Abstract
Background
Esophageal diseases (ED) are a kind of common diseases of upper digestive tract. Previous studies have proved that metabolic disorders are closely related to the occurrence and development of ED. However, there is a lack of evidence for causal relationships between metabolites and ED, as well as between metabolite ratios representing enzyme activities and ED. Herein, we explored the causality of genetically determined metabolites (GDMs) on ED through Mendelian Randomization (MR) study.
Methods
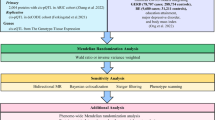
Two-sample Mendelian randomization analysis was used to assess the causal effects of genetically determined metabolites and metabolite ratios on ED. A genome-wide association analysis (GWAS) encompassing 850 individual metabolites along with 309 metabolite ratios served as the exposures. Meanwhile, the outcomes were defined by 10 types of ED phenotypes, including Congenital Malformations of Esophagus (CME), Esophageal Varices (EV), Esophageal Obstructions (EO), Esophageal Ulcers (EU), Esophageal Perforations (EP), Gastroesophageal Reflux Disease (GERD), Esophagitis, Barrett's Esophagus (BE), Benign Esophageal Tumors (BETs), and Malignant Esophageal Neoplasms (MENs). The standard inverse variance weighted (IVW) method was applied to estimate the causal relationship between exposure and outcome. Sensitivity analyses were carried out using multiple methods, including MR-Egger, Weighted Median, MR-PRESSO, Cochran's Q test, and leave-one-out analysis. P < 0.05 was conventionally considered statistically significant. After applying the Bonferroni correction for multiple testing, a threshold of P < 4.3E-05 (0.05/1159) was regarded as indicative of a statistically significant causal relationship. Furthermore, metabolic pathway analysis was performed using the web-based MetaboAnalyst 6.0 software.
Results
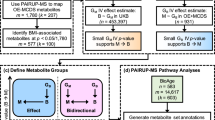
The findings revealed that initially, a total of 869 candidate causal association pairs (\({P}_{ivw}\) < 0.05) were identified, involving 442 metabolites, 145 metabolite ratios and 10 types of ED. However, upon applying the Bonferroni correction for multiple testing, only 36 pairs remained significant, involving 28 metabolites (predominantly lipids and amino acids), 5 metabolite ratios and 6 types of ED. Sensitivity analyses and reverse MR were performed for these 36 causal association pairs, where the results showed that the pair of EV and 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (p-16:0/18:2) did not withstand the sensitivity tests, and Hexadecenedioate (C16:1-DC) was found to have a reverse causality with GERD. The final 34 robust causal pairs included 26 metabolites, 5 metabolite ratios and 5 types of ED. The involved 26 metabolites predominantly consisted of methylated nucleotides, glycine derivatives, sex hormones, phospholipids, bile acids, fatty acid dicarboxylic acid derivatives, and N-acetylated amino acids. Furthermore, through metabolic pathway analysis, we uncovered 8 significant pathways that played pivotal roles in five types of ED conditions.
Conclusions
This study integrated genomics with metabolomics to assess causal relationships between ED and both metabolites and metabolite ratios, uncovering several key metabolic features in ED pathogenesis. These findings have potential as novel biomarkers for ED and provide insights into the disease's etiology and progression. However, further clinical and experimental validations are necessary
Similar content being viewed by others
Background
Esophageal diseases (ED) represent a prevalent category of upper gastrointestinal diseases, such as Congenital Malformations of Esophagus (CME), Esophageal Varices (EV), Esophageal Obstructions (EO), Esophageal Ulcers (EU), Esophageal Perforations (EP), Gastroesophageal Reflux Disease (GERD), Esophagitis, Barrett's Esophagus (BE), Benign Esophageal Tumors (BETs), and Malignant Esophageal Neoplasms (MENs). Among these, GERD is the most common. According to epidemiological data, GERD is estimated to affect between 15% and 21% of the population in Europe, while globally, approximately 14% of individuals experience reflux symptoms on a weekly basis or more frequently [1, 2]. The evolution of GERD often culminates in a host of secondary complications, such as esophageal inflammation, ulcers, strictures, perforations, bleeding, and the development of Barrett's esophagus, a recognized precursor lesion for esophageal adenocarcinoma, which is the eighth most frequently diagnosed malignancy globally and the sixth leading cause of cancer-associated deaths [3, 4]. The increasing burden of healthcare costs renders the in-depth exploration of the fundamental physiological mechanisms underlying ED particularly essential. However, the complex interplay among diverse elements such as environmental exposures, lifestyle factors, genetic components, and biochemistry continues to confound the mechanistic underpinnings and the risk factors associated with ED entities.
In recent years, an increasing body of research has demonstrated a strong correlation between ED and metabolic dysregulation, notably including diabetes, arterial hypertension, dyslipidemia, non-Alcoholic fatty liver disease (NAFLD), and obesity [5,6,7,8,9,10]. Metabolites, embodying the substrates and end-products derived from the intertwined metabolic activities of the host and its resident microbiome, offer a direct snapshot of the organism's metabolic milieu. The application of metabolomics methodologies, designed to measure minute fluctuations in the metabolite profile either quantitatively or qualitatively, enables the elucidation of biological phenotypes, disease trajectories, and adaptive responses to extrinsic environmental factors. As such, metabolomics represents a powerful instrument to identify and characterize biomarkers [11]. Several investigations highlight the prospective diagnostic utility of metabolites in relation to ED, where these molecules are predominantly found in urinary, plasma, and serum specimens [12,13,14,15,16,17]. Despite the utilization of metabolites as proxies for evaluating risk stratification and diagnostic potential in ED, a significant hurdle in establishing their credibility as definitive biomarkers involves contending with substantial inter-individual variation in metabolomic signatures, which is influenced by both genetic determinants and environmental exposure [18, 19]. Moreover, the causal relationship between these variables and the metabolic alterations remain incompletely understood.
Currently, the rapid advancements in high-throughput technologies have facilitated large-scale implementation of Genome-Wide Association Studies (GWAS) in epidemiology and genetics, enabling the identification of a vast number of single nucleotide polymorphisms (SNPs) associated with metabolites, consequently providing a necessary condition for the establishment of genetically determined metabolites (GDMs) databases[18, 20]. Alternatively, Mendelian randomization (MR) represents a genetically grounded approach to causal inference that utilizes SNPs endowed at conception as instrumental variables (IVs) to probe the causal linkages between specific exposures, e.g., biomarkers, environmental exposures, or behavioral traits, and disease outcomes, effectively mitigating the risks of confounding and reverse causation inherent to traditional observational epidemiologic research [21]. To our knowledge, no studies have inferred a causal relationship between GDMs and ED yet. Thus, in this study, we adopted a two-sample Mendelian randomization design to assess the causal impact of GDMs on ED, aiming to provide empirical support for the underlying pathological mechanisms involved in ED.
Methods
MR study design
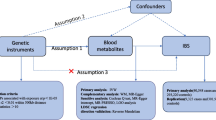
The validity of the MR approach depends on adherence to three core assumptions, which in the context of this study are specifically articulated as follows: (1) The Relevance Assumption dictates that IVs must exhibit a strong and consistent statistical association with GDMs. (2) The Independence Assumption requires that IVs should be independent of any confounding factors influencing ED, apart from their relationship with the GDMs. (3) The Exclusion Assumption posits that the IVs only affect ED outcomes indirectly through their influence on the GDMs, without any additional direct effects, thus negating the presence of pleiotropy, which refers to the phenomenon where a single genetic variant influences multiple seemingly unrelated traits or outcomes. The schematic overview of the study design is depicted in Fig. 1. The study methods were compliant with the STROBE-MR checklist [22].
GWAS data for metabolites and metabolite ratios
GWAS data of 1,091 metabolites and 309 metabolite ratios are available from the study of Chen et al. [20], which is more comprehensive than the 486 metabolite GWAS data in the study of Shin et al. [18] and is the most comprehensive metabolite-related GWAS data set to date. The GWAS summary statistics were deposited to GWAS catalog (https://www.ebi.ac.uk/gwas/). The dataset was derived from 8299 individuals from The Canadian Longitudinal Study on Aging (CLSA) cohort. In this study, the levels of 1,458 metabolites were quantified in plasma samples by Metabolon using the ultrahigh performance liquid chromatography-tandem mass spectroscopy (UPLC-MS/MS) platform which is also known as Metabolon HD4 platform. For metabolite ratios, researchers identified 309 metabolite pairs that share enzymes or transporters using the HMDB [23]. Then, the metabolite ratio was calculated for each pair of metabolites by dividing the batch normalized measurement value of one metabolite by the measurement of the other metabolite in the same individual. The metabolite ratios were then trimmed (retaining those within three standard deviations) and inverse rank normal transformed. Of the 1,091 metabolites tested, 850 had known identities across eight superpathways (that is, lipid, amino acid, xenobiotics, nucleotide, cofactor and vitamins, carbohydrate, peptide and energy). The remaining 241 were categorized as unknown or ‘partially’ characterized molecules. In order to ensure the interpretability of the results, 241 uncertain metabolites were discarded, and only 850 known metabolites and 309 metabolite ratios were retained.
GWAS data for esophageal diseases
Summary GWAS data for 10 types of ED were obtained from UK Biobank (UKB) and FinnGen consortium R9 release data, including BE (1,123 cases and 320,387 controls), BETs (295 cases and 376,982 controls), CME (110 cases and 376,152 controls), EO (1,157 cases and 360,037 controls), EP (114 cases and 320,387 controls), Esophagitis (19,905 casesand 341,289 controls), EU (1,157 cases and 360,037 controls), EV (cases and controls), GERD (29,975 cases and 331,219 controls), MENs (566 cases and 287,137 controls) (Table 1). To minimize bias introduced by population stratification, our analysis focused on the European population. The UK Biobank is a large-scale biomedical database and research resource housing in-depth genetic and health information derived from over 500,000 volunteer participants across the United Kingdom, all aged between 40-69 years at recruitment and having contributed a comprehensive array of personal data encompassing their lifestyles, environments, and medical histories [24]. The FinnGen project capitalizes on the unique genetic characteristics of the Finnish population and its extensive national health registry data, demonstrating remarkable advantages in the field of genetic research. Due to historical isolation and the founder effect within the Finnish population, harmful genetic variations tend to cluster among a limited number of low-frequency variants, which is particularly advantageous for uncovering rare but potentially high-impact genetic mutations [25]. The FinnGen project initiative aims to collect and analyze genomic data from 500,000 Finns along with their corresponding health records, and as of now, over 224,000 participants have already undergone both genotype and phenotype assessments. The definition of ED in the datasets were based on clinical diagnosis, and the tenth edition of the International Classification of Diseases (ICD-10).
Selection of genetic instrumental variables
The selection of IVs must satisfy the three assumptions mentioned above, namely, Relevance Assumption, Independence Assumption and Exclusion Assumption. To satisfy Relevance Assumption, SNPs associated with each metabolite at genome-wide significance (P < 1E-05) were first screened as IVs. Secondly, we utilized the SumStatsRehab software tool [26] to supplement missing variant IDs for IVs. In contrast to the conventional approach of removing SNPs with missing variant IDs, our method maximized retention of all available IVs, thereby ensuring the completeness and accuracy of the results. Thirdly, the clumping procedure was done by linkage disequilibrium analysis with \(r^2\) threshold < 0.1 and a kilobase (kb) window > 10,000 in the European 1000 Genomes Project Phase 3 reference panel [21]. Finally, we evaluated the strength of all IVs by calculating the explained variance \((R^2)\) and F statistical parameters for each IV. As a general consensus, IVs with F > 10 were considered to be strong instruments and would be used in subsequent MR analysis. Please refer to Table S1 for calculation formulas of \(R^2\) and F statistical parameters.
MR statistical analysis
Under the premise of satisfying the three assumptions inherent to MR, the standard inverse variance weighted (IVW) method is considered the most effective and reliable approach for consistently estimating the causal relationship between exposure and outcome, as it delivers consistent estimates of the causal effect of the exposure [27]. Therefore, we preferred IVW to identify causal associations between metabolites and ED. However, the IVW method relies on strict satisfaction of the IV assumptions, in particular to exclude the bias due to the presence of horizontal pleiotropy, so we also need to perform the following sensitivity analysis to satisfy Independence Assumption and Exclusion Assumption: (1) MR-Egger [28] and Weighted Median [29] methods were used to strengthen and supplement the results of IVW and increase the robustness of the results, ensuring the satisfaction of the Exclusion Assumption. The MR-Egger method can detect violations of the IV assumptions and provide effect estimates that remain unaltered by such deviations. The Weighted Median method allows for unbiased estimation of causal effects even when some IVs potentially violate the IV assumptions, as long as at least half of the valid instruments satisfy the relevant assumptions. (2) The horizontal pleiotropy of IVs was estimated according to the intercept of MR-Egger regression and MR-PRESSO [30] to ensure that SNPs was independently correlated with exposure and outcome, ensuring the satisfaction of the Independence Assumption. (3) Cochran's Q test for IVW and MR-Egger were used to detect heterogeneity to ensure that there was no heterogeneity in the results [31] and the satisfaction of the Exclusion Assumption. (4) Leave-one-out analysis were used to assess the possibility that if a single SNP had an impact on the overall causal results, ensuring the satisfaction of the Independence Assumption. All analyses were conducted using R software (version 4.2.3), where the “ieugwasr” and “plinkbinr” packages were employed for the removal of linkage disequilibrium, while the “TwoSampleMR” package was utilized for two-sample MR analysis, and the “MRPRESSO” package was used for MR-PRESSO analysis. P < 0.05 was conventionally considered statistically significant. After applying the Bonferroni correction for multiple testing, a threshold of P < 4.3E-05 (0.05/1159) was regarded as indicative of a statistically significant causal relationship. The statistical power in this study was calculated using R code [32].
Metabolic pathway analysis
To further understand the biological processes and disease mechanisms of metabolites in ED, we employed MetaboAnalyst 6.0 ( https://www.metaboanalyst.ca/) [33] to conduct pathway analysis on metabolites demonstrating significant causal relationships within ED (\(P_{IVW}\) <0.05). The Small Molecule Pathway Database (SMPDB) [34] and the Kyoto Encyclopedia of Genes and Genomes (KEGG) Database [35]were used in the functional enrichment analysis module and pathway analysis module.
Results
Selection of genetic instrumental variables
After P threshold filtering of 850 metabolite and 309 metabolite ratio SNPs (P < 1E-05), variant ID completion, linkage disequilibrium removal, and weak instrument removal, we obtained 306,240 valid IVs that satisfy Relevance Assumption (Table S2). The number of IVs for each metabolite or metabolite ratio ranged from 9 to 113 (Table S3), explaining 0.235-23.262% of the variance (Table S2). In addition, the F statistic values of all IVs were greater than 10 (19.503-2297.785), indicating that there were no weak IVs (Table S2).
Causal effects of metabolites and metabolite ratios on esophageal diseases
(1) An overview of the number of causal associations: In this study, we used IVW to assess causality in 850 metabolites and 309 metabolite ratios for ED. The results showed that there were a total of 869 pairs of potential causal associations (\({P}_{ivw}\) < 0.05), involving 442 metabolites and 145 metabolite ratios, with 101 pairs for BE, 57 pairs for BETs, 74 pairs for CME, 99 pairs for EO, 83 pairs for EP, 85 pairs for Esophagitis, 119 pairs for EU, 93 pairs for EV, 110 pairs for GERD, and 48 pairs for MENs (Table S3). Next, we applied the Bonferroni correction method to adjust for multiple testing of the 869 pairs of causal associations. The results showed that a total of 36 pairs passed the correction, involving 28 metabolites and 5 metabolites ratios, of which GERD was the most, followed by EU (Fig. 2).
(2) Statistically significant causal effects results: For BE, there were 2 pairs, including Paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil ratio (\({OR}_{IVW}\) = 0.879 95% CI: 0.829-0.933 \({P}_{IVW-adjusted}\) = 0.023), and N-acetylputrescine (\({OR}_{IVW}\) = 1.263 95% CI: 1.131-1.411 \({P}_{IVW-adjusted}\) = 0.042).
For EO, there were 5 pairs, namely Glycine to phosphate ratio (\({OR}_{IVW}\) = 1.001 95% CI: 1.001-1.001 \({P}_{IVW-adjusted}\) = 5.36E-06), Gamma-glutamylglycine (\({OR}_{IVW}\) = 1.001 95% CI: 1.001-1.001 \({P}_{IVW-adjusted}\) = 1.08E-04), Glycine (\({OR}_{IVW}\) = 1.001 95% CI: 1.000-1.001 \({P}_{IVW-adjusted}\) = 3.91E-04) , Glycine to alanine ratio (\({OR}_{IVW}\) = 1.001 95% CI: 1.001-1.001 \({P}_{IVW-adjusted}\) = 0.001), and Glycine to pyridoxal ratio (\({OR}_{IVW}\) = 1.001 95% CI: 1.001-1.001 \({P}_{IVW-adjusted}\) = 0.003).
For EP, there were 2 pairs, namely 2'-o-methyluridine (\({OR}_{IVW}\) = 0.562 95% CI: 0.435-0.725 \({P}_{IVW-adjusted}\) = 0.011), and 2'-o-methylcytidine (\({OR}_{IVW}\) = 0.567 95% CI: 0.439-0.733 \({P}_{IVW-adjusted}\) = 0.017).
For EU, there were 8 pairs, namely Andro steroid monosulfate C19H28O6S (1) (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.024), Bilirubin (Z,Z) to androsterone glucuronide ratio (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.022), 1-oleoyl-GPG (18:1) (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.026), Taurodeoxycholic acid 3-sulfate (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.049), Octadecadienedioate (C18:2-DC) (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.023), 16a-hydroxy DHEA 3-sulfate (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.029), Octadecenedioate (C18:1-DC) (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.045), and Octadecenedioylcarnitine (C18:1-DC) (\({OR}_{IVW}\) = 0.999 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.011).
For EV, there was only 1 pair, namely 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (p-16:0/18:2) (\({OR}_{IVW}\) = 0.999 95% CI: 0.999-1.000 \({P}_{IVW-adjusted}\) = 0.045).
For GERD, there were 18 pairs, namely Glycocholenate sulfate (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.003 \({P}_{IVW-adjusted}\) = 3.05E-05), Hexadecenedioate (C16:1-DC) (\({OR}_{IVW}\) = 1.002 95% CI: 1.002-1.003 \({P}_{IVW-adjusted}\) = 1.59E-04), 1-linoleoyl-GPG (18:2) (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 0.020), N-acetyltyrosine (\({OR}_{IVW}\) = 0.997 95% CI: 0.997-0.998 \({P}_{IVW-adjusted}\) = 9.84E-07), N-acetyl-1-methylhistidine (\({OR}_{IVW}\) = 0.998 95% CI: 0.997-0.999 \({P}_{IVW-adjusted}\) = 2.18E-04), Hexadecanedioate (C16-DC) (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 1.09E-04), Glycochenodeoxycholate glucuronide (1) (\({OR}_{IVW}\) = 1.002 95% CI: 1.001-1.003 \({P}_{IVW-adjusted}\) = 0.005), N-acetylarginine (\({OR}_{IVW}\) = 0.998 95% CI: 0.998 - 0.999 \({P}_{IVW-adjusted}\) = 0.013), Tetradecanedioate (C14-DC) (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 0.001), 1-oleoyl-GPG (18:1) (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 0.014), Glycodeoxycholate 3-sulfate (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 1.38E-04), Taurodeoxycholic acid 3-sulfate (\({OR}_{IVW}\) = 1.002 95% CI: 1.001-1.004 \({P}_{IVW-adjusted}\) = 0.046), N-acetylasparagine (\({OR}_{IVW}\) = 0.998 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.042), Deoxycholic acid 12-sulfate (\({OR}_{IVW}\) = 1.003 95% CI: 1.001-1.004 \({P}_{IVW-adjusted}\) = 0.037), N-acetylkynurenine (2) (\({OR}_{IVW}\) = 0.998 95% CI: 0.997-0.999 \({P}_{IVW-adjusted}\) = 0.026), N-acetylcitrulline (\({OR}_{IVW}\) = 0.998 95% CI: 0.998-0.999 \({P}_{IVW-adjusted}\) = 0.007), Taurocholenate sulfate (\({OR}_{IVW}\) = 1.003 95% CI: 1.001-1.004 \({P}_{IVW-adjusted}\) = 0.023), Octadecenedioylcarnitine (C18:1-DC) (\({OR}_{IVW}\) = 1.003 95% CI: 1.002-1.004 \({P}_{IVW-adjusted}\) = 5.71E-04).
(3) Statistical summary and characteristics overview of metabolite categories with a causal association to ED: There were 17 lipids and 8 amino acids among the 28 metabolites, which accounted for 89.28% of the metabolite entries and the sub pathways mainly involved in these metabolites were “Fatty Acid, Dicarboxylate” and “Secondary Bile Acid Metabolism” (Fig. 2).
Strong causal effects results: We found that the causal relationships between BE and Paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil ratio (\({OR}_{IVW}\) = 0.879 95% CI: 0.829-0.933 \({P}_{IVW-adjusted}\) = 0.023), BE and N-acetylputrescine (\({OR}_{IVW}\) = 1.263 95% CI: 1.131-1.411 \({P}_{IVW-adjusted}\) = 0.042) , EP and 2'-o-methyluridine (\({OR}_{IVW}\) = 0.562 95% CI: 0.435-0.725 \({P}_{IVW-adjusted}\) = 0.011), EP and 2'-o-methylcytidine (\({OR}_{IVW}\) = 0.567 95% CI: 0.439-0.733 \({P}_{IVW-adjusted}\) = 0.017) were stronger than other causal associations.
The reverse MR results: We then performed a reverse MR analysis for 36 causal association pairs, which showed that only GERD had a significant causal relationship with Tetradecanedioate (C14-DC) (\({OR}_{IVW}\) = 0.024 95% CI: 0.001-0.559 \({P}_{IVW-adjusted}\) = 0.020) (Table S4).
Sensitivity analysis
In order to eliminate the bias effect of weak IVs on IVW results, we performed sensitivity analysis to evaluate potential horizontal pleiotropy and heterogeneity, so as to ensure the satisfaction of Independence Assumption and Exclusion Assumption. Firstly, we augmented the IVW results by applying both the MR-Egger and Weighted Median methods to validate the Exclusion Assumption. The subsequent findings revealed that there were four instances of causal relationships for which consistent statistical significance was not maintained across these approaches, namely N-acetylputrescine on BE (\({P}_{MR-Egger}\)= 0.099), Paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil ratio on BE (\({P}_{MR-Egger}\) = 0.076), 1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (p-16:0/18:2) on EV (\({P}_{MR-Egger}\) = 0.536),1-(1-enyl-palmitoyl)-2-linoleoyl-GPE (p-16:0/18:2) levels on EV (\({P}_{Weighted Median}\) = 0.102), and Glycine to pyridoxal ratio on EO (\({P}_{MR-Egger}\) = 0.142) (Fig. S1). However, we could still consider the potential causal relationship between the N-acetylputrescine on BE, Paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil ratio on BE and Glycine to pyridoxal ratio on EO, as these three pairs of associations passed all sensitivity tests except for the MR-Egger analysis. We further screened out possible horizontal pleiotropy in all associations by MR-Egger’s intercept term, MR-PRESSO’s global test, scatter plots, forest plots, and funnel plots to conduct Independence Assumption validation (Fig. S2-S13). The results showed that except the causal association between Taurocholenate sulfate and GERD and the causal association between 1-oleoyl-GPG (18:1) and GERD failed to pass MR-Egger's intercept term test, no horizontal pleiotropy was found in other causal relationships (\({P}_{MR-PRESSO}\) > 0.05 and \({P}_{MR-Egger{\prime}s {\prime}intercept}\) > 0.05) (Table 2). However, given that both causal associations passed MR-PRESSO's global test, we still consider the level of horizontal pleiotropy in these two causal relationships to be limited. In addition, the results of the leave-one-out method showed that a single SNP did not have a significant effect on the causality outcome, suggesting that the Independence Assumption is satisfied (Fig. S14-S17). Finally, the results of Cochran’s Q test for IVW and MR-Egger showed that there was no heterogeneity, suggesting that the Exclusion Assumption is satisfied (Table 2).
Metabolic pathway analysis
Figure 3 summarized the pathways identified as important in various types of ED (P < 0.01), and the results showed that "Caffeine metabolism" played a pivotal role in BE (P = 8.07E-05), while "Glutathione metabolism" was significantly associated with EO (P = 0.008). For EP, "Butanoate metabolism" (P = 0.005) and "Pentose and glucuronate interconversions" (P = 0.008) were identified as the most critical pathways. For GERD, "Glycine, serine and threonine metabolism" (P = 2.77E-04) standed out as a key pathway. For all ED, "Caffeine metabolism" (P = 0.003), "Arginine and proline metabolism" (P = 0.006), "Pyrimidine metabolism" (P = 0.008), and "Arginine biosynthesis" (P = 0.010) were recognized as crucial pathways. Detailed results of Metabolic pathway analysis are available for reference Table S5.
Discussion
In this study, we employed a two-sample MR approach using the IVW method to investigate causal relationships between 850 metabolites and 309 metabolite ratios, respectively, with 10 distinct ED phenotypes, successfully identifying 36 causal associations encompassing 28 individual metabolites, 5 metabolite ratios, and 6 distinct ED phenotypes. Next, we performed sensitivity analyses, reverse MR, and Bonferroni correction for multiple testing. The results indicated that EV did not withstand sensitivity analysis, while for Hexadecenedioate (C16:1-DC) and GERD, a potential reverse causality was suggested, indicating uncertainty regarding their actual relationship, necessitating further research for validation. Below, we will discuss in detail the remaining 34 causal associations thus identified.
(1) Causal associations involving BE: N-acetylputrescine is a derivative of biogenic amines that, in the context of our study, emerged as a risk factor for BE. Although a direct association between N-acetylputrescine and BE has not been definitively established, there are reports indicating its upregulation in cases of esophagitis, which aligns with our present findings [36]. Caffeine undergoes metabolism in the liver via the enzyme CYP1A2, approximately 84% of which is converted into paraxanthine. The ratio of paraxanthine to caffeine in plasma serves as an index to evaluate CYP1A2 activity, while in urine, the ratio of (1-methylxanthine + 1-methylurate + 5-acetylamino-6-formylamino-3-methyluracil) to 1,7-dimethylurate can be used to assess CYP1A2 activity. Notably, CYP1A2 has been identified as a potential therapeutic target in treating the development of esophageal strictures resulting from chronic esophagitis [37]. Therefore, it can be inferred that the ratio of paraxanthine to 5-acetylamino-6-formylamino-3-methyluracil in plasma is closely associated with CYP1A2 activity, and this correlation may play a protective role during the formation of BE.
(2) Causal associations involving EP: 2'-O-methylcytidine and 2'-O-methyluridine are methylated nucleotides, and although no studies have directly implicated them in an intrinsic relationship with the esophagus, RNA modifications have been demonstrated to mediate gene regulation in the development and progression of BE and esophageal adenocarcinoma (EAC) [38]. Thus, we hypothesize that they may also have a close connection with the occurrence and progression of EP.
(3) Causal associations involving EO: For EO, four out of the five causal factors are directly related to glycine, and Gamma-glutamylglycine is composed of glycine. While there is currently no direct literature implicating a specific close relationship between glycine and the esophagus, studies have reported that Glycine-extended gastrin may promote cell survival rather than apoptosis in BE and EAC cells, thereby exerting a detrimental influence on the progression of BE or the development of EAC [39]. Consequently, we suppose that dysregulation in glycine metabolism might contribute to the development of certain ED, including EO, which aligns with the five causal elements related to glycine. BE and EAC are more prevalent in males, suggesting a possible intimate association between sex hormones and ED.
(4) Causal associations involving EU: Androsterone Monosulfate C19H28O6S (1) is a sulfate conjugate of an androgenic steroid, representing a major metabolite of male sex hormones in the human body. It is derived from testosterone through a series of biochemical transformations and ultimately exists in the form of a sulfate conjugate in blood and urine [40]. Consequently, we consider that Androsterone Monosulfate C19H28O6S (1) as a sex hormone metabolite could potentially play a role in the development of BE and EAC. Analogous to BE, EU is also a complication associated with GERD. Hence, we suppose that Androsterone Monosulfate C19H28O6S (1) may also influence the occurrence and progression of EU. Bilirubin (Z,Z) is a stereoisomer of bilirubin, while Androsterone glucuronide is a conjugate of an endogenous androgenic metabolite, formed when the liver enzymatically attaches a glucuronic acid moiety to the androsterone molecule. Existing research has revealed an inverse association between bilirubin levels and the risk of EAC [41]. However, there are currently no published reports establishing a link between Androsterone glucuronide and ED, and the ratio between these two substances requires further validation. 1-oleoyl-GPG (18:1) is a phospholipid, and while there are currently no studies directly implicating its inherent relationship with ED, existing research has shown that aberrant phospholipid synthesis is closely associated with the onset and progression of esophageal cancer [42]. As an early esophageal lesion, the occurrence of EU might potentially be related to 1-oleoyl-GPG (18:1). however, in this study, 1-oleoyl-GPG (18:1) appears as a protective factor, which contradicts existing research and necessitates further verification. Taurodeoxycholic acid 3-sulfate is a bile acid derivative, wherein the hydroxyl group at the 3-position of Taurodeoxycholic acid is sulfated to form a 3-sulfate ester. Current research indicates that Taurodeoxycholic acid impairs normal esophageal barrier function in the early stages of ED [43]. However, other studies have shown that TDCA plays a positive role in inflammatory responses [44]. Considering the results of the present study, it suggests that Taurodeoxycholic acid 3-sulfate may predominantly exhibit a more beneficial effect in the context of EU. Octadecadienedioate (C18:2-DC), Octadecenedioate (C18:1-DC), and Octadecenedioylcarnitine (C18:1-DC) are three metabolites for which there are currently no reported associations with ED. Octadecadienedioate (C18:2-DC) and Octadecenedioate (C18:1-DC) belong to the category of fatty acid dicarboxylic acid derivatives, while Octadecenedioylcarnitine (C18:1-DC) is an acylcarnitine derivative, also stemming from fatty acids. In our findings, these three metabolites demonstrate a protective effect against EU. 16a-hydroxy DHEA 3-sulfate is a hormonal compound belonging to the class of Androgenic Steroids. Currently, there are no reports linking it directly to ED. However, we inferred that its mechanism may be similar to Andro steroid monosulfate C19H28O6S (1) in relation to EU.
(5) Causal associations involving GERD: Glycocholic acid sulfate, Glycochenodeoxycholic acid glucuronide (1), Glycodeoxycholic acid 3-sulfate, Taurodeoxycholic acid 3-sulfate, Deoxycholic acid 12-sulfate, and Taurocholic acid sulfate are all belong to bile acid metabolism. Bile acid reflux has been established as a significant risk factor for GERD [43, 45, 46]. Studies have shown that bile acid perfusion experiments can elicit heartburn sensations. Additionally, another research has revealed that exposing rabbit esophageal mucosa to weakly acidic solutions containing bile acids leads to increased permeability and the formation of dilated intercellular spaces (DIS), a pathological mechanism considered essential for provoking heartburn symptoms [47]. A recent study found that bile reflux is not only associated with esophageal mucosal injury but also with symptom development [48]. These findings align with our results, suggesting that bile acids are indeed high-risk factors in the initiation and progression of GERD. Hexadecanedioate (C16-DC) and Tetradecanedioate (C14-DC) are both fatty acid derivatives containing dicarboxylic acids, whereas Octadecenedioylcarnitine (C18:1-DC) belongs to the class of acylcarnitines, also being a derivative of fatty acids. To date, there have been no reported studies associating these metabolites with GERD. Our findings indicate that all three are risk factors for GERD. This outcome contradicts the protective effects previously observed for fatty acid dicarboxylic acid derivatives on EU, necessitating further validation. As previously mentioned, 1-linoleoyl-GPG (18:2) and 1-oleoyl-GPG (18:1) are both classified as phospholipids [42]. Research has shown that abnormal phospholipid synthesis is closely linked to the development and progression of esophageal cancer. Chronic GERD can lead to the onset of BE, which is a precancerous condition for esophageal cancer. Our findings support the notion that phospholipids may serve as contributing factors to GERD, and over the course of disease progression, eventually culminate in the development of esophageal cancer. N-acetyltyrosine, N-acetyl-1-methylhistidine, N-acetylarginine, N-acetylasparagine, N-acetylkynurenine (2), and N-acetylcitrulline are all N-acetylated amino acids. Currently, there are no reported associations between N-acetylated amino acids and ED. However, research has demonstrated that N-acetylated amino acids can maintain the hydrocarbon chain packing structure of intercellular lipids through electrostatic repulsion, thereby preserving skin barrier function. We infer that the six N-acetylated amino acids discovered in our study may potentially enhance esophageal mucosal barrier function through a similar mechanism, offering protection to the esophageal mucosa.
(6) The characteristics overview of metabolite categories with a causal association to ED: In summary, we have found that the metabolic compounds causally associated with ED mainly include methylated nucleotides, glycine derivatives, sex hormones, phospholipids, bile acids, fatty acid dicarboxylic acid derivatives, and N-acetylated amino acids. These discoveries highlight the multifactorial nature of esophageal pathologies and suggest potential targets for further investigation and intervention in disease prevention and management.
(7) The metabolic pathway analysis results: Moreover, we also identified through metabolic pathway analysis 8 critical pathways in 5 different types of ED, with some of these findings already substantiated. "Pyrimidine metabolism" is a complex enzymatic network that integrates nucleoside salvage, de novo nucleotide synthesis, and catalytic degradation of pyrimidines [49]. There are currently no reports indicating perturbations in "Pyrimidine metabolism" in the context of esophageal diseases. "Caffeine metabolism" is the process by which caffeine is consumed and converted into other compounds that are eventually excreted [50]. Research has shown that caffeine exhibits certain chemopreventive effects in individuals with precancerous lesions and heightened risk of developing cancer in the esophageal region [51]. Arginine is converted into nitric oxide (NO) within endothelial cells through the catalysis by nitric oxide synthase (NOS), which under normal circumstances helps to maintain smooth muscle relaxation and blood supply in the esophagus. In cases of esophagitis or disorders of esophageal motility, the production of NO may be compromised, thereby affecting the normal physiological functions of the esophagus [52]. Therefore, "Arginine biosynthesis" may affect the normal physiological function of the esophagus. Research has indicated that there is a relationship between the arginine and proline polymorphism of the p53 gene and HPV infection status during the occurrence and development of esophageal squamous cell carcinoma, with the presence of the arginine allele potentially elevating an individual's risk for HPV-associated esophageal squamous cell carcinoma, which may indicate that "Arginine and proline metabolism" plays a important role in ED [53]. "Pentose and glucuronate interconversions" are limited in studies in ED and require further clinical and experimental studies. Recent research has highlighted the significant role played by the NEK (Never in mitosis A (NIMA)-related kinase) gene family of serine/threonine kinases in the development of EAC [54]. Coupled with the findings regarding glycine, it is reliable to infer that the "Glycine, Serine, and Threonine Metabolism" pathway exerts a substantial impact on ED. Daysha et al. found that glutathione S-transferase theta 2 (GSTT2) protects esophageal squamous cells from DNA damage caused by genotoxic stress, which is consistent with our discovery of the key to "Glutathione metabolism" [55]. Analysis of the association between "Butanoate metabolism" and ED is limited. However, considering the strong anti-inflammatory effect of butyric acid, it can maintain intestinal barrier function, promote the proliferation and differentiation of intestinal epithelial cells, and reduce the probability of intestinal inflammation [56]. Therefore, we think that "Butanoate metabolism" might play a role in maintaining esophageal barrier function.
(8) Advantages and limitations: The present study has several key strengths: Firstly, it is the first study to report causal associations between ED and metabolites, as well as metabolite ratios, based on both genomic and metabolomic data. Secondly, this research encompasses a comprehensive range of 10 different ED phenotypes and utilizes a vast number of samples sourced from internationally recognized public databases such as UKB and FinnGen. Thirdly, the scope of metabolites studied is extensive and includes metabolite ratios, thereby allowing for the assessment not only of causal relationships between individual metabolites and ED but also between enzyme activity-representative metabolite ratios and ED. Fourthly, strict sensitivity analyses and multiple testing correction have been employed to ensure robustness of the findings. Notably, in selecting IVs, we have completed missing variant IDs, which contrasts with traditional approaches that discard IVs with unknown IDs, thereby rendering our results more complete and reliable. However, the study also has several limitations. Primarily, our analysis is confined to a sample set derived solely from the European population, necessitating validation of our findings using datasets from additional ethnicities. Secondly, we did not fully exclude SNPs that may correspond to confounders of the metabolites and metabolite ratios, which could potentially introduce slight biases into the results. Thirdly, the preliminary causal relationship between identified metabolites and ED may require further analysis using genetic methods such as Colocalization Analysis and PrediXcan, to uncover deeper genetic relationships between the two. Lastly, our findings reveal some conflicting causal associations and previously unreported ones, which call for further experimental validation to illustrate the underlying physiological mechanisms in detail.
Conclusion
In conclusion, this is the first study to integrate genomics and metabolomics in order to summarize the causal relationships between ED and both metabolites as well as metabolite ratios. In this research, we identified 34 pairs of causal associations between metabolic features and ED, through the use of IVW analysis and multiple sensitivity assessments. Furthermore, we uncovered 8 pivotal pathways within 6 categories of ED conditions. These findings provide new reference evidence for elucidating the pathogenesis and progression of ED diseases and offer valuable insights into the diagnosis and treatment strategies for ED in subsequent research. However, further clinical and experimental validations are necessary.
References
Eusebi LH, Ratnakumaran R, Yuan Y, Solaymani-Dodaran M, Bazzoli F, Ford AC. Global prevalence of, and risk factors for, gastro-oesophageal reflux symptoms: a meta-analysis. Gut. 2018;67(3):430–40.
Taraszewska A. Risk factors for gastroesophageal reflux disease symptoms related to lifestyle and diet. Rocz Panstw Zakl Hig. 2021;72(1):21–8.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistic4s 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. Cancer J Clinic. 2021;71(3):209–49.
Iyer PG, Katzka DA. Nonendoscopic Detection of Barrett Esophagus and Esophageal Adenocarcinoma: Recent Advances and Implications. Ann Intern Med. 2021;174(7):1006–7.
Yuan S, Larsson SC. Adiposity, diabetes, lifestyle factors and risk of gastroesophageal reflux disease: a Mendelian randomization study. Eur J Epidemiol. 2022;37(7):747–54.
Cameron AJ, Lagergren J, Henriksson C, Nyren O, Locke GR 3rd, Pedersen NL. Gastroesophageal reflux disease in monozygotic and dizygotic twins. Gastroenterology. 2002;122(1):55–9.
Camilleri M, Malhi H, Acosta A. Gastrointestinal Complications of Obesity. Gastroenterology. 2017;152(7):1656–70.
Steffen A, Huerta JM, Weiderpass E, Bueno-de-Mesquita HB, May AM, Siersema PD, Kaaks R, Neamat-Allah J, Pala V, Panico S, et al. General and abdominal obesity and risk of esophageal and gastric adenocarcinoma in the European Prospective Investigation into Cancer and Nutrition. Int J Cancer. 2015;137(3):646–57.
Richter JE. Oesophageal motility disorders. Lancet (London, England). 2001;358(9284):823–8.
Elliott JA, Reynolds JV. Visceral Obesity, Metabolic Syndrome, and Esophageal Adenocarcinoma. Front Oncol. 2021;11:627270.
Johnson CH, Patterson AD, Idle JR, Gonzalez FJ. Xenobiotic metabolomics: major impact on the metabolome. Annu Rev Pharmacol Toxicol. 2012;52:37–56.
Wang L, Chen J, Chen L, Deng P, Bu Q, Xiang P, Li M, Lu W, Xu Y, Lin H, et al. 1H-NMR based metabonomic profiling of human esophageal cancer tissue. Mol Cancer. 2013;12:25.
Zhang J, Bowers J, Liu L, Wei S, Gowda GA, Hammoud Z, Raftery D. Esophageal cancer metabolite biomarkers detected by LC-MS and NMR methods. PLoS ONE. 2012;7(1):e30181.
Zhang X, Xu L, Shen J, Cao B, Cheng T, Zhao T, Liu X, Zhang H. Metabolic signatures of esophageal cancer: NMR-based metabolomics and UHPLC-based focused metabolomics of blood serum. Biochem Biophys Acta. 2013;1832(8):1207–16.
Hasim A, Ma H, Mamtimin B, Abudula A, Niyaz M, Zhang LW, Anwer J, Sheyhidin I. Revealing the metabonomic variation of EC using 1H-NMR spectroscopy and its association with the clinicopathological characteristics. Mol Biol Rep. 2012;39(9):8955–64.
Ikeda A, Nishiumi S, Shinohara M, Yoshie T, Hatano N, Okuno T, Bamba T, Fukusaki E, Takenawa T, Azuma T, et al. Serum metabolomics as a novel diagnostic approach for gastrointestinal cancer. Biomedical chromatography : BMC. 2012;26(5):548–58.
Yang Y, Wang L, Wang S, Liang S, Chen A, Tang H, Chen L, Deng F. Study of metabonomic profiles of human esophageal carcinoma by use of high-resolution magic-angle spinning 1H NMR spectroscopy and multivariate data analysis. Anal Bioanal Chem. 2013;405(10):3381–9.
Shin SY, Fauman EB, Petersen AK, Krumsiek J, Santos R, Huang J, Arnold M, Erte I, Forgetta V, Yang TP, et al. An atlas of genetic influences on human blood metabolites. Nat Genet. 2014;46(6):543–50.
Hamdalla MA, Ammar RA, Rajasekaran S. A molecular structure matching approach to efficient identification of endogenous mammalian biochemical structures. BMC Bioinformatics. 2015;16 Suppl 5(Suppl 5):S11.
Chen Y, Lu T, Pettersson-Kymmer U, Stewart ID, Butler-Laporte G, Nakanishi T, Cerani A, Liang KYH, Yoshiji S, Willett JDS, et al. Genomic atlas of the plasma metabolome prioritizes metabolites implicated in human diseases. Nat Genet. 2023;55(1):44–53.
Hemani G, Zheng J, Elsworth B, Wade KH, Haberland V, Baird D, Laurin C, Burgess S, Bowden J, Langdon R et al: The MR-Base platform supports systematic causal inference across the human phenome. eLife. 2018;7.
Skrivankova VW, Richmond RC, Woolf BAR, Yarmolinsky J, Davies NM, Swanson SA, VanderWeele TJ, Higgins JPT, Timpson NJ, Dimou N, et al. Strengthening the Reporting of Observational Studies in Epidemiology Using Mendelian Randomization: The STROBE-MR Statement. JAMA. 2021;326(16):1614–21.
Wishart DS, Feunang YD, Marcu A, Guo AC, Liang K, Vázquez-Fresno R, Sajed T, Johnson D, Li C. Karu N et al: HMDB 40: the human metabolome database for 2018. Nucleic Acids Res. 2018;46(D1):D608-d617.
Sudlow C, Gallacher J, Allen N, Beral V, Burton P, Danesh J, Downey P, Elliott P, Green J, Landray M, et al. UK biobank: an open access resource for identifying the causes of a wide range of complex diseases of middle and old age. PLoS Med. 2015;12(3):e1001779.
Kurki MI, Karjalainen J, Palta P, Sipilä TP, Kristiansson K, Donner KM, Reeve MP, Laivuori H, Aavikko M, Kaunisto MA, et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature. 2023;613(7944):508–18.
Matushyn M, Bose M, Mahmoud AA, Cuthbertson L, Tello C, Bircan KO, Terpolovsky A, Bamunusinghe V, Khan U, Novković B, et al. SumStatsRehab: an efficient algorithm for GWAS summary statistics assessment and restoration. BMC Bioinformatics. 2022;23(1):443.
Burgess S, Butterworth A, Thompson SG. Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol. 2013;37(7):658–65.
Bowden J, Davey Smith G, Burgess S. Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol. 2015;44(2):512–25.
Bowden J, Davey Smith G, Haycock PC, Burgess S. Consistent Estimation in Mendelian Randomization with Some Invalid Instruments Using a Weighted Median Estimator. Genet Epidemiol. 2016;40(4):304–14.
Verbanck M, Chen CY, Neale B, Do R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat Genet. 2018;50(5):693–8.
Cohen JF, Chalumeau M, Cohen R, Korevaar DA, Khoshnood B, Bossuyt PM. Cochran’s Q test was useful to assess heterogeneity in likelihood ratios in studies of diagnostic accuracy. J Clin Epidemiol. 2015;68(3):299–306.
Burgess S. Sample size and power calculations in Mendelian randomization with a single instrumental variable and a binary outcome. Int J Epidemiol. 2014;43(3):922–9.
Ewald JD, Zhou G, Lu Y, Kolic J, Ellis C, Johnson JD, Macdonald PE, Xia J. Web-based multi-omics integration using the Analyst software suite. Nat Protoc. 2024;19(5):1467–97.
Jewison T, Su Y, Disfany FM, Liang Y, Knox C, Maciejewski A, Poelzer J, Huynh J, Zhou Y, Arndt D, et al. SMPDB 20: big improvements to the Small Molecule Pathway Database. Nucleic Acids Res. 2014;42(Database issue):D478-484.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Moye LM, Liu Y, Coarfa C, Putluri N, Rhoads JM. Plasma Urea Cycle Metabolites May Be Useful Biomarkers in Children With Eosinophilic Esophagitis. Front Pediatr. 2018;6:423.
Wu R, Fu M, Tao HM, Dong T, Fan WT, Zhao LL, Fan ZN, Liu L. Benign esophageal stricture model construction and mechanism exploration. Sci Rep. 2023;13(1):11769.
Zou K, Dong H, Li M, Zhang Y, Zhang K, Song D, Chu C. Comprehensive analysis of transcriptome-wide N6-methyladenosine methylomes in the Barrett’s esophagus in rats. Genomics. 2023;115(5): 110687.
Beales IL, Ogunwobi OO. Glycine-extended gastrin inhibits apoptosis in Barrett’s oesophageal and oesophageal adenocarcinoma cells through JAK2/STAT3 activation. J Mol Endocrinol. 2009;42(4):305–18.
Xie SH, Fang R, Huang M, Dai J, Thrift AP, Anderson LA, Chow WH, Bernstein L, Gammon MD, Risch HA, et al. Association Between Levels of Sex Hormones and Risk of Esophageal Adenocarcinoma and Barrett’s Esophagus. Clinical gastroenterology and hepatology : the official clinical practice journal of the American Gastroenterological Association. 2020;18(12):2701-2709.e2703.
Seyed Khoei N, Wagner KH, Carreras-Torres R, Gunter MJ, Murphy N, Freisling H. Associations between Prediagnostic Circulating Bilirubin Levels and Risk of Gastrointestinal Cancers in the UK Biobank. Cancer. 2021;13(11):2749.
Ma S, Zhou B, Yang Q, Pan Y, Yang W, Freedland SJ, Ding LW, Freeman MR, Breunig JJ, Bhowmick NA, et al. A Transcriptional Regulatory Loop of Master Regulator Transcription Factors, PPARG, and Fatty Acid Synthesis Promotes Esophageal Adenocarcinoma. Can Res. 2021;81(5):1216–29.
Gadelha KKL, Batista-Lima FJ, de Oliveira DMN, Carvalho EF, Sifrim D, Santos AAD, Magalhães PJC. Impairment of rat oesophageal muscle contractility associated with experimental non-erosive oesophageal mucosal damage. Exp Physiol. 2019;104(2):199–208.
Quante M, Iske J, Uehara H, Minami K, Nian Y, Maenosono R, Matsunaga T, Liu Y, Azuma H, Perkins D, et al. Taurodeoxycholic acid and valine reverse obesity-associated augmented alloimmune responses and prolong allograft survival. Am J Transplant Off J Am Soc Transplant Am Soc Transplant Surg. 2022;22(2):402–13.
Fass R, Boeckxstaens GE, El-Serag H, Rosen R, Sifrim D, Vaezi MF. Gastro-oesophageal reflux disease. Nat Rev Dis Primers. 2021;7(1):55.
Bredenoord AJ. Mechanisms of reflux perception in gastroesophageal reflux disease: a review. Am J Gastroenterol. 2012;107(1):8–15.
Farré R, van Malenstein H, De Vos R, Geboes K, Depoortere I, Vanden Berghe P, Fornari F, Blondeau K, Mertens V, Tack J, et al. Short exposure of oesophageal mucosa to bile acids, both in acidic and weakly acidic conditions, can impair mucosal integrity and provoke dilated intercellular spaces. Gut. 2008;57(10):1366–74.
de Bortoli N, Gyawali CP, Frazzoni M, Tolone S, Frazzoni L, Vichi E, Visaggi P, Bellini M, Marabotto E, Penagini R, et al. Bile reflux in patients with nerd is associated with more severe heartburn and lower values of mean nocturnal baseline impedance and chemical clearance. Neurogastroenterol Motil. 2020;32(12):e13919.
Siddiqui A, Ceppi P. A non-proliferative role of pyrimidine metabolism in cancer. Molecular metabolism. 2020;35:100962.
Mazzafera P. Catabolism of caffeine in plants and microorganisms. Front Biosci. 2004;9:1348–59.
Conney AH. Enzyme induction and dietary chemicals as approaches to cancer chemoprevention: the Seventh DeWitt S. Goodman Lecture Cancer research. 2003;63(21):7005–31.
Conklin JL, Du C, Murray JA, Bates JN. Characterization and mediation of inhibitory junction potentials from opossum lower esophageal sphincter. Gastroenterology. 1993;104(5):1439–44.
Kawaguchi H, Ohno S, Araki K, Miyazaki M, Saeki H, Watanabe M, Tanaka S, Sugimachi K. p53 polymorphism in human papillomavirus-associated esophageal cancer. Can Res. 2000;60(11):2753–5.
Chen L, Ballout F, Lu H, Hu T, Zhu S, Chen Z, Peng D. Differential Expression of NEK Kinase Family Members in Esophageal Adenocarcinoma and Barrett's Esophagus. Cancer. 2023;15(19):4821.
Ferrer-Torres D, Nancarrow DJ, Steinberg H, Wang Z, Kuick R, Weh KM, Mills RE, Ray D, Ray P, Lin J, et al. Constitutively Higher Level of GSTT2 in Esophageal Tissues From African Americans Protects Cells Against DNA Damage. Gastroenterology. 2019;156(5):1404–15.
Chen G, Ran X, Li B, Li Y, He D, Huang B, Fu S, Liu J, Wang W. Sodium Butyrate Inhibits Inflammation and Maintains Epithelium Barrier Integrity in a TNBS-induced Inflammatory Bowel Disease Mice Model. EBioMedicine. 2018;30:317–25.
Acknowledgements
We thank all the teams that contributed to the GWAS data involved in this study.
Funding
This work was supported by the Nurture projects for basic research of Shanghai Chest Hospital (No: 2022YNJCQ14).
Author information
Authors and Affiliations
Contributions
HLY, YLW, CQC and WJY were responsible for the study design. HLY and YLW were responsible for GWAS data acquisition and data analysis. HLY and WJY drafted the manuscript. YWZ, and LQC contributed to interpretation and editing of the manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This article contains human participants collected by several studies performed by previous studies. All participants gave informed consent in all the corresponding original studies, as described in the Methods. Here, our study is based on the large-scale GWAS datasets, and not the individual-level data. Hence, ethical approval was not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Yang, H., Wang, Y., Zhao, Y. et al. Causal effects of genetically determined metabolites and metabolite ratios on esophageal diseases: a two-sample Mendelian randomization study. BMC Gastroenterol 24, 310 (2024). https://doi.org/10.1186/s12876-024-03411-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12876-024-03411-8