Abstract
Background
Pagiophloeus tsushimanus (Coleoptera: Curculionidae), an emerging forest pest exclusively infesting camphor trees, has recently caused severe ecological and economic damage in localized areas in China. Its population outbreak depends largely on the capacity to overcome the pressure of terpenoid-derived metabolites (e.g. linalool) from camphor trees. At present, the molecular basis of physiological adaptation of P. tsushimanus to dietary linalool is poorly understood, and there is no available reference genome or transcriptome.
Results
Herein, we constructed the transcriptome profiling of P. tsushimanus larvae reared on linalool-infused diets using RNA sequencing and single-molecule real-time sequencing. A total of 20,325 high-quality full-length transcripts were identified as a reference transcriptome, of which 14,492 protein-coding transcripts including 130 transcription factors (TFs), and 5561 long non-coding RNAs (lncRNAs) were detected. Also, 30 alternative splicing events and 8049 simple sequence repeats were captured. Gene ontology enrichment of differential expressed transcripts revealed that overall up-regulation of both cytochrome P450s (CYP450s) and cuticular proteins (CPs), was the primary response characteristic against dietary linalool. Other physiological effects possibly caused by linalool exposure, such as increase in Reactive Oxygen Species (ROS) and hormetic stimulation, were compensated by a handful of induced genes encoding antioxidases, heat shock proteins (HSPs), juvenile hormone (JH) epoxide hydrolases, and digestive enzymes. Additionally, based on co-expression networks analysis, a diverse array of hub lncRNAs and TFs co-expressed with CYP450s and CPs were screened as the potential gene regulators. Temporal expression of candidate transcripts determined by quantitative real-time PCR also indicated a cooperative relationship between the inductions of CYP450s and CPs upon exposure to linalool.
Conclusions
Our present study provides an important transcriptome resource of P. tsushimanus, and lays a valuable foundation for understanding how this specialist pest copes with chemical challenges in its specific host environments.
Similar content being viewed by others
Background
It is well established that plants and herbivores are in a constant battle for survival [1,2,3,4]. The former keeps adapting their chemistry to defend against the latter, which sparks the rise of extensive specialized metabolites (mainly secondary metabolites) currently found in plants. Among these, terpenoids are the largest and most diverse plant metabolites and have been widely known as the defensive vehicles in conifers protection [5,6,7,8,9,10]. But actually some broad-leaved trees, as exemplified by camphor trees Cinnamomum camphora, are also equipped with a suite of terpenoid-derived metabolites. Different parts of camphor trees are abundant in monoterpenoid oxygenated derivatives such as D-camphor and linalool [11]. The evidence strongly suggests that these specialized metabolites have toxic and/or repellent effects on pests and diseases with the potential as antimicrobial or insecticidal agents [12, 13]. In general, terpenoids as important chemical mediators in plant-herbivore interactions work to the benefit of the plants and to the detriment of herbivores.
In the face of a diverse range of phytochemicals, specialization could lead to a competitive advantage by developing more fine-tuned metabolic features for digestion and detoxification of host plants [14, 15]. In this context, classic examples are that specialist herbivores feeding on a single or a few plants with terpenoid-based defenses have evolved adaptation strategies, as mainly demonstrated for pine-feeding bark beetles [16, 17]. Some bark beetles can use cytochrome P450 monooxygenases (CYP450s) to detoxify or even biotransform pine-derived terpenoids into pheromones by both sexes [18]. Progress in the molecular functions of CYP450 genes has deciphered the roles of genes belonging to the CYP6 family in response to phytochemicals including several terpenoids [19]. It also must be cautioned that exposure to terpenoids appears to disrupt the insects’ cuticular development because of their strong volatility and penetrability in analogy to mechanisms of contact insecticides or fumigants [20]. In contrast, the resistant strains of target insects have evolved to deploy a class of structural components, cuticular proteins (CPs), in order to enable the cuticle thickening that minimizes penetration of xenobiotics (e.g. contact insecticides or fumigants) into their bodies [20]. Penetration resistance referring to reduced insecticide penetration through the cuticle has been reported in a variety of insect species [21,22,23]. However, whether some insect pests of camphor trees have co-evolved similar or distinct mechanisms to counter monoterpenoid oxygenated derivatives (e.g. D-camphor and linalool) remains elusive so far.
More recently, an emerging insect pest, Pagiophloeus tsushimanus (Coleoptera: Curculionidae) has been found to persist exclusively on camphor trees, and locally cause serious damage in China [24, 25]. It evokes our great interest that how this insect pest tolerates host-specific terpenoid defenses from camphor trees without incurring apparent fitness costs. In our previous study, we tested the sensitivity of P. tsushimanus eggs and larvae to camphor oil and several monoterpenoids including D-camphor and linalool, suggesting that this weevil possesses various levels of tolerance to different monoterpenoids [26]. Subsequently, more work place emphasis on the mechanism underlying the tolerance of this weevil to D-camphor, and the role of this compound as a key semiochemical in host recognition (unpublished data). But also, linalool is undeniably an important source of chemical pressure when the larvae fed on the phloem of camphor trees. Published work indicated a broad spectrum of toxicity or repellent activity of linalool to some stored-product pests [13, 26,27,28], while little is known about how this specialist weevil overcomes the detrimental effects of linalool exposure.
Next-generation sequencing has attained considerable momentum so far, and RNA sequencing (RNA-seq) is an indispensable tool for exploring the mRNA expression profiles of target genes, and further predicting their physiological functions [29]. Studies investigating the transcriptomic responses of insect pests to insecticides and plant metabolites (including a small fraction of terpenoids) exposures have been well-established [30,31,32,33,34]. Single-Molecule Real-Time (SMRT) sequencing as an emerging technology overcomes the limitation of short-read sequences, and provides a full-length reference transcriptome for RNA-seq analysis of non-model insects without reference genomes [35]. Also, SMRT sequencing contributes to accurately analyzing the long non-coding RNA (lncRNA) and the structural information of sequences, such as alternative splicing (AS) and simple sequence repeat (SSR) [35]. The combination of RNA-seq and SMRT sequencing has been used in various studies to obtain more complete transcriptome information reflecting the physiological state of insects under stress environments [36,37,38].
Therefore, in this study we employed an integrated RNA-seq and SMRT sequencing approach to a general view of the response of P. tsushimanus larvae to a long-term linalool exposure (from 1st- to 3th-instar) in the dietary. AS and SSR prediction, expression analysis of lncRNA, transcription factor, and functional genes (mRNA) were then performed to determine the mechanism driving the tolerance to dietary linalool in P. tsushimanus larvae. Understanding of how P. tsushimanus adapts to a linalool-rich dietary will provide a valuable clue for the physiological mechanisms of its dietary specialization and outbreak. Our findings will aid in screening potential molecular targets for this pest management from the perspective of plant chemical defenses.
Results
Summary of the transcriptome data output from single-molecule real-time and Illumina sequencing
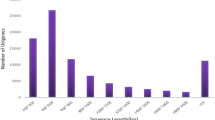
To obtain a comprehensive transcriptome database of Pagiophloeus tsushimanus, one library with 1–6 K cDNA insert size was generated from the pooled RNA samples (three male adults, three female adults, sixty eggs, three newly molted 1st- to 5th-instar larvae, and three pupae) using the Pacific Sequel platform. As shown in Table S1, a total of 18.31 Gb nucleotides with 16,263,362 subreads were obtained after the initial filtering process. These subreads were then subjected to circular consensus sequence (CCS) analysis, and 113,436 CCSs with a full-length non-chimera (FLNC) reads of 68,407 were produced. 26,937 consensus isoforms were screened out after the Iterative clustering of error correction (ICE). Finally, a total of 20,325 high-quality full-length transcripts with a mean length of 1194.31 bp were established as a reference transcriptome to correct the reads produced by Illumina sequencing. In addition, detailed information on the length distribution of these reads were shown in Fig. S1. For Illumina sequencing, six RNA samples (CK: A1, A2, A3; Linalool_LC15: B1, B2, B3) were separately sequenced on the Illumina HiSeq 2000 platform. We obtain 50,560,795 raw reads and 50,265,983 clean reads per sample with an average of Q20 of 98.60%, Q30 of 95.56% and GC content of 43.20%, which indicated a high accuracy and quality of the sequenced data for further analysis (Table S2).
Structural analysis of transcripts
For alternative splicing (AS) analysis, only 30 AS events were detected without determining their types, since so far no reference genome is available for transcriptome assembly in P. tsushimanus. For simple sequence repeat (SSR) analysis, total of 8049 SSRs were identified from 5728 SSR-containing sequences among the 15,148 tested transcripts. Specifically, all SSR repeat types were divided into six SSR motif units, including Mono-nucleotide, Di-nucleotide, Tri-nucleotide, Tetra-nucleotide, and Penta-nucleotide motifs. Among these, the highest number of SSR motif units is Mono-nucleotide motif (7201, 89.46%), as shown in Table S3, followed by Tri-nucleotide motif (551, 6.85%) and Di-nucleotide motif (265, 3.29%).
Functional annotation and enrichment of differentially expressed transcripts
In total, 20,325 full-length transcripts were performed into functional annotation analysis, of which 14,492 transcripts (71.30%) were annotated in at least one database (Table S4). Homologous species information in the annotation of Nr database showed that 56.6% of transcripts had the highest scoring blast matches to Dendroctonus ponderosae, followed by Sitopjilus oryzae (22.5%) (Fig. S2a). Based on GO and KEGG annotation, a total of 8623 and 12,305 transcripts were respectively classified into three main GO categories (i.e. biological processes, cellular components, and molecular functions) and six KEGG categories (i.e. human diseases, metabolism, organismal systems, cellular processes, environmental information processing, and genetic information processing), which are detailed in Fig. S2b & c.
To explore the functional genes involved in response to dietary linalool, we evaluated the differences in transcription levels between the linalool-exposed and control groups. The results showed that 565 differentially expressed (DE) transcripts were observed in the larvae reared on the linalool-contained diets, including 449 up-regulated transcripts and 116 down-regulated transcripts (Fig. 1a & b). Furthermore, 10 top-most up-regulated transcripts predominantly consisted of transcripts encoding cytochrome P450s (CYP450s) and cuticular proteins (CPs), while tropomyosin-associated transcripts were mainly suppressed in the exposed larvae (Table 1). GO enrichment analysis consistently revealed that the pathways related to oxidoreductase activity (GO: 0016705) and cuticular structural constituents (GO: 0005198 and GO: 0042302) were significantly activated by linalool exposure (Table 2, Fig. 1c). A total of 11 CYP450 and 69 CP transcripts that were differentially expressed in the linalool-exposed group were further identified, and most of them exhibited an up-regulation expression in varying degrees (Fig. 2). Furthermore, other inducible transcripts of interest, such as skin secretory proteins, digestive enzymes, odorant binding proteins, antioxidases, 5-aminolevulinate synthases, ATP-binding cassette, heat shock proteins, juvenile hormone (JH) epoxide hydrolases, and esterase, were targeted as functional mRNAs engaging in responses to dietary linalool (Fig. 2). Detailed information on their expression levels was provided in Table S5.
Identification and enrichment analysis of differentially expressed transcripts. (a) Number of significantly down-regulated and up-regulated transcripts in Linalool_LC15 group when compared with those in CK group. (b) Volcano plot of differentially expressed transcripts. Dotted horizontal and vertical lines indicate P-value thresholds and ± 2-fold change, respectively. Red and blue dots indicate significantly up-regulated and down-regulated transcripts, respectively. Names of top 10 up-regulated and down-regulated transcripts are shown. (c) Bubble chart of significantly functionally enriched Gene Ontology (GO) terms in up-regulated and down-regulated transcripts. Dotted horizontal line indicates P-value thresholds. X-axis represents Z-score [(No. up-regulated transcripts - No. down-regulated transcripts) / SQRT (No. up-regulated transcripts + No. down-regulated transcripts)]. GO terms highlighted in red are further discussed in subsequent analysis
Heatmap analysis of differentially expressed transcripts of interest. Data are shown as the average values of expression for each group, and are Z-score normalized across each row. Red bars indicate up-regulated transcripts while blue bars indicate down-regulated transcripts. Detailed information on the transcript levels is listed in Table S5
Co-expression networks between lncRNA and target mRNA, and TF and target mRNA
As an important component of full-length transcriptome, lncRNAs were identified using four coding potential analysis methods (i.e. CNCI, Pfam, PLEK, and CPC). As shown in Fig. 3a, given that intersection of four databases, a total of 5561 lncRNAs were yielded to further DE analysis, and only 95 lncRNAs were found to be differentially expressed in the larvae exposed to dietary linalool. More details of DE lncRNAs were provided in Table S6. To determine the potential relevance of biological functions between lncRNAs and target mRNAs (i.e, 11 CYP450 and 69 CP transcripts), we constructed lncRNA-mRNA co-expression networks on the basis of their expression correlations (Fig. 3b & d). In the networks, the top-ranked lncRNAs in terms of the degree values were considered as hub lncRNAs. There were inconsistent responses occurring in the top 10 hub lncRNAs related to the expression of CYP450s, where six of them were up-regulated but the other four displayed down-regulated expression (Fig. 3c). In addition, all of 20 hub lncRNAs related to the expression of CPs were markedly inducible in the treatment group (Fig. 3e). Also, 130 TF transcripts were identified using the animal TFDB version 3.0 database. Among them, 71 TF transcripts belonged to Zinc-finger family, accounting for the highest percentage (Fig. 4a). More details of TFs were provided in Table S7. Subsequently, TF-mRNA co-expression networks were established to explore the transcriptional regulatory of TFs on the target mRNAs (Fig. 4b & d). The degree values of TFs were calculated and ranked to determine the top 8 hub TFs. Moreover, the expression levels of TF111 and TF102 significantly increased in the Linalool_LC15 group, and consequently the two TFs were regarded as key regulators mediating the transcriptional expression of target mRNAs (Fig. 4c & e).
Identification of differentially expressed lncRNAs involved in regulation of cytochrome P450 (CYP450) and cuticular protein (CP) genes. (a) Venn diagram of showing the number of lncRNAs predicted by four methods, Coding Potential Calculator (CPC, score < 0.5), Coding-Non-Coding Index (CNCI, score < 0), PLEK (score < 0), and Pfam protein structure domain analysis (no annotation). A total of 95 differentially expressed lncRNAs (DELncRNAs) were identified for subsequent tests. (b, d) Co-expression networks between DELncRNA and CYP450 genes, DELncRNA and CP genes using Pearson correlation coefficient. Rectangle and circular nodes indicate target mRNAs (i.e. CYP450 or CP) and DELncRNAs, respectively. Red and blue lines indicate positive and negative correlations, respectively. The lncRNAs with top-ranked degrees are colored red. (c, e) Heatmap of hub lncRNAs ranked by degrees. Data are shown as the average values of expression for each group, and are Z-score normalized across each row. Red bars indicate up-regulated lncRNAs while blue bars indicate down-regulated lncRNAs
Identification of transcription factors (TFs) involved in regulation of cytochrome P450 (CYP450) and cuticular protein (CP) genes. (a) Statistics of TF family. (b, d) Co-expression networks between TFs and CYP450 genes, TFs and CP genes using Pearson correlation coefficient. Rectangle and circular nodes indicate target mRNAs (i.e. CYP450 or CP) and DELncRNAs, respectively. Red and blue lines indicate positive and negative correlations, respectively. The TFs with top-ranked degrees are colored red. (c, e) Heatmap of hub TFs ranked by degrees. Data are shown as the average values of expression for each group, and are Z-score normalized across each row. Red bars indicate up-regulated TFs while blue bars indicate down-regulated TFs
Temporal expression of candidate lncRNAs, TFs, and mRNAs involved in response to dietary linalool
Based on samples for Illumina sequencing, we determined the relative expression levels of 16 interesting transcripts (including two lncRNAs, two TFs, six CYP450s, and six CPs) using RT-qPCR. These tested transcripts showed a positive correlation between RNA-seq and RT-qPCR data (Pearson’s r = 0.744, P < 0.001), which suggests the sequencing results are highly reliable (Fig. S3).
To further investigate the roles of these candidate transcripts involving in response to dietary linalool, a long-lasting monitor on their expression levels from 1st- to 3rd instar larvae was performed. As shown in Fig. 5, lncRNA77 and TF111, as potential regulators of CYP450 transcription, showed a contradictory trend that the expression of the former continued to be suppressed with the increasing of larval instar, while the latter was significantly induced by linalool treatment, with 1.56- to 2.15-fold higher than the control. In regard to six CYP450 transcripts, varying degrees of up-regulation were observed when the larvae chronically fed on linalool-contained diets, with fold-change values ranging from 1.81 to 20.37. More interestingly, there was a bell-shaped relationship between the fold changes and developmental stages occurring in the majority of tested CYP450s. In addition, six transcripts encoding cuticular proteins showed a global up-regulation during the later stages of treatment (i.e. 2nd to 3rd instar), and the fold-change values varied from two to 30 (Fig. 6). lncRNA80, one hub lncRNA related to the expression of CPs, was slightly up-regulated. However, there is no significant changes occurring in the mRNA level of TF102 that was considered as one of hub TFs (Fig. 6).
Temporal expression of the tested lncRNA, transcription factor, cytochrome P450 (CYP450) genes under Linalool exposure using RT-qPCR. Data are shown as the mean ± SE. The values with red or blue background indicate the fold changes between CK and Linalool_LC15 groups that reach a significant level according to one-way ANOVA followed by independent sample t-test (P < 0.05). L1-D1, day 1-1st instar larvae; L1-D3, day 3-1st instar larvae; L2-D3, day 3-2nd instar larvae; L3-D3, day 3-3rd instar larvae
Temporal expression of the tested lncRNA, transcription factor, cuticular protein (CP) genes under Linalool exposure using RT-qPCR. Data are shown as the mean ± SE. The values with red or blue background indicate the fold changes between CK and Linalool_LC15 groups that reach a significant level according to one-way ANOVA followed by independent sample t-test (P < 0.05). L1-D1, day 1-1st instar larvae; L1-D3, day 3-1st instar larvae; L2-D3, day 3-2nd instar larvae; L3-D3, day 3-3rd instar larvae
Discussion
A global picture of Pagiophloeus tsushimanus transcriptome
To take advantage of RNA-seq and SMRT sequencing to offset the disadvantages of each alone, the combination of the two techniques has been increasingly applied to produce more complete transcripts [38, 39]. In this study, we adopted this hybrid-sequencing strategy to successfully construct the full-length transcriptome database of the weevil pest, Pagiophloeus tsushimanus. To be concise, we obtained 20,325 full-length transcripts with 1194 bp of average length and 1759 bp of N50 length. In other Coleoptera insects, transcriptome data of red palm weevil Rhynchophorus ferrugineus has been generated with a high quality in read length (average length 2964 bp and N50 length 3547 bp) [37], while P. tsushimanus transcriptome obtained in the present study shared a similar sequence length with the full-length transcriptome of flea beetle Agasicles hygrophila (average length 1684 bp and N50 length 2331 bp) [37]. We speculated that differences in read length were closely related to insect species and sequencing depth. In some studies, multiple cDNA libraries with different sizes (e.g. 1–2, 2–3, and 3–6 kb) were established, and using several SMRT cells to sequence these libraries [40, 41]. However, in this work, only one SMRT cell was used for one pooled cDNA library (1–6 kb in length), which results in a relatively short read length. Occurrence of alternative splicing (AS) events in the post-transcriptional stage increases the complexity and diversity of the transcriptome, and plays a role in response to environmental cues [42]. Herein, only 30 AS events were captured from P. tsushimanus transcriptome, while other information on AS events was not available due to a lack of reference genome. Simple sequence repeats (SSRs), referring to tandem repeats of one to six nucleotides, have been widely applied to analysis of genetic structure and diversity [43]. The abundance levels of different SSR repeat types usually vary among different insect species. In analogy to our results, mono-nucleotide repeat motif is dominating in most insects [44]. However, the largest abundance of tri-nucleotide repeat motif occurs in some other insects, such as Tomicus yunnanensis and Anoplophora chinensis [45, 46]. We also found tri-nucleotide repeat motif was the second largest abundance. One possibility is that tri-nucleotide rarely produces sliding mutations of the coding frame, and thus may be more stable than other repeat types in the protein-coding region.
Moreover, a very high percentage of P. tsushimanus transcriptome was similar to that of the mountain pine beetle, Dendroctonus ponderosae (Fig. S2), which implies that a closely linked genetic background allows them to survive in their harsh and terpenoid-rich host environment. Notably, high-quality genomes of Dendroctonus spp. recently published provide valuable references for interaction studies between the Curculionoidea superfamily and their host terpenoids [47, 48]. In this context, some rapidly expanded and positively selected genes related to metabolic capacity, including cytochrome P450 and glutathione S-transferase gene families, were thought to promote Dendroctonus adaptations to oleoresin terpenoids in conifers [48, 49]. Consequently, in the future, genome sequence of P. tsushimanus deserves in-depth studies to perform a comparative analysis with genomes of Dendroctonus spp. Despite the lack of genome resource, full-length P. tsushimanus transcriptome in this study facilitates the functional study and decipher mechanism underlying host adaptations.
Transcriptional responses underlying the tolerance of Pagiophloeus tsushimanus larvae to dietary linalool.
To understand the functional molecules recruited by P. tsushimanus larvae to counteract the specific xenobiotics of their unique host plant (i.e. a suit of monoterpenoid oxygenated derivatives from camphor trees), differential expression profile of the larvae upon exposure to dietary linalool were investigated in this study. The remarkable finding is that the pathways related to oxidoreductase activity and cuticular structural constituents were in an especially active stage, as characterized by a cluster of up-regulated transcripts encoding cytochrome P450s (CYP450s) and cuticular proteins (CPs) (Fig. 2). Other physiological effects possibly caused by linalool exposure, such as increase in Reactive Oxygen Species (ROS) and hormetic stimulation, were compensated by a handful of induced genes encoding antioxidases, heat shock proteins (HSPs), juvenile hormone (JH) epoxide hydrolases, and digestive enzymes (Fig. 2). Therefore we proposed a mode for mechanism underlying tolerance to dietary linalool primarily involves co-expression of CYP450s and CPs in P. tsushimanus larvae (Fig. 7).
From the perspective of detoxification metabolism, CYP450s as a key phase I detoxification enzyme is widely known to metabolize a variety of allelochemicals via its monooxygenation reaction [50]. Ample evidence has accumulated in the last few years indicating the induction of CYP450s by specific phytochemicals [51]. Our present results confirm that linalool is a phytochemical inducer of CYP450s in P. tsushimanus larvae. It is presumed that the biological activity of linalool against the tested larvae may be predominantly reduced by the oxidation of CYP6A subfamily. It is of valuable reference that well-established studies of CYP6 subfamily functioning in terpene detoxification have been conducted in bark beetles [18]. In these reports, a number of functionally-characterized CYP450s have been thought to hydroxylate the terpenes in conifers to some alcohol products as pheromone precursors or kairomones [17, 52,53,54]. However, the metabolic fate of some terpenoid derivatives from broad-leaved tree species (e.g. D-camphor and linalool from camphor trees) in insects remains poorly understood. Similarly, a bank of CYP450 transcripts in our former investigations were up-regulated after D-camphor exposure [55]. We expected that there is a parallel CYP450-mediated metabolic process occurring in the detoxification of D-camphor and linalool since they are a group of isomers. Furthermore, it is alarming that to what extent the induction of CYP450s represents their metabolic functions is still elusive. Thus we will soon further decipher the biological roles of these inducible CYP450s in the metabolism of host-specific terpenoids using the heterologous expression of insect P450 enzymes and RNAi experiments [56,57,58]. Besides, overexpression of 5-aminolevulinate synthases indicated that production of 5-aminolevulinate, as the precursor of an indispensable cofactor (called heme), assists in the metabolic reaction of CYP450s [59]. We also found that a fraction of up-regulated odorant binding proteins (OBPs) potentially capture the linalool molecule as signals to activate CYP450s or other downstream responses.
Unlike other chemicals, terpenoids, particularly mono- and sesquiterpenes, used in most studies belong to a class of plant volatiles that can be ingested by contact with the cuticle or fumigation [10]. The insect cuticle is the first and major barrier to prevent the penetration of exogenous compounds [60]. In the current study we revealed a great accumulation of transcripts coding for structural cuticular proteins (CPs) and skin secretory proteins in the treated larvae, suggesting their role in decreasing the penetration of linalool. Consistently, some of transcriptomic studies on the insects’ adaptation towards host plants also identified the differential expression of cuticle-related genes [61]. Meanwhile, the down-regulation of chitinase-related genes was found in the treated larvae. We speculate this negative regulation allows excessive chitin to be accumulated to achieve thickening of the cuticle.
Another is that CP family also participates in the developmental process of insect molting and metamorphosis, and consequently is sensitive to hormonal regulations [62]. Insect pheromones control the production of cuticle formation and mating behaviour. As found by Sureshan et al. [63], a number of genes related to pheromone biosynthesis and CP genes were differentially expressed when Oxycarenus laetus fed on gossypol soaked cotton seeds. That is reasonable to hypothesize that gossypol may be directly or indirectly involved in the excessive pheromone production, leading to the enhanced cuticle production in O. laetus. In the present work, two genes encoding JH epoxide hydrolases correspondingly exhibited a significant upregulation, suggesting that the ingestion of exogenous linalool may also directly or indirectly stimulate the excessive pheromone production, resulting ultimately in the differential response of CP family. To our knowledge, terpenes are widely considered as a precursor for the pheromone system of bark beetles [64], and JH induction for pheromone synthesis among bark beetles is well documented [53, 54, 65]. Therefore this potential cascade caused by the hormetic stimulation merits further investigation.
Finally, the intake of linalool may also affect digestive process of nutrients from the host plant, as characterized by up-regulation of some genes encoding digestive enzymes, and consequently regulate the larval growth and development. A similar case has shown that low dose of gossypol acts as a feeding stimulus to promote the growth of Helicoverpa armigera larvae by increasing food consumption rate [66]. More evolutionary explanation for these beneficial effects of specific plant defensive compounds on the specialist herbivores remain to further investigate. Conversely, the detrimental effects of phytochemicals, typically as enhanced ROS production, pose a great challenge to insects survivorship [67, 68]. Herein, we also observed the enhanced expression of several antioxidase and HSP genes, which may play key roles in regulating the intracellular ROS balance and preventing protein denaturation to alleviate oxidative damage [69, 70].
Conclusion
There is a long-standing consensus in plant-insect interactions that specialist insects possess more specific and sometimes more efficient detoxification systems when compared to generalist ones [14, 71, 72]. In this context, the evolution of specific tolerance mechanisms propelled by selective pressure of host chemicals has been witnessed in many examples [73, 74]. In our previous work, an emerging specialist pest, Pagiophloeus tsushimanus, feeding exclusively on camphor trees Cinnamomum camphora, has been reported that its tolerance performance on different host monoterpenoids including D-camphor and linalool varied [26]. Furthermore, a potential mechanism underlying the tolerance of this specialist to D-camphor exposure has been proposed [55]. In this work, we obtained a more comprehensive transcriptome of P. tsushimanus using a combination of RNA-seq and SMRT sequencing, and emphasized the functional transcripts involved in response to dietary linalool. Based on 20,325 identified high-quality full-length transcripts, a narrow, targeted set of differentially expressed (DE) transcripts predominantly consisting of cytochrome P450 (CYP450) and cuticular protein (CP) families were considered to perform important physiological functions in overcoming the pressure of host-specific chemical (i.e. linalool). According to the results of co-expression networks, some DE lncRNAs and transcription factors strongly associated with the expressions of CYP450 and CP families were respectively determined as key regulators to activate the transcriptional responses of the two types of target mRNAs. Likewise, we further monitored the transcription dynamics of candidate CYP450 and CP transcripts under the chronical exposure of dietary linalool, suggesting that the specific adaptation to linalool is embodied in a rapid and moderate induction of CYP450 transcripts and a cumulative and massive induction of CP transcripts. Overall, the combined findings of this study will provide a new idea for future works on the adaptation traits of specialists responding to their specific host chemicals, and also facilitate better pest management.
Material and methods
Insect culture, plant material and reagents
A laboratory colony of Pagiophloeus tsushimanus was established from adults in Cinnamomum camphora plantations in Songjiang district, Shanghai city, China (30°56′6.15″N, 121°12′32.76″E) in May 2021. A complete breeding system for P. tsushimanus in the laboratory was described in our previous study [25]. In brief, the adults were allowed to feed, mate randomly and oviposit on fresh branches of camphor tree in a plastic cage. The eggs were kept in a Petri dish (5 cm i.d. × 1 cm) with moist cotton for incubation. The neonate larvae were maintained into a six-well Petri dish (3.5 cm i.d. × 1.5 cm each well) with fresh semi-artificial diets (50% distilled water, 40% powder of camphor tree bark, 5% agar, 3% sucrose, 1% yeast, 0.5% sorbic acid and 0.5% sodium benzoate) until pupation. The pupae were collected and deposited in glass tubes (2.0 cm i.d. × 5.0 cm) for emergence. All insects were kept in an environmental incubator (MTI-201B, Tokyo Rikakikai, Japan) at 28 ± 1 °C with a 70 ± 5% relative humidity and a photoperiod of 16: 8 h (L: D). Finally, the plant material authorized and used in insect culture was collected from the campus of Nanjing Forestry University (32°4′43″N/118°48′30″E), China, and authenticated by Prof. Zengfang Yin from Nanjing Forestry University, China. The voucher specimen (NF2002553) was deposited at Dendrological Herbarium, Nanjing Forestry University (Institution code from Chinese Virtual Herbarium: NF). All reagents for semi-artificial diets were purchased from Sangon, Shanghai, China. Linalool (95% purity, CAS: 78–70-6) was purchased from Sigma-Aldrich, Shanghai, China.
Sample preparation and RNA extraction
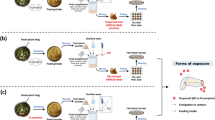
The procedure of sample preparation for SMRT sequencing and Illumina RNA-seq was visualized in Fig. 8.
For SMRT sequencing, three newly emerged males and females, sixty newly laid eggs, three newly molted 1st- to 5th-instar larvae, and three pupae aged 1–3 days were collected and frozen in liquid nitrogen for RNA extraction. Total RNA of each SMRT sequencing sample was respectively isolated using TRIzol Reagent (TaKaRa, Japan) according to the manufacturer’s protocol. RNA quantity was checked using a Nanodrop 2000 (Thermo Scientific, Waltham, MA, United States). RNA integrity was monitored on 1% agarose gels. An equal amount of total RNA from each sample was mixed in order to increase the sequence coverage of SMRT sequencing.
For Illumina RNA-seq, the neonate larvae were separated into two experimental groups. On the one hand larvae in the treatment group were reared on linalool-contained diets at the lethal concentration killing 15% of the population (19.7 mg·g− 1) (unpublished data) until they reached the 3th-instar stage (approximately 20 days old) (group name: Linalool_LC15), on the other larvae in the control group were reared on linalool-free diets until they reached the 3th-instar stage (group name: CK). Vigorous larvae from each group were selected and frozen in liquid nitrogen for subsequent tests. Three biological replicates with five larvae per replicate (3 × 5 individuals) were used for RNA extraction. Total RNA of each replicate was isolated and checked as described above. Six RNA samples (2 groups × 3 replicated samples) were used for Illumina RNA-seq.
Library construction and sequencing
For SMRT sequencing, a mixed RNA sample (3 μg) as described above was used to generate a Full-length cDNA library according to the manufacturer’s protocol of SMARTerTM PCR cDNA Synthesis Kits (Clontech, USA). One SMRT cell (1–6 kb) was sequenced using a PacBio Sequel platform (Majorbio, Shanghai, China). The raw SMRT data were uploaded to the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) with accession CRA007259.
For Illumina RNA-seq, six cDNA libraries (2 groups × 3 replicated samples; 1 μg of RNA used per sample) were generated with NEBNext UltraTM RNA Library Prep Kits (NEB, Beverly, MA, United States) following the manufacturer’s protocol. The high quality of the libraries was confirmed using a high-sensitivity DNA chip on an Agilent Bioanalyzer 2100 (Agilent Technologies, Palo Alto, CA, United States). Paired-end sequencing was conducted on the Illumina HiSeq 2000 platform (Majorbio, Shanghai, China). The raw Illumina data were uploaded to the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) with accession CRA007253.
Processing of SMRT sequencing data and Illumina data
To obtain the clean reads, the raw SMRT data were polished via filtering out short length (< 50 bp) and low quality (< 0.9) of polymerase reads using SMRTlink version 6.0 software. Then, a circular consensus sequence (CCS) was generated after the clean reads were processed into error-corrected reads of inserts (ROIs) with parameters of full passes of ≥3 and accuracy of > 0.9, and CCS.BAM files were output. CCS.BAM files were divided into full-length non-chimeric (FLNC) and non-full-length (NFL) reads according to whether the reads contained the 5′ primer, 3′ primer, and poly A tail. Subsequently, to obtain polished consensus isoforms, FLNC reads were clustered using the Iterative clustering of error correction (ICE) program and NFL reads were clustered using Quiver algorithm. High-quality isoforms with a post-correction accuracy of > 99% were identified as full-length transcripts. Ultimately, the final full-length transcripts were generated for further analysis after removing the redundancies by CD-HIT-EST program with default parameters.
The raw Illumina data were filtered by removing reads containing adapters, > 10% ambiguous ‘N’ nucleotides, and > 20% of the bases having quality scores < 10 in SeqPrep software (https://github.com/jstjohn/SeqPrep). The Q20, Q30 and GC content of the clean reads were calculated to assess sequencing quality using fastx_toolkit_0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/). The obtained full-length transcripts from SMRT data were applied as a reference transcriptome to Illumina data correction. We used Bowtie2 version 2.2.9 to map the clean reads from Illumina sequencing to the reference transcriptome with default parameters [75].
Prediction of alternative splicing, simple sequence repeat, long non-coding RNA, and transcription factor
Alternative splicing (AS) events were predicted using pair-wise BLAST analysis based on the full-length transcript sequences. The alignment results were considered as candidate AS events according to the following criteria: 1) Both tested sequences lengths were more than 1000 bp, and there should be two High-scoring Segment Pairs in the alignment. 2) The alternative splicing gap was more than 100 bp, with at least 100 bp distance to the 3′/5′ end. 3) All alternatively-spliced transcripts allowed 5 bp overlap.
Transcripts > 500 bp were subjected to simple sequence repeat (SSR) analysis using MISA version 2.3.6. Seven SSR types, including mononucleotide, dinucleotide, trinucleotide, tetra-nucleotide, pentanucleotide, hexanucleotide, and compound SSRs (mixed microsatellites, two SSRs with > 100 bp distance), were detected.
long non-coding RNA (lncRNA) was predicted using Coding Potential Calculator (CPC, score < 0.5), Coding-Non-Coding Index (CNCI, score < 0), PLEK (score < 0), and Pfam protein structure domain analysis (no annotation). The prediction of transcription factors (TFs) was performed by the animal TFDB version 3.0 database.
Functional annotation of full-length transcripts
To annotate the obtained full-length transcripts, six public databases, namely, NCBI nonredundant proteins (Nr, e-value ≤1.0 × 10− 5), Swiss-Prot (e-value ≤1.0 × 10− 5), Protein families (Pfam, e-value ≤1.0 × 10− 5), Clusters of Orthologous Groups (COG, e-value ≤1.0 × 10− 5), (Gene Ontology (GO, e-value ≤1.0 × 10− 5), and Kyoto Encyclopedia of Genes and Genomes (KEGG, e-value ≤1.0 × 10− 10) databases, were searched [76].
Differential expression analysis and GO enrichment analysis of full-length transcripts
The expression level of each transcript from six RNA-seq samples was calculated and normalized as TPM (transcripts per million) values by RSEM version 1.3.1. Then, we used DESeq2 version 1.24.0 for differential expression analyses between CK and Linalool_LC15 groups. The criteria for determining the differentially expressed (DE) transcripts was P-value < 0.05 (false discovery rate ≤ 0.01) and |log2 (fold change)| ≥ 1. For significantly up-regulated and down-regulated transcripts, GO enrichment analysis was separately performed using Goatools program (https://github.com/tanghaibao/GOatools), and an adjusted P-value < 0.05 was the threshold value for determining the significantly enriched terms. Heatmap of differentially expressed transcripts of interest were present using Chiplot online tools (https://www.chiplot.online/tvbot.html).
lncRNA-mRNA and TF-mRNA co-expression networks construction
According to the results of differential expression analysis and GO enrichment analysis, the DE transcripts encoding cytochrome P450s (CYP450s) and cuticular protein (CPs) were identified as target genes (mRNAs) that involved in tolerance to dietary linalool. To screen for the regulating factors associated with target mRNAs, the correlation coefficients between target mRNAs and lncRNAs, target mRNAs and TF were evaluated on the basis of TBM value using the Pearson correlation method in Origin version 2021b. The correlation coefficients of > 0.8 (positive) or < 0.8 (negative) between target mRNAs and lncRNAs, target mRNAs and TF were considered as co-expressed genes. Data of correlation analysis were put into Cytoscape 3.8.2 to generate co-expression networks.
Expression pattern of interesting transcripts using real-time quantitative PCR
We selected two lncRNAs, two TFs, six CYP450 transcripts, and six CP transcripts to explore their transcriptional responses to linalool exposure using Real-time quantitative PCR (RT-qPCR). The sampling scheme was as follows: Day 1, 1st instar larvae (L1-D1), Day 3, 1st instar larvae (L1-D3), Day 3, 2nd instar larvae (L2-D3), and Day 3, 3rd instar larvae (L3-D3) were respectively collected from the treatment and CK groups referring to the protocols for Illumina RNA-Seq samples. Three biological replicates with five larvae per replicate (3 × 5 individuals) were used for each time point. All samples were frozen in liquid nitrogen for RNA extraction. cDNA was synthesized using a HiScript 1st Strand cDNA Synthesis Kit according to the manufacturer’s protocol (Vazyme Biotech, Nanjing, China). RT-qPCR experiments were performed using SYBR Premix Ex Taq II (TaKaRa, Japan) on an Applied Biosystem 7500 Real-Time PCR System (Thermo Fisher, Massachusetts, United States). PCR conditions were as follows: 5 min at 95 °C, followed by 40 cycles of 10 s at 95 °C and 40 s at 60 °C. Specific primers were designed using Primer Premier version 5.0 (Table S8) and synthesized from Sangon, Shanghai, China. Relative expression levels of each transcript were determined by the 2−△△Ct method with Ribosomal protein L10 (RPL10) and TBP (TATA-binding protein) as the housekeeping genes. Statistical significance of gene expression was determined using one-way ANOVA followed by independent sample t-test (P < 0.05) in SPSS version 20.0. To verify the reliability of RNA-Seq data, Pearson’s correlation between the results of RT-qPCR (Log2 2−△△Ct) and RNA-Seq (Log2 fold change) was analysed in SPSS version 20.0.
Availability of data and materials
The raw SMRT data generated during the current study were uploaded to the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) with accession CRA007259. The raw Illumina data generated during the current study were uploaded to the Genome Sequence Archive (GSA) (https://ngdc.cncb.ac.cn/gsa/) with accession CRA007253. The qPCR data were uploaded to figshare (https://figshare.com/account/home). Li, Shouyin (2022): Data for qPCR. figshare. Dataset. https://doi.org/10.6084/m9.figshare.20941435
References
Petschenka G, Agrawal AA. How herbivores coopt plant defenses: natural selection, specialization, and sequestration. Curr Opin Insect Sci. 2016;14:17–24.
Stahl E, Hilfiker O, Reymond P. Plant-arthropod interactions: who is the winner? Plant J. 2018;93(4):703–28.
Erb M, Reymond P. Molecular interactions between plants and insect herbivores. Annu Rev Plant Biol. 2019;70:527–57.
Wari D, Aboshi T, Shinya T, Galis I. Integrated view of plant metabolic defense with particular focus on chewing herbivores. J Integr Plant Biol. 2022;64(2):449–75.
Keeling CI, Bohlmann J. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006;170(4):657–75.
Zulak KG, Bohlmann J. Terpenoid biosynthesis and specialized vascular cells of conifer defense. J Integr Plant Biol. 2010;52(1):86–97.
Ashour M, Wink M, Gershenzon J. Biochemistry of terpenoids: monoterpenes, sesquiterpenes and diterpenes. Annu Rev Plant Biol. 2018;40:258–303.
Whitehill J, Bohlmann J. A molecular and genomic reference system for conifer defence against insects. Plant Cell Environ. 2019;42(10):2844–59.
Whitehill J, Yuen M, Henderson H, Madilao L, Kshatriya K, Bryan J, et al. Functions of stone cells and oleoresin terpenes in the conifer defense syndrome. New Phytol. 2019;221(3):1503–17.
Boncan D, Tsang S, Li C, Lee I, Lam HM, Chan TF, et al. Terpenes and terpenoids in plants: Interactions with environment and insects. Int J Mol Sci. 2020;21(19):7382.
Poudel DK, Rokaya A, Ojha PK, Timsina S, Satyal R, Dosoky NS. The chemical profiling of essential oils from different tissues of Cinnamomum camphora L. and their antimicrobial activities. Molecules. 2021;26(17):5132.
Jiang H, Wang J, Song L, Cao X, Yao X, Tang F. GCxGC-TOFMS analysis of essential oils composition from leaves, twigs and seeds of Cinnamomum camphora L. Presl and their insecticidal and repellent activities. Molecules. 2016;21(4):423.
Fouad HA, Tavares WDS, Zanuncio JC. Toxicity and repellent activity of monoterpene enantiomers to rice weevils (Sitophilus oryzae). Pest Manag Sci. 2021;77(7):3500–7.
Govind G, Mittapalli O, Griebel T, Allmann S, Böcker S, Baldwin IT. Unbiased transcriptional comparisons of generalist and specialist herbivores feeding on progressively defenseless Nicotiana attenuata plants. PLoS One. 2010;5(1):e8735.
Bras A, Roy A, Heckel DG, Anderson P, Karlsson GK. Pesticide resistance in arthropods: ecology matters too. Ecol Lett. 2022;25:1746–59.
Seybold SJ, Huber DPW, Lee SJ, Graves AD, Bohlmann J. Pine monoterpenes and pine bark beetles: a marriage of convenience for defense and chemical communication. Phytochem Rev. 2006;5:143–78.
Verónica T, Gabriel O, Soto-Robles LV, Arnulfo A, María FL, Gerardo Z. Gut transcriptome of two bark beetle species stimulated with the same kairomones reveals molecular differences in detoxification pathways. Comput Struct Biotec. 2022;20:3080–95.
Blomquist GJ, Tittiger C, MacLean M, Keeling CI. Cytochromes P450: terpene detoxification and pheromone production in bark beetles. Curr Opin Insect Sci. 2021;43:97–102.
Vandenhole M, Dermauw W, Van Leeuwen T. Short term transcriptional responses of P450s to phytochemicals in insects and mites. Curr Opin Insect Sci. 2021;43:117–27.
Balabanidou V, Grigoraki L, Vontas J. Insect cuticle: a critical determinant of insecticide resistance. Curr Opin Insect Sci. 2018;27:68–74.
Koganemaru R, Miller DM, Adelman ZN. Robust cuticular penetration resistance in the common bed bug (Cimex lectularius L.) correlates with increased steady-state transcript levels of CPR-type cuticle protein genes. Pestic Biochem Phys. 2013;106:190–7.
Fang F, Wang W, Zhang D, Yuan L, Zhou D, Ma L, et al. The cuticle proteins: a putative role for deltamethrin resistance in Culex pipiens pallens. Parasitol Res. 2015;114:4421–9.
Yahouedo GA, Chandre F, Rossignol M, Ginibre C, Balaban MN, et al. Contributions of cuticle permeability and enzyme detoxification to pyrethroid resistance in the major malaria vector Anopheles gambiae. Sci Rep. 2017;7(1):11091.
Chen C, Zhang CC, Li SY, Zhu H, Fan BQ, Su P, et al. Biological traits and life history of Pagiophloeus tsushimanus (Coleoptera: Curculionidae), a weevil pest on camphor trees in China. J For Res. 2021;32(5):1979–88.
Li SY, Chen C, Jia ZY, Li Q, Tang ZZ, Zhong MF, et al. Offspring performance and female preference of Pagiophloeus tsushimanus (coleoptera curculionidae) on three Lauraceae tree species: a potential risk of host shift caused by larval experience. J Appl Entomol. 2021;145(6):530–42.
Li SY, Wang JT, Chen C, Li H, Hao DJ. Tolerance, biochemistry and related gene expression in Pagiophloeus tsushimanus (Coleoptera: Curculionidae) exposed to chemical stress from headspace host-plant volatiles. Agric For Entomol. 2022;22(2):189–203.
Eljazi JS, Bachrouch O, Salem N, Msaada K, Aouini J, Hammami M, et al. Chemical composition and insecticidal activity of essential oil from coriander fruit against Tribolium castaenum, Sitophilus oryzae, and Lasioderma serricorne. Int J Food Prop. 2017;20:S2833–45.
Liu TT, Chao LK, Hong KS, Huang YJ, Yang TS. Composition and insecticidal activity of essential oil of Bacopa caroliniana and interactive effects of individual compounds on the activity. Insects. 2019;11(1):23.
Stark R, Grzelak M, Hadfield J. RNA sequencing: the teenage years. Nat Rev Gene. 2019;20(11):631–56.
Vogel H, Musser RO, Celorio-Mancer MDLP. Transcripome responses in herbivorous insects towards host plant and toxin feeding. Annu Rev Plant Biol. 2014;47:197–233.
Rand E, Smit S, Beukes M, Apostolides Z, Pirk C, Nicolson SW. Detoxification mechanisms of honey bees (Apis mellifera) resulting in tolerance of dietary nicotine. Sci Rep. 2015;5:11779.
Roy A, Walker WB III, Vogel H, Chattington S, Larsson MC, Anderson P, et al. Diet dependent metabolic responses in three generalist insect herbivores Spodoptera spp. Insect Biochem Mol Biol. 2016;71:91–105.
Huang Y, Liao M, Yang QQ, Xiao JJ, Hu ZY, Zhou LJ. Transcriptome profiling reveals differential gene expression of detoxfication enzymes in Sitophilus zeamais responding to terpinen-4-olfumigation. Pestic Biochem Phys. 2018;149:44–53.
Zheng X, Liu FD, Shi M, Li S, Xie XF, Li GN, et al. Transcriptome analysis of the reproduction of silkworm (Bombyx mori) under dimethoate stress. Pestic Biochem Phys. 2022;183:105081.
Rhoads A, Au KF. PacBio sequencing and its applications. Genom Proteom Bioinf. 2015;13(5):278–89.
Jia D, Wang Y, Liu Y, Hu J, Guo Y, Gao L. SMRT sequencing of full-length transcriptome of flea beetle Agasicles hygrophila (Selman and Vogt). Sci Rep. 2018;8(1):2197.
Yang H, Xu D, Zhuo Z, Hu J, Lu B. SMRT sequencing of the full-length transcriptome of the Rhynchophorus ferrugineus (Coleoptera: Curculionidae). PeerJ. 2020;8:e9133.
Wang X, Xu X, Ullah F, Qian D, Gao XW, Desneux N, et al. Comparison of full-length transcriptomes of different imidacloprid-resistant strains of Rhopalosiphum padi (L.). Entomol Gen. 2021;41:289–304.
Xu D, Yang H, Zhuo Z, Lu B, Hu J, Yang F. Characterization and analysis of the transcriptome in Opisina arenosella from different developmental stages using single-molecule real-time transcript sequencing and RNA-seq. Int J Biol Macromol. 2021;169:216–27.
Chen H, Lin L, Xie M, Zhong Y, Zhang G, Su W. Survey of the Bradysia odoriphaga transcriptome using PacBio single-molecule long-read sequencing. Genes. 2019;10(6):481.
Chen T, Sun Q, Ma Y, Zeng W, Liu R, Qu D, et al. A transcriptome atlas of silkworm silk glands revealed by PacBio single-molecule long-read sequencing. Mol Gen Genomics. 2020;295(5):1227–37.
Vuong CK, Black DL, Zheng S. The neurogenetics of alternative splicing. Nat Rev Neurosci. 2016;17(5):265–81.
Buschiazzo E, Gemmell NJ. The rise, fall and renaissance of microsatellites in eukaryotic genomes. BioEssays. 2006;28(10):1040–50.
Ouyang H, Wang X, Zheng X, Lu W, Qin F, Chen C. Full-length SMRT transcriptome sequencing and SSR analysis of Bactrocera dorsalis (Hendel). Insects. 2021;12(10):938.
Yuan Y, Zhang L, Wu G, Zhu J. High-throughput discovery microsatellites in Tomicus yunnanensis (Coleoptera: Scolytinae). J Environ Entomol. 2014;36:166–70 (In Chinese).
Han X, Wang Y, Lu C, Lin H, Shi Y, He H, et al. Characteristics of the SSR loci in the Anoplophora chinensis transcriptome. Chi J Appl Entomol. 2019;56:1799–805 (in Chinese).
Keeling CI, Campbell EO, Batista PD, Shegelski VA, Trevoy SA, Huber DP, et al. Chromosome-level genome assembly reveals genomic architecture of northern range expansion in the mountain pine beetle, Dendroctonus ponderosae Hopkins (Coleoptera: Curculionidae). Mol Ecol Resour. 2022;22:1149–67.
Liu ZD, Xing LS, Huang WL, Liu B, Wan FH, Raffa KF, et al. Chromosome-level genome assembly and population genomic analyses provide insights into adaptive evolution of the red turpentine beetle. BMC Biol. 2022;20:190.
Keeling CI, Yuen MM, Liao NY, Docking TR, Chan SK, Taylor GA, et al. Draft genome of the mountain pine beetle, Dendroctonus ponderosae Hopkins, a major forest pest. Genome Biol. 2013;14:R27.
Nauen R, Bass C, Feyereisen R, Vontas J. The role of cytochrome P450s in insect toxicology and resistance. Annu Rev Plant Biol. 2022;67:105–24.
Lu K, Song Y, Zeng R. The role of cytochrome P450-mediated detoxification in insect adaptation to xenobiotics. Curr Opin Insect Sci. 2021;43:103–7.
Keeling CI, Henderson H, Li M, Dullat HK, Ohnishi T, Bohlmann J. CYP345E2, an antenna-specific cytochrome P450 from the mountain pine beetle, Dendroctonus ponderosae Hopkins, catalyses the oxidation of pine host monoterpene volatiles. Insect Biochem Mol Biol. 2013;43(12):1142–51.
Chiu CC, Keeling CI, Bohlmann J. Monoterpenyl esters in juvenile mountain pine beetle and sex-specific release of the aggregation pheromone trans-verbenol. Proc Natl Acad Sci U S A. 2018;115(14):3652–7.
Ramakrishnan R, Roy A, Kai M, Svatoš A, Jirošová A. Metabolomics and transcriptomics of pheromone biosynthesis in an aggressive forest pest Ips typographus. Insect Biochem Molec. 2022;140:103680.
Li SY, Li H, Wang JT, Chen C, Hao DJ. Hormetic response and co-expression of cytochrome P450 and cuticular protein reveal the tolerance to host-specific terpenoid defences in an emerging insect pest, Pagiophloeus tsushimanus (Coleoptera: Curculionidae). J Pest Sci. 2022. https://doi.org/10.1007/s10340-022-01509-0.
Mao YB, Cai WJ, Wang JW, Hong GJ, Tao XY, Wang LJ. Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat Biotechnol. 2007;25(11):1307–13.
Wang RL, Xia QQ, Baerson SR, Ren Y, Wang J, Su YJ, et al. A novel cytochrome P450 CYP6AB14 gene in Spodoptera litura (Lepidoptera: Noctuidae) and its potential role in plant allelochemical detoxification. J Insect Physiol. 2015;75:54–62.
Nauen R, Zimmer CT, Vontas J. Heterologous expression of insect P450 enzymes that metabolize xenobiotics. Curr Opin Insect Sci. 2021;43:78–84.
Hrycay EG, Bandiera SM. In: Hrycay EG, Bandiera SM, editors. Monooxygenase, peroxidase and peroxygenase properties and reaction mechanisms of cytochrome P450 enzymes. Cham: Springer International Publishing; 2015. p. 1–61.
Tajiri R. Cuticle itself as a central and dynamic player in shaping cuticle. Curr Opin Insect Sci. 2017;19:30–5.
Birnbaum S, Abbot P. Gene expression and diet breadth in plant-feeding insects: summarizing trends. Trends Ecol Evol. 2020;35(3):259–77.
Charles JP. The regulation of expression of insect cuticle protein genes. Insect Biochem Mol Biol. 2010;40(3):205–13.
Sureshan SC, Mohideen HS. Ramya M differential expression profiling of Oxycarenus laetus Kirby (Hemiptera: Lygaeidae) upon exposure to gossypol. Mol Biol Rep. 2022;49:4727–35.
Chiu CC, Bohlmann J. Mountain pine beetle epidemic: an interplay of terpenoids in host defense and insect pheromones. Annu Rev Plant Biol. 2022;73:475–94.
Bearfield JC, Henry AG, Tittiger C, Blomquist GJ, Ginzel MD. Two regulatory mechanisms of monoterpenoid pheromone production in Ips spp. of bark beetles. J Chem Ecol. 2009;35:689–97.
Celorio-Mancera MP, Ahn S, Vogel H, Heckel DG. Transcriptional responses underlying the hormetic and detrimental effects of the plant secondary metabolite gossypol on the generalist herbivore Helicoverpa armigera. BMC Genomics. 2011;12:575.
Lu K, Li YM, Cheng YB, Li WR, Zeng BX, Gu CZ, et al. Activation of the ROS/CncC and 20-Hydroxyecdysone signaling pathways is associated with xanthotoxin-induced tolerance to λ-Cyhalothrin in Spodoptera litura. J Agric Food Chem. 2021;69:13425–35.
Cen YJ, Zou XP, Li LB, Chen SN, Lin YG, Liu L, et al. Inhibition of the glutathione biosynthetic pathway increases phytochemical toxicity to Spodoptera litura and Nilaparvata lugens. Pestic Biochem Phys. 2020;168:104630.
Corona M, Robinson GE. Genes of the antioxidant system of the honey bee: annotation and phylogeny. Insect Mol Biol. 2006;15:687–701.
King AM, MacRae TH. Insect heat shock proteins during stress and diapause. Annu Rev Entomol. 2015;60:59–75.
Scriber MJ, Feeny P. Growth of herbivorous caterpillars in relation to feeding specialization and to the growth form of their food plants. Ecology. 1979;60:829–50.
Rothwell EM, Holeski LM. Phytochemical defences and performance of specialist and generalist herbivores: a meta-analysis. Ecol Entomol. 2020;45(3):396–405.
Ali JG, Agrawal AA. Specialist versus generalist insect herbivores and plant defense. Trends Plant Sci. 2012;17(5):293–302.
Beran F, Petschenka G. Sequestration of plant defense compounds by insects: from mechanisms to insect-plant coevolution. Annu Rev Entomol. 2022;67:163–80.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Acknowledges
We acknowledge Mr. Liang Chen and Mr. Yanbo Sun for their assistance with the collection of tested insects, and Prof. Zengfang Yin for her assistance with authentication of plant material. Also thanks for the financial support of the Forest Station of Shanghai and Jiangsu Education Department.
Funding
This study was supported by the Science and Technology Commission of Shanghai Municipality (18391903200), and Postgraduate Research and Practice Innovation Program of Jiangsu Province (KYCX22_1111).
Author information
Authors and Affiliations
Contributions
S.L. and D.H. designed the experiment. S.L. performed the experiments and analyzed the data. S.L., H.L., C.C., and D.H. wrote and reviewed the manuscript. H.L. and D.H. provided the reagents and materials. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The plant material used in this study was collected from the campus of Nanjing Forestry University, China (32°4′43″N/118°48′30″E), and in compliance with the appropriate permissions and/or licenses for the collection of plant specimens. Experimental research on plants (either cultivated or wild), including the collection of plant material, comply with relevant institutional, national, and international guidelines and legislation.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Li, S., Li, H., Chen, C. et al. Tolerance to dietary linalool primarily involves co-expression of cytochrome P450s and cuticular proteins in Pagiophloeus tsushimanus (Coleoptera: Curculionidae) larvae using SMRT sequencing and RNA-seq. BMC Genomics 24, 34 (2023). https://doi.org/10.1186/s12864-023-09117-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-023-09117-7