Abstract
Background
An increasing number of small nucleolar RNA host genes (SNHGs) have been revealed to be dysregulated in lung cancer tissues, and abnormal expression of SNHGs is significantly correlated with the prognosis of lung cancer. The purpose of this study was to conduct a meta-analysis to explore the correlation between the expression level of SNHGs and the prognosis of lung cancer.
Methods
A comprehensive search of six related databases was conducted to obtain relevant literature. Relevant information, such as overall survival (OS), progression-free survival (PFS), TNM stage, lymph node metastasis (LNM), and tumor size, was extracted. Hazard ratios (HRs) and 95% confidence intervals (CIs) were pooled to evaluate the relationship between SNHG expression and the survival outcome of lung cancers. Sensitivity and publication bias analyses were performed to explore the stability and reliability of the overall results.
Results
Forty publications involving 2205 lung cancer patients were included in this meta-analysis. The pooled HR and 95% CI values indicated a significant positive association between high SNHG expression and poor OS (HR: 1.890, 95% CI: 1.595–2.185), disease-free survival (DFS) (HR: 2.31, 95% CI: 1.57–3.39) and progression-free survival (PFS) (HR: 2.01, 95% CI: 0.66–6.07). The pooled odds ratio (OR) and 95% CI values indicated that increased SNHG expression may be correlated with advanced TNM stage (OR: 1.509, 95% CI: 1.267–1.799), increase risk of distant lymph node metastasis (OR: 1.540, 95% CI: 1.298–1.828), and large tumor size (OR: 1.509, 95% CI: 1.245–1.829). Sensitivity analysis and publication bias results showed that each result had strong reliability and robustness, and there was no significant publication bias or other bias.
Conclusion
Most SNHGs are upregulated in lung cancer tissues, and high expression of SNHGs predicts poor survival outcomes in lung cancer. SNHGs may be potential prognostic markers and promising therapeutic targets.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Cancer is a major threat to human health [1, 2]. Thousands of people die of cancer every year, which brings an enormous economic burden to the whole world [3]. According to reports, in 2020, there was an estimated 19.8 million new cancer cases and nearly 10 million cancer deaths worldwide [2]. Among cancers, lung cancer ranks first in incidence among men and second among women [4, 5]. Various treatment modalities, such as surgery, radiotherapy, chemotherapy, targeted therapy and immunotherapy, have been applied for cancer treatment, and patient survival has improved. However, many patients are already in the middle and advanced stages of the disease when they are diagnosed [6], and the five-year survival rate of lung cancer is still not optimistic [7, 8]. An increasing number of researchers are trying to find new therapeutic targets [9,10,11].
With the advancement of cancer research at the level of molecular biology, long noncoding RNAs have been reported by many scientists to be significant factors in the progression of lung cancer [12,13,14]. Although they have no protein coding ability, long noncoding RNAs can directly act on downstream genes or signaling pathways and intervene in the proliferation, migration, invasion and drug resistance of lung cancer cells [15]. For instance, Guo et al. uncovered that linc00261 could suppress the proliferation, migration and invasion of lung cancer cells by increasing FOXO1 expression by downregulating miR-1269a [16]. Xu et al. revealed that linc00473 contributes to the invasion, migration and proliferation of NSCLC cells by sponging and downregulating miR-497-5p [17].
The expression of many SNHGs has been revealed by researchers to be dysregulated in lung cancer tissues and to be closely involved in the occurrence and development of lung cancer. SNHG can directly regulate the downstream genes or signaling pathways of lung cancer cells or act as a molecular sponge of microRNAs and then indirectly regulate downstream signaling cascades to affect the proliferation, migration, invasion and apoptosis of tumor cells [18, 19]. For instance, Wang et al. suggested that SNHG12 promotes the migration and invasion of NSCLC cells by interacting with the Slug/ZEB2 signaling pathway by serving as a sponge of miR-218 [20]. Zhao et al. discovered that SNHG3 facilitates the invasion, proliferation, and migration and inhibits the apoptosis of NSCLC cells through the upregulation of nuclear factor IX (NFIX) by sponging and downregulating miR-1343-3p [21]. An increasing number of studies have reported that SNHGs are upregulated and significantly related to the prognosis of lung cancer, while other studies have obtained the opposite results. Considering that the sample size of single studies on this topic are insufficient, and the conclusions of different studies are not completely consistent, the purpose of this study was to conduct a meta-analysis to comprehensively explore the correlation between the expression level of SNHG and the prognosis of lung cancer.
Materials and methods
Literature search strategy
Based on the Preferred Reporting Items for Systematic Reviews and Meta-Analyses, a comprehensive search of six related electronic databases, including PubMed, Embase, Web of Science, Cochrane Library, Google Scholar and China National Knowledge Infrastructure (CNKI), was performed. The detailed search terms were as follows: (“small nucleolar RNA host gene” OR “Long noncoding RNA SNHG” OR “SNHG” OR “lnc SNHG”) AND (“non-small cell lung cancer” OR “lung cancer” OR “Lung adenocarcinoma” OR “NSCLC” OR “prognosis” OR “survival” OR “outcome”). The references of the included literature were also read in detail to avoid omitting relevant literature as much as possible.
Inclusion and exclusion criteria
Based on the reporting specification of The Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) for meta-analysis, the original documents included in this study met the following inclusion and exclusion criteria. Inclusion criteria: (1) There is a clear detection method to detect the expression level of SNHG in tumor tissues, such as real-time fluorescence quantitative polymerase chain reaction (qRT‒PCR). (2) Based on the expression level of SNHG, patients were divided into a high-expression SNHG group and a low-expression SNHG group. (3) The literature mainly evaluates the correlation between the expression level of SNHG and the prognosis of lung cancer. (4) The original documents provide sufficient data for statistics. Exclusion criteria: (1) The original literature did not evaluate the correlation between the expression level of SNHG and the prognosis of lung cancer. (2) Insufficient or unavailable data. (3) The research objects are not humans but animals. (4) Written in a language other than English.
Quality evaluation of included literature
For each publication included in this meta-analysis, the quality assessment according to the Newcastle‒Ottawa Scale (NOS) score was independently conducted by two researchers, which included three major items: selection method of case group and control group, comparability of case group and control group, and contact exposure assessment method. According to the star rating system, the three items have a total of 9 points; the lower the score is, the worse the quality of the literature research. The literature with a score below 6 will be excluded, and the literature with a score of 6–9 is considered suitable for inclusion in this study [22].
Data extraction
Two researchers independently obtained usable or related data, such as the name of the first author, year of publication, sample size, cutoff value, detection methods, and follow-up month. The number of occurrences and the total number of events were extracted to evaluate the correlation between SNHG expression levels and various clinicopathological features, such as TNM stage, LNM, DM, tumor size, and histological grade. Hazard ratios (HRs) with 95% confidence intervals (CIs) were obtained to evaluate the relationship between SNHG expression and the survival outcomes of lung cancer, including OS, progression-free survival (PFS), disease-free survival (DFS) and relapse-free survival (RFS). If the literature did not directly give the HR value but contained a survival curve and the number of people with high and low expression of SNHG, we obtained the HR value and its 95% confidence interval according to the software Enguage version 4.0 [23].
Statistical analysis
Stata version 12.0 software (Stata Corporation, College Station, TX) and Review Manager 5.4.0 (Cochrane Collaboration) were utilized in this meta-analysis. Pooling HR with 95% CI was carried out to assess the association between SNHG expression and cancer prognosis. Pooling OR with 95% CI was performed to explore the relationship between SNHG expression and clinicopathological features of lung cancers. For the heterogeneity of each result, the fix-effect model was performed for small heterogeneity (I2 < 50%, p ≥ 0.05). If the heterogeneity was significant (I2 ≥ 50%, p < 0.05), the random-effect model was used, and subgroup analysis was conducted based on the SNHG expression, follow-up month, number of patients, NOS score and so on.
Results
The basic characteristics of the included studies
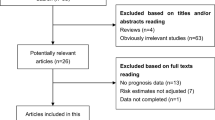
After the comprehensive search of related databases, 975 articles were initially obtained, 174 duplicate publications were excluded, and 696 studies were discarded because they did not assess the correlation between the expression level of SNHG and the prognosis of lung cancer. In addition, 22 meta-analyses, 17 reviews and 26 articles with insufficient data were removed. Finally, 40 studies with 2205 lung cancer patients were enrolled in this meta-analysis [20, 21, 24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60] (Fig. 1). All patients were from China, and the expression level of SNHG was mostly detected by clear detection methods, such as real-time fluorescent quantitative polymerase chain reaction (qRT‒PCR), and there were clear reference genes (Table 1). Based on the NOS scale, the research quality of the 41 original documents was no less than 6 points (Table 2).
The association between SNHG expression and overall survival
Thirty studies with 1748 patients were enrolled to assess the association between SNHG expression and the prognosis of lung cancer. The pooled HR with 95% CI indicated a significant positive relationship between high SNHG expression and poor OS (HR: 1.890, 95% CI: 1.595–2.185) (Fig. 2). In consideration of the inconsistent cutoff values, follow-up time, NOS scores and HR sources between different primary studies, the results of the subgroup analysis show that elevated SNHG expression implies worse OS in the mean value cutoff subgroup (HR: 2.262, 95% CI: 1.790–2.734), the median-value cutoff subgroup (HR: 1.643, 95% CI: 1.262–2.024), the subgroup of multivariate analysis (HR: 2.793, 95% CI: 1.631–3.955) and univariate analysis (HR: 1.828, 95% CI: 1.523–2.133). At the same time, the pooled HR and 95% CI values revealed that high SNHG expression predicted poor disease-free survival (DFS) (HR: 2.31, 95% CI: 1.57–3.39) (Fig. 3A) and progression-free survival (PFS) (HR: 2.01, 95% CI: 0.66–6.07) (Fig. 3B and Table 3).
The association between SNHG expression and TNM stage
Twenty-eight studies comprising 1589 patients were included in this study to explore the relationship between SNHG expression and TNM stage. Pooled OR and 95% CI values showed that high SNHG expression predicted advanced TNM stage (OR: 1.509, 95% CI: 1.267–1.799) (Fig. 4). The results of subgroup analysis indicated advanced TNM stage of lung cancers was correlated with high SNHG expression (OR: 1.650, 95% CI: 1.374–1.980), NOS score ≥ 9 (OR: 2.043, 95% CI: 1.215–3.435) and NOS score < 9 (OR: 1.451, 95% CI: 1.204–1.748). However, in the low SNHG expression subgroup, high SNHG expression implies a favorable TNM stage (OR: 0.424, 95% CI: 0.207–0.867) (Table 4).
The association between SNHG expression and LNM
Twenty-nine publications involving 1662 patients were enrolled to evaluate the relationship between SNHG expression and LNM. Pooled OR and 95% CI results suggested a significant association between increased SNHG expression and distant lymph node metastasis (OR: 1.540, 95% CI: 1.298–1.828) (Fig. 5). Based on the subgroup analysis, we found an increased risk of distant lymph node metastasis of lung cancer cells in the subgroups with high SNHG expression (OR: 1.681, 95% CI: 1.406–2.008), NOS score ≥ 9 (OR: 1.857, 95% CI: 1.203–2.865) and NOS score < 9 (OR: 1.488, 95% CI: 1.235–1.792). In addition, there was a lower likelihood of distant lymph node metastasis in the subgroup with low SNHG expression (OR: 0.435, 95% CI: 0.216–0.876) (Table 4).
The association between SNHG expression and other clinicopathological parameters
Pooled OR and 95% CI values also revealed a marked positive correlation between high SNHG expression and large tumor size (OR: 1.509, 95% CI: 1.245–1.829) (Fig. 6). The correlations between SNHG expression and histological grade (OR: 1.248, 95% CI: 0.938–1.661) (Fig. 7A), depth of invasion (OR: 1.029, 95% CI: 0.297–3.566), DM (OR: 0.933, 95% CI: 0.463–1.882) (Fig. 7B), age (OR: 1.045, 95% CI: 0.879–1.242) and sex (OR: 0.970, 95% CI: 0.806–1.166) were nonsignificant (Table 4).
Sensitivity and publication bias analyses
The results of the sensitivity analysis showed that removing any one study did not significantly change the overall results, supporting the reliability and stability of this meta-analysis (Fig. 8). The results of publication bias analysis showed that there was no significant publication bias for each outcome, suggesting that none of the individual studies contribute significant statistical bias or other types of bias (Fig. 9).
Discussion
An increasing number of long noncoding RNAs have been shown to be significantly involved in cancer progression and are clearly associated with cancer prognosis. Many noncoding RNAs have been reported to affect the occurrence and development of cancer by affecting cell biological behaviors such as tumor cell proliferation, migration, invasion, apoptosis, drug resistance or immune escape. In recent years, lung cancer-associated long noncoding RNAs have been gradually revealed. The biological behavior of lung cancer cells is significantly regulated by long noncoding RNAs. For example, Xu et al. reported that linc00473 may contribute to the proliferation, migration, invasion and inhibition of apoptosis of NSCLC cells by activating the ERK/p38 and MAPK signaling axes by sponging and downregulating miR-497-5p [17]. Zhong et al. revealed that lncRNA TTN-AS1 enhances the invasion and migration of NSCLC cells by increasing ZEB1 expression by suppressing miR-4677-3p [61]. Moreover, the prognosis of lung cancer patients has been reported to be significantly correlated with dysregulation of long noncoding RNAs [62].
The SNHG long noncoding RNA family, a class of small molecules without protein coding function, includes dozens of family members. An increasing number of studies have reported that the expression of SNHGs is dysregulated in lung tissue. Abnormally expressed SNHGs can affect the occurrence and development of lung cancer by affecting a series of biological behaviors of lung cancer cells, such as proliferation, migration, apoptosis, immune escape and drug resistance. Differential expression of SNHGs is significantly correlated with the prognosis of lung cancer. In this study, by pooling HR values from different studies, high expression of SNHGs was found to be positively correlated with a poor prognosis for lung cancer. SNHG2, SNHG3, and SNHG10 are expressed at low levels in lung cancer tissues, while other SNHGs are upregulated in lung cancer tissues. Considering these findings, we conducted a subgroup analysis based on the expression level of SNHGs in lung cancer, and the results showed that low expression of SNHG2 and SNHG3 predicted a poor prognosis for lung cancer, while high expression predicted a poor prognosis for lung cancer. Due to different cutoff values (mean and median), different analysis methods (univariate analysis and multivariate analysis), and different follow-up times and sample sizes among different studies, we performed subgroup analysis to compare the results of these types of studies. The results showed that among studies employing multivariate analysis, univariate analysis, the mean cutoff value and the median cutoff value, high expression of SNHG predicted poor prognosis of lung cancer. In addition, the combined HR value results showed that high expression of SNHG was significantly positively correlated with unsatisfactory progression-free survival and disease-free survival. The pooled OR value and 95% CI results showed that high expression of SNHG predicted advanced TNM stage, increased risk of lymph node metastasis and distant metastasis, larger tumor diameter, and worse histological grade. Considering the differences in research quality and inconsistent cutoff values among original studies, we also conducted subgroup analysis, and the results showed that high expression of SNHG still predicted poor survival outcomes among different subgroups.
An increasing number of studies have reported the molecular biological mechanism by which SNHG affects the progression of lung cancer (Table 5). First, SNHG can directly act on downstream genes or signaling pathways to affect a series of biological behaviors of lung cancer cells; for example, Zhang et al. reported that SNHG1 may contribute to the migration and invasion of NSCLC cells by upregulating zinc finger E-box-binding homeobox 1 (ZEB1) [56]. Shi et al. revealed that SNHG3 could drive the proliferation and migration of lung cancer cells by interacting with the IL-6/JAK2/STAT3 pathway [49]. Guo et al. discovered that DANCR (also named SNHG13) facilitates the proliferation, migration, invasion and EMT process of tumor cells through the upregulation of the p21 gene [34]. Second, SNHG could serve as a competing endogenous RNA and indirectly regulate downstream genes or signaling pathways by sponging microRNAs. For instance, Cui et al. suggested that SNHG1 may induce the proliferation and cell cycle and suppress the apoptosis of lung cancer cells by upregulating Wnt/β-catenin signaling by sponging and downregulating miR-101-3p [28]. Zhao et al. demonstrated that SNHG3 may promote the proliferation, migration, and invasion and inhibit the apoptosis of NSCLC cells through the upregulation of nuclear factor IX (NFIX) by sponging miR-1343-3p [21]. Wang et al. demonstrated that SNHG12 could facilitate the migration and EMT process of tumor cells by interacting with the Slug/ZEB2 signaling pathway by decreasing miR-218 [20]. In addition, some family members of SNHG can also regulate the drug resistance of NSCLC cells; for example, Wei et al. revealed that SNHG1 may reduce the cisplatin sensitivity of A549/DDP cells by increasing Rho-associated coiled-coil containing protein kinase 2 (ROCK2) expression by sponging and downregulating miR-101-3p [54]. Yang et al. reported that growth arrest specific 5 (GAS5, also named SNHG2) could reduce H1299/DDP cell migration, invasion and EMT and reduce cisplatin resistance through the upregulation of phospholysine phosphohistidine inorganic pyrophosphate phosphatase (LHPP) by sponging miR-217 [55]. Wang et al. discovered that SNHG5 was downregulated in NSCLC tissues, and high SNHG5 expression may enhance the sensitivity of A549 cells to gefitinib by interacting with the miR377/CASP1 axis [63]. In addition, SNHG may interfere with the immune escape of lung cancer cells; for instance, Huang et al. revealed that SNHG12 facilitates the immune escape of H1299 cells by interacting with the HuR/PD-L1/USP8 axis [37].
There are inevitably some limitations to this study. First, all the included studies were from China, so the conclusions of this study may only be applicable to patients in China or East Asia. Second, among all the included original studies, some studies directly provided HR values, while others only provided survival curves. We could only obtain HR values indirectly through Enguage software, which makes the combined OS value of this study somewhat inaccurate. statistical bias. However, this study is the first meta-analysis to explore the correlation between the expression level of the SNHG family and the prognosis of lung cancer. In addition, this study comprehensively summarizes the molecular biological mechanism of each member of the SNHG family affecting the occurrence and development of lung cancer.
Conclusion
Most SNHGs are upregulated in lung cancer, and only some SNHGs are downregulated in lung cancer. High SNHG expression predicts poor overall survival and disease-free survival in lung cancer. SNHG may be a potential prognostic marker and a promising therapeutic target.
Availability of data and materials
All data generated or analyzed during this study are included in this published article or are available from the corresponding author upon reasonable request.
References
Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin. 2023;73(1):17–48. https://doi.org/10.3322/caac.21763.
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: globocan estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–49. https://doi.org/10.3322/caac.21660.
Bhakta N, Force LM, Allemani C, Atun R, Bray F, Coleman MP, et al. Childhood cancer burden: a review of global estimates. Lancet Oncol. 2019;20(1):e42–53. https://doi.org/10.1016/s1470-2045(18)30761-7.
Wong MCS, Lao XQ, Ho KF, Goggins WB, Tse SLA. Incidence and mortality of lung cancer: global trends and association with socioeconomic status. Sci Rep. 2017;7(1):14300. https://doi.org/10.1038/s41598-017-14513-7.
Khanmohammadi S, Saeedi Moghaddam S, Azadnajafabad S, Rezaei N, Esfahani Z, Rezaei N, et al. Burden of tracheal, bronchus, and lung cancer in North Africa and middle east countries, 1990 to 2019: results from the Gbd study 2019. Front Oncol. 2022;12:1098218. https://doi.org/10.3389/fonc.2022.1098218.
Manser R, Lethaby A, Irving LB, Stone C, Byrnes G, Abramson MJ, et al. Screening for lung cancer. Cochrane Database Syst Rev. 2013;2013(6):Cd001991. https://doi.org/10.1002/14651858.CD001991.pub3.
Torre LA, Siegel RL, Jemal A. Lung cancer statistics. Adv Exp Med Biol. 2016;893:1–19. https://doi.org/10.1007/978-3-319-24223-1_1.
Bray FI, Weiderpass E. Lung cancer mortality trends in 36 european countries: secular trends and birth cohort patterns by sex and region 1970–2007. Int J Cancer. 2010;126(6):1454–66. https://doi.org/10.1002/ijc.24855.
Nooreldeen R, Bach H. Current and future development in lung cancer diagnosis. Int J Mol Sci. 2021;22(16):8661. https://doi.org/10.3390/ijms22168661.
Sears CR, Mazzone PJ. Biomarkers in lung cancer. Clin Chest Med. 2020;41(1):115–27. https://doi.org/10.1016/j.ccm.2019.10.004.
Seijo LM, Peled N, Ajona D, Boeri M, Field JK, Sozzi G, et al. Biomarkers in lung cancer screening: achievements, promises, and challenges. J Thorac Oncol. 2019;14(3):343–57. https://doi.org/10.1016/j.jtho.2018.11.023.
Xu K, Zhang C, Du T, Gabriel ANA, Wang X, Li X, et al. Progress of exosomes in the diagnosis and treatment of lung cancer. Biomed Pharmacother. 2021;134:111111. https://doi.org/10.1016/j.biopha.2020.111111.
Entezari M, Ghanbarirad M, Taheriazam A, Sadrkhanloo M, Zabolian A, Goharrizi M, et al. Long non-coding rnas and exosomal lncrnas: potential functions in lung cancer progression, drug resistance and tumor microenvironment remodeling. Biomed Pharmacother. 2022;150:112963. https://doi.org/10.1016/j.biopha.2022.112963.
Li Y, Jiang T, Zhou W, Li J, Li X, Wang Q, et al. Pan-cancer characterization of immune-related Lncrnas identifies potential oncogenic biomarkers. Nat Commun. 2020;11(1):1000. https://doi.org/10.1038/s41467-020-14802-2.
Wang J, Tan L, Yu X, Cao X, Jia B, Chen R, et al. Lncrna Znrd1-As1 promotes malignant lung cell proliferation, migration, and angiogenesis via the Mir-942/Tns1 axis and is positively regulated by the M(6)a reader Ythdc2. Mol Cancer. 2022;21(1):229. https://doi.org/10.1186/s12943-022-01705-7.
Guo C, Shi H, Shang Y, Zhang Y, Cui J, Yu H. Lncrna Linc00261 overexpression suppresses the growth and metastasis of lung cancer via regulating mir-1269a/Foxo1 Axis. Cancer Cell Int. 2020;20:275. https://doi.org/10.1186/s12935-020-01332-6.
Xu SH, Bo YH, Ma HC, Zhang HN, Shao MJ. Lncrna Linc00473 promotes proliferation, migration, invasion and inhibition of apoptosis of non-small cell lung cancer cells by acting as a sponge of Mir-497-5p. Oncol Lett. 2021;21(6):429. https://doi.org/10.3892/ol.2021.12690.
Nie Z, Zhang K, Li Z, Bing X, Jin S, Li M. Human papillomavirus 16 E6 promotes angiogenesis of lung cancer via Snhg1. Cell Biochem Biophys. 2023. https://doi.org/10.1007/s12013-022-01121-0.
Zhang K, Chen J, Li C, Yuan Y, Fang S, Liu W, et al. Exosome-mediated transfer of Snhg7 enhances docetaxel resistance in lung adenocarcinoma. Cancer Lett. 2022;526:142–54. https://doi.org/10.1016/j.canlet.2021.10.029.
Wang Y, Liang S, Yu Y, Shi Y, Zheng H. Knockdown of Snhg12 suppresses tumor metastasis and epithelial-mesenchymal transition via the Slug/Zeb2 signaling pathway by targeting mir-218 in Nsclc. Oncol Lett. 2019;17(2):2356–64. https://doi.org/10.3892/ol.2018.9880.
Zhao S, Gao X, Zhong C, Li Y, Wang M, Zang S. Snhg3 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through regulating Mir-216a/Zeb1 Axis. Onco Targets Ther. 2020;13:11327–36. https://doi.org/10.2147/ott.S263637.
Stang A. Critical evaluation of the newcastle-ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol. 2010;25(9):603–5. https://doi.org/10.1007/s10654-010-9491-z.
Tierney JF, Stewart LA, Ghersi D, Burdett S, Sydes MR. Practical methods for incorporating summary time-to-event data into meta-analysis. Trials. 2007;8: 16. https://doi.org/10.1186/1745-6215-8-16.
Chen C, Zhang Z, Li J, Sun Y. Snhg8 is identified as a key regulator in non-small-cell lung cancer progression sponging to mir-542-3p by targeting Ccnd1/Cdk6. Onco Targets Ther. 2018;11:6081–90. https://doi.org/10.2147/ott.S170482.
Chen X, Song P, Yao Y, Yang Y. Long non-coding rna Snhg14 regulates Spin1 expression to accelerate tumor progression in non-small cell lung cancer by sponging Mir-382-5p. Cancer Manage Res. 2020;12:9113–23. https://doi.org/10.2147/cmar.S250893.
Chen Z, Chen X, Chen P, Yu S, Nie F, Lu B, et al. Long non-coding rna Snhg20 promotes non-small cell lung cancer cell proliferation and migration by epigenetically silencing of P21 expression. Cell Death Dis. 2017;8(10):e3092. https://doi.org/10.1038/cddis.2017.484.
Cui HX, Zhang MY, Liu K, Liu J, Zhang ZL, Fu L. Lncrna Snhg15 promotes proliferation and migration of lung cancer via targeting microrna-211-3p. Eur Rev Med Pharmacol Sci. 2018;22(20):6838–44. https://doi.org/10.26355/eurrev_201810_16152.
Cui Y, Zhang F, Zhu C, Geng L, Tian T, Liu H. Upregulated Lncrna Snhg1 contributes to progression of non-small cell lung cancer through inhibition of Mir-101-3p and activation of Wnt/Β-catenin signaling pathway. Oncotarget. 2017;8(11):17785–94. https://doi.org/10.18632/oncotarget.14854.
Dong YZ, Meng XM, Li GS. Long non-coding rna Snhg15 indicates poor prognosis of non-small cell lung cancer and promotes cell proliferation and invasion. Eur Rev Med Pharmacol Sci. 2018;22(9):2671–9. https://doi.org/10.26355/eurrev_201805_14963.
Dong Z, Liu H, Zhao G. Long noncoding rna Snhg6 promotes proliferation and inhibits apoptosis in non-small cell lung cancer cells by regulating Mir-490-3p/Rsf1 Axis. Cancer Biother Radiopharm. 2020;35(5):351–61. https://doi.org/10.1089/cbr.2019.3120.
Fan H, Yuan J, Li Y, Jia Y, Li J, Wang X, et al. Mkl1-Induced Lncrna Snhg18 drives the growth and metastasis of non-small cell lung cancer via the Mir-211-5p/Brd4 Axis. Cell Death Dis. 2021;12(1):128. https://doi.org/10.1038/s41419-021-03399-z.
Gao N, Ye B. Spi1-induced upregulation of Lncrna Snhg6 promotes non-small cell lung cancer via Mir-485-3p/Vps45 Axis. Biomed Pharmacother. 2020;129:110239. https://doi.org/10.1016/j.biopha.2020.110239.
Geng H, Li S, Xu M. Long noncoding rna Snhg6 functions as an oncogene in non-small cell lung cancer via modulating Ets1 signaling. Onco Targets Ther. 2020;13:921–30. https://doi.org/10.2147/ott.S235336.
Guo L, Gu J, Hou S, Liu D, Zhou M, Hua T, et al. Long non-coding Rna Dancr promotes the progression of non-small-cell lung cancer by inhibiting P21 expression. Onco Targets Ther. 2019;12:135–46. https://doi.org/10.2147/ott.S186607.
Han P, Zhao J, Gao L. Increased serum exosomal long non-coding Rna Snhg15 expression predicts poor prognosis in non-small cell lung cancer. J Clin Lab Anal. 2021;35(11).https://doi.org/10.1002/jcla.23979.
Han W, Du X, Liu M, Wang J, Sun L, Li Y. Increased expression of long non-coding rna Snhg16 correlates with tumor progression and poor prognosis in non-small cell lung cancer. Int J Biol Macromol. 2019;121:270–8. https://doi.org/10.1016/j.ijbiomac.2018.10.004.
Huang Y, Xia L, Tan X, Zhang J, Zeng W, Tan B, et al. Molecular mechanism of Lncrna Snhg12 in immune escape of non-small cell lung cancer through the Hur/Pd-L1/Usp8 Axis. Cell Mol Biol Lett. 2022;27(1):43. https://doi.org/10.1186/s11658-022-00343-7.
Huang YF, Zhang Y, Fu X. Long non-coding Rna Dancr promoted non-small cell lung cancer cells metastasis via modulating of Mir-1225-3p/Erbb2 signal. Eur Rev Med Pharmacol Sci. 2021;25(2):758–69. https://doi.org/10.26355/eurrev_202101_24637.
Jin B, Jin H, Wu HB, Xu JJ, Li B. Long non-coding rna Snhg15 promotes Cdk14 expression Via Mir-486 to accelerate non-small cell lung cancer cells progression and metastasis. J Cell Physiol. 2018;233(9):7164–72. https://doi.org/10.1002/jcp.26543.
Kang B, Qiu C, Zhang Y. The effect of Lncrna Snhg3 overexpression on lung adenocarcinoma by regulating the expression of Mir-890. J Healthc Eng. 2021;2021: 1643788. https://doi.org/10.1155/2021/1643788.
Li L, Ye D, Liu L, Li X, Liu J, Su S, et al. Long noncoding rna Snhg7 accelerates proliferation, migration and invasion of non-small cell lung cancer cells by suppressing Mir-181a-5p through Akt/Mtor signaling pathway. Cancer Manage Res. 2020;12:8303–12. https://doi.org/10.2147/cmar.S258487.
Li X, Zheng H. Lncrna Snhg1 influences cell proliferation, migration, invasion, and apoptosis of non-small cell lung cancer cells via the Mir-361-3p/Frat1 axis. Thorac Cancer. 2020;11(2):295–304. https://doi.org/10.1111/1759-7714.13256.
Li Y, Jiang L, Zhu Z, Fu B, Sun X, Jiao Y. Long noncoding rna Snhg16 regulates the growth of human lung cancer cells by modulating the expression of aldehyde dehydrogenase 2 (Aldh2). J Oncol. 2022;2022:2411642. https://doi.org/10.1155/2022/2411642.
Liang R, Xiao G, Wang M, Li X, Li Y, Hui Z, et al. Snhg6 functions as a competing endogenous Rna to regulate E2f7 expression by Sponging Mir-26a-5p in lung adenocarcinoma. Biomed Pharmacother. 2018;107:1434–46. https://doi.org/10.1016/j.biopha.2018.08.099.
Lingling J, Xiangao J, Guiqing H, Jichan S, Feifei S, Haiyan Z. Snhg20 knockdown suppresses proliferation, migration and invasion, and promotes apoptosis in non-small cell lung cancer through acting as a Mir-154 sponge. Biomed Pharmacother. 2019;112:108648. https://doi.org/10.1016/j.biopha.2019.108648.
Liu S, Yang N, Wang L, Wei B, Chen J, Gao Y. Lncrna Snhg11 promotes Lung cancer cell proliferation and migration via activation of Wnt/Β-catenin signaling pathway. J Cell Physiol. 2020;235(10):7541–53. https://doi.org/10.1002/jcp.29656.
Ma XR, Xu YL, Qian J, Wang Y. Long non-coding rna Snhg15 accelerates the progression of non-small cell lung cancer by absorbing Mir-211-3p. Eur Rev Med Pharmacol Sci. 2019;23(4):1536–44. https://doi.org/10.26355/eurrev_201902_17112.
Pang L, Cheng Y, Zou S, Song J. Long noncoding rna Snhg7 contributes to cell proliferation, migration, invasion and epithelial to mesenchymal transition in non-small cell lung cancer by regulating Mir-449a/Tgif2 Axis. Thorac Cancer. 2020;11(2):264–76. https://doi.org/10.1111/1759-7714.13245.
Shi J, Li J, Yang S, Hu X, Chen J, Feng J, et al. Lncrna Snhg3 is activated by E2f1 and promotes proliferation and migration of non-small-cell lung cancer cells through activating Tgf-Β pathway and Il-6/Jak2/Stat3 pathway. J Cell Physiol. 2020;235(3):2891–900. https://doi.org/10.1002/jcp.29194.
Wang F, Quan Q. The long non-coding rna Snhg4/Microrna-Let-7e/Kdm3a/P21 pathway is involved in the development of non-small cell lung cancer. Mol Ther Oncolytics. 2021;20:634–45. https://doi.org/10.1016/j.omto.2020.12.010.
Wang R, Chen C, Kang W, Meng G. Snhg9 was upregulated in Nsclc and Associated with ddp-resistance and poor prognosis of Nsclc patients. Am J Translational Res. 2020;12(8):4456–66.
Wang S, Jiang M. The long non-coding Rna-Dancr exerts oncogenic functions in non-small cell lung cancer Via Mir-758-3p. Biomed Pharmacother. 2018;103:94–100. https://doi.org/10.1016/j.biopha.2018.03.053.
Wang X, Gu G, Zhu H, Lu S, Abuduwaili K, Liu C. Lncrna Snhg20 promoted proliferation, invasion and inhibited cell apoptosis of lung adenocarcinoma via sponging Mir-342 and upregulating Ddx49. Thorac Cancer. 2020;11(12):3510–20. https://doi.org/10.1111/1759-7714.13693.
Wei L, Yang N, Sun L, Zhang L, Li Z, Li D, et al. Lncrna Snhg1 upregulates rock2 to reduce cisplatin sensitivity of Nsclc cells by targeting Mir-101-3p. Transl Cancer Res. 2019;8(5):2141–50. https://doi.org/10.21037/tcr.2019.09.24.
Yang X, Meng L, Zhong Y, Hu F, Wang L, Wang M. The long intergenic noncoding rna Gas5 reduces cisplatin-resistance in non-small cell lung cancer through the Mir-217/Lhpp Axis. Aging. 2021;13(2):2864–84. https://doi.org/10.18632/aging.202352.
Zhang HY, Yang W, Zheng FS, Wang YB, Lu JB. Long non-coding rna Snhg1 regulates zinc finger E-box binding homeobox 1 expression by interacting with Tap63 and promotes cell metastasis and invasion in lung squamous cell carcinoma. Biomed Pharmacother. 2017;90:650–8. https://doi.org/10.1016/j.biopha.2017.03.104.
Zhang N, Jiang W. Long non–coding rna Dancr promotes Hmga2–mediated invasion in lung adenocarcinoma cells. Oncol Rep. 2019;41(2):1083–90. https://doi.org/10.3892/or.2018.6897.
Zhang Z, Wang Y, Zhang W, Li J, Liu W, Lu W. Long non-coding rna Snhg14 exerts oncogenic functions in non-small cell lung cancer through acting as an Mir-340 sponge. Biosci Rep. 2019;39(1): BSR20180941. https://doi.org/10.1042/bsr20180941.
Zhang Z, Yan Y, Zhang B, Ma Y, Chen C, Wang C. Long non-coding rna Snhg17 promotes lung adenocarcinoma progression by targeting the Microrna-193a-5p/Neto2 Axis. Oncol Lett. 2021;22(6):818. https://doi.org/10.3892/ol.2021.13079.
Zhao L, Song X, Guo Y, Ding N, Wang T, Huang L. Long non–coding rna Snhg3 promotes the development of non–small cell lung cancer via the Mir–1343–3p/Nfix pathway. Int J Mol Med. 2021;48(2):1–2. https://doi.org/10.3892/ijmm.2021.4980.
Zhong Y, Wang J, Lv W, Xu J, Mei S, Shan A. Lncrna Ttn-As1 drives invasion and migration of lung adenocarcinoma cells via modulation of Mir-4677-3p/Zeb1 axis. J Cell Biochem. 2019;120(10):17131–41. https://doi.org/10.1002/jcb.28973.
Pan J, Fang S, Tian H, Zhou C, Zhao X, Tian H, et al. Lncrna Jpx/Mir-33a-5p/Twist1 Axis regulates tumorigenesis and metastasis of lung cancer by activating Wnt/Β-Catenin signaling. Mol Cancer. 2020;19(1):9. https://doi.org/10.1186/s12943-020-1133-9.
Wang Z, Pan L, Yu H, Wang Y. The long non-coding rna snhg5 regulates gefitinib resistance in lung adenocarcinoma cells by targetting Mir-377/Casp1 Axis. Biosci Rep. 2018;38(4): BSR20180400. https://doi.org/10.1042/bsr20180400.
Liang M, Wang L, Cao C, Song S, Wu F. Lncrna Snhg10 Is Downregulated in Non-Small Cell Lung Cancer and Predicts Poor Survival. BMC Pulm Med. 2020;20(1):273. https://doi.org/10.1186/s12890-020-01281-w.
Zhang Z, Nong L, Chen ML, Gu XL, Zhao WW, Liu MH, et al. Long Noncoding Rna Snhg10 Sponges Mir-543 to Upregulate Tumor Suppressive Sirt1 in Nonsmall Cell Lung Cancer. Cancer Biother Radiopharm. 2020;35(10):771–5. https://doi.org/10.1089/cbr.2019.3334.
Funding
Not applicable.
Author information
Authors and Affiliations
Contributions
Author ContributionsYi-Zhang and Guoyi-Shen design the project; Rong-zhi Huang and Shao-bin Yang searched databases and performed literature screen; Rong-qiang Shen and Jian-li Gao extracted and analyzed the data, analysis; Yi-Zhang and Guoyi-Shen evaluated the quality of included literature; Yi-Zhang, Guoyi-Shen, Rong-zhi Huang and Shao-bin Yang contributed to writing the manuscript. Final draft was approved by all the authors.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
No clinical patient study was conducted for this article, so no ethical statement is needed. The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Shen, Gy., Huang, Rz., Yang, Sb. et al. High SNHG expression may predict a poor lung cancer prognosis based on a meta-analysis. BMC Cancer 23, 1243 (2023). https://doi.org/10.1186/s12885-023-11706-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-023-11706-4