Abstract
Background
Endonucleases play a crucial role in plant growth and stress response by breaking down nuclear DNA. However, the specific members and biological functions of the endonuclease encoding genes in wheat remain to be determined.
Results
In this study, we identified a total of 26 TaENDO family genes at the wheat genome-wide level. These genes were located on chromosomes 2 A, 2B, 2D, 3 A, 3B, and 3D and classified into four groups, each sharing similar gene structures and conserved motifs. Furthermore, we identified diverse stress-response and growth-related cis-elements in the promoter of TaENDO genes, which were broadly expressed in different organs, and several TaENDO genes were significantly induced under drought and salt stresses. We further examined the biological function of TaENDO23 gene since it was rapidly induced under drought stress and exhibited high expression in spikes and grains. Subcellular localization analysis revealed that TaENDO23 was localized in the cytoplasm of wheat protoplasts. qRT-PCR results indicated that the expression of TaENDO23 increased under PEG6000 and abscisic acid treatments, but decreased under NaCl treatment. TaENDO23 mainly expressed in leaves and spikes. A kompetitive allele-specific PCR (KASP) marker was developed to identify single nucleotide polymorphisms in TaENDO23 gene in 256 wheat accessions. The alleles with TaENDO23-HapI haplotypes had higher grain weight and size compared to TaENDO23-HapII. The geographical and annual frequency distributions of the two TaENDO23 haplotypes revealed that the elite haplotype TaENDO23-HapI was positively selected in the wheat breeding process.
Conclusion
We systematically analyzed the evolutionary relationships, gene structure characteristics, and expression patterns of TaENDO genes in wheat. The expression of TaENDO23, in particular, was induced under drought stress, mainly expressed in the leaves and grains. The KASP marker of TaENDO23 gene successfully distinguished between the wheat accessions, revealing TaENDO23-HapI as the elite haplotype associated with improved grain weight and size. These findings provide insights into the evolution and characteristics of TaENDO genes at the genome-wide level in wheat, laying the foundation for further biological analysis of TaENDO23 gene, especially in response to drought stress and grain development.
Similar content being viewed by others
Introduction
Wheat (Triticum aestivum L.) is one of the world’s most crucial cereal crops and is cultivated widely covering more than 200 million hectares, with an annual production of approximately700 million tons. This crop is essential for global food security. However, drought and heat stresses have substantially reduced national cereal production by 9–10% [1]. Thus, identifying key genes associated with grain yield and drought tolerance is crucial for developing high-yielding, drought-resistant wheat varieties through molecular marker-assisted breeding [2].
Plant nucleases are involved in nearly every stage of the plant life cycle through the programmed cell death (PCD) pathway. They play essential roles in various processes, such as endosperm degradation, aerenchyma formation, leaf senescence, tapetum destruction during pollen development, and immune responses [3,4,5,6,7]. These nucleases function as bifunctional enzymes capable of efficiently degrading single-stranded DNA or RNA, but not double-stranded DNA [3, 4, 8]. During seed or grain development, the degradation of DNA by nucleases is crucial for the proper growth of organs. For instance, nucleases are activated in the formation of the suspensor and degradation of the aleurone layer [9, 10]. In Arabidopsis (Arabidopsis thaliana), embryos lacking poly (A) -specific 3’-5’ ribonuclease or with decreased enzymatic activity, exhibit retarded development, ultimately arresting at the bent-cotyledon stage [11]. In addition, nucleases are implicated in the regulation of abiotic stress responses. In Arabidopsis, mutations in certain members of the CAF1 family affect cellular mRNA levels, thereby influencing responses to diverse abiotic stresses [12]. In barley (Hordeum vulgare L.), the nuclease-encoding gene Bnuc1 is induced by salt stress and exogenous abscisic acid (ABA) treatment [13]. These findings underscore the vital role of plant nucleases in plant growth and stress response mechanisms.
Plant nucleases can be categorized into exonuclease and endonuclease based on their substrate specificity, particularly at the 5 ‘or 3’ terminus. Endonucleases are further divided into two groups depending on their cofactors, Zn2+ and Ca2+. Zn2+-dependent endonucleases play crucial roles in plant development, differentiation, and PCD, whereas Ca2+-dependent endonucleases are primarily involved in plant defense [14]. The enzymatic properties of nucleases are influenced by various factors, such as divalent cations, optimal pH, and variations in amino acid sequences [6], which can cause functional differences of these enzymes in plant development and their responses to abiotic or biotic stresses. Although the functions of ENDO genes have been extensively studied in Arabidopsis, their evolutionary relationships and biological roles in wheat remain largely unknown. In Arabidopsis, at least five endonuclease-encoding genes have been identified and classified into three groups: ENDO1/BFN1 in subclade I; ENDO3, ENDO4, and ENDO5 in subclade II; and ENDO2 in subclade III [15]. Among these, ENDO1 has been widely studied and reported to be involved in various biological processes related to cell death [16, 17]. ENDO1 expression was detected during leaf and stem senescence, flower and seed development, and abscission zone formation [18]. ENDO2 expressed in leaves and was reported to respond to various biotic and abiotic stresses, ENDO3 primarily expressed in flowers, and ENDO5 specifically expressed in roots [6].
Although several studies have reported the role of plant nucleases in seed development and abiotic stress responses, our understanding of endonuclease—largely derived from studies in Arabidopsis—remains limited. In this study, we identified ENDO family genes in the wheat genome and analyzed their gene structure, conserved domains, and collinearity. We also examined their expression patterns using RNA sequencing (RNA-seq) data. Furthermore, we amplified the gene TaENDO23 to analyze its biological function. We transiently expressed TaENDO23-GFP fusion proteins in wheat protoplasts to determine the subcellular localization of TaENDO23. In addition, we analyzed the expression of the TaENDO23 gene across different organs and its response to salt and drought stresses. Finally, a kompetitive allele-specific PCR (KASP) marker was developed to distinguish different haplotypes in 256 wheat accessions. This marker was used to identify the elite haplotypes of TaENDO23 gene. Our study provides a theoretical foundation for understanding the evolution and potential biological functions of TaENDO family genes, especially TaENDO23, in drought stress responses and grain development.
Results
Identification and characteristics of TaENDO family members
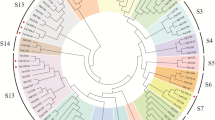
A total of 26 TaENDO family genes were identified and named TaENDO1 to TaENDO26 on the basis of their chromosomal positions (Table 1). The theoretical molecular weights of their encoded proteins ranged from 23.21 KDa (TaENDO19) to 34.67 KDa (TaENDO1), with isoelectric points varying between 5.35 (TaENDO19) and 8.29 (TaENDO5) (Table 1). Subcellular localization prediction indicated that TaENDO proteins were distributed across various cellular compartments, including the nucleus, cytoplasm, mitochondrion, endoplasmic reticulum, vacuole, and chloroplast. This finding suggests that TaENDO proteins have diverse roles in cellular processes (Table 1). To explore the phylogenetic relationships among ENDO proteins, the amino acid sequences from five AtENDOs, four OsENDOs, four ZmENDOs, and 26 TaENDOs were aligned and used to construct a phylogenetic tree (Fig. 1). The analysis classified ENDO proteins into five groups (Fig. 1). Notably, subclade III contained only three Arabidopsis ENDO proteins and lacked monocotyledon ENDO proteins. By contrast, subclades IV and V were composed exclusively of monocotyledon ENDOs (Fig. 1), which exhibited distinct subcellular localization (Table 1). This diversity in localization suggests that TaENDO family members within these subclades participate in various regulation pathways.
Gene structure and conserved motif analysis of TaENDO family members
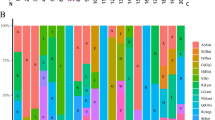
To investigate the structural diversity of TaENDO proteins, conserved motifs were identified using the MEME database. A total of 20 conserved motifs were discovered, designated as motifs 1 through 20 (Fig. 2A). Motifs 1 to 3 were found in all TaENDO proteins, suggesting a high degree of functional conservation (Fig. S1). In addition, TaENDO proteins within the same subgroup exhibited similar motif compositions and distribution patterns. For example, members of subgroup I all contained motifs 1, 2, 3, 6, and 10, whereas motif 11 was found only in subgroup II. To further explore the TaENDO family, gene structures were analyzed using GSDS software (Fig. 2B). The analysis revealed that most TaENDO genes consisted of eight exons and seven introns, with family members within the same subfamily sharing similar exon/intron positions, indicating a high level of conservation among TaENDO family genes (Fig. 2B). However, TaENDO2 and TaENDO4 in subclade I had only seven and six exons, respectively, leading to the loss of several conserved motifs in their encoded sequences (Fig. 2B). This structural variation may contribute to functional differentiation during the evolution of TaENDO genes.
Conserved motifs of TaENDO family proteins and gene structures of their encoding gene. A. Conserved motifs in TaENDO proteins. Colored boxes represent different motifs. B. Exon-intron structures of TaENDO genes. Blue boxes indicate untranslated regions, yellow boxes represent exons, and black lines represent introns. The phylogenetic tree was constructed as in Fig. 1, focusing only on ENDO proteins in wheat
Chromosomal localization, gene duplication, and collinearity analysis of TaENDO family genes
To gain a deeper understanding of the expansion and evolution of TaENDO family genes, we analyzed the chromosomal distribution of the 26 TaENDO genes. The results revealed that TaENDO genes were localized exclusively on chromosomes 2 A, 2B, 2D, 3A, 3B, and 3D (Fig. S2). Seventeen pairs of TaENDO genes were identified as products of segmental duplication, whereas no tandem duplications were observed (Fig. 3A). The Ka/Ks ratio calculated to assess the selection pressure on duplicated genes indicated that the majority of duplicated TaENDO gene pairs were under purifying selection, given that the ratios were all less than 1 (Table S1). In addition, we conducted a collinearity analysis to explore the evolutionary relationships between ENDO genes in wheat, rice, and maize (Fig. 3B). The analysis showed that wheat ENDO genes shared six homologous genes with rice and nine with maize, suggesting functional conservation of ENDO genes among different species.
Collinearity analysis of TaDNDO family genes. A. Duplicated gene pairs of TaDNDO genes in the wheat genome. B. Collinearity analysis of TaDNDO genes with the genomes of rice and maize. Duplicated gene pairs are linked by different lines. Blue lines indicate collinear blocks between the genome and other species
Analysis of cis-acting elements of TaENDO genes
To investigate the regulatory elements in the promoter regions of TaENDO family genes, we analyzed the 2000-bp upstream sequences from the initiation codon by using the PlantCARE platform. A total of 30 cis-acting elements were indentified and categorized into four groups: plant hormone response elements (7), light response elements (7), growth and development elements (6), and stress regulation elements (10) (Fig. S3). The analysis revealed that most TaENDO genes contained ABA-responsive elements (ABRE), suggesting their involvement in the ABA regulation pathway. In addition, several stress response elements (MYB, STRE, LTR) and growth-related elements (CAT-box, sa-1, CCGTCC-box) were identified in the promoters of TaENDO genes. These results indicated the potential involvement of TaENDO family genes in diverse regulatory pathways.
The expression level of TaENDO genes in different organs and under different abiotic stress treatments
The expression levels of TaENDO family genes were analyzed in various organs—roots, stems, leaves, spikes, and grains— of wheat variety Chinese Spring at different growth stages by using RNA-seq data derived from the WheatOmics 1.0 platform. The transcripts per kilobase of exon model per million mapped reads (TPM) values were normalized by TBtools software to quantify gene expression across these organs. The analysis revealed that TaENDO genes within the same subclade exhibited similar expression patterns (Fig. 4A). Specifically, TaENDO genes in subclades I and V exhibited broad expression across different organs, whereas those in subclade II specifically expressed in young grains. TaENDO genes in subclade IV displayed diverse expression patterns, with genes such as TaENDO3, 5, 9, 14, and 16 having high expression in roots, and TaENDO1, 7, and 12 being predominantly expressed in spikes. The high expression of certain TaENDO genes in spikes and grains suggests their potential role in regulating grain development.
Expression pattern analysis of TaDNDO family genes. (A) Heatmap of TaDNDO genes expressions at different growth stages and in various organs of Chinese Spring wheat plants. (B) Expression Heatmap of TaDNDO expressions under NaCl treatment. (C) Heatmap of TaDNDO expressions under PEG treatment. Growth stages: Z10 One-leaf stage, Z13 Three-leaf stage, Z30 Booting stage, Z32 Early jointing stage, Z39 Late jointing stage, Z65 Mid-flowering stage, Z71 2 d after flowering, Z75 10 d after flowering, and Z85 30d after flowering. The phylogenetic tree was constructed as in Fig. 1, focusing only on ENDO proteins in wheat
To further explore the expression patterns of TaENDO genes under abiotic stress, we analyzed RNA-seq data derived from the WheatOmics 1.0 platform. The results indicated that most TaENDO genes were significantly down-regulated under salt stress compared with the control, except for three genes, TaENDO20, 21, and 24, which were up-regulated (Fig. 4B). In response to PEG6000 treatment, several TaENDO genes (TaENDO13, 15, 19, 22, 23, and 25) were rapidly induced within 2 h compared with that under 0 h PEG6000 treatment, whereas only a few genes, such as TaENDO1, 4, 7, and 12, remained insensitive (Fig. 4C). The rapid induction of certain TaENDO genes under PEG6000 treatment suggests their crucial roles in responding to drought stress.
To further validate these findings, we selected three TaENDO genes, TaENDO13, 21, and 24, to examine their response to drought stress and whether they were highly expressed in spikes or grains. The transcription levels of these genes in different organs and under PEG6000 treatment were validated using qRT-PCR with the winter wheat cultivar Jinmai 47 (JM47), a drought-resistant and high-yielding variety widely cultivated in the dryland regions of northwest China. Consistent with the RNA-seq data from the WheatOmics 1.0 platform, the qRT-PCR results demonstrated that TaENDO13, 21, and 15 were rapidly induced under 20% PEG6000 treatment and exhibited high expression in young spikes (Fig. S4). These results further conformed that several TaENDO genes may participate in responding to drought stress and regulating grain development.
The expression pattern of TaENDO23 gene and the subcellular localization of its encoding protein
Given that TaENDO23 was one of the TaENDO family genes rapidly induced under PEG6000 treatment (PEG6000-2 h) and exhibited broad expression across various organs, particularly in spikes and grains, the biological function of TaENDO23 was further analyzed. TaENDO23 gene contains 8 exons and 7 introns, with 11 conserved motifs possessed in its encoding protein. The phenotype response of wheat seedling was observed after treatment with 20% PEG6000 and 200 mM NaCl for both 0 and 1 day, respectively. No significant effect on wheat leaves was found after 1 day of PEG 000 or NaCl treatment (Fig. 5A). On the basis of these findings, multiple time points within a day were selected to analyze the expression levels of TaENDO23 gene under salt and drought stress conditions. The TaENDO23 expression rapidly induced after 1 h of PEG6000 treatment, peaking at 3 h; the expression levels were nine times higher than those in untreated samples (0-h) (Fig. 5B). After NaCl treatment, the expression of TaENDO23 gene quickly dropped to 0.3 times after 1 h and then continued down-regulated compared with the 0-h treatment (Fig. 5C). These results were generally consistent with RNA-seq data. Because the ABA signaling pathway is often involved in the response to diverse abiotic stresses, the expression level of the TaENDO23 gene under ABA treatment was also examined. Following ABA treatment, TaENDO23 transcription levels steadily increased, reaching the peak at 12 h (Fig. 5D). These results indicated that TaENDO23 gene participates in the response to drought stress through an ABA-dependent pathway.
qRT-PCR analysis of TaDNDO23 expression under PEG 6000, NaCl, and ABA treatments. (A) Phenotypes of 12-day-old JM47 seedling under 200 mM NaCl and 20% PEG6000 treatments. Bar = 4 cm. (B) Expression patterns of TaDNDO23 following PEG6000 treatment. (C) Expression patterns of TaDNDO23 following NaCl treatment. (D) Expression patterns of TaDNDO23 following ABA treatment. The relative expression levels in control samples (0 h) were normalized to 1. Means and standard deviations (SDs) were calculated from three biological and three technical replicates. Different letters denote significant differences between means (P < 0.05)
The qRT-PCR results also indicated that TaENDO23 gene expressed mainly in the leaves and spikes (Fig. 6A). Although TaENDO23 did not show high expression in grain as indicated in RNA-seq data (Figs. 4A and 6A), its expression in grains gradually increased over time (Fig. 6A), suggesting a potential role in grain development. To determine the subcellular localization of the TaENDO23 protein in wheat cells, the coding sequence (CDS) of TaENDO23 was amplified and fused into a 35 S::eGFP vector, which was then introduced into wheat protoplasts. The results revealed that green fluorescence signals were present in the nucleus, cytoplasm, and cytomembrane of wheat protoplasts transfected with the empty 35 S::eGFP vector. By contrast, the TaENDO23-eGFP fusion protein emitted green fluorescence only in the cytoplasm (Fig. 6B). These findings reveal that the TaENDO23 protein is predominantly localized in the cytoplasm.
Expression level of TaDNDO23 in different organs and subcellular localization of TaDNDO23 protein in wheat protoplasts. (A) qRT-PCR analysis of TaDNDO23 expression pattern in different wheat organs. TaActin was used as a reference gene. Means and standard deviations (SDs) were calculated from three biological and three technical replicates. DAA, days after anthesis. Different letters denote significant differences between means (P < 0.05). (B) Subcellular localization of TaDNDO23 protein by transiently expressing TaDNDO23-GFP in the wheat protoplast. Bar = 10 μm
The KASP marker development and relationship between two haplotypes of TaENDO23 gene and grain-related traits in wheat
The TaENDO23 gene is highly expressed in spikes at booting stage, suggesting its crucial role in spike and grain development. To explore this further, polymorphisms in the promoter and coding regions of TaENDO23 were analyzed using genomic re-sequencing data from 681 varieties obtained from the Wheat Union Database (Table S2). This analysis revealed three single-nucleotide polymorphisms (SNPs) in the promoter region and four SNPs in the coding region (Fig. 7A). Based on these SNPs, two haplotypes of TaENDO23 were identified, TaENDO23-HapI and TaENDO23-HapII (Fig. 7A). A KASP marker was successfully developed at 2,869 bp (C/G) in the coding region of TaENDO23 to differentiate wheat natural populations into two genotypes (Fig. 7B; Table S3). This population included 256 wheat accessions from various regions of China, which were planted in three farm stations at multiple years (five environments) in this study to measure their grain weight and size. The association between the different TaENDO23 haplotypes and grain-related traits was then examined in this wheat natural population. The results showed that TaENDO23-HapI had significantly higher thousand kernel weight (TKW) and kernel thickness (KT) than TaENDO23-HapII in four growth environments, whereas kernel length (KL) and kernel width (KW) remained similar between the haplotypes in all five environments (Fig. 7C, Table S4). These results suggested that TaENDO23-HapI is an elite haplotype for improving grain yield in wheat.
Effects of TaDNDO23 haplotypes on grain phenotypic traits. (A) Distribution of SNP sites in the TaDNDO23 gene structure. Red boxes and dotted lines represent exons and introns, respectively. The polymorphic SNP site (in red color) at position 2,869 bp from the ATG of TaDNDO23 was used to develop a KASP molecular marker. (B) The TaDNDO23-KASP was used to identify different haplotypes of TaDNDO23 among 256 wheat accessions. (C) Association of two TaDNDO23 haplotypes with thousand kernel weight (TKW), kernel length (KL), kernel width (KW), and kernel thickness (KT) across five environments. E1-E5 indicate Tongwei in 2021, Tongwei in 2022, Zhuanglang in 2022, Zhuanglang in 2023, and Zhongliang in 2023. * represents P < 0.05; ** represents P < 0.01; ns means denotes not significant
Selection of TaENDO23 haplotypes in the wheat breeding process
To assess whether the elite haplotype TaENDO23-HapI has undergone positive selection, we evaluated the geographical distribution of the two TaENDO23 haplotypes by using 256 accessions from the natural population and 302 accessions from the Wheat Union Database, which have well-documented growth regions (Table S5). The results revealed that TaENDO23-HapI was the dominant haplotype in all detected provinces of China, particularly in key wheat-producing regions, such as Henan (73.2%), Shandong (75%), Shanxi (93.6%), and Hebei (63.6%) (Fig. 8A; Table S5). In addition, the selection of allele variants of TaENDO23 gene in wheat during historical breeding was analyzed using 194 accessions from the natural population and 82 accessions from the Wheat Union Database, with clear historical growing periods (Table S6). The frequency of the favorable haplotype TaENDO23-HapI increased gradually from 71% in the pre-1980 period to 83% in the post-2010 period (Fig. 8B; Table S6). These findings suggest that TaENDO23-HapI was positively selected in the history of wheat breeding. The spatiotemporal distribution results indicated that TaENDO23-HapI was preferentially selected during wheat domestication to enhance grain traits in China.
Spatial and temporal distribution of two TaDNDO23 haplotypes. (A) Geographic distribution of wheat accessions with different TaDNDO23 haplotypes in China. The map was downloaded from the Standard Map Service System (http://bzdt.ch.mnr.gov.cn/). (B) Frequencies of TaDNDO23 allelic variation in Chinese wheat breeding programs in different decades
Discussion
PCD is a vital process in multicellular organisms, including animals and plants. In plants, endonucleases, which are involved in DNA degradation during PCD, play crucial roles in various biological processes, such as growth, development, and tolerance to biotic and abiotic stress [19]. Members of the endonuclease family have been identified in various species, including Arabidopsis, barley, and tomato [4, 6, 20]. However, the members and biological functions of the endonuclease family have not yet been comprehensively analyzed in wheat. In the present study, we identified 26 TaENDO genes from the wheat genome and characterized their molecular features using bioinformatic approaches. In addition, we selected the gene TaENDO23 for a detailed analysis of its biological functions. Our findings suggest that TaENDO family genes, particularly TaENDO23, play critical roles in grain development.
Five AtENDO genes have been identified in Arabidopsis to date. However, more ENDO genes were identified in the wheat genome, reflecting wheat’s status as an allohexaploid plant with a much larger genome than Arabidopsis and rice. A phylogenetic tree was constructed using five AtENDOs, four OsENDOs, four ZmENDOs, and 26 TaENDOs proteins. The classification results of the five AtENDO proteins was consistent with that reported in a previous study [15]. During evolution, gene duplication events often occur, leading to the expansion of gene family. This expansion results in increased gene numbers, functional differentiation, and redundancy, which allow genomes to adapt to diverse environments [21]. In the present study, 17 pairs of collinear TaENDO genes were identified on the A, B, and D chromosomes of wheat (Fig. 3A), indicating that the TaENDO family genes have been highly conserved throughout evolution. Collinearity analysis of ENDO genes in rice, wheat, and maize revealed close genetic relationships among these species (Fig. 3B). Furthermore, all members of the TaENDO family proteins contain three conserved motifs—motif 1, motif 2, and motif 3 (Fig. 2A) —implying that ENDO proteins may possess conserved biological functions in wheat. The presence of introns, which are noncoding regions within a gene, substantially increases the genetic diversity of higher organisms through alternative splicing. We observed that the members of the same subfamily within the TaENDO family generally have similar exon/intron structures, whereas the number of exons and introns varies across different subfamilies (Fig. 2B). This finding suggests that some exons or introns may have been lost during evolution as an adaptation to the environmental conditions, leading to arise new gene function.
Nuclease activity was observed to be induced in grains [22]. In barley (Hordeum vulgare), the nuclease BEN1 is secreted from the aleurone layers of seeds and play a role in the development of seed endosperm [3]. In Arabidopsis, the ENDO1/BFN1 gene is broadly expressed in leaves, flowers, and seeds [18], whereas the ENDO3 gene is mainly expressed in flower organs [6]. Furthermore, the expression of ENDO2 can be triggered by various biotic and abiotic stresses [6]. According to RNA-seq data derived from the WheatOmics 1.0 platform, several TaENDO genes were found to be highly expressed in spikes and grains and significantly induced under drought stress in wheat (Fig. 4A and C). These findings suggest that ENDO genes play crucial roles in seed development and in the response to various abiotic stresses. Transcription factors participate in many physiological and biochemical processes, such as hormone response, abiotic stress response, and development, by directly binding to different cis-acting elements [23]. Understanding the transcriptional regulation and potential function of genes requires identifying the cis-acting elements in their promoter regions. In this study, TaENDO family genes were found to contain numerous stress-responsive (MYB/STRE/ABRE) and growth-related elements (CAT-box/as-1/CCGTCC-box) in their promoter regions (Fig. S3) [24,25,26,27,28]. Notably, CCGTCC-box is commonly found in the promoters of auxin-induced genes, whereas MYB, ABA-responsive elements (ABRE), and LTR are typically involved in various abiotic stresses [29,30,31,32]. For instance, ABRE and MYB both respond to drought treatment and play critical roles in different plants [33]. These results indicate that TaENDO genes may have pivotal roles in regulating drought stress response in wheat. Both RNA-seq data and our qRT-PCR results demonstrated that the TaENDO23 gene not only expressed in spikes and grains but also widely expressed in nutritive organs (Figs. 4A and 6A), suggesting that TaENDO23 participates in regulating other biological processes in wheat. For example, the high expression of TaENDO23 in leaves and its rapid response to PEG6000 treatment (PEG6000-2 h) suggest that it enhances drought tolerance by regulating the function of wheat leaves.
The subcellular localization of a protein is crucial in determining its biological function within the cell [34]. Analyzing this localization is therefore essential for understanding the function of the target protein. Our results indicated that the TaENDO23-GFP fusion protein was primarily localized in the cytoplasm of wheat protoplasts, and green fluorescence was also observed around the nucleus (Fig. 6B). A previous study demonstrated that the initial localization of nuclease BFN-1 in tobacco protoplasts was in filamentous structures throughout the cytoplasm and eventually gathered around the nucleus during protoplast senescence [35]. This finding suggests that TaENDO23 similarly participates in the PCD process. In cereal crops, the endosperm primarily synthesizes and stores starch and storage proteins. PCD occurs in endosperm cells during development [36]. Understanding the mechanisms of PCD during endosperm development and using this knowledge to prolong the grain-filling period could be effective strategies for improving grain yield and quality in cereal crops. Previous studies have shown that PCD first appears in maize endosperm cells 16 days after anthesis, as observed using the Evans Blue staining method. PCD then spreads from the middle and upper endosperm layers to the edges, with nearly all endosperm cells dying by 40 days after anthesis [36]. Similarly, PCD was observed in wheat endosperm cells at 16 days after anthesis [37]. Our study found that several TaENDO genes were highly expressed in spikes and grains (Fig. 4A). In addition, TaENDO23 gene exhibits two haplotypes among 256 wheat accessions (Fig. 7B). During domestication, superior haplotypes tend to accumulate through artificial selection, leaving a distinct genomic imprint. TaENDO23-HapI was found to have higher TKW and KT than TaENDO23-HapII and was positively selected in wheat breeding history (Figs. 7C and 8). Taken together, TaENDO23 gene may play a role in grain development through the PCD pathway via its endonuclease activity.
Twelve SNPs were identified in the promoter and coding regions of TaENDO23 gene. Among these, an SNP at 2,869 bp (C/G) in the coding region was used to develop a KASP marker, which successfully distinguished 256 wheat accessions into two genotypes. Our results revealed that TaENDO23-HapI is an elite haplotype associated with higher grain weight and size. Molecular markers, such as TaDA1-A-HapI, TaGS5-3 A-T, and TaSus2-2 A-HapA, which are significantly associated with grain weight, have been widely used in molecular-assisted breeding in wheat [38,39,40]. Similarly, the molecular marker for TaENDO23 gene developed in this study can be applied in molecular-assisted breeding to identify and select the elite haplotype TaENDO23-HapI in wheat.
Conclusion
In this study, 26 TaENDO genes were identified in the wheat genome and grouped into four subfamilies based on structural similarities. Several TaENDO genes were highly expressed in spikes and grains and significantly induced under drought stress, suggesting their involvement in grain development and drought stress response. Notably, TaENDO23 was highly expressed in leaves and grains, with its protein primarily localized in the cytoplasm. To identify SNP in the TaENDO23 gene across 256 wheat accessions, a KASP marker was developed. The results revealed that TaENDO23-HapI is an elite haplotype associated with higher grain weight and size in wheat. These findings provide valuable insights into the biological functions of TaENDO genes in wheat, and the use of the developed molecular marker and elite haplotype in future wheat breeding programs can be promising.
Materials and methods
Plant materials and growth conditions
The winter wheat cultivar Jinmai 47 (JM47), a drought-resistant and high-yielding variety which has been widely cultivated in the dryland area of northwest China, was used to analyze TaENDO23 expression levels and clone the gene. Seeds were grown in an incubator at 25 °C with a 16-h light/8-h dark cycle. To identify nucleotide polymorphisms in TaENDO23 and perform an association analysis of its different haplotypes with grain-related traits, 256 wheat accessions bred in various regions of China were used (Table S3). These accessions were planted at the Tongwei farm station (35°11′N, 105°19′E, altitude 1750 m) during the 2020–2021 (E1) and 2021–2022 (E2) seasons, at the Zhuanglang farm station (35°12′N, 106°2′E, altitude 1790 m) during the 2021–2022 (E3) and 2022–2023 (E4) seasons, and at the Zhongliang farm station (34°36′N, 105°39′E, altitude 1540 m) during the 2022–2023 (E5) season. Each line was planted in a plot consisting of six rows, with a 20-cm spacing between rows and 30 seeds sown per row in a 1-m plot.
Measurement of wheat grain size and weight
At maturity, spikes from 15 randomly selected plants in the middle of each plot were sampled to analyze kernel length (KL), kernel width (KW), thousand kernel weight (TKW), and kernel thickness (KT). After air drying, 200 seeds from each line were randomly selected to measure KL, KW, and TKW using an SC-G image analysis system (Hangzhou Wanshen Detection Technology Co., Ltd., Hangzhou, China). KT was measured using a vernier caliper.
Identification of TaENDO family genes in wheat
Wheat genomic data, including protein sequences and annotation files, were obtained from the Ensembl Plants database (http://plants.ensembl.org). To identify candidate proteins, the ENDO conserved domain HMMER file (PF03145) was downloaded from the InterPro database (https://www.ebi.ac.uk/interpro/) [41]. The HMMER 3.0 software (https://www.ebi.ac.uk/Tools/hmmer/search/hmmsearch) was used to search for protein sequences across the entire genome, using the ENDO conserved domain with a threshold E value of < 1e-5 using the conserved domain of ENDO [42]. The candidate wheat TaENDO proteins were further verified using NCBI-CDD (https://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi), PFAM, and SMART (http://smart.embl-heidelberg.de/) [42,43,44]. The final identified protein sequences of TaENDO family members were submitted to ExPASY (http://web.expasy.org/protparam/) to predict their physical and chemical properties such as molecular weight and isoelectric point. Finally, Gpos-mPLoc (http://www.csbio.sjtu.edu.cn/bioinf/Gpos-multi/) was used to predict the subcellular localization of TaENDO proteins [34].
Phylogenetic tree construction of TaENDO family members and analysis of their gene structures and conservated motifs
Multiple sequence alignment of ENDO proteins from wheat, Arabidopsis, rice, and maize was performed using MEGA11.0 software (https://www.megasoftware.net/). A phylogenetic tree was then constructed using the Neighbor-Joining algorithm (NJ) with 1,000 bootstrap replicates [45]. The gene structures of TaENDO family members were analyzed using the GSDS database (http://gsds.cbi.pku.edu.cn/) based on the genome and CDS sequences of TaENDO genes. Conserved motifs within TaENDO proteins were identified using the MEME platform (http://meme.nbcr.net/meme) and visualized with TBtools software [46]. In addition, Weblogo (http://weblogo.berkeley.edu/) was sued to visualize the amino acids sequences of the conserved motifs.
Chromosomal location, gene duplication, and collinearity analysis of TaENDO genes in wheat
The chromosomal locations of TaENDO genes were determined using the wheat genome annotation information (gff3) from the Ensembl Plants database (http://ftp.ebi.ac.uk/ensemblgenomes/pub/release-51/plants/gff3/triticum_aestivum/). Collinearity analysis was performed using the One Step MCScanX module in TBtools to analyze the rate of sequence evolution. The synonymous substitution rates (Ks) and non-synonymous substitution (Ka) rates of duplicate gene pairs were calculated using the Multiple Synteny Plot module of TBtools. A Ka/Ks ratio greater than 1 indicates positive selection, a ratio equal to 1 suggests neutral selection, and a ratio less than 1 represents purifying selection [47].
Analysis of cis-acting elements and expression patterns of TaENDO genes
The DNA sequences of 2000 bp upstream of the initiation codon (ATG) of the TaENDO genes in the Chinese Spring wheat variety were extracted from the WheatOmics 1.0 database using TBtools software. The cis-acting elements within these sequences were then predicted using the PlantCARE database (http://bioinformatics.psb.ugent.be/webtools/plancare/). RNA-seq data from different wheat organs and under various abiotic stress treatments for the Chinese Spring wheat variety were downloaded from WheatOmics1.0 (http://202.194.139.32/expression/wheat.html) [48]. The expression patterns of TaENDO genes were assessed based on TPM values (Table S7), which were then normalized and mapped using TBtools software. The expression levels of each gene were displayed in different colors corresponding to their TPM values.
qRT-PCR analysis of the expression levels of TaENDO23 in different organs and under abiotic stress treatment
To analyze the expression levels of TaENDO23 gene in different organs of the wheat cultivar JM47, samples were collected from the roots, stems, leaves, developing spikes at the anthesis stage, spikes at the booting and young stages, and spikes and grains at 5, 10, 15, 20, and 25 days after anthesis. To assess the expression level of TaENDO23 gene under different abiotic stress treatments, grains of the JM47 cultivar were initially cultured in 1/2 Hoagland nutrient solution. After 12 days, the solution was replaced with 1/2 Hoagland nutrient solution supplemented with 200 mM NaCl, 20% PEG6000, or 100 μm ABA, respectively. The first leaves from three wheat seedlings were then collected at 0, 6, 12, 24, and 48 h post-treatment. qRT-PCR analysis was performed with three biological replicates.
Total RNA was extracted using the TIANGEN® Plant Tissue RNA Rapid Extraction Kit, and RNA concentration was determined using an ultra-microphotometer. The first strand of cDNA was synthesized using FastKing gDNA Dispelling RT SuperMix (Beijing). The expression of TaENDO23 was detected through qRT-PCR using FastReal qPCR PreMix (SYBR Green). TaActin1 [49] and TaTubulin [50] were used as reference genes for analyzing the TaENDO23 expression levels under abiotic stress and in different tissues, respectively. Relative transcript expression levels of TaENDO23 were calculated using the 2−ΔΔCt method [51]. The primers used for qRT-PCR are listed in Table S8.
Subcellular localization analysis of TaENDO23 protein in wheat protoplast
Specific primers (Table S8) were designed using Primer 5.0 software to amplify the CDS sequence of TaENDO23 gene from JM47 cDNA. The amplified CDS sequence was then inserted into the pEarlyGate101 vector by using homologous recombinase, creating the 35 S:: TaENDO23-GFP construct. Wheat protoplasts were isolated and transfected as described previously [52]. The TaENDO23-GFP protein was transiently expressed in the wheat protoplasts, and the green fluorescence signal was observed using a laser confocal scanning microscope with an excitation wavelength of 488 nm.
Development of KASP marker in TaENDO23 genomic sequence
SNP sites within the TaENDO23 gene were identified in 681 wheat accessions by using resequencing data from the Wheat Union database (http://wheat.cau.edu.cn/WheatUnion/c_5/). A specific SNP (C/G) located at 2,869 bp downstream from the initiation codon of TaENDO23 was converted into a KASP marker for genotyping 256 wheat accessions from different regions. The KASP marker (Table S8) was designed using the WheatOmics 1.0 platform (http://wheatomics.sdau.edu.cn/). The KASP assay was performed in a 96-well plate, with a total reaction volume of 4 µl containing 1 µl of SNP primer mix (4×), 2 µl of 2× KASP master mix, and 1 µl of DNA template. PCR conditions were set as follows: 94 °C for 15 min; 10 touchdown cycles (94 °C for 20 s, touchdown at 61 °C, decreasing by 0.6 °C per cycle for 40 s); followed by 35 amplification cycles (94 °C for 20 s and 55 °C for 45 s). Fluorescence signal were detected and analyzed using Kluster Caller software on the FLUOstar Omega instrument (LGC Genomics Ltd., Hangzhou, China).
Genomic polymorphism analysis of TaENDO23 genes and the association with grain-related traits
Two distinct TaENDO23 haplotypes from 256 wheat accessions were analyzed for grain weights and sizes by using Excel software (Table S4). Significant differences between the phenotypes of the two haplotypes alleles were evaluated using the t-test. The geographical distribution of the two TaENDO23 haplotypes in China was studied using 256 accessions from the natural population and 302 accessions from the Wheat Union Database (http://wheat.cau.edu.cn/WheatUnion/c_5/), which have well-documented growth regions (Table S5). Breeding selection of the two TaENDO23 haplotypes in China was further analyzed using 194 accessions from the natural population and 82 accessions from the Wheat Union Database, with clear historical growing periods (Table S6).
Data availability
The relevant data and additional information are available in the supplementary files.
Abbreviations
- ABA:
-
Abscisic acid
- ABRE:
-
Abscisic acid-responsive elements
- CDS:
-
Coding sequence
- KASP:
-
Kompetitive allele-specific PCR
- KL:
-
Kernel length
- KT:
-
Kernel thickness
- KW:
-
Kernel width
- MW:
-
Molecular weights
- PCD:
-
Programmed cell death
- RNA-seq:
-
RNA sequencing
- TKW:
-
Thousand Grain Weight
- TPM:
-
Transcripts per kilobase of exon model per million mapped reads
References
Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529(7584):84–7.
Xiao J, Liu B, Yao Y, Guo Z, Jia H, Kong L, et al. Wheat genomic study for genetic improvement of traits in China. Sci China Life Sci. 2022;65(9):1718–75.
Aoyagi S, Sugiyama M, Fukuda H. BEN1 and ZEN1 cDNAs encoding S1-type DNases that are associated with programmed cell death in plants. FEBS Lett. 1998;429:134–8.
Granot G, Morgenstern Y, Khan A, Givaty Rapp Y, Pesok A, Nevo E, et al. Internucleosomal DNA fragmentation in wild emmer wheat is catalyzed by S1-type endonucleases translocated to the nucleus upon induction of cell death. Biochim Biophys Acta. 2015;1849:239–46.
Podzimek T, Matoušek J, Lipovová P, Poučková P, Spiwok V, Šantrůček J. Biochemical properties of three plant nucleases with anticancer potential. Plant Sci. 2011;180:343–51.
Givaty-Rapp Y, Yadav NS, Khan A, Grafi G. S1-type endonuclease 2 in dedifferentiating Arabidopsis protoplasts: translocation to the nucleus in senescing protoplasts is associated with de-glycosylation. PLoS ONE. 2017;12:e0170067.
He X, Kermode AR. Nuclease activities and DNA fragmentation during programmed cell death of megagametophyte cells of white spruce (Picea glauca) seeds. Plant Mol Biol. 2003;51(4):509–21.
Ito J, Fukuda H. ZEN1 is a key enzyme in the degradation of nuclear DNA during programmed cell death of tracheary elements. Plant Cell. 2002;14:3201–11.
Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Mol Biol. 1999;39:915–26.
Lombardi L, Ceccarelli N, Picciarelli P, Lorenzi R. DNA degradation during programmed cell death in Phaseolus coccineus suspensor. Plant Physiol Biochem. 2007;45:221–7.
Reverdatto SV, Dutko JA, Chekanova JA, Hamilton DA, Belostotsky DA. mRNA deadenylation by PARN is essential for embryogenesis in higher plants. RNA. 2004;10:1200–14.
Walley JW, Kelley DR, Nestorova G, Hirschberg DL, Dehesh K. Arabidopsis deadenylases AtCAF1a and AtCAF1b play overlapping and distinct roles in mediating environmental stress responses. Plant Physiol. 2010;152:866–75.
Muramoto Y, Watanabe A, Nakamura T, Takabe T. Enhanced expression of a nuclease gene in leaves of barley plants under salt stress. Gene. 1999;234:315–21.
Sugiyama M, Ito J, Aoyagi S, Fukuda H, Endonucleases. Plant Mol Biol. 2000;44(3):387–97.
Triques K, Sturbois B, Gallais S, Dalmais M, Chauvin S, Clepet C, et al. Characterization of Arabidopsis thaliana mismatch specific endonucleases: application to mutation discovery by TILLING in pea. Plant J. 2007;51:1116–25.
Fendrych M, Van Hautegem T, Van Durme M, Olvera-Carrillo Y, Huysmans M, Karimi M, et al. Programmed cell death controlled by ANAC033/SOMBRERO determines root cap organ size in Arabidopsis. Curr Biol. 2014;24:931–40.
Sakamoto W, Takami T. Nucleases in higher plants and their possible involvement in DNA degradation during leaf senescence. J Exp Bot. 2014;65:3835–43.
Farage-Barhom S, Burd S, Sonego L, Perl-Treves R, Lers A. Expression analysis of the BFN1 nuclease gene promoter during senescence, abscission, and programmed cell death-related processes. J Exp Bot. 2008;59:3247–58.
Daneva A, Gao Z, Van Durme M, Nowack MK. Functions and regulation of programmed cell death in plant development. Annu Rev Cell Dev Biol. 2016;32:441–68.
Lesniewicz K, Karlowski WM, Pienkowska JR, Krzywkowski P, Poreba E. The plant s1-like nuclease family has evolved a highly diverse range of catalytic capabilities. Plant Cell Physiol. 2013;54:1064–78.
Cannon SB, Mitra A, Baumgarten A, Young ND, May G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004;4:10.
Ingle J, Hageman RH. Metabolic changes associated with the germination of corn. II. Nucleic acid metabolism. Plant Physiol. 1965;40:48–53.
Zhang Y, Lu Y, Sayyed H, El, Bian J, Lin J, Li X. Transcription factor dynamics in plants: insights and technologies for in vivo imaging. Plant Physiol. 2022;189:23–36.
Song S, Xu Y, Huang D, Miao H, Liu J, Jia C, et al. Identification of a novel promoter from banana aquaporin family gene (MaTIP1;2) which responses to drought and salt-stress in transgenic Arabidopsis thaliana. Plant Physiol Biochem. 2018;128:163–9.
Ebeed HT. Genome-wide analysis of polyamine biosynthesis genes in wheat reveals gene expression specificity and involvement of STRE and MYB-elements in regulating polyamines under drought. BMC Genomics. 2022;23(1):734.
Alabd A, Cheng H, Ahmad M, Wu X, Peng L, Wang L, et al. ABRE-BINDING FACTOR3-WRKY DNA-BINDING PROTEIN44 module promotes salinity-induced malate accumulation in pear. Plant Physiol. 2023;192(3):1982–96.
Wu L, Xie Z, Li D, Chen Y, Xia C, Kong X, et al. TaMYB72 directly activates the expression of TaFT to promote heading and enhance grain yield traits in wheat (Triticum aestivum L). J Integr Plant Biol. 2024;66(7):1266–9.
Khan M, Dahro B, Wang Y, Wang M, Xiao W, Qu J, et al. The transcription factor ERF110 promotes cold tolerance by directly regulating sugar and sterol biosynthesis in citrus. Plant J. 2024. https://doi.org/10.1111/tpj.16925.
Lv G, Han R, Shi J, Chen K, Liu G, Yu Q, et al. Genome-wide identification of the TIFY family reveals JAZ subfamily function in response to hormone treatment in Betula platyphylla. BMC Plant Biol. 2023;23(1):143.
Tran LSP, Nakashima K, Sakuma Y, Simpson SD, Fujita Y, Maruyama K, et al. Isolation and functional analysis of Arabidopsis stress-inducible NAC transcription factors that bind to a drought-responsive cis-element in the early responsive to dehydration stress 1 promoter. Plant Cell. 2004;16:2481–98.
Cao B, Cui Y, Lou K, Luo D, Liu Z, Zhou Q. Genome-wide identification and expression analysis of the Dof gene family in Medicago sativa L. under various abiotic stresses. DNA Cell Biol. 2020;39:1976–89.
Xiao J, Hu R, Gu T, Han J, Qiu D, Su P, et al. Genome-wide identification and expression profiling of trihelix gene family under abiotic stresses in wheat. BMC Genomics. 2019;20(1):287.
Roychoudhury A, Sengupta DN. The promoter-elements of some abiotic stress-inducible genes from cereals interact with a nuclear protein from tobacco. Biol Plant. 2009;53(3):583.
Chou KC, Shen H, Bin. Plant-mPLoc: a top-down strategy to augment the power for predicting plant protein subcellular localization. PLoS ONE. 2010;5(6):e11335.
Farage-Barhom S, Burd S, Sonego L, Mett A, Belausov E, Gidoni D, et al. Localization of the Arabidopsis senescence-and cell death-associated BFN1 nuclease: from the ER to fragmented nuclei. Mol Plant. 2011;4:1062–73.
Young TE, Gallie DR. Programmed cell death during endosperm development. Plant Mol Biol. 2000;44(3):283–301.
Young TE, Gallie DR. Analysis of programmed cell death in wheat endosperm reveals differences in endosperm development between cereals. Plant Mol Biol. 1999;39(5):915–26.
Ma L, Li T, Hao C, Wang Y, Chen X, Zhang X. TaGS5-3A, a grain size gene selected during wheat improvement for larger grain and yield. Plant Biotechnol J. 2016;14:1269–80.
Liu H, Li H, Hao C, Wang K, Wang Y, Qin L, et al. TaDA1, a conserved negative regulator of grain size, has an additive effect with TaGW2 in common wheat (Triticum aestivum L). Plant Biotechnol J. 2020;18:1330–42.
Hou J, Jiang Q, Hao C, Wang Y, Zhang H, Zhang X. Global selection on sucrose synthase haplotypes during a century of wheat breeding. Plant Physiol. 2014;164:1918–29.
Paysan-Lafosse T, Blum M, Chuguransky S, Grego T, Pinto BL, Salazar GA, et al. InterPro in 2022. Nucleic Acids Res. 2023;51:418–27.
Finn RD, Mistry J, Schuster-Böckler B, Griffiths-Jones S, Hollich V, Lassmann T, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:247–51.
Yang M, Derbyshire MK, Yamashita RA, Marchler-Bauer A. NCBI’s conserved domain database and tools for protein domain analysis. Curr Protoc Bioinf. 2020;69(1):e90.
Letunic I, Khedkar S, Bork P, SMART. Recent updates, new developments and status in 2020. Nucleic Acids Res. 2021;49:458–60.
Tamura K, Stecher G, Kumar S. MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol. 2021;38:3022–7.
Chen C, Wu Y, Li J, Wang X, Zeng Z, Xu J, et al. TBtools-II: a one for all, all for one bioinformatics platform for biological big-data mining. Mol Plant. 2023;16:1733–42.
Wang Y, Li J, Paterson AH. MCScanX-transposed: detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics. 2013;29:1458–60.
Ma S, Wang M, Wu J, Guo W, Chen Y, Li G, et al. WheatOmics: a platform combining multiple omics data to accelerate functional genomics studies in wheat. Mol Plant. 2021;14:1965–8.
Mei F, Chen B, Du L, Li S, Zhu D, Chen N, et al. A gain-of-function allele of a DREB transcription factor gene ameliorates drought tolerance in wheat. Plant Cell. 2022;34:4472–94.
An X, Zhao S, Luo X, Chen C, Liu T, Li W, et al. Genome-wide identification and expression analysis of the regulator of chromosome condensation 1 gene family in wheat (Triticum aestivum L). Front Plant Sci. 2023;14:1124905.
Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–8.
Luo G, Li B, Gao C. Protoplast isolation and transfection in wheat. Methods Mol Biol. 2022;2464:131–41.
Acknowledgements
We thank the scientists for providing a large amount of resequencing data in the Wheat Union database.
Funding
This work was financially supported by the Research Program Sponsored by the State Key Laboratory of Aridland Crop Science of China (GSCS-2023-07), the Key Sci & Tech Special Project of Gansu Province (22ZD6NA009), the Key Cultivation Project of University Research and Innovation Platform of Gansu Province (2024CXPT-01), the Sci &Tech Plan Project of Gansu Province (24JRRA633), the Breakthrough Project in Seed Industry of Gansu Province (GYGG-2024-2), the Industrial Support Plan of Colleges and Universities in Gansu Province (2022CYZC-44), and the Development Fund Project of National Guiding Local Science and Technology (23ZYQA0322).
Author information
Authors and Affiliations
Contributions
TC conducted analysis and wrote the draft of the manuscript. TC, LZ, YZ, WG, PZ, and LG performed data analysis. DY conceived and designed the experiments, and reviewed and edited the writing of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Chen, T., Zhang, L., Zhang, Y. et al. Genome-wide identification of the endonuclease family genes implicates potential roles of TaENDO23 in drought-stressed response and grain development in wheat. BMC Genomics 25, 919 (2024). https://doi.org/10.1186/s12864-024-10840-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-024-10840-y