-8749.2008.03197.x.
Abugessaisa I, Ramilowski JA, Lizio M, Severin J, Hasegawa A, Harshbarger J, Kondo A, Noguchi S, Yip CW, Ooi JL, Tagami M. FANTOM enters 20th year: expansion of transcriptomic atlases and functional annotation of non-coding RNAs. Nucleic Acids Res. 2021;49(D1):D892–8.
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12(6):996–1006. https://doi.org/10.1101/gr.229102.
Parikh N, Boyd S. Proximal algorithms. Found Trends® Optim. 2014;1(3):127–239.
Ho LS, Tran G. 2021 Adaptive Group Lasso Neural Network Models for Functions of Few Variables and Time-Dependent Data. arXiv preprint arXiv:2108.10825.
Kingma DP, Ba J. Adam: A method for stochastic optimization. arXiv preprint arXiv:1412.6980. 2014 Dec 22.
Basu SN, Kollu R, Banerjee-Basu S. AutDB: a gene reference resource for autism research. Nucleic Acids Res. 2009;37(suppl_1):D832–6. https://doi.org/10.1093/nar/gkn835.
Marshall CR, Howrigan DP, Merico D, Thiruvahindrapuram B, Wu W, Greer DS, Antaki D, Shetty A, Holmans PA, Pinto D, Gujral M. Contribution of copy number variants to schizophrenia from a genome-wide study of 41,321 subjects. Nat Genet. 2017;49(1):27–35. https://doi.org/10.1038/ng.3725.
Mosci S, Rosasco L, Santoro M, Verri A, Villa S. Solving structured sparsity regularization with proximal methods. InMachine Learning and Knowledge Discovery in Databases: European Conference, ECML PKDD 2010, Barcelona, Spain, September 20-24, 2010, Proceedings, Part II 21 2010 (pp. 418-433). Springer Berlin Heidelberg
Acknowledgements
This study makes use of data generated by the DECIPHER community. A complete list of centers that contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project [15] was provided by Welcome [grant number WT223718/Z/21/Z]. Those who carried out the original analysis and collection of the Data in the DECIPHER project bear no responsibility for the analysis or interpretation of the analyses provided in this study. Analysis was made possible with computational resources provided by the UNSW BioMedical Machine Learning Laboratory (BML) Servers with funding from the UNSW Scientia Program Fellowship.
Funding
This work was supported by the UNSW Scientia Program Fellowship and the Australian Research Council Discovery Early Career Researcher Award (DECRA) under Grant No. DE220101210 to HAR. This study makes use of data generated by the DECIPHER community. A complete list of centers that contributed to the generation of the data is available from https://deciphergenomics.org/about/stats and via email from contact@deciphergenomics.org. Funding for the DECIPHER project was provided by Welcome, Grant No. WT223718/Z/21/Z. Those who carried out the original analysis and collection of the data in the DECIPHER project bear no responsibility for the analysis or interpretation of the analyses provided in this study. Analysis was made possible with computational resources provided by the UNSW BioMedical Machine Learning Laboratory (BML) Servers with funding from the UNSW Scientia Program Fellowship. The funders had no role in study design, data collection and analysis, the decision to publish, or the preparation of the manuscript. HRR was partially supported by the IR National Science Foundation (INSF), Grant No. 96006077.
Author information
Authors and Affiliations
Contributions
HRR, ZR, and HAR did the conceptualization of the study. ZR did the data curation. ZR and HRR performed the Formal Analysis. ZR did the investigation. ZR and HRR developed the methodology. HRR did the project administration and supervision. ZR did the software development. ZR and HRR did the validation. ZR did the visualization. ZR wrote the manuscript. HRR and HAR reviewed and edited the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethics approval and consent to participate.
Not applicable.
Consent for publication
Not applicable.
Informed consent.
The consent approval was not needed; all the data used in this study are publicly available.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Rahaie, Z., Rabiee, H.R. & Alinejad-Rokny, H. CNVDeep: deep association of copy number variants with neurocognitive disorders. BMC Bioinformatics 25, 283 (2024). https://doi.org/10.1186/s12859-024-05874-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12859-024-05874-8