Abstract
Background
Fusarium head blight (FHB), caused by Fusarium graminearum, is a major disease of wheat in North America. FHB infection causes fusarium damaged kernels (FDKs), accumulation of deoxynivalenol (DON) in the grain, and a reduction in quality and grain yield. Inheritance of FHB resistance is complex and involves multiple genes. The objective of this research was to identify QTL associated with native FHB and DON resistance in a ‘D8006W’/’Superior’, soft white winter wheat population.
Results
Phenotyping was conducted in replicated FHB field disease nurseries across multiple environments and included assessments of morphological and FHB related traits. Parental lines had moderate FHB resistance, however, the population showed transgressive segregation. A 1913.2 cM linkage map for the population was developed with SNP markers from the wheat 90 K Infinium iSelect SNP array. QTL analysis detected major FHB resistance QTL on chromosomes 2D, 4B, 5A, and 7A across multiple environments, with resistance from both parents. Trait specific unique QTL were detected on chromosomes 1A (visual traits), 5D (FDK), 6B (FDK and DON), and 7D (DON). The plant height and days to anthesis QTL on chromosome 2D coincided with Ppd-D1 and were linked with FHB traits. The plant height QTL on chromosome 4B was also linked with FHB traits; however, the Rht-B1 locus did not segregate in the population.
Conclusions
This study identified several QTL, including on chromosome 2D linked with Ppd-D1, for FHB resistance in a native winter wheat germplasm.
Similar content being viewed by others
Background
Wheat (Triticum aestivum L.) is a major staple food crop. Fusarium head blight (FHB) caused by Fusarium graminearum Schwabe is one of the major diseases affecting wheat produced around the world, including North America [1]. Fusarium head blight causes premature bleaching of spikelets, rachis discoloration, and shriveled grain known as Fusarium-damaged kernels (FDKs), and accumulation of the mycotoxin deoxynivalenol (DON) in the grain [2, 3]. FHB has been reported as a threat to wheat yield, and end-use quality, in addition to causing economic losses in many countries, including Canada [4]. There are also strict regulatory limits for DON levels which are 2 ppm in adult and 1 ppm in baby foods in Canada [5]. Ensuring DON concentrations in grain do not exceed regulatory limits and reducing the high yield penalty associated with FHB are challenges for Canadian wheat growers.
An integrated approach is required to control FHB and may include growing cultivars with higher FHB resistance, use of fungicides, crop rotation, use of biological control, and other agronomic practices [4]. Among the FHB management practices, genetic resistance is a very crucial component. The wheat-Fusarium graminearum pathosystem is complex due to multiple types of host resistance; type I (resistance to initial infection), type II (resistance against fungal spread within spike), type III (resistance to kernel infection/FDK), type IV (tolerance), and type V (resistance to mycotoxin/DON accumulation in grain) [6, 7]. Environment also plays a significant role on FHB disease development [1]. As a result, breeding for FHB resistance is a challenging task.
Numerous FHB resistance quantitative trait loci (QTL) mapping studies have been conducted using different populations and more than 52 QTL have been reported across all of the wheat chromosomes [7, 8]. However, many of these QTL are based on Sumai-3 or related Asian sources of resistance, which are mainly present on chromosomes 3B and 6B [9,10,11,12,13]. Frontana, a Brazilian based source of FHB resistance, is also commonly used by breeders and resistance from this source has been identified on chromosomes 3A [14] and 5A [15, 16]. There are few studies on QTL mapping for FHB resistance in Canadian wheat and those that have been conducted mainly showed either Sumai-3 (3B and 6B) based resistance [17, 18] or association of FHB reaction with the semi-dwarfing loci on chromosomes 4B and 4D [19, 20]. Despite mapping of major FHB loci such as Fhb1 and Fhb2, resistance is not complete and multiple genes are required to improve FHB resistance. The genetic background of winter wheat in North America is quite different from spring wheat, and FHB resistance in winter wheat has been considered a challenging puzzle.
This research is based on soft white winter wheat which is mainly used for cakes, biscuits, and pastries, and has been facing FHB as a major threat [21, 22]. Superior has been widely grown as a soft white winter wheat variety in Eastern Canada. Superior does not have a known major FHB resistance background such as Sumai-3 or Frontana (Tamburic-llincic, personal communication). D8006W is a breeding line from Michigan State University and possesses an intermediate reaction to FHB (Tamburic-llincic, preliminary trials). The detection of QTL associated with FHB resistance in this population may be unique and could be used in marker-assisted selection in winter wheat breeding programs as an alternative to Sumai-3 based FHB resistance. The objective of this research was to identify QTL associated with FHB resistance, plant height and days to anthesis in a D8006W/Superior doubled haploid (DH) population.
Methods
Plant materials
The doubled haploid (DH) winter wheat population used in this study was developed using the wheat-maize pollination and embryo rescue method [23]. The parental cross was developed by Dr. Tamburic-Ilincic, and DHs were created from the F1 generation of a cross between two soft white winter wheat genotypes, ‘D8006W’ (a breeding line from Michigan State University) and ‘Superior’ (a cultivar from Ontario, Canada). ‘Superior’ has the pedigree ‘Rebecca’/’Harus’ and ‘D8006W’ has the pedigree ‘2555’/’Lowell’ (Plant Gene Resource of Canada, Agriculture and Agri-Food Canada). The DH population consisted of 105 winter wheat lines. Both parents have native, intermediate resistance to FHB [24], and lack backgrounds related to Sumai-3, Frontana, or other major known FHB resistance sources (24, Neupane, Tamburic-Ilinic, and McCartney unpublished haplotype analysis).
Disease phenotyping
FHB symptom evaluation was conducted in eight site years. The environments included two sites in Western Canada and one in Eastern Canada in 2016 and 2017, one site in Germany in 2017, and one site in Western Canada in 2018. The Western Canada sites were located at Winnipeg and Carman, Manitoba and the Eastern Canada site was at Ridgetown, Ontario. The Germany site was in Hohenheim.
A randomized complete block design with three replications was used in each site year. The DH population, parents and checks were tested for FHB reaction at each site. The check lines used in the Manitoba trials were ‘FHB 148’, ‘32 C*17’, and ‘Emerson’ as resistant checks, ‘Hanover’ and ‘Caledonia’ as susceptible checks, and ‘Freedom’ and ‘43I*18’ as intermediate FHB checks. The checks used in the Ontario trials were ‘Ava’ and ‘AC Morley’ (moderately resistant), and ‘Emmit’ (moderately susceptible). Different checks were used in the two Canadian provinces based on adaptation of the checks in the respective climatic conditions. Each plot was a single row and were 1 m long in Manitoba and 2m long in Ontario. The plots in Germany were double row microplots (0.42 × 1.25m2). The checks used in Germany were ‘Venture’ (moderately susceptible), ‘Gallus’ (moderately resistant), and ‘Priesley’ (moderately resistant).
The Julian calendar date, in which 50% of spikes in each plot were at anthesis (referred to as days to anthesis and abbreviated DA), was recorded by visually assessing the flowering spikes in each plot. In the Manitoba trials, each plot was inoculated at 50% anthesis and then again three days later, using a mixture of two 15-acetyldeoxynivalenol (15-ADON) F. graminearum isolates (M7-07-1, M9-07-1), and two 3-acetyldeoxynivalenol (3-ADON) F. graminearum isolates (M1-07-2, M3-07-2). The isolates were provided by Dr. Jeannie Gilbert of the former Cereal Research Centre of Agriculture and Agri-Food Canada. The inoculum for each isolate was prepared with slight modification to research article from von der Ohe et al. [25]. Briefly, each isolate was grown in Spezieller Nahrstoffarmer agar (SNA) media plates and transferred to Carboxymethyl Cellulose (CMC) flasks. The isolates were mixed in equal concentrations, and the final spore concentration was 50,000 macroconidia/ml. Four milliliters of Tween 20 (Uniqema Americas LLC) were added into each 2-liter mixture solution to increase conidia adhesion. Each plot was spray inoculated with 50 ml of the F. graminearum macroconidia suspension. Spraying was done using a CO2 backpack sprayer with 30 psi pressure. Through the ten hours period after field inoculation, a misting system was operated for ten minutes every hour to maintain high humidity for disease development. Similar inoculation procedures were followed in Ontario as described in Tamburic-Ilincic et al. [26]. Eighteen to twenty-one days after the initial inoculation, each plot was visually assessed for disease incidence and severity. The FHB disease incidence (DI) was estimated as a percentage of spikes in the plot that were infected and disease severity (DS) as the percentage of the infected spikelets in the infected spikes. FHB index was calculated as disease incidence x disease severity /100. Plant height (PHT) was recorded, in each plot, using the mean of three measurements from the soil surface to the tip of the main spike (excluding awns).
At maturity, each plot was hand harvested, and spikes were threshed with a small plot stationary combine with reduced wind speed to reduce loss of FDK. FDK were measured from a 50 g of sample, from each line, using a composite of replicates. After the FDK measurement, the sample was ground into powder that was fine enough to pass through 20 mesh sieves. Ten grams of flour were sampled for DON quantification with the enzyme linked immunosorbent assay (ELISA) using DON kit (Neogen Corporation, Lansing, MI). DON quantification limit was 5ppm. The samples with over 5ppm of DON were kept within limit using dilutions of filtrate.
The D8006W/Superior population was also tested in Germany for FHB resistance, by Dr. Thomas Meidner’s program [27]. The spray inoculation was performed using an aggressive F. culmorum isolate at a spore suspension concentration of 200,000 macroconidia/ml applied with an agricultural field sprayer. All genotypes were inoculated three to four times, with two days interval at mid anthesis, to ensure successful inoculation based on flowering time. The FHB rating was conducted based on Miedaner et al. [28] using both the number of infected spikes per plot and number of infected spikelets per spike in one rating scale [25, 29]. The percentage of infected spikelet’s was rated (0-100) for each plot which is the product of percentage of infected spikes per plot and percentage of infected spikelets per spike. Additionally, anthesis date was recorded at 50% anthesis and anther extrusion was recorded on a 1–4 scale (1 being the least extruded anthers and 4 being the most extruded anthers). The post-harvest measurements, including FDK, and DON analysis were not conducted for trials from Germany.
Genotyping
The DH and parental lines were grown on sterilized cotton balls placed on Deep Inserts trays using 4–5 seeds per line. The Deep Inserts contained 48 cells with each cell dimension of 2” x 2.25” x 3.25” and were placed over flat trays (https://www.greenhousemegastore.com/containers-trays/trays-flats/deep-inserts). Cotton balls were moistened regularly with distilled water for optimal germination and growth. Trays were placed in normal room temperature under 12 h light and grown until seedlings reached the one to two-leaf stage. Tissue samples were collected and freeze dried and stored at -78 °C until DNA extraction could be performed. The DNA extraction of D8006W/Superior population was done using Qiagen DNeasy kit (Qiagen, Toronto, Canada). DNA was quantified using PicoGreen stain (Molecular Probes, Inc., Eugene, Oregon, USA) and the working DNA concentration was adjusted to 50 ng/µl. Genotyping of the DH population was performed using the wheat 90 K Infinium iSelect SNP array (Illumina, San Diego, CA, USA) [30]. Genome Studio V2011.1 software (Illumina, San Diego, CA, USA) was used to analyze the raw data generated from the genotyping platform. Genome Studio identified clusters, which were then converted to A (female parent), B (male parent), or missing data. Markers with greater than 10% missing data or strong segregation distortion, (based on X2 value at P < 0.001), were removed from the dataset. The final set of high quality markers was used for linkage map construction and QTL analysis. The DH population was also genotyped using the Fhb1 Kompetitive allele specific PCR (KASP) marker ‘wMAS0008’ to check for the presence of Fhb1 on chromosome 3B [31, 32]. In addition, the photoperiod gene Ppd-D1 on chromosome 2D was analyzed using the KASP marker ‘wMAS00024’ [33].
Linkage and QTL analysis
Polymorphic markers were used to create a linkage map using MapDisto version 1.7.7 [34]. Linkage map construction was similar to the method described in McCartney et al. [35]. Briefly, the first linkage groups were created using the Automap function in MapDisto with the Kosambi mapping function. Initially, a stringent LOD score of 4 and recombination fraction (RF) of 0.20 cM were used to identify the number of linkage groups and later a LOD score of 3 and RF of 0.3 cM were used to relax the criteria and finalize linkage groups. To order the markers, “sum of adjacent recombination fraction” (SARF) was used as an objective function along with “Branch and Bound II” and “Seriation II”, as the fitting criteria. All marker data were checked, and possible error candidates were replaced with missing data. All linkage groups were assigned to respective chromosomes using available wheat reference maps [30, 36].
The QTL analysis method was similar to McCartney et al. [35] and Thambugala et al. [37]. For QTL analysis, markers were sorted based on position and the most informative marker (i.e., least missing data) was retained per linkage bin. QTL analysis was conducted using software QTL IciMapping version 4.0.6.0 [38] using the interval mapping (IM) and inclusive composite interval mapping (ICIM) functions. The QTL analysis was conducted with 0.1 cM scanning steps, and QTL were identified as significant based on a 5% experiment-wise LOD significance threshold based upon 10,000 permutations. The LOD threshold was 3.10 for additive QTL analysis for IM and 3.06 for ICIM. The QTL confidence interval was estimated based upon one-LOD drop from QTL peak position. QTL were reported if they were significant in at least two environments. For those significant QTL, other environments QTL data were also reported for QTL with minimum LOD 2.5. The QTL that fit the above criteria were considered QTL of interest in this research and were further discussed. The additional QTL were further discussed if they were associated to multiple traits. The physical position of SNP markers flanking the important QTL, were checked with the International Wheat Genome Consortium (IWGSC) Chinese Spring RefSeq v1.0 (https://wheat-urgi.versailles.inra.fr/Seq-Repository/Assemblies) and potential candidate genes were analyzed with the basic local alignment tool (BLAST) available in different bioinformatics platform (https://urgi.versailles.inra.fr/blast_iwgsc/?dbgroup=wheat_iwgsc_refseq_v1_chromosomes&program=blastn; https://plants.ensembl.org ; https://wheat.triticeaetoolbox.org).
Statistical analysis
All statistical analyses were performed using SAS 9.4. Analysis of variance was performed using the proc mixed function with treatments as a fixed effect, and site year and block as random effects. Least-squares means were calculated for each phenotypic trait for each environment as well as the combined dataset. The normality of the datasets was tested using the proc univariate test, and homogeneity of the variance for the combined dataset was checked using the Levene test in SAS. Data were homogenous across environments, and thus combined across multiple site years. Correlations between all phenotypic variables were calculated using the Pearson correlation and variance components were estimated using the proc var procedure in SAS. Heritability was calculated, for each trait, using the broad sense heritability equation:
h2 = 𝜎2G ∕ (𝜎2G + 𝜎2GS/S+ 𝜎2e/SR); where h2 = heritability of traits, 𝜎2 = variance component, G = genotypes, e = error variance, S = site years, S x E = genotype x site years interaction, R = replicates/blocks.
Results
Phenotypic analysis
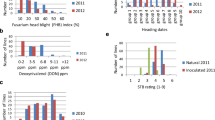
The DH population showed transgressive segregation of genotypes, with DH lines containing phenotypic trait values higher or lower than the parents used in study, for all traits measured (Fig. 1, Table S1). ‘D8006W’ was shorter and had earlier anthesis than ‘Superior’. The mean plant height of ‘D8006W’ parent was 75.3 cm, while Superior was 86.2 cm. The mean days to anthesis for ‘D8006W’ was 161.4, and for ‘Superior’ was 163.4. Although both parents had an intermediate FHB resistance, ‘D8006W’ had slightly higher DI, DS, FHB index, FDK and DON than ‘Superior’ (Fig. 1, Table S1). The mean DI, DS, FHB index, FDK, and DON for ‘D80006W’ was 32.9%, 37.1%, 12,8%, 8.2%, and 15.7 ppm, respectively, while for ‘Superior’ was 26.0%, 36.3%, 9.8%, 8.0%, and 13.5 ppm, respectively.
The frequency distribution of plant height (A), days to anthesis (B), FHB disease incidence (C), FHB disease severity (D), FHB index (E), Fusarium-damaged kernels (FDK) (F), and deoxynivalenol (DON) content (G) of D8006W/Superior doubled haploid (DH) population (parents and 105 DH lines) based on combined site years of data. Means of parental lines are indicated for each traits (A-G). Pearson correlation coefficients and statistical significance levels among phenotypic traits are presented (bottom). The significant correlations are referred as ****p < 0.0001; ***p < 0.001; **p < 0.01; *p < 0.05 and non-significant correlation referred to as ns. Phenotypic data were collected across seven site years (Winnipeg 2016, Carman 2016, Ridgetown 2016, Winnipeg 2017, Carman 2017, Ridgetown 2017, Winnipeg 2018) except plant height and days to anthesis which were based on six site years (Ridgetown 2017 excluded)
Analysis of variance showed that both genotype and genotype x site year interaction were significant for all phenotypic traits (Table 1). The broad sense heritability (h2) calculation showed that plant height (85%) and days to anthesis (89%), had higher heritability than FHB related traits, DI (77%), DS (76%), FHB index (75%), FDK (71%) and DON (75%) (Table 1). All variables were significantly correlated with each other, except plant height and days to anthesis with disease incidence (Fig. 1). However, plant height and days to anthesis were strongly correlated with each other and to DS, FHB index, FDK and DON. There was also strong correlation between FHB symptoms (DI, DS) and harvested grain data (FDK, DON).
Linkage mapping
In total, 5195 SNP polymorphic markers were used for marker grouping into linkage groups, and after further refining, 1254 high quality, informative SNPs were used to generate the final linkage map (Supplementary Table S4). The linkage map of the ‘D8006W’/’Superior’ DH population consisted of 31 linkage groups (LG) with a total map length of 1913.17 cM and 1254 high quality informative SNP marker loci (Table 2, Supplementary Table S5). The majority of the markers mapped to the B genome (about half of the SNP markers), followed by the A genome containing one third of the SNP markers, and the D genome contained the least amount of total SNP markers (Table 2). The low number of SNP markers in the D genome was expected based on the literature and nature of polyploidization in hexaploid wheat. The average spacing per SNP marker loci was 1.5 cM in the genetic map and 9.4 Mb in the physical map, giving the overall genetic and physical map ratio of 6.2 Mb/cM. The physical map and genetic map were fitted in expected order based on reference maps (Fig. S1, Table S5) [30, 36].
QTL analysis
The major QTL for each measured phenotypic trait were detected in different environments as well as the combined environment using interval mapping (IM) and composite inclusive interval mapping methods (ICIM) of QTL IciMapping version 4.0.6.0 [38]. QTL were detected across multiple chromosomes and linkage groups (LG) within chromosomes. The QTL results using IM and ICIM are presented in Table 3 and results were discussed based on ICIM data, and its consensus to IM data.
Plant height QTL
Six plant height QTL were reported on chromosomes 2B, 2D, 3B, 4B, 5A (LG: 5A.3) and 7B (LG: 7B.2) which were named as QPht.ufg-2B, QPht.ufg-2D, QPht.ufg-3B, QPht.ufg-4B, QPht.ufg-5A, and QPht.ufg-7B respectively (Table 3). QPht.ufg-2D was a major plant height QTL that explained 30.0-47.5% of the phenotypic variation across individual environments and 50.6% variation for the combined environments using ICIM. QPht.ufg-2D was located in the 30-35 cM region of the chromosome 2D genetic map, however, the IM method also showed a shoulder peak approximately at 23 cM across individual and combined environments (Fig. 2; Table 3). QPht.ufg-4B explained 8.4–10.4%, and 9.8% of the phenotypic variation across individual and combined environments, respectively, using ICIM. QPht.ufg-7B explained 8.7–12.5%, and 6.4% of the phenotypic variation across individual and combined environments, respectively, using ICIM. The effect of QPht.ufg-2B, QPht.ufg-3B, and QPht.ufg-5A were minor, reported in one or two environments, and explained 7.4–13.6%, 5.7–5.8%, and 6.4% of the phenotypic variation across one or two environment(s), respectively, using ICIM. The additive effect was negative for five of the six plant height QTL, including QPht.ufg-2D, indicating that the ‘D8006W’ alleles decreased plant height. However, the allelic effect was positive for QPht.ufg-4B, indicating that the ‘Superior’ allele decreased plant height at this locus.
Days to anthesis QTL
Eight QTL associated with days to anthesis were reported on chromosomes 1B, 2A, 2D, 4A, 5A (LG: 5A.2), 7A, and 7D which were named as QDa.ufg-1B, QDa.ufg-2A, QDa.ufg-2D.1, QDa.ufg-2D.2, QDa.ufg-4A, QDa.ufg-5A, QDa.ufg-7A, and QDa.ufg-7D, respectively (Table 3). Two QTL were detected on chromosome 2D, QDa.ufg-2D.1 and QDa.ufg-2D.2, which were in the 30-35 cM, and 55-60 cM regions of chromosome 2D, respectively. QDa.ufg-2D.1 was consistently expressed across most of the environments, and accounted for 16.0–54.0% of the phenotypic variation using ICIM. QDa.ufg-2D.2 was expressed in fewer environments. QDa.ufg-2D.1 was a major days to anthesis QTL, and the ‘D80006W’ allele decreased days to anthesis on this locus. QDa.ufg-2D.1 collocated with a plant height QTL QPht.ufg-2D at 30-35 cM. The KASP marker ‘wMAS00024’ for Ppd-D1 located at 31.70 cM on chromosome 2D showed that the population segregated for Ppd-D1 and this locus lies in the region identified for plant height and anthesis QTL (30–35 cM) (Fig. 2). The remaining six QTL: QDa.ufg-1B, QDa.ufg-2A, QDa.ufg-4A, QDa.ufg-5A, QDa.ufg-7A, and QDa.ufg-7D were detected in fewer environments and explained less phenotypic variation compared to QDa.ufg-2D.1. QDa.ufg-1B, QDa.ufg-2A, QDa.ufg-4A, QDa.ufg-5A, and QDa.ufg-7A explained 4.2–49.7%, 0.7–7.8%, 2.1%, 9.6–48.9%, and 1.6% of the phenotypic variation across individual environment(s), respectively using ICIM. The additive effects were negative across six out of eight of the QTL, indicating that the ‘D8006W’ alleles contributed to early anthesis. However, the additive effect was positive for QDa.ufg-7A, and QDa.ufg-7D indicating that the ‘Superior’ allele decreased the days to anthesis at these loci, but this effect was minimal compared to other QTL.
Anther extrusion QTL
Two QTL associated with anther extrusion were reported on chromosomes 5A (LG: 5A.3), and 7A which were named as QAE.ufg-5A, and QAE.ufg-7A, respectively, based on one German site year of data (Table 3). QAE.ufg-5A (LG: 5A.3), and QAE.ufg-7A explained 17.1% and 17.4% of the phenotypic variation, respectively, using ICIM. The additive effects were positive for QAE.ufg-5A and negative for QAE.ufg-7A which means that the ‘D8006W’ and ‘Superior’ alleles contributed to higher anther extrusion, respectively, on those two QTL.
FHB resistance QTL
Disease incidence, disease severity, FHB index, FDK and DON content (collectively referred as FHB related traits) are measures of FHB and DON resistance in this study. QTL related to DI, DS and FHB Index, which were based on direct phenotypic measurement, were grouped under the same QTL designation as QFhb. QTL related to FDK and DON were kept distinct as QFdk and QDon, respectively (Table 3). The QTL analysis detected common QTL across multiple FHB traits along with a few unique QTL specific for each trait (Table 3; Table S3).
FHB incidence, FHB severity, and FHB Index
A total of eight FHB resistance QTL were detected on chromosomes 1A.1 (DI and FHB index), 1B (DI and DS), 2D (DI, DS, and FHB index), 4B (DS, and FHB Index), 5A.2 (DI, DS, and FHB index), 5A.3 (DI, DS, and FHB index), and 7A (DI, DS, FHB index) and were named as QFhb.ufg-1A, QFhb.ufg-1B, QFhb.ufg-2D.1, QFhb.ufg-2D.2, QFhb.ufg-4B, QFhb.ufg-5A.1 (LG: 5A.2), QFhb.ufg-5A.2 (LG: 5A.3), and QFhb.ufg-7A, respectively (Table 3).
QFhb.ufg-1A explained 11.5%, and 4.1% of the phenotypic variation for DI and FHB index, respectively, across individual environments using ICIM (Table 3). QFhb.ufg-1B explained 9.6%, and 9.2% of the phenotypic variation for DI and DS, respectively, across individual environments using ICIM (Table 3). For both QFhb.ufg-1A and QFhb.ufg-1B, the additive effect was negative, indicating that resistance was contributed from the ‘D8006W’ parent. However, these QTL were detected in only a few environments and explained less phenotypic variation compared to other QTL.
The major FHB resistance QTL on chromosome 2D were classified into two groups QFhb.ufg-2D.1 and QFhb.ufg-2D.2, based on genetic position within the chromosome. QFhb.ufg-2D.1 explained phenotypic variation of 14.4%, 16.3%, and 13.5–17.0% for DI, DS, and FHB index, respectively, across individual and combined environments using ICIM (Table 3). The phenotypic variation explained by QFhb.ufg-2D.2 were 10.7–15.6%, 17.0-23.9%, and 14.0–20.0% for DI, DS, and FHB index, respectively, across individual and combined environments using ICIM. The additive effect was negative for QFhb.ufg-2D.1 and QFhb.ufg-2D.2 indicating that ‘D8006W’ decreased DI, DS, and FHB index and acted as the allelic source of resistance.
The QTL located on two linkage groups (LG) of chromosome 5A; 5A.2 and 5A.3, were major FHB resistance QTL which were detected across multiple environments and multiple FHB traits (Table 3). The QFhb.ufg-5A.1 (LG: 5A.2) explained phenotypic variation of 11.6–18.4%, 13.6%, and 15.9–16.6% for DI, DS and FHB index, respectively, across individual environments using ICIM. The phenotypic variation explained by QFhb.ufg-5A.1 (LG: 5A.2) were 7.4%, 9.2%, and 11.1% for DI, DS, and FHB index, respectively, across combined environments using ICIM. The QFhb.ufg-5A.2 (LG: 5A.3) explained 10.2–34.3%, 8.7–32.4%, and 33.7–34.1% of phenotypic variation across individual environments for DI, DS, and FHB Index, respectively using ICIM. The phenotypic variation explained by QFhb.ufg-5A.2 (LG: 5A.3) across combined environments were 35.9%, 22.4%, and 27.1% for DI, DS, and FHB index, respectively using ICIM. The additive effect was negative for all of the QFhb.ufg-5A.1 and QFhb.ufg-5A.2 QTL indicating that ‘D8006W’ decreases DI, DS, and FHB index and was an allelic source of resistance.
QFhb.ufg-7A was the third most important FHB QTL in this study which was detected for DI and FHB index using ICIM (Table 3). QFhb.ufg-7A explained phenotypic variation of 7.0-22.9%, and 20.9%, for DI and FHB index, respectively across individual environment(s) using ICIM. QFhb.ufg-7A explained 12.8% of phenotypic variation for DI across combined environment using ICIM. The additive effects were positive for QFhb.ufg-7A indicating that ‘Superior’ decreases DI, DS, and FHB index and was the allelic source of resistance.
QFhb.ufg-4B was another important QTL which was detected using DS and FHB index. The QFhb.ufg-4B explained phenotypic variation of 12.9–17.8%, and 12.3–14.2% for DS and FHB index, respectively, for individual environments using ICIM (Table 3). The phenotypic variation explained by QFhb.ufg-4B across combined environments was 12.4%, and 6.9% for DS, and FHB index, respectively using ICIM. The additive effect was positive for QFhb.ufg-4B, indicating that the ‘Superior’ allele decreased DI and, FHB index, and acted as allelic source of resistance.
The German site also detected significant FHB QTL on chromosomes 1B, 2D, 5 A.3 and 6B (Table 3). QFhb.ufg-1B, QFhb.ufg-5A.2 (LG: 5A.3), and QFhb.ufg-6B explained 9.8%, 25.1%, and 10.9% of phenotypic variation for FHB index, respectively using ICIM (Table 3). The additive effects of those QTL were negative, indicating ‘D8006W’ acted as a resistance allele. QFhb.ufg-5A.2 (LG:5A.3) collocated with anther extrusion QAE.ufg-5A at the 48 cM position of the 5A.3 linkage group. Phenotypically, both parents also showed intermediate FHB resistance based on Germany data (Table S2).
FDK and DON content
Seven major QTL were detected for FDK on chromosomes 2B, 2D, 4B, 5A (LG: 5A.2), 5D (LG: 5D.2), and 7A which were named as QFdk.ufg-2B, QFdk.ufg-2D.1, QFdk.ufg-2D.2, QFdk.ufg-4B, QFdk.ufg-5A, QFdk.ufg-5D, and QFdk.ufg-7A, respectively (Table 3). The QTL on chromosome 2D were classified into two groups QFdk.ufg-2D.1 and QFdk.ufg-2D.2 based on genetic position within the chromosome. QFdk.ufg-2B, QFdk.ufg-2D.1, QFdk.ufg-2D.2, and QFdk.ufg-5A explained 6.2%, 8.2–15.9%, 13.6–21.7%, and 10.4–30.7% of phenotypic variation for FDK, respectively, across individual environment(s) using ICIM. QFdk.ufg-2B, QFdk.ufg-2D.1, and QFdk.ufg-5A explained 7.4%, 15.9%, and 19.9% of phenotypic variation across combined environments, respectively using ICIM. The additive effect of those three QTL was negative, indicating that resistance was contributed from ‘D8006W’ parent. The additive effects of QFdk.ufg-4B, QFdk.ufg-5D, and QFdk.ufg-7A were positive, indicating that resistance was contributed from ‘Superior’ parent for this three QTL. QFdk.ufg-4B explained 8.7%, and 13.5% of phenotypic variation for FDK across individual and combined environment(s), respectively using ICIM. QFdk.ufg-5D, and QFdk.ufg-7A explained 13.5% and 23.1% of phenotypic variation, respectively, in individual environment using ICIM. Two additional minor QTL related to FDK were detected in single environment on chromosomes 1D (QFdk.ufg-1D) and 6B (QFdk.ufg-6B) where ‘D8006W’ carried the resistant allele.
Four major QTL were detected for DON content on chromosomes 2D, 4D (LG: 4D.1), 5 A (LG: 5A.2) and 7 A which were named as QDon.ufg-2D.1, QDon.ufg-4D, QDon.ufg-5A, and QDon.ufg-7A, respectively (Table 3). The QTL on chromosome 2D were classified into two groups QDon.ufg-2D.1 and QDon.ufg-2D.2 based on genetic position within the chromosome, but QDon.ufg-2D.2. was only detected using IM. QDon.ufg-2D.1 and QDon.ufg-5A explained 25.6%, and 13.5–18.6% of phenotypic variation across individual and combined environment(s), respectively using ICIM. The additive effect of those two QTL was negative, indicating that lower DON content was contributed from the ‘D8006W’ parent. QDon.ufg-4D, and QDon.ufg-7 A explained 10.5%, and 17.9% of phenotypic variation across individual environments, respectively using ICIM. The phenotypic variation explained by QDon.ufg-4D, and QDon.ufg-7A across combined environments were 16.4% and 5.2%, respectively using ICIM. The additive effect of those two QTL was positive, indicating that lower DON content was contributed from the ‘Superior’ parent. A few additional minor QTL related to DON content were detected in single environments on chromosomes 1B, 1D, and 6B (D8006W as a resistance allele), and 4B, and 7D (Superior carried the resistant allele).
QTL common across FHB traits
The QTL on chromosomes 2D, 4B, 5A.2 and 7A contained combinations of four or more FHB related traits; DI, DS, FHB index, FDK, and DON (Table 3). QFhb.ufg-2D.1, QFdk.ufg-2D.1 and QDon.ufg-2D.1 collocated approximately at 8 cM in the ‘D8006W’/’Superior’ 2D linkage map (Table 3). Similarly, QFhb.ufg-2D.2, QFdk.ufg-2D.2 and QDon.ufg-2D.2 collocated at 40–60 cM in the ‘D8006W’/’Superior’ 2D linkage map. The additive effect was negative, for both groups, on chromosome 2D QTL indicating that ‘D8006W’ decreased all FHB related traits and acted as the allelic source of resistance. QFhb.ufg-5 A.1 (LG: 5 A.2), QFdk.ufg-5 A (LG: 5A.2) and QDon.ufg-5A (LG: 5A.2) collocated at 8–13 cM of the ‘D8006W’/’Superior’ 5 A.2 linkage map. The additive effect was negative for those QTL indicating that ‘D8006W’ decreases all FHB related traits and acted as allelic source of resistance. The QFhb.ufg-7A, QFdk.ufg-7A and QDon.ufg-7A were collocated approximately at 92–112 cM of the ‘D8006W’/’Superior’ 7 A linkage map. The additive effect was positive for those QTL indicating that ‘Superior’ decreases all FHB traits and acted as allelic source of resistance. Likewise, QFhb.ufg-4B, QFdk.ufg-4B and QDon.ufg-4B were also detected at 17–27 cM of 4B linkage map. The additive effect was positive for those 4B QTL, indicating that ‘Superior’ allele decreases DS, FHB index, FDK and DON content and acted as allelic source of resistance. A few minor QTL were also associated with multiple FHB traits. QFhb.ufg-1B and QDon.ufg-1B were located within 5 cM distance with the resistance allele contributed from ‘D8006W’ parent. Similarly, the QFdk.ufg-1D and QDon.ufg-1D were located within 5 cM distance with the resistance allele, contributed from ‘D8006W’. QFdk.ufg-6B and QDon.ufg-6B were located in the 7–11 cM vicinity with the resistance allele from ‘D8006W’.
Discussion
Phenotypic attributes of the DH population
This study reported on inheritance of FHB visual symptoms, FDK and DON resistance, and related morphological traits based on a biparental mapping population. The analysis of variance with significant genotype x site year interaction illustrates the effect of multiple environments on FHB traits. The phenotypic variation for FHB traits showed continuous distribution patterns and heritability (Table 1) of the trait was quite high (75% using FHB index, 72% using FDK, and 76% using DON). This phenotypic performance illustrated that enough trait variation existed in the population; parents differed in genes associated with FHB resistance; and multiple genes were associated with FHB resistance. Similar results were illustrated in a previous study [39]. The phenotypic correlation between FHB variables were strong and supported the QTL analysis results.
Morphological traits QTL and relationship with FHB QTL
Plant height, and days to anthesis were key morphological traits studied in this study. The major plant height QTL QPht.ufg-2D collocated with the days to anthesis QTL QDa.ufg-2D.1 approximately at 34 cM on chromosome 2D (Table 3). The photoperiod gene Ppd-D1, based on ‘wMAS00024’KASP marker, mapped at 31.7 cM on 2D, which corresponds to QPht.ufg-2D and QDa.ufg-2D.1 region with the flanking SNP marker ‘D_GA8KES402GRIFZ_148’-23.2 cM and ‘Excalibur_c51559_220’-34.4 cM (Fig. 2). The physical location of Ppd-D1 was also compared using IWGSC Chinese Spring RefSeq v1.0 map and it appeared in the same physical region of QDa.ufg-2D.1 flanking SNP markers, between 27.980 and 36.114 Mbp based on the physical map (Fig. S1, Table S5). This is the expected position of Ppd-D1 [40]. Therefore, the days to anthesis QTL QDa.ufg-2D.1 is due to the effect of Ppd-D1. Few studies reported that Rht8 and Ppd-D1 are closely linked [40,41,42]. The Rht8 allele, based on the ‘Rht8-KASP-V3’ marker, didn’t segregate in population suggesting a pleiotropic effect of the Ppd-D1 loci to both the days to anthesis and plant height. The additive effect was negative for both QPht.ufg-2D and QDa.ufg-2D.1 implying that ‘D8006W’ decreases the values (shorter plant height and early days to anthesis). This agrees with the phenotypic differences between the parents for these traits; the ‘D8006W’ parent was shorter and also reached anthesis earlier than ‘Superior’. Surprisingly, the alleles for shorter plant height and earlier anthesis at this locus was associated with greater FHB resistance, which were mainly due to Ppd-D1 effect. The FHB QTL QFhb.ufg-2D.1 for DI, DS, FHB index, which was also located in vicinity of QPht.ufg-2D and QDa.ufg-2D.1, showed negative additive effects, meaning that the ‘D8006W’ alleles for this QTL also account for a decrease in FHB value, which is unique to this study. Additionally, the additive effects of QFdk.ufg-2D.1 and QDon.ufg-2D.1 were negative indicating that all FHB (DI, DS, FHB index, FDK and DON) QTL were correlated and also linked with QPht.ufg-2D and QDa.ufg-2D.1. Similarly, QDa.ufg-2D.2 was in vicinity with QFhb.ufg-2D.2, QFdk.ufg-2D.2 and QDon.ufg-2D.2 with all QTL containing negative additive effects, meaning that the ‘D8006W’ alleles for those QTL also account for decrease in days to anthesis and FHB related traits.
Another plant height QTL of interest was on chromosome 4B (QPht.ufg-4B) which was collocated along with QTL for FHB traits (QFhb.ufg-4B, QFdk.ufg-4B, and QDon.ufg-4B) where ‘Superior’ allele decreased both plant height and FHB level. Previous studies reported important dwarfing gene, Rht-B1, located on chromosome 4B, was associated with FHB susceptibility [43]. In our study, the ‘wMAS00001’ Rht-B1 KASP marker was monomorphic for the population and QPht.ufg-4B was detected a distance away from the expected location of Rht-B1 indicating that the effect of QPht.ufg-4B was not due to the major dwarfing gene. The effect QPht.ufg-4B was also minor compared to the plant height QTL QPht.ufg-2D.
In addition to QDa.ufg-2D, there were days to anthesis QTL on chromosomes 5A and 7A which accounted for less phenotypic variation than the QDa.ufg-2D (Table 3). The QDa.ufg-5A QTL collocated with QFhb.ufg-5A.1, QFdk.ufg-5A.1, and QDon.ufg-5A.1 with negative additive effects for all these QTL on chromosome 5 A (LG: 5A.2). This suggests that early anthesis was also associated with lower FHB index, FDK and DON. This phenomenon is dependent on anthesis coinciding with weather conditions that are conducive to disease development. This association will be useful for breeders that want to combine shorter plants and early maturity with FHB resistance. Previous studies also reported association of favourable QTL for plant height, days to anthesis, and FHB related traits [19, 20], however, further study is required to confirm the effect of the Ppd-D1 gene in FHB resistance/susceptibility in this study. Other minor effect morphological traits QTL, such as QPht.ufm-3B, QPht.ufm-7B, QPht.ufm-2A, and QDa.ufm-4A were not associated with FHB traits.
Anther extrusion (AE) was another morphological trait for discussion based on data from Germany. QFhb.ufg-5A.2 and QAE.ufg-5A.2 were collocated at 48 cM on chromosome 5 A (LG: 5A.3) (Table 3) where ‘D80006W’ allele increased anther extrusion and conferred FHB resistance. Likewise, QAE.ufg-7A was detected within the same region of QFhb.ufg-7A, with the ‘Superior’ allele contributing to higher anther extrusion and lower FHB values. Anther extrusion data was not evaluated in field environments in Canada; however, QTL for AE and FHB were associated based on Germany data. Further research is required to better understand the role of AE in FHB resistance in D8006W/Superior population. Previous studies also reported association of AE with FHB resistance [16, 43,44,45]. Steiner et al. [45] fine mapped FHB resistance QTL on chromosome 5A, Qfhs.ifa-5A, in Fhb5 region, in two resistance QTL Qfhs.ifa-5As and Qfhs.ifa-5Ac which were associated with anther extrusion. The Qfhs.ifa-5A interval corresponds to physical position 46.6-364.4 Mbp flanked by Xbarc186 and Xwmc805 on IWGSC Chinese Spring RefSeq v1.0 map. QFhb.ufg-5A.2 (LG: 5A.3) and QAE.ufg-5A.2 (LG: 5A.3) in this study mapped on the 48 cM position of the linkage map and approximately at 700.4 Mbp based on IWGSC Chinese Spring RefSeq v1.0 map, indicating that QAE.ufg-5A.2 (LG: 5A.3) is not genetically linked with Fhb5 and it is likely that this QTL is novel to D8006W/Superior population.
Relationship among QTL for FHB related traits (DI, DS, FHB index, FDK and DON)
Both parents contributed favourable alleles for FHB visual traits, FDK and DON content. ‘D8006W’ contributed FHB resistance alleles on chromosomes 1 A, 1B, 2D, 5 A.2, and 5 A.3, while ‘Superior’ contributed FHB resistance alleles on chromosomes 4B, 7 A (Table 3 and S3). The QTL for the three field disease measurements DI, DS and FHB Index were similar to the FDK and DON QTL. The FHB related QTL on chromosomes 2D; QFhb.ufg-2D.1, and QFhb.ufg-2D.2 were in proximity of QFdk.ufg-2D.1/QDon.ufg-2D.1 and QFdk.ufg-2D.2/QDon.ufg-2D.2 respectively. Similarly, QFhb.ufg-5 A.1 (LG: 5 A.2) and QFhb.ufg-7 A were in proximity of QFdk.ufg-5 A.1 (LG: 5 A.2)/QDon.ufg-5 A.1 (LG: 5 A.2), and QFdk.ufg-7 A/ QDon.ufg-7 A, respectively. For these QTL, it is likely that the same gene(s) controlling all FHB traits (DI, DS, FHB index, FDK and DON). The strong phenotypic correlation between these traits may be explained, in part, by QTL that control the same traits, or by the biological relationship between these traits (Table 3 and S3). However, there were some QTL unique to FDK and DON, QFdk.ufg-5D, QFdk.ufg-6B, QDon.ufg-6B, and QDon.ufg-7D, indicating that FDK and DON resistance should be considered in addition to field FHB evaluations in wheat breeding programs. Similarly, some QTL such as QFhb.ufg-5 A.2 (LG: 5 A.3), and QFhb.ufg-1 A were not detected for FDK and DON, signifying the importance of field resistance.
Possible source of FHB resistance QTL in D8006W/Superior population
FHB resistance QTL QFhb.ufg-2D.1, QFhb.ufg-2D.2, QFhb.ufg-4B, QFhb.ufg-5 A.2, and QFhb.ufg-7 A were detected across multiple environments, and collocated with FDK or DON QTL, thus are considered for discussion of the source of native FHB resistance. Other important QTL were QFhb.ufg-1 A, QFhb.ufg-1B, QFhb.ufg-5 A.3, QFdk.ufg-2B, and QDon.ufg-4D which were detected in at least two environments.
The major FHB resistance QTL (3B) used in North America is primarily derived from Sumai-3 and derivatives [12, 17], but is not present in native sources of resistance from North America. The marker test with the ‘wMAS00008’ KASP marker for segregation of the major FHB resistance gene from Sumai-3, Fhb1 on chromosome 3B [31, 32, 46], confirmed that the Fhb1 background was lacking in D8006W/Superior population. Further, the haplotype analysis for FHB resistance in our study (data not presented) showed that both parents ‘D8006W’ and ‘Superior’ do not share Fhb1 haplotypes. There is also no direct source of Asian lines in the genetic background based on pedigree information of the two parents (https://pgrc-rpc.agr.gc.ca/gringlobal/search).
The QTL on chromosome 2D associated with all FHB related traits, plant height and days to anthesis was the most prominent QTL detected in thus study. ‘D8006W’ contributed the favourable alleles for FHB resistance, shorter plants, and earlier days to anthesis. Most previous studies have reported FHB QTL on chromosome 2D with negative association among plant height and FHB resistance [43]. McCartney et al. [35] reported FHB QTL on chromosome 2D in ‘Kenyon’/’86ISMN’ population, where 86IMSN2137 had the early maturity and short alleles, but increased FHB susceptibility. The FHB QTL on chromosome 2D was also reported in a winter wheat cross ‘Vienna’/’25R47’ [20] and a cross between Asian sources ‘Wuhan-1’/’NyuBai’ [15]. The DH population in this study does not share a relationship with any of those populations. The source of FHB resistance might be linked to Ppd-D1 on chromosome 2D since plant height and days to anthesis QTL were also detected in this study. The QTL on chromosome 4B was associated with DS, FHB index, FDK, and DON with the resistance allele contributed by ‘Superior’. Multiple studies have reported FHB QTL on chromosome 4B [20, 47, 48]. Rht-B1 did not segregate in the D8006W/Superior population, thus QFhb.ufg-4B could not be explained by allelic variation for Rht-B1. Therefore, QFhb.ufg-4B is unique to this population and it will be interesting to discover the possible source of resistance associated with QFhb.ufg-4B in this population.
Another major FHB resistance alleles in this study was on chromosome 5A.2 (QFhb.ufg-5A.1) for all FHB related traits and on 5A.3 for field visual traits (QFhb.ufg-5A.2) with the resistance allele contributed by ‘D8006W’. The FHB QTL on chromosome 5A was reported in studies involving ‘Frontana’ with a Brazilian background, and studies with European wheat backgrounds in Steiner et al. [45]. QTL on chromosome 5A were also reported in other studies including the Asian line ‘Wangshuibai’ [49], European wheat cultivar ‘Arina’ [50] and the US cultivars ‘Everest’ [51] and ‘Ernie’ [52]. Fhb5 is one of the major FHB resistance genes and is associated with FHB QTL on chromosome 5A based on previous studies [45, 53]. The physical position of Fhb5 located between 46.6 and 364.4 Mbp of IWGSC Chinese Spring RefSeq v1.0 map, based on Steiner et al. [45]. This region also corresponds to QFhb.ufg-5A.1 (LG: 5A.2) located at 0–10 cM on chromosome 5A.2 (Fig. S1). Therefore, the QFhb.ufg-5A.1 (LG: 5A.2) is likely Fhb5. However, no known pedigree information was available to further validate Fhb5 in D8006W/Superior population. QFhb.ufg-5A.2 (LG: 5A.3) is a different locus than Fhb5 since it is in a different linkage group and QTL position, mapped at 48 cM and approximately at 700.4 Mbp of the IWGSC Chinese Spring RefSeq v1.0 map. Based on Steiner et al. [45], the Fhb5 locus is located between 46.6 and 364.4 Mbp of IWGSC Chinese Spring RefSeq v1.0 map. Thus, QFhb.ufg-5A.2 (LG: 5A.3) is not associated with Fhb5 and it is likely that this QTL is novel to D8006W/Superior population.
Another QTL for FHB traits (DI, DS, FHB index, FDK and DON) was on chromosome 7A with ‘Superior’ as a source of resistance allele (Table 3 and S3). FHB related QTL on chromosome 7A were reported in previous studies with Asian based germplasm [54,55,56]. Skinnes et al. [57] mapped FHB resistance QTL on the 7AL chromosomal region using European winter wheat ‘Arina’. QFhb.ufg-7A was also mapped in the long arm of chromosome 7A, however, it is not clear whether these QTL are the same or genetically linked based on pedigree information and marker information.
QFhb.ufg-1A was detected for visual FHB triats (DI, DS and Index) in this study with ‘D8006W’ as a source of the resistance allele. QFhb.ufg-1B was detected for DI, DS and DON and ‘D8006W’ was the source of the resistance allele. QFdk.ufg-2B and QDon.ufg-4D had ‘D8006W’ and ‘Superior’ as the source of the resistance allele, respectively. FHB QTL on chromsomes 1 A, 1B, 2B and 4D have been also previously reported in Buerstmayr et al. studies [7, 43].
Conclusion
This study used two winter wheat parents which showed intermediate resistance to FHB, FDK and DON. The segregating DH population contained genotypes with better FHB related traits resistance than both parents. Both parents contributed FHB resistance alleles, however ‘D8006W’ parent contributed more resistance alleles. The QTL analysis showed common QTL across measured phenotypic traits, in addition to unique QTL for each trait. QTL for FHB visual traits were detected consistently across multiple environments on chromosomes 2D, 4B, 5A and 7A. Additional QTL specific for FDK and DON resistance were detected on chromosomes 1D, 5D and 6B. The major QTL on chromosome 2D, which is related with shorter plants, earlier anthesis and resistant to all FHB related traits, brings couples useful alleles for use in wheat breeding programs. These findings contrast with past studies in which shorter genotypes are associated with FHB susceptibility [35, 43]. Further, QTL on chromosomes 4B, 5A and 7A would be useful sources of resistance as they exist in adapted backgrounds and would reduce the potential for linkage drag often associated with using unadapted sources of resistance. This would also provide an alternative to ‘Sumai-3’ based resistance, which is often associated with a negative impact on yield and other agronomic traits. In addition, all the significant QTL can be used in the marker-assisted selection-based winter wheat breeding programs. QTL bringing forward in the breeding program should be validated and the phenotypic effect examined using other populations for effective introgression.
Data availability
The datasets generated and/or analyzed during the current study are available in the public repository; DOI: https://doi.org/10.6084/m9.figshare.26485174 [58]. The phenotypic evaluation and genetic mapping data are presented in article main contents, and supplementary files. Please contact authors to obtain the plant materials used in study.
Abbreviations
- QTL:
-
Quantitative Trait Loci
- FHB:
-
Fusarium Head Blight
- FDK:
-
Fusarium Damaged Kernels
- DON:
-
Deoxynivalenol
- DH:
-
Doubled haploid
- SNP:
-
Single Nucleotide Polymorphism
- KASP:
-
Kompetitive allele specific PCR
- IWGSC:
-
International Wheat Genome Consortium
- BLAST:
-
Basic Local Alignment Tool
- LG:
-
Linkage Group
References
McMullen M, Jones R, Gallenberg D. Scab of wheat and barley: a re-emerging disease of devastating impact. Plant Dis. 1997;81:1340–8.
Puri KD, Zhong S. The 3ADON population of Fusarium Graminearum found in North Dakota is more aggressive and produces a higher level of DON than the prevalent 15ADON population in spring wheat. Phytopathology. 2010;100:1007–14. https://doi.org/10.1094/phyto-12-09-0332.
Amarasinghe CC, Tittlemier SA, Fernando WGD. Nivalenol-producing Fusarium cerealis associated with fusarium head blight in winter wheat in Manitoba, Canada. Plant Pathol. 2014;988–95. https://doi.org/10.1111/ppa.12329.
Gilbert J, Tekauz a. Review: recent developments in research on fusarium head blight of wheat in Canada. Can J Plant Pathol. 2000;22:1–8. https://doi.org/10.1080/07060660009501155.
Health Canada. (2020). Maximum levels for deoxynivalenol (DON) in food. https://www.canada.ca/en/health-canada/services/food-nutrition/food-safety/food-contaminants-chemicals/deoxynivalenol.html (Accessed 31 Mar 2021).
Schroeder HW, Christensen JJ. Factors affecting resistance of wheat to scab caused by Gibberella zeae. Phytopathol. 1963;53(7):831–8.
Buerstmayr H, Ban T, Anderson JA. QTL mapping and marker-assisted selection for Fusarium head blight resistance in wheat: a review. Plant Breed. 2009;128:1–26. https://doi.org/10.1111/j.1439-0523.2008.01550.x.
Liu S, Hall MD, Griffey CA, McKendry AL. Meta-analysis of QTL associated with fusarium head blight resistance in wheat. Crop Sci. 2009;49:1955–68. https://doi.org/10.2135/cropsci2009.03.0115.
Bai G, Kolb FL, Shaner G, Domier LL. Amplified fragment length polymorphism markers linked to a major quantitative trait locus controlling scab resistance in wheat. Phytopathol. 1999;89:343–8.
Anderson JA, Stack RW, Liu S, et al. DNA markers for Fusarium head blight resistance QTLs in two wheat populations. Theor Appl Genet. 2001;102:1164–8. https://doi.org/10.1007/s001220000509.
Buerstmayr H, Lemmens M, Hartl L, et al. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. I. Resistance to fungal spread (type II resistance). Theor Appl Genet. 2002;104:84–91. https://doi.org/10.1007/s001220200009.
Cuthbert PA, Somers DJ, Thomas J, et al. Fine mapping Fhb1, a major gene controlling fusarium head blight resistance in bread wheat (Triticum aestivum L). Theor Appl Genet. 2006;112:1465–72. https://doi.org/10.1007/s00122-006-0249-7.
Zhou MP, Hayden MJ, Zhang ZY, et al. Saturation and mapping of a major Fusarium head blight resistance QTL on chromosome 3BS of Sumai 3 wheat. J Appl Genet. 2010;51:19–25. https://doi.org/10.1007/BF03195706.
Steiner B, Lemmens M, Griesser M, Scholz U, Schondelmaier J, Buerstmayr H. Molecular mapping of resistance to Fusarium head blight in the spring wheat cultivar Frontana. Theor Appl Genet. 2004;109:215–24.
Somers DJ, Fedak G, Savard M. Molecular mapping of novel genes controlling Fusarium head blight resistance and deoxynivalenol accumulation in spring wheat. Genome. 2003;46:555–64. https://doi.org/10.1139/g03-033.
Buerstmayr H, Steiner B, Hartl L, et al. Molecular mapping of QTLs for Fusarium head blight resistance in spring wheat. II. Resistance to fungal penetration and spread. Theor Appl Genet. 2003;107:503–8. https://doi.org/10.1007/s00122-003-1272-6.
Cuthbert PA, Somers DJ, Brulé-Babel A. Mapping of Fhb2 on chromosome 6BS: a gene controlling Fusarium head blight field resistance in bread wheat (Triticum aestivum L). Theor Appl Genet. 2007;114:429–37. https://doi.org/10.1007/s00122-006-0439-3.
Tamburic-Ilincic L. Effect of 3B, 5A and 3A QTL for Fusarium head blight resistance on agronomic and quality performance of Canadian winter wheat. Plant Breed. 2012;131:722–7. https://doi.org/10.1111/pbr.12005.
McCartney CA, Somers DJ, Fedak G, et al. The evaluation of FHB resistance QTLs introgressed into elite Canadian spring wheat germplasm. Mol Breed. 2007;20:209–21. https://doi.org/10.1007/s11032-007-9084-z.
Tamburic-Ilincic L, Barcellos Rosa S. Alleles on the two dwarfing loci on 4B and 4D are main drivers of FHB-related traits in the Canadian winter wheat population Vienna × 25R47. Plant Breed. 2017;136:799–808. https://doi.org/10.1111/pbr.12527.
Canadian Grain Commission. (2020). Soft white wheat. Retrieved from https://www.grainscanada.gc.ca/en/grain-quality/soft-white-wheat.html (Accessed 31 Mar 2021).
Tamburic-Ilincic L. 2008 survey for fusarium head blight of winter wheat in Ontario. Can plant Dis Surv = Inven des Mal des plantes au Canada; 2009.
Devaux P, Pickering R. Haploids in the improvement of Poaceae. Haploids in crop improvement II. Berlin, Heidelberg: Springer Berlin Heidelberg; 2005. pp. 215–42.
OCCC (Ontario Cereal Crops Committee). (2020). Ontario wheat performance trial results. Agriculture and Agri-Food Canada. Retrieved from http://www.gocereals.ca/wp-content/uploads/2021/02/OWPT20.pdfat (Acessed on 2 Feb 2021).
von der Ohe C, Gauthier V, Tamburic-Ilincic L, et al. A comparison of aggressiveness and deoxynivalenol production between Canadian Fusarium graminearum isolates with 3-acetyl and 15-acetyldeoxynivalenol chemotypes in field-grown spring wheat. Eur J Plant Pathol. 2010;127:407–17. https://doi.org/10.1007/s10658-010-9607-z.
Tamburic-Ilincic L, Schaafsma AW, Falk DE. Indirect selection for lower deoxynivalenol (DON) content in grain in a winter wheat population. Can J Plant Sc Oct. 2007;1:87(4):931–6. https://doi.org/10.4141/P06-024.
Nisidis P. (2017) Segregation variance for Fusarium head blight and yellow rust resistance and important agronomic traits in two Canadian winter wheat populations. Master Thesis. University of Hohenheim, June 2017.
Miedaner T, Kalih R, Großmann MS, Maurer HP. Correlation between Fusarium head blight severity and DON content in triticale as revealed by phenotypic and molecular data. Plant Breed. 2016;135:31–7. https://doi.org/10.1111/pbr.12327.
Miedaner T, Wilde F, Steiner B, et al. Stacking quantitative trait loci (QTL) for Fusarium head blight resistance from non-adapted sources in an European elite spring wheat background and assessing their effects on deoxynivalenol (DON) content and disease severity. Theor Appl Genet. 2006;112:562–9. https://doi.org/10.1007/s00122-005-0163-4.
Wang S, Wong D, Forrest K, et al. Characterization of polyploid wheat genomic diversity using a high-density 90 000 single nucleotide polymorphism array. Plant Biotechnol J. 2014;12:787–96. https://doi.org/10.1111/pbi.12183.
Liu S, Pumphrey MO, Gill BS, et al. Toward positional cloning of fhb1, a major qtl for fusarium head blight resistance in wheat. Cereal Res Commun Akadémiai Kiadó. 2008;195–201. https://doi.org/10.1556/crc.36.2008.suppl.b.15. https://doi-org.uml.idm.oclc.
Bernardo AN, Ma H, Zhang D, Bai G. Single nucleotide polymorphism in wheat chromosome region harboring Fhb1 for Fusarium head blight resistance. Mol Breed. 2012;29:477–88. https://doi.org/10.1007/s11032-011-9565-y.
Beales J, Turner A, Griffiths S, et al. A Pseudo-response Regulator is misexpressed in the photoperiod insensitive Ppd-D1a mutant of wheat (Triticum aestivum L). Theor Appl Genet. 2007;115:721–33. https://doi.org/10.1007/s00122-007-0603-4.
Lorieux M. MapDisto: fast and efficient computation of genetic linkage maps. Mol Breed. 2012;30(2):1231–5.
McCartney CA, Brûlé-Babel AL, Fedak G, et al. Fusarium Head Blight Resistance QTL in the Spring Wheat Cross Kenyon/86ISMN 2137. Front Microbiol. 2016;7:1542. https://doi.org/10.3389/fmicb.2016.01542.
Lukaszewski AJ, Alberti A, Sharpe A, et al. A chromosome-based draft sequence of the hexaploid bread wheat (Triticum aestivum) genome. Sci (80-). 2014;345. https://doi.org/10.1126/science.1251788.
Thambugala D, Brûlé-Babel AL, Blackwell BA, Fedak G, Foster AJ, MacEachern D, Gilbert J, Henriquez MA, Martin RA, McCallum BD, Spaner D, Iqbal M, Pozniak CJ, N’Diaye A, McCartney CA. Genetic analyses of native Fusarium head blight resistance in two spring wheat populations identifies QTL near the B1, Ppd-D1, Rht-1, Vrn-1, Fhb1, Fhb2, and Fhb5 loci. Theor Appl Gene. 2020;133(10):2775–96. https://doi.org/10.1007/s00122-020-03631-y.
Li H, Ribaut JM, Li Z, Wang J. Inclusive composite interval mapping (ICIM) for digenic epistasis of quantitative traits in biparental populations. Theor Appl Genet. 2008;116:243–60. https://doi.org/10.1007/s00122-007-0663-5.
Somers DJ, Thomas J, DePauw R, et al. Assembling complex genotypes to resist Fusarium in wheat (Triticum aestivum L). Theor Appl Genet. 2005;111:1623–31. https://doi.org/10.1007/s00122-005-0094-0.
Gasperini D, Greenland A, Hedden P, et al. Erratum: genetic and physiological analysis of Rht8 in bread wheat: an alternative source of semi-dwarfism with a reduced sensitivity to brassinosteroids. J Exp Bot. 2014;65:6760. https://doi.org/10.1093/jxb/ers138.
McCartney CA, Brûlé-Babel AL, Lamari L, Tekauz A, Mamer DF. Identification of Fusarium head blight resistance QTL in two hexaploid wheat populations. Theor Appl Genet. 2005;111:753–60. https://doi.org/10.1007/s00122-005-2021-9.
Chai G, Zhang H, Hu X, Ma Y, Qi X, Wang X, Peng Y, Sun Q. Fhb7 and Fhb1 are two major quantitative trait loci (QTLs) conferring Fusarium head blight resistance in wheat. J Genet Genomics. 2018;45(5):307–13. https://doi.org/10.1016/j.jgg.2018.04.005.
Buerstmayr M, Steiner B, Buerstmayr H. Breeding for Fusarium head blight resistance in wheat—progress and challenges. Plant Breed. 2020;139:429–54. https://doi.org/10.1111/pbr.12797.
Steiner B, Buerstmayr M, Michel S, et al. Breeding strategies and advances in line selection for Fusarium head blight resistance in wheat. Trop Plant Pathol. 2017;42:165–74. https://doi.org/10.1007/s40858-017-0127-7.
Steiner B, Buerstmayr M, Wagner C, et al. Fine-mapping of the Fusarium head blight resistance QTL Qfhs.ifa-5A identifies two resistance QTL associated with anther extrusion. Theor Appl Genet. 2019;132:2039–53. https://doi.org/10.1007/s00122-019-03336-x.
Marker Assisted Selection in Wheat/MASWheat. (2020). https://www.maswheat.ucdavis.edu. (Accessed 1 October 2020).
He X, Singh PK, Dreisigacker S, Singh S. Dwarfing genes Rht-B1b and Rht-D1b are Associated with both type I FHB susceptibility and low Anther Extrusion in two Bread Wheat populations. PLoS ONE. 2016;11(9):e0162499. https://doi.org/10.1371/journal.pone.0162499.
Prat N, Guilbert C, Prah U, et al. QTL mapping of Fusarium head blight resistance in three related durum wheat populations. Theor Appl Genet. 2017;130:13–27. https://doi.org/10.1007/s00122-016-2785-0.
Lin F, Xue SL, Zhang ZZ, et al. Mapping QTL associated with resistance to Fusarium head blight in the Nanda2419 x Wangshuibai population. II: type I resistance. Theor Appl Genet. 2006;112:528–35. https://doi.org/10.1007/s00122-005-0156-3.
Paillard S, Schnurbusch T, Tiwari R, et al. QTL analysis of resistance to Fusarium head blight in Swiss winter wheat (Triticum aestivum L). Theor Appl Genet. 2004;109:323–32. https://doi.org/10.1007/s00122-004-1628-6.
da Silva CL, Fritz A, Clinesmith M, et al. QTL mapping of Fusarium head blight resistance and deoxynivalenol accumulation in the Kansas wheat variety ‘Everest’. Mol Breed. 2019;39(3):1–21. https://doi.org/10.1007/s11032-019-0937-z.
Liu S, Abate ZA, Lu H, et al. QTL associated with Fusarium head blight resistance in the soft red winter wheat Ernie. Theor Appl Genet. 2007;115:417–27. https://doi.org/10.1007/s00122-007-0577-2.
Xue S, Xu F, Tang M, Zhou Y, Li G, An X, Lin F, Xu H, Jia H, Zhang L, Kong Z. Precise mapping Fhb5, a major QTL conditioning resistance to Fusarium infection in bread wheat (Triticum aestivum L). Theor Appl Genet. 2011;123:1055–63. https://doi.org/10.1007/s00122-011-1647-z.
Zhang M, Zhang R, Yang JZ, Luo PG. Identification of a new QTL for Fusarium head blight resistance in the wheat genotype Wang shui-bai. Molecul Biol Rep. 2010;37:1031–5. https://doi.org/10.1007/s11033-009-9809-7.
Jayatilake DV, Bai GH, Dong YH. A novel quantitative trait locus for Fusarium head blight resistance in chromosome 7A of wheat. Theor Appl Genet. 2011;122:1189–98.
Li T, Bai GH, Wu SY, Gu SL. Quantitative trait loci for resistance to Fusarium head blight in the Chinese wheat landrace Huangfangzhu. Euphytica. 2012;185:93–102. https://doi.org/10.1007/s00122-011-1549-0.
Skinnes H, Semagn K, Tarkegne Y, Marøy AG, Bjørnstad Å. The inheritance of anther extrusion in hexaploid wheat and its relationship to Fusarium head blight resistance and deoxynivalenol content. Plant Breed. 2010;129(2):149–55.
Neupane A. (2024). QTL analysis of native Fusarium head blight and deoxynivalenol resistance in ‘D8006W’/’Superior’, soft white winter wheat population. figshare. Dataset. https://doi.org/10.6084/m9.figshare.26485174
Acknowledgements
The research project was conducted in collaboration between three institutions: University of Manitoba, University of Guelph Ridgetown Campus, and Agriculture and Agri-Food Canada. The author would like to thank Maria Stoimenova, Mary Meleshko, Zesong Ye, Yang Lin, Eppie Austria, John Patterson, Todd Phibbs and numerous summer students for assistance in collecting phenotypic data for this study. We are also grateful to National Research Council, Saskatoon and staff Kerry Boyle for genotyping of population and Agriculture and Agri-Food Canada Morden research center and AAFC staffs (Leslie Bezte, Suzanne Enns, Linda Langille) for assisting in in lab experiments associated with this research. We would like to acknowledge Thomas Miedaner, University of Hohenheim, Germany for phenotyping population in Germany. The first author acknowledges Western Grain Research Fund for providing Graduate scholarship for support in PhD program.
Funding
The funding of this project was provided by AAFC Growing Forward-two and Western Grain Research Fund.
Author information
Authors and Affiliations
Contributions
AN, ABB and LTI designed the research experiment. LTI developed the DH population used in research. ABB, LTI, and CM supervised the research work. AN performed the experiments, analyze data, and wrote manuscript. ABB, LTI and CM edited and revised the manuscripts. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Neupane, A., Tamburic-llincic, L., Brûlé-Babel, A. et al. QTL analysis of native Fusarium head blight and deoxynivalenol resistance in ‘D8006W’/’Superior’, soft white winter wheat population. BMC Plant Biol 24, 852 (2024). https://doi.org/10.1186/s12870-024-05536-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12870-024-05536-0