Abstract
Background
Cisplatin (CDDP) remains a key agent in the treatment of muscle-infiltrating bladder carcinoma (MIBC). However, a proportion of MIBC patients do not respond to chemotherapy, which may be caused by the increased repair of CDDP-induced DNA damage. The purpose of this study was to explore the prognostic value of proteins involved in nucleotide excision repair (NER) and translesion DNA synthesis (TLS) in MIBC patients.
Methods
This is a retrospective analysis of 86 MIBC patients. The XPA, XPF, XPG, ERCC1, POLI, POLH and REV3L proteins were stained in primary bladder tumors and their levels were analyzed both in the total cohort and in a subgroup with metastatic urothelial carcinoma (mUC) that received gemcitabine and CDDP as a first-line therapy. Both cohorts were divided by percentage of cancer cells stained positive for each protein into subgroups with high and low expression. In the same manner, the combined expression of NER (XPA + ERCC1 + XPF + XPG) and TLS (POLI + POLH + REV3L), as the whole pathways, was analyzed.
Results
Mortality was 89.5% at the median follow-up of 120.2 months. In the total cohort, patients with tumors stained positive for XPA, XPG and POLI had significantly worse overall survival (OS) compared to those with negative staining [hazard ratio (HR) = 0.60, 0.62 and 0.53, respectively]. Both XPG and POLI were independent prognostic factors in multivariate analyses (MVA). In addition, an increase in NER and TLS pathway expression was significantly associated with worse OS in the total cohort (HR = 0.54 and 0.60, respectively). In the mUC subgroup, high POLI expression was associated with significant deterioration of OS (HR = 0.56) in univariate analyses, and its independent prognostic value was shown in MVA.
Conclusions
Our study showed significant correlations between the tumor expression of XPG and POLI, as well as NER and TLS as the whole pathways, and inferior OS. Hence, they could constitute prognostic biomarkers and potentially promising therapeutic targets in MIBC. However, a prospective trial is required for further validation, thereby overcoming the limitations of this study.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Muscle-infiltrating bladder carcinoma (MIBC) remains a lethal disease in many cases, with a five-year overall survival (OS) of approximately 50% [1, 2]. For locally advanced or metastatic forms of this disease, the OS is reduced to 10-15% [3], highlighting the need for new therapies. Irrespective of the disease stage, cisplatin (CDDP)-based chemotherapy, mostly in combination with gemcitabine (GC), is the standard of care in both neoadjuvant and metastatic settings [4, 5]. As the main pharmacological target of CDDP is DNA, DNA damage represents the key factor determining CDDP toxicity. CDDP-induced DNA damage, mainly CDDP-DNA intra-strand cross-links (IaCLs) and inter-strand cross-links (ICLs), activates DNA damage response (DDR) pathways involving the complex coordination of several DNA repair mechanisms, including nucleotide excision repair (NER) and translesion DNA synthesis (TLS) [6, 7]. Schematic representation of these pathways is shown in Fig. 1.
Simplified representation of NER and TLS pathways. Only proteins involved in this study are highlighted by their names. In NER (left panel), XPA is involved in DNA damage recognition, while XPF-ERCC1 (XPF and ERCC1 form a heterodimeric complex) and XPG are nucleases responsible for dual incision nearby the lesion [8, 9]. In TLS (right panel), POLH and POLI DNA polymerases are considered the primary inserter DNA polymerases because they incorporate one or a few nucleotides opposite the damaged site, while Pol ζ through its REV3L catalytic subunit mediates extension step from the improperly paired DNA [10, 11]. Abbreviations: NER, nucleotide excision repair; TLS, translesion DNA synthesis; XPA, xeroderma pigmentosum group A protein; XPF, xeroderma pigmentosum group F protein; ERCC1, excision repair cross-complementation group 1; XPG, xeroderma pigmentosum group G protein; POLH, DNA polymerase H; POLI, DNA polymerase I; REV3L, REV3-like DNA-directed polymerase ζ catalytic subunit
NER contributes to the repair of CDDP-induced DNA damage by mediating lesion recognition and incision [8, 9]. Central to DNA lesion recognition is the xeroderma pigmentosum complementation group A (XPA) protein, which also functions in DNA lesion verification and the assembly of NER incision complexes [12, 13, 14]. Incision during NER is mediated by the two structure-specific nucleases, xeroderma pigmentosum complementation group F and G (XPF and XPG, respectively), the former forming a complex with the excision repair cross-complementation group 1 (ERCC1) protein. ERCC1 per se is catalytically inactive and serves to target XPF to different substrates, thereby regulating its availability and activity [15,16,17,18]. XPF and XPG cleave damaged DNA 5' and 3' to the site of the lesion, respectively [19], with XPG also having a non-enzymatic role in 5' incision by the XPF-ERCC1 heterodimer [20]. Importantly, this heterodimer is recruited to the DNA lesion by the XPA protein through an XPA-ERCC1 interaction [21].
In cell, CDDP-induced DNA damage can disrupt DNA replication by arresting replicative DNA polymerases. Stalled DNA replication forks can collapse into DNA double-strand breaks (DSBs), a potentially lethal event. To prevent such scenario, CDDP-induced DNA damage can be bypassed by TLS [22, 23]. TLS requires specialized polymerases of the Y and B families with a broad catalytic site lacking proofreading activity, enabling them to bypass DNA lesions [24]. DNA polymerase eta (POLH, Pol η) and iota (POLI, Pol ι) belong to the Y family, while REV3L [REV3-like DNA-directed polymerase zeta (Pol ζ) catalytic subunit] belongs to the B family of TLS DNA polymerases. Biochemically, POLH can efficiently bypass a wide variety of DNA lesions and has been shown to incorporate nucleotides opposite to CDDP-induced IaCLs with high catalytic efficiency [25, 26, 27, 28, 29, 30]. POLI is highly error-prone when replicating undamaged DNA, exhibiting an unusual misincorporation spectrum [31]. It can also facilitate mutagenic replication past a wide range of DNA lesions, including minor groove-adducted purines, bulky major groove-adducted purines, and other types of DNA damage [32]. REV3L has a pivotal and irreplaceable role in TLS across several DNA lesions [24, 30], including CDDP-DNA lesions [30].
Both NER and TLS have been shown to play a role in acquired resistance to chemotherapeutic agents, including CDDP. NER directly repairs DNA damage caused by this drug [33], while TLS tolerates CDDP-induced DNA damage by mediating replication of CDDP-damaged DNA templates. The importance of these two pathways in development of CDDP resistance is highlighted by the fact that CDDP-treated cells defective in NER and TLS accumulate DNA damage, unfilled post-replicative gaps, collapsed replication forks and DSBs, all potentially lethal events [34, 35]. Altered or aberrant expression of many genes encoding the NER and TLS factors has been found in various types of cancers, where it represents a valuable prognostic and predictive biomarker [24, 36, 37, 38, 39].
Recently, we have examined the expression of three NER proteins (ERCC1, XPF and XPA) in germ cell tumor (GCT) patients and correlated it with their prognosis [40]. We showed that GCT patients with lower XPA expression had significantly better OS than patients with higher expression. In addition, XPA expression was increased in the non-seminomatous histological subtype, in patients with poor prognosis according to International Germ Cell Cancer Collaborative Group classification, in patients with increasing S stage, as well as in patients with lung, liver and non-pulmonary visceral metastases. Therefore, we proposed that XPA expression, and NER as a whole, represented an additional independent prognostic biomarker for stratifying GCT patients, allowing for improvements in decision-making on treatment for those at high risk of refractoriness or relapse. In addition, we suggested that XPA could serve as a novel therapeutic target in GCTs [40].
The purpose of the present retrospective study was to extend our previous results and to review the prognostic power of NER in another genitourinary malignancy, MIBC. This malignancy was of particular interest, as it is known to present with a higher level of DNA alterations than any other adult cancer [41, 42]. Like other carcinogen-induced tumors, the somatic mutation burden in MIBC is quite high, with a median of nearly eight mutations per megabase [41]. Therefore, DNA repair pathways are believed to have a significant impact on this malignity, as documented by the genomic landscape data [41, 42, 43]. Importantly, these pathways not only play a role in process of carcinogenesis, but they also govern chemotherapy response. In line with this, numerous mutations in DNA repair pathways have been found in MIBC. Interestingly, unlike some other cancers, in which most mutations affecting DNA repair involve the homologous recombination pathway, MIBC tumors have alterations across several DNA repair pathways, including higher rates of NER alterations than other tumor types [43]. Therefore, we examined the expression level of the key NER factors, ERCC1, XPA, XPF and XPG, in chemotherapy-naïve primary bladder tumors of patients with urothelial MIBC and in cohort of patients with metastatic urothelial carcinoma (mUC) treated by GC in first line. To examine the role of TLS in CDDP response, we also included three TLS factors, POLH, POLI and REV3L, in this study, strengthening its novelty.
In summary, high proportion of patients with urothelial MIBC do not respond to CDDP-based chemotherapy. This could be caused by an aberrant repair of induced DNA damage. Should NER and TLS pathways show significant prognostic power, they could become biomarkers for stratifying patients with MIBC to optimize their management. Such findings could initiate testing the NER and TLS inhibitors, the biology of which has already been investigated for a while, for use in the clinical trials. It is believed that NER and TLS inhibitors could be successful in overcoming the resistance to CDDP.
Methods
Patients
Patients ≥ 18 years of age with MIBC of urothelial origin including its variants and an Eastern Cooperative Oncology Group (ECOG) performance status of 0 to 2 with available formalin-fixed, paraffin-embedded (FFPE) tissue were eligible for this study. Subjects with chemotherapy prior to transurethral resection of bladder tumor (TUR BT) due to MIBC or second primary malignancy were excluded. All candidates were selected from a local database of the National Cancer Institute (NCI), Bratislava, Slovakia.
Study design
This is a retrospective study on biomarkers, reflecting single-center, real-world experience, which provides the correlation between DNA damage repair/tolerance factors and survival. All patients included in the sub-group analysis were treated in metastatic setting with at least one cycle of GC, CDDP 70 mg/m2 on day 1 with gemcitabine 1,000 mg/m2 days 1 and 8, new cycle day 22. The flowchart showing study design is depicted in Fig. 2.
Study design flowchart. The database of the National Cancer Institute in Bratislava, Slovakia was used for selection of patients with MIBC. The total study population consisted of patients with MIBC who met the inclusion and exclusion criteria. The subgroup with mUC, treated with GC chemotherapy, comprised the patients with de novo mUC and progressive mUC. Abbreviations: I/E, inclusion/exclusion; MIBC, muscle-infiltrating bladder cancer; N, number of patients; GC, gemcitabine + cisplatin; mUC; metastatic urothelial carcinoma
This study was approved by the Ethical Committees of St. Elisabeth Cancer Institute (SECI), Bratislava, Slovakia, on February 3, 2022 (6-2022/EK), and NCI on April 24, 2023 (UC-SK002/2023).
Study assessments and end points
FFPE samples from pre-treatment primary tumors of individual cases selected from the archive of the Department of Pathology of the SECI were stained with hematoxylin and eosin; and reviewed by two pathologists (KK and CB). One representative sample was selected from each case and subjected to immunohistochemical (IHC) analysis for the following DNA damage repair and tolerance factors: ERCC1, XPA, XPF, XPG, POLH, POLI and REV3L. Basic characteristics of the primary antibodies are shown in Table 1.
Slides for IHC analysis were deparaffinized and rehydrated. Automated antigen staining was performed in Autosteiner Link 48 (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). EnVisionTM FLEX/HRP, High pH kit (K8000, Dako; Agilent Technologies, Inc., Santa Clara, CA, USA) was used for visualization of antibodies according to manufacturer's instructions. Antigen retrieval was induced by incubation of the sections with EnVisionTM FLEX Target Retrieval Solution High pH (pH 9.0) for 20 min at 97-98°C in PT Link (Dako; Agilent Technologies, Inc., Santa Clara, CA, USA). Endogenous peroxidases were blocked with EnVision FLEX Peroxidase-Blocking Reagent.
Primary mouse and rabbit antibodies were diluted in Antibody Diluent with Background Reducing Components (S3022, Dako Agilent Technologies, Inc., Santa Clara, CA, USA), as shown in Table 1, and slides were incubated with primary antibodies for 20 or 30 min. Subsequently, sections were incubated with Mouse or Rabbit Linker in the case of primary antibodies against ERCC1, XPA and XPG.
After washing with EnVision FLEX Wash Buffer, the sections were incubated with secondary antibody EnVision FLEX/HRP for 20 min. Visualization of the antibody complexes was achieved by incubation with EnVision FLEX buffered substrate and 3,3'-diaminobenzidine (DAB) chromogen for 10 min. EnVision FLEX Hematoxylin was used for counterstaining. After drying, the sections were mounted with solakryl.
Tissue samples of tonsil were used as positive control for ERCC1, XPF and XPG, skin samples for XPA and POLI, and liver samples for POLH and REV3L.
IHC-stained slides were evaluated by 4 independent observers (KK, CB, AH and PM) and a consensus opinion was taken for equivocal cases. Only nuclear positivity was considered, and the expression of individual markers was evaluated semi-quantitatively, i.e., by the percentage expression of positively stained cells. The examined cases were divided into 4 categories: I – 0-25% of positive cells, II – 26-50% of positive cells, III – 51-75% of positive cells and IV – more than 75% of positive cells. Subsequently, the cases were divided into two groups according to the percentage of positive cells, one group with low expression (up to 75% of positive cells) was considered as negative and the second group with high expression (> 75% of positive cells) was regarded as positive. Moreover, XPG was evaluated by using a cut-off of 50% positive cells for the low proportion of patients in the positive group.
The combined NER and TLS expression levels were scored by (weighted) histoscore (HS), accounting for both the extent of cell staining and the intensity. The percentage of positively stained cells was calculated and the average intensity (percentage) of positively stained cells was given a score from 0 to 4 (0 = no staining1 = 0-25%; 2 = 26-50%; 3 = 51-75%; 4 = 76-100%). The HS for individual DNA repair pathways (NER and TLS, respectively) was then calculated as a sum of individual protein scores and median value for each pathway was used as a cut-off value (13 NER; 8 TLS). The expression level for each DNA repair pathway was graded as “low” (< 13, NER; < 8, TLS) and “high” (≥ 13, NER; ≥ 8, TLS).
All data were entered by the investigators into electronic data files, and their accuracy was validated for each patient by an independent investigator. For the total cohort, OS was calculated from the day of TUR BT with muscle-infiltrating UC histology until the last follow-up or mortality for any reason. In the sub-group of mUC patients, progression-free survival (PFS) was calculated from day 1 of the first GC cycle until disease progression, last follow-up, or mortality for any reason. OS was calculated from day 1 of the first GC cycle until mortality for any reason or last follow-up. Objective response (OR) to chemotherapy was evaluated by the Response Evaluation Criteria in Solid Tumors (RECIST) criteria, version 1.1.
Statistical analysis
All statistical analyses were performed using NCSS 23 Statistical Software, version 23.0.1, Kaysville, Utah, USA. Data were summarized by the median ± standard deviation (SD) or range for continuous variables. For the median follow-up calculation, we applied the reversed Kaplan-Meier estimator. The Kaplan-Meier method was used to estimate PFS and OS. The differences between the groups were determined by the log rank test. Estimated hazard ratios (HR), 95% confidence intervals (CI), and p values were calculated from the Cox proportional hazard regression models.
Multivariate analysis (MVA) was carried out by the Cox regression model. In the total cohort, MVA included disease distribution (locally and locally advanced v metastatic disease) and positively correlated experimental factors in univariate analyses. In the mUC sub-group, MVA included a performance status by ECOG, metastatic status by Bajorin [44], and the experimental factors positively correlated in univariate analyses.
The association between biomarker tumor staining and OR was carried out by the one-way analysis of variance (ANOVA). Two-sided p value < 0.05 was considered statistically significant.
Results
Patient selection
From the local database, a total of 86 patients with MIBC with complete clinical data and available tumor tissues showing smooth bladder muscle infiltration with urothelial carcinoma (UC) were selected for this study.
Patient, tumor and treatment characteristics
All patients had high-grade urothelial MIBC, including 11 subjects (12.8%) with variants (3 squamous-cell, 2 sarcomatous, 2 nested, 2 micro-papillary, 1 glandular and 1 clear-cell). Further characteristics are shown in Table 2.
Correlations between the expression of selected NER and TLS factors and survival
At data cut-off (June 26, 2023), the median follow-up was 120.2 months (95% CI :101.5-139.0 months). OS was significantly worse in patients with metastatic disease compared to local/locally advanced disease [median OS (mOS) 14.4 vs 29.5 months, HR = 0.50, 95% CI: 0.28-0.89, p = 0.0032].
The total cohort
In the total cohort, 58 patients had primary bladder tumor stained positive for ERCC1, 74 for XPF, 8 for XPG (> 75%), 11 for POLH, and 46 for REV3L. mOS was shorter in the ERCC1 positive compared to negative subgroup (18.6 vs 29.0 months, HR = 0.69, 95% CI: 0.44-1.09, p = 0.1318), XPF positive vs negative (18.9 vs 29.0 months, HR = 0.73, 95% CI: 0.40-1.31, p = 0.34), XPG positive (76% to 100%) vs negative (15.7 vs 21.1 months, HR = 0.50, 95% CI: 0.19-1.33, p = 0.0541, Fig. 3B), POLH positive vs negative (20.0 vs 21.1 months, HR = 0.89, 95% CI: 0.44-1.78, p = 0.7256), and longer in REV3L positive vs negative (21.1 vs 17.3 months, HR = 0.91, 95% CI: 0.58-1.42, p = 0.6618) (Table 3).
Kaplan-Meier estimates of OS for XPA (A), XPG (B and C) and POLI (D) in MIBC patients. The XPG was evaluated by two different cut-off values (75% and 50%; B and C, respectively) for the low proportion of patients in the positive group where high expression was regarded as > 75% of positive cells. Abbreviations: OS, overall survival; XPA, xeroderma pigmentosum group A protein; XPG, xeroderma pigmentosum group G protein; POLI, DNA polymerase I; MIBC, muscle-infiltrating bladder cancer; N, number of patients; HR, hazard ratio; CI, confidence interval; p, probability
Positive staining in primary bladder tumors for XPA, XPG (> 50%) and POLI was detected in 59, 24 and 25 patients, respectively. Significantly shorter mOS was experienced in XPA positive vs negative populations (18.7 vs 30.5 months, HR = 0.60, 95% CI: 0.38-0.94, p = 0.0354, Fig. 3A), XPG positive vs negative (≤ 50%) subgroup (18.7 vs 21.1 months, HR = 0.62, 95% CI: 0.36-1.07, p = 0.0465, Fig. 3C), and POLI positive vs negative populations (15.7 vs 23.0 months, HR = 0.53, 95% CI: 0.30-0.93; p = 0.0066; Fig. 3D). Representative images of positive and negative staining for these proteins are provided in Fig. 4.
IHC detection of XPA, XPG and POLI in primary tumor cells of high-grade urothelial MIBC. HE (A), XPA (B), XPG (C) and POLI (D) staining. Upper panels show representative images of positive XPA, XPG and POLI staining, while the lower panels illustrate negative staining for these proteins. Magnification 400x. Scale applies for all images. Abbreviations: IHC, immunohistochemical; XPA, xeroderma pigmentosum group A protein; XPG, xeroderma pigmentosum group G protein; POLI, DNA polymerase I; MIBC, muscle-infiltrating bladder cancer; HE, hematoxylin and eosin
In multivariate analysis, the stage of disease (metastatic vs local/locally advanced), XPG (> 75%) and POLI (> 50%) positive staining were identified as independent predictive factors for OS [risk ratio (RR) = 1.99, 95% confidence limit (CL): 1.18-3.36, p = 0.0103; RR = 1.67, 95% CL:1.01-2.78, p = 0.048 and RR = 1.95, 95% CL: 1.17-3.26, p = 0.0103, respectively).
In addition to correlation of expression of individual NER and TLS proteins with prognosis of patients with MIBC, we analyzed the roles of these two DNA damage repair/tolerance pathways in predicting patient survival. This was achieved by combining the expression of their individual factors (for more details, see the Methods section). Interestingly, an increase in NER expression was significantly associated with worse OS (HR = 0.54, 95% CI: 0.34-0.84, p = 0.0105) (Fig. 5A). Similarly, higher levels of TLS led to shorter OS (HR = 0.60, 95% CI: 0.38-0.94, p = 0.0224) (Fig. 5B).
Kaplan-Meier estimates of OS for NER (A) and TLS (B) in MIBC patients. The histoscore for NER and TLS pathways was calculated as a sum of individual protein scores. Based on median value for each pathway (13 and 8 for NER and TLS, respectively), the expression level for each DNA repair pathway was graded as low and high. Abbreviations: OS, overall survival; NER, nucleotide excision repair; TLS, translesion DNA synthesis; MIBC, muscle-infiltrating bladder cancer; N, number of patients; HR, hazard ratio; CI, confidence interval; p, probability
In multivariate analysis, the stage of disease (metastatic vs local/locally advanced), but not NER and TLS, was identified as independent predictive factor for OS (RR = 1.84, 95% CL: 1.09-3.11, p = 0.0233; RR = 1.67, 95% CL: 0.99-2.83, p = 0.0548 and RR = 1.26, 95% CL: 0.77-2.06, p = 0.3670, respectively).
mUC cohort
In the mUC cohort treated with first-line GC, 17 patients were identified as ECOG 2, while a positive metastasis status, by Bajorin criteria, was identified in 43 subjects. mOS was significantly worse in patients with ECOG 2 compared to ECOG 0 to 1 (4.4 vs 14.0 months, HR = 0.20, 95% CI: 0.07-0.52, p < 0.0001) and in patients with positive vs negative metastasis status (9.3 vs 19.8 months, HR = 0.43, 95% CI: 0.26-0.73, p = 0.0025) (Table 4).
The bladder tumors staining positive for ERCC1, XPA, XPF, XPG (< 75% and < 50%), POLH, POLI and REV3L were found in 40, 43, 52, 8 and 20, 8, 18, and 36 patients, respectively. mOS was shorter in ERCC1 positive vs negative (10.2 vs 14.0 months, HR = 0.73, 95% CI: 0.43-1.24, p = 0.2573), XPA positive vs negative (10.5 vs 12.3 months, HR = 0.84, 95% CI: 0.48-1.46, p = 0.5377), XPF positive vs negative (10.9 vs 15.2 months, HR = 0.81, 95% CI: 0.39-1.69, p = 0.5987), XPG positive (51 to 100%) vs negative (10.2 vs 11.7 months, HR = 0.73, 95% CI: 0.41-1.30, p = 0.2391), and longer in POLH positive vs negative (11.7 vs 10.9 months, HR = 0.67, 95% CI: 0.34-1.32, p = 0.3061) and REV3L positive vs negative subgroup (11.7 vs 9.3 months, HR = 0.96, 95% CI: 0.57-1.64, p = 0.8911) (Table 4).
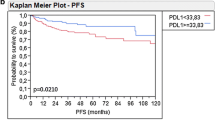
Significantly worse mOS was detected in POLI positive vs negative population (9.3 vs 12.6 months, HR = 0.56, 95% CI: 0.30-1.07, p = 0.0347) (Fig. 6) and XPG positive (≤ 75%) vs negative (8.4 vs 11.7 months, HR = 0.48, 95% CI: 0.18-1.31, p = 0.0478); however, the number of subjects in the XPG negative sub-group was too low (Table 4). In contrast to total population, neither NER nor TLS combined expression was significantly associated with OS in the mUC cohort (HR = 0.70, 95% CI: 0.41-1.22, p = 0.2379 and HR = 1.20, 95% CI: 0.70-2.05, p = 0.4976, respectively).
Multivariate analysis found performance status by ECOG 2, positive metastasis status by Bajorin and POLI positive (76% to 100%) staining to be the independent predictive factors for OS (RR = 6.51, 95% CL: 3.24-13.08, p < 0.0001; RR = 2.13, 95% CL: 1.11-4.08, p = 0.023; RR = 2.14, 95% CL: 1.16-3.95, p = 0.0156, respectively).
PFS was significantly worse in patients with ECOG 2 compared to ECOG 0 to 1 [median PFS (mPFS) 2.4 vs 10.0 months, HR = 0.19, 95% CI: 0.07-0.52, p < 0.0001] and with positive vs negative visceral metastasis status by Bajorin (mPFS 7.0 vs 16.1 months, HR = 0.42, 95% CI: 0.25-0.70, p = 0.0015) (Table 5).
mPFS was shorter in tumors stained ERCC1 positive vs negative (7.3 vs 8.4 months, HR = 0.89, 95% CI: 0.51-1.53, p = 0.6706), XPA positive vs negative (7.3 vs 8.9 months, HR = 0.99, 95% CI: 0.55-1.77, p = 0.9723), XPG positive (51 to 100%) vs negative (7.0 vs 8.1 months, HR = 0.68, 95% CI: 0.38-1.22; p = 0.1532), POLI positive vs negative (7.0 vs 8.1 months, HR = 0.63, 95% CI: 0.34-1.18, p = 0.0970), and longer in XPF positive vs negative (8.0 vs 4.7 months; HR = 0.82, 95% CI: 0.35-1.93, p = 0.6205), POLH positive vs negative (8.4 vs 7.8 months, HR = 0.67, 95% CI: 0.34-1.32, p = 0.3079) and REV3L positive vs negative subgroup (8.0 vs 7.3 months, HR = 0.89, 95% CI: 0.52-1.52, p = 0.664) (Table 5).
Significantly worse PFS was detected in XPG positive (75 to 100%) vs negative subgroup (median PFS 6.8 vs 8.1 months, HR = 0.47, 95% CI: 0.17-1.28; p = 0.0347); however, more patients with negative XPG staining would need to be analyzed to confirm this trend (Table 5).
Correlations of OR to GC with expression of selected NER and TLS factors in mUC sub-group
Patients that did not achieve OR to first-line GC compared to those who responded had identical, or very similar, IHC staining for ERCC1 (median 4.0 ± 0.8 vs 4.0 ± 0.8, p < 0.5659), XPA (median 4.0 ± 0.6 vs 4.0 ± 1.1, p < 0.4809), XPF (median 4.0 ± 0.3 vs 4.0 ± 0.7, p < 0.8810), XPG (median 2.0 ± 1.3 vs 2.0 ± 0.9, p < 0.3590), POLH (median 1.5 ± 1.3 vs 2.0 ± 1.4, p < 0.5567), POLI (median 2.5 ± 1.3 vs 3.0 ± 1.2, p < 0.3185) and REV3L (median 4.0 ± 0.9 vs 4.0 ± 1.1, p < 0.7790).
In summary, our data show that positive staining for XPA, XPG, and POLI in tumors significantly correlates with shorter OS in the total cohort of patients with MIBC. Interestingly, expression of NER and TLS DNA damage repair/tolerance pathways (assessed by the combined expression of XPA + ERCC1 + XPF + XPG and POLI + POLH + REV3L, respectively) also inversely correlates with OS. In the multivariate analyses, the stage of disease, as well as XPG and POLI positive staining were found to be the independent predictive factors for OS of patients with MIBC. In the mUC cohort, significantly worse OS was revealed in POLI and XPG, and PFS in XPG positive population, although XPG findings may be inconclusive due to a low number of patients in the XPG negative sub-group. Neither NER, nor TLS expression was significantly associated with OS. In the multivariate analyses, performance status, metastasis status by Bajorin and POLI positive staining were shown to have an independent predictive value for OS.
Discussion
Unfavorable response to CDDP represents a serious problem for cancer patient management as it often leads to poor prognosis. CDDP manifests its toxic effects through induction of DNA damage, and therefore cellular response to this drug strongly involves DNA damage repair/tolerance pathways. Central to the repair of CDDP-induced DNA lesions is the Fanconi anemia pathway, a highly complex DDR pathway that coordinates the action of a plethora of DNA damage repair/tolerance mechanisms, with NER and TLS being of primary significance. In addition to CDDP, these two pathways are known to remove/tolerate DNA adducts induced by many other anti-cancer chemotherapeutic agents, highlighting their clinical importance.
Previously, we have correlated expression of the three NER proteins, ERCC1, XPF and XPA, with prognosis of GCT patients. We demonstrated that XPA expression was higher in patients with more advanced disease and poor prognostic features. We proposed that the expression level of XPA, and NER as an entire pathway, represent additional prognostic biomarkers for stratifying GCT patients to optimize their disease management [40]. The purpose of the present study was to extend our previous work by studying another genitourinary malignancy, MIBC, and a higher number of DNA damage repair/tolerance factors. Here, we determined the expression levels of the key NER and TLS factors, ERCC1, XPF, XPG, XPA, POLH, POLI and REV3L, and correlated them with prognosis of MIBC patients. Both the total cohort and a cohort of subjects with mUC that received GC as a first-line therapy were examined.
Of all the DNA damage repair/tolerance factors involved in the present study, only ERCC1 has been examined for its prognostic value in bladder cancer (BC) to date. The first report to show that ERCC1 expression at the mRNA level inversely correlates with OS in advanced BC patients treated with CDDP-based regimens was described by Bellmunt et al. [45]. This association was later validated at the protein level in populations with advanced or recurrent UC [46] and mUC [47]. However, other studies failed to show a predictive value for ERCC1 expression in chemotherapy-naïve primary bladder tumor specimens of metastatic BC patients treated with GC chemotherapy [48] or specimens obtained from cystectomy or nephroureterectomy of advanced UC patients treated by first-line GC combination [49]. ERCC1 expression also predicted PFS in a locally advanced BC cohort treated by CDDP-based regimens in adjuvant setting [50]. High ERCC1 positivity, defined as HS calculated by multiplying IHC nuclear staining intensity by percentage of positive cells, had a negative prognostic significance in patients observed after radical cystectomy due to UC and its variants. In the sub-group treated by adjuvant GC-based chemotherapy, high ERCC1 HS was associated with shorter OS [51]. Furthermore, urothelial MIBC patients following radical cystectomy with ERCC1 positive specimens had significantly better disease-free and cancer-specific survivals than those with ERCC1 negative tumors. ERCC1 status itself did not affect the outcome of adjuvant CDDP-based chemotherapy [52]. ERCC1 positivity, defined as weak to strong staining of the entire nucleus in >10% of tumor cells, was a predictive factor for shorter survival in patients with predominantly urothelial bladder carcinoma treated in neoadjuvant setting with CDDP-based regimens, followed by radical cystectomy [53]. ERCC1 nuclear staining correlated with the efficacy of chemoradiation therapy for MIBC in a small cohort [54]. ERCC1 positive locally advanced MIBC patients, who received CDDP-based chemoradiotherapy following TUR BT, exhibited significantly improved disease-specific survival rates [55]. These studies suggest that the ERCC1 expression could represent a potential prognostic biomarker for UC, although its status may have different prognostic and predictive implications across clinical contexts. Our results show that ERCC1 would not be a suitable prognosticator for OS and PFS neither in the overall urothelial MIBC patient cohort, nor in the mUC subgroup treated with GC in first-line setting.
Interestingly, examination of prognostic relationship of NER and pyrimidine synthesis factors in urothelial bladder carcinoma showed that CAD/POLD2 gene expression was associated with poor OS and CDDP-based chemotherapy resistance [56]. The CAD gene encodes a trifunctional protein which is associated with the enzymatic activities of the first 3 enzymes in de novo pyrimidine biosynthesis [57]. Therefore, this protein represents a target for treating tumors, as its role in pyrimidine synthesis can be manipulated to destroy cancer cells that are generally in a high need of nucleotides due to intensive proliferation [58]. POLD2 is an indispensable subunit of the DNA polymerase delta (POLD, Pol δ) complex, the major DNA replication enzyme in eukaryotic cells that is also involved in many forms of DNA repair processes, including NER [59]. Notably, POLD2 was shown to associate with the IaCLs bypass process [60, 61]. Simultaneously increased expression of CAD and POLD2 could lead to bypassing of CDDP-induced DNA damage by bladder UC cells and establishment of the chemo-resistant phenotype, leading to poor OS. In a similar manner, poor prognosis of MIBC patients with high POLI expression observed here might result from a similar process, as both POLD2 and POLI contribute to TLS in bypassing IaCLs. In this regard, it would be interesting to find out whether CAD is co-expressed with POLI.
Similarly to ERCC1 expression, we demonstrate that XPF, POLH and REV3L expressions were not associated with OS and/or PFS in the total MIBC population and the mUC sub-population treated with GC in first-line setting. In contrast, XPA, XPG and POLI expression inversely correlated with OS in the total population and, in multivariate analysis, XPG and POLI emerged as independent factors of survival. Moreover, POLI expression was associated with OS, but not PFS, in the sub-group of mUC receiving first-line systemic therapy. In the mUC cohort, POLI represented an independent predictor of OS, as shown by multivariate analysis. This is the first study to suggest that XPG and POLI are prognostic factors for MIBC, with POLI also being a prognostic factor for mUC subgroup.
Alongside the analysis of individual proteins, the expression of whole NER and TLS pathways was also correlated with prognosis of MIBC patients. Similarly to GCTs [40], expression of NER is a prognostic factor in MIBC. In MIBC, however, TLS also acts as valuable prognostic factor. Worse prognosis for cancer patients with high expression of NER and TLS pathways signifies that the efficacy of CDDP-combined chemotherapy could, in general, be potentiated by inhibition of these pathways. Inhibitors of these DNA damage repair/tolerance pathways could improve prognosis for MIBC patients. Importantly, great progress has been made in the development of tools to inhibit NER expression or activity, mainly through XPA component and its essential interaction with ERCC1 [62, 63, 64, 65]. Of the agents designed to target TLS, the most promising are small molecule inhibitors targeting the REV7-binding surface of the REV1 C-terminal domain (CTD), which is responsible for the recruitment of TLS polymerases POLK, POLI, POLH and REV3L through interaction with REV7. Recently, a specific small molecule inhibitor inducing dimerization of the REV1 CTD at its REV7-binding surface and blocking the REV1-REV7 interaction, was reported. Importantly, its co-administration with CDDP lead to significantly stronger inhibition of drug-resistant tumors in mice [66].
Single nucleotide polymorphisms (SNPs) in numerous NER genes have been found in MIBC. Based on their germline or somatic origin, they have been associated with the risk of developing disease, its stage and grade, progression and reoccurrence, response to chemotherapy, and patient survival [41, 67, 68, 69, 70]. Regarding treatment response, it has been hypothesized that suboptimal levels of NER in the MIBC tumors might lead to decreased removal of DNA lesions induced by CDDP, and thus increased clinical response and survival after chemotherapy [69, 71]. Different levels of NER might thus explain interindividual differences among MIBC patients in response to, and prognosis after, chemotherapy. Such levels can also be determined by SNPs, as already shown for certain NER factors [72, 73]. Interestingly, to predict response to chemotherapy, a model using SNPs rather than clinical characteristics per se has been suggested [74]. This model accommodates the combined effect of multiple SNPs that is more beneficial in modulating NER capacity than the investigation of single SNP in single gene [73]. Therefore, a whole pathway-based approach is preferred to study the effects of SNPs on clinical outcomes [70]. One can expect that SNPs are, at least in part, responsible for the different levels of individual NER, and potentially TLS factors, including the whole pathway activity observed in this study. Such variant genotypes, when mutually combined with each other as well as with well-known cases from previous studies, could constitute a platform for improving current diagnostic (germline origin) and treatment (somatic origin) protocols, the latter potentially forming the basis for more directed, personalized medicine.
We are aware of limitations to our study that include its retrospective nature and single-center setting; however, no prospective trials covering this subject have been published to date. Hence, our future tasks will be to validate our biomarkers prospectively in a larger sample size (once confirmed, they can become candidate targets for pharmaceutical intervention); and to disclose possible association between the levels of NER and TLS factors and SNPs in the corresponding genes. As several NER inhibitors are in development, their testing in BC cell lines, animal models, and finally MIBC patients could represent the future directions.
Conclusions
In this study, we have revealed a significant deterioration in OS of MIBC patients with high expression of the XPA, XPG and POLI proteins. High POLI expression was similarly associated with worse survival in mUC sub-group. In addition, we demonstrated that NER and TLS as whole pathways have prognostic values in the total MIBC population. Based on our results, we propose that XPA, XPG and POLI expression individually, and, by inference, NER and TLS, represent potential prognostic biomarkers for stratifying MIBC patients to optimize disease management. We believe that MIBC is an ideal model for therapies targeting these two DNA damage repair/tolerance pathways with a view of overcoming resistance to CDDP and, therefore, improving prognosis of patients with this malignancy. Further research is, however, urgently needed to validate these findings and to overcome limitations of the present study. Once validated and tested in clinical trials, integration of NER and TLS inhibitors, which have been under thorough investigation for the past few years, into practice is highly advised. We are convinced that NER and TLS inhibitors have potential to improve MIBC patient outcomes in future.
Availability of data and materials
All data generated or analyzed during this study are included in the published article.
Abbreviations
- BC:
-
Bladder cancer
- CDDP:
-
Cisplatin
- CI:
-
Confidence interval
- CL:
-
Confidence limit
- CTD:
-
C-terminal domain
- DDR:
-
DNA damage response
- ECOG:
-
Eastern Cooperative Oncology Group
- ERCC1:
-
Excision repair cross-complementation group 1
- FFPE:
-
Formalin-fixed, paraffin-embedded
- GC:
-
Gemcitabine + cisplatin
- GCT:
-
Germ cell tumor
- HE:
-
Hematoxylin and eosin
- HS:
-
Histoscore
- HR:
-
Hazard ratio
- IaCLs:
-
Intra-strand cross-links
- ICLs:
-
Inter-strand cross-links
- IHC:
-
Immunohistochemical
- MIBC:
-
Muscle-infiltrating bladder carcinoma
- mUC:
-
Metastatic urothelial carcinoma
- N:
-
Number of patients
- NER:
-
Nucleotide excision repair
- OR:
-
Objective response
- OS:
-
Overall survival
- P:
-
Probability
- PFS:
-
Progression-free survival
- POLD:
-
DNA polymerase D
- POLI:
-
DNA polymerase I
- POLH:
-
DNA polymerase H
- REV3L:
-
REV3-like DNA-directed polymerase ζ catalytic subunit
- SNPs:
-
Single nucleotide polymorphisms
- TLS:
-
Translesion DNA synthesis
- TUR BT:
-
Transurethral resection of bladder tumor
- UC:
-
Urothelial carcinoma
- XPA:
-
Xeroderma pigmentosum group A protein
- XPF:
-
Xeroderma pigmentosum group F protein
- XPG:
-
Xeroderma pigmentosum group G protein
References
Catto JW, Yates DR, Rehman I, Azzouzi AR, Patterson J, Sibony M, Cussenot O, Hamdy FC. Behavior of urothelial carcinoma with respect to anatomical location. J Urol. 2007;177:1715–20. https://doi.org/10.1016/j.juro.2007.01.030. <PMID: 17437794>.
Stein JP, Lieskovsky G, Cote R, Groshen S, Feng AC, Boyd S, Skinner E, Bochner B, Thangathurai D, Mikhail M, Raghavan D, Skinner DG. Radical cystectomy in the treatment of invasive bladder cancer: long-term results in 1,054 patients. J Clin Oncol. 2001;19:666–75. https://doi.org/10.1200/JCO.2001.19.3.666. <PMID: 11157016>.
von der Maase H, Sengelov L, Roberts JT, Ricci S, Dogliotti L, Oliver T, Moore MJ, Zimmermann A, Arning M. Long-term survival results of a randomized trial comparing gemcitabine plus cisplatin, with methotrexate, vinblastine, doxorubicin, plus cisplatin in patients with bladder cancer. J Clin Oncol. 2005;23:4602–8. https://doi.org/10.1200/JCO.2005.07.757. <PMID: 16034041>.
Yin M, Joshi M, Meijer RP, Glantz M, Holder S, Harvey HA, Kaag M, Fransen van de Putte EE, Horenblas S, Drabick JJ. Neoadjuvant chemotherapy for muscle-invasive bladder cancer: a systematic review and two-step meta-analysis. Oncologist. 2016;21:708–15. https://doi.org/10.1634/theoncologist.2015-0440. <PMID: 27053504>.
von der Maase H, Hansen SW, Roberts JT, Dogliotti L, Oliver T, Moore MJ, Bodrogi I, Albers P, Knuth A, Lippert CM, Kerbrat P, Sanchez Rovira P, Wersall P, Cleall SP, Roychowdhury DF, Tomlin I, Visseren-Grul CM, Conte PF. Gemcitabine and cisplatin versus methotrexate, vinblastine, doxorubicin, and cisplatin in advanced or metastatic bladder cancer: results of a large, randomized, multinational, multicenter, phase III study. J Clin Oncol. 2000;18:3068–77. https://doi.org/10.1200/JCO.2000.18.17.3068. <PMID: 11001674>.
Hashimoto S, Anai H, Hanada K. Mechanisms of interstrand DNA crosslink repair and human disorders. Genes and Environ. 2016;38:9. https://doi.org/10.1186/s41021-016-0037-9. <PMID: 27350828>.
Semlow DR, Walter JC. Mechanisms of vertebrate DNA interstrand cross-link repair. Annu Rev Biochem. 2021;90:107–35. https://doi.org/10.1146/annurev-biochem-080320-112510. <PMID: 33882259>.
Schärer OD. Nucleotide excision repair in eukaryotes. Cold Spring Harb Perspect Biol. 2013;5:a012609. https://doi.org/10.1101/cshperspect.a012609. <PMID: 24086042>.
Spivak G. Nucleotide excision repair in humans. DNA Repair (Amst). 2015;36:13–8. https://doi.org/10.1016/j.dnarep.2015.09.003. <PMID: 26388429>.
Shilkin ES, Boldinova EO, Stolyarenko AD, et al. Translesion DNA Synthesis and Carcinogenesis. Biochemistry (Mosc). 2020;85:425–35. https://doi.org/10.1134/S0006297920040033. <PMID: 32569550>.
Menck CF, Munford V. DNA repair diseases: What do they tell us about cancer and aging? Genet Mol Biol. 2014;37(1 Suppl):220–33. https://doi.org/10.1590/s1415-47572014000200008.%3cPMID:24764756%3e.
Fadda E. Role of the XPA protein in the NER pathway: A perspective on the function of structural disorder in macromolecular assembly. Comput Struct Biotechnol J. 2015;14:78–85. https://doi.org/10.1016/j.csbj.2015.11.007. <PMID: 26865925>.
Borszéková Pulzová L, Ward TA, et al. XPA: DNA Repair Protein of Significant Clinical Importance. Int J Mol Sci. 2020;21:2182. https://doi.org/10.3390/ijms21062182. <PMID: 32235701>.
Krasikova YS, Lavrik OI, Rechkunova NI. The XPA Protein-Life under Precise Control. Cells. 2022;11:3723. https://doi.org/10.3390/cells11233723. <PMID: 36496984>.
Schärer OD. XPG: its products and biological roles. Adv Exp Med Biol. 2008;637:83–92. https://doi.org/10.1007/978-0-387-09599-8_9. <PMID: 19181113>.
Manandhar M, Boulware KS, Wood RD. The ERCC1 and ERCC4 (XPF) genes and gene products. Gene. 2015;569:153–61. https://doi.org/10.1016/j.gene.2015.06.026. <PMID: 26074087>.
Faridounnia M, Folkers GE, Boelens R. Function and Interactions of ERCC1-XPF in DNA Damage Response. Molecules. 2018;23:3205. https://doi.org/10.3390/molecules23123205. <PMID: 30563071>.
Pal R, Paul N, Bhattacharya D, et al. XPG in the nucleotide excision repair and beyond: a study on the different functional aspects of XPG and its associated diseases. Mol Biol Rep. 2022;49:7995–8006. https://doi.org/10.1007/s11033-022-07324-1. <PMID: 35596054>.
Aboussekhra A, Biggerstaff M, Shivji MK, et al. Mammalian DNA nucleotide excision repair reconstituted with purified protein components. Cell. 1995;80:859–68. https://doi.org/10.1016/0092-8674(95)90289-9. <PMID: 7697716>.
Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030–4. https://doi.org/10.1074/jbc.272.25.16030. <PMID: 9188507>.
Park CH, Sancar A. Formation of a ternary complex by human XPA, ERCC1, and ERCC4(XPF) excision repair proteins. Proc Natl Acad Sci U S A. 1994;91:5017–21. https://doi.org/10.1073/pnas.91.11.5017. <PMID: 8197175>.
Sale JE. Competition, collaboration and coordination-determining how cells bypass DNA damage. J Cell Sci. 2012;125:1633–43. https://doi.org/10.1242/jcs.094748. <PMID: 22499669>.
Roos WP, Kaina B. DNA damage-induced cell death: from specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013;332:237–48. https://doi.org/10.1016/j.canlet.2012.01.007. <PMID: 22261329>.
Lange SS, Takata K, Wood RD. DNA polymerases and cancer. Nat Rev Cancer. 2011;11:96–110. https://doi.org/10.1038/nrc2998. <PMID: 21258395>.
Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–4. https://doi.org/10.1038/21447. <PMID: 10385124>.
Johnson RE, Kondratick CM, Prakash S, Prakash L. hRAD30 mutations in the variant form of xeroderma pigmentosum. Science. 1999;285:263–5. https://doi.org/10.1126/science.285.5425.263. <PMID: 10398605>.
Haracska L, Yu SL, Johnson RE, Prakash L, Prakash S. Efficient and accurate replication in the presence of 7,8-dihydro-8-oxoguanine by DNA polymerase eta. Nat Genet. 2000;25:458–61. https://doi.org/10.1038/78169. <PMID: 10932195>.
Vaisman A, Masutani C, Hanaoka F, Chaney SG. Efficient translesion replication past oxaliplatin and cisplatin GpG adducts by human DNA polymerase eta. Biochemistry. 2000;39:4575–80. https://doi.org/10.1021/bi000130k. <PMID: 10769112>.
Johnson RE, Washington MT, Prakash S, Prakash L. Fidelity of human DNA polymerase eta. J Biol Chem. 2000;275:7447–50. https://doi.org/10.1074/jbc.275.11.7447. <PMID: 10713043>.
Shachar S, Ziv O, Avkin S, Adar S, Wittschieben J, Reissner T, Chaney S, Friedberg EC, Wang Z, Carell T, Geacintov N, Livneh Z. Two-polymerase mechanisms dictate error-free and error-prone translesion DNA synthesis in mammals. EMBO J. 2009;28:383–93. https://doi.org/10.1038/emboj.2008.281. <PMID: 19153606>.
Tissier A, McDonald JP, Frank EG, Woodgate R. Poliota, a remarkably error-prone human DNA polymerase. Genes Dev. 2000;14:1642–50 <PMID: 10887158>.
Washington MT, Carlson KD, Freudenthal BD, Pryor JM. Variations on a theme: eukaryotic Y-family DNA polymerases. Biochim Biophys Acta. 2010;1804:1113–23. https://doi.org/10.1016/j.bbapap.2009.07.004. <PMID: 19616647>.
Kirschner K, Melton DW. Multiple roles of the ERCC1-XPF endonuclease in DNA repair and resistance to anticancer drugs. Anticancer Res. 2010;30:3223–32 <PMID: 20944091>.
Chen YW, Cleaver JE, Hanaoka F, Chang CF, Chou KM. A novel role of DNA polymerase eta in modulating cellular sensitivity to chemotherapeutic agents. Mol Cancer Res. 2006;4:257–65. https://doi.org/10.1158/1541-7786.MCR-05-0118. <PMID: 16603639>.
Ummat A, Rechkoblit O, Jain R, Roy Choudhury J, Johnson RE, Silverstein TD, Buku A, Lone S, Prakash L, Prakash S, Aggarwal AK. Structural basis for cisplatin DNA damage tolerance by human polymerase η during cancer chemotherapy. Nat Struct Mol Biol. 2012;19:628–32. https://doi.org/10.1038/nsmb.2295. <PMID: 22562137>.
Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol. 2006;37:225–38. https://doi.org/10.1007/s10735-006-9041-x. <PMID: 16855787>.
Makridakis NM, Reichardt JK. Translesion DNA polymerases and cancer. Front Genet. 2012;3:174. https://doi.org/10.3389/fgene.2012.00174. <PMID: 22973298>.
Marteijn JA, Lans H, Vermeulen W, Hoeijmakers JH. Understanding nucleotide excision repair and its roles in cancer and ageing. Nat Rev Mol Cell Biol. 2014;15:465–81. https://doi.org/10.1038/nrm3822. <PMID: 24954209>.
Anand J, Chiou L, Sciandra C, Zhang X, Hong J, Wu D, Zhou P, Vaziri C. Roles of trans-lesion synthesis (TLS) DNA polymerases in tumorigenesis and cancer therapy. NAR Cancer. 2023;5:zcad005. https://doi.org/10.1093/narcan/zcad005. <PMID: 36755961>.
Cierna Z, Miskovska V, Roska J, Jurkovicova D, Pulzova LB, Sestakova Z, Hurbanova L, Machalekova K, Chovanec M, Rejlekova K, Svetlovska D, Kalavska K, Kajo K, Babal P, Mardiak J, Ward TA, Mego M, Chovanec M. Increased levels of XPA might be the basis of cisplatin resistance in germ cell tumours. BMC Cancer. 2020;20:17. https://doi.org/10.1186/s12885-019-6496-1. <PMID: 31906898>.
Cancer Genome Atlas Research Network. Comprehensive molecular characterization of urothelial bladder carcinoma. Nature. 2014;507(7492):315–22. https://doi.org/10.1038/nature12965. <PMID: 24476821>.
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortés ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature. 2013;499(7457):214–8. https://doi.org/10.1038/nature12213. <PMID: 23770567>.
Mouw KW. DNA repair pathway alterations in bladder cancer. Cancers (Basel). 2017;9(4):28. https://doi.org/10.3390/cancers9040028. <PMID: 28346378>.
Bajorin DF, Dodd PM, Mazumdar M, Fazzari M, McCaffrey JA, Scher HI, Herr H, Higgins G, Boyle MG. Long-term survival in metastatic transitional-cell carcinoma and prognostic factors predicting outcome of therapy. J Clin Oncol. 1999;17:3173–81. https://doi.org/10.1200/JCO.1999.17.10.3173. <PMID: 10506615>.
Bellmunt J, Paz-Ares L, Cuello M, Cecere FL, Albiol S, Guillem V, Gallardo E, Carles J, Mendez P, de la Cruz JJ, Taron M, Rosell R, Baselga J; Spanish Oncology Genitourinary Group. Gene expression of ERCC1 as a novel prognostic marker in advanced bladder cancer patients receiving cisplatin-based chemotherapy. Ann Oncol. 2007,18:522–8. https://doi.org/10.1093/annonc/mdl435. <PMID: 17229776>.
Kim KH, Do IG, Kim HS, Chang MH, Kim HS, Jun HJ, Uhm J, Yi SY, Lim DH, Ji SH, Park MJ, Lee J, Park SH, Kwon GY, Lim HY. Excision repair cross-complementation group 1 (ERCC1) expression in advanced urothelial carcinoma patients receiving cisplatin-based chemotherapy. APMIS. 2010;118:941–8. https://doi.org/10.1111/j.1600-0463.2010.02648.x. <PMID: 21091775>.
Mullane SA, Werner L, Guancial EA, Lis RT, Stack EC, Loda M, Kantoff PW, Choueiri TK, Rosenberg J, Bellmunt J. Expression levels of DNA damage repair proteins are associated with overall survival in platinum-treated advanced urothelial carcinoma. Clin Genitourin Cancer. 2016;14:352–9. https://doi.org/10.1016/j.clgc.2015.12.029. <PMID: 26778300>.
Matsumura N, Nakamura Y, Kohjimoto Y, Inagaki T, Nanpo Y, Yasuoka H, Ohashi Y, Hara I. The prognostic significance of human equilibrative nucleoside transporter 1 expression in patients with metastatic bladder cancer treated with gemcitabine-cisplatin-based combination chemotherapy. BJU Int. 2011;E110–6. https://doi.org/10.1111/j.1464-410X.2010.09932.x. <PMID: 21166756>.
Kim M, Ku JH, Kwak C, Kim HH, Lee E, Keam B, Kim TM, Heo DS, Lee SH, Moon KC. Predictive and prognostic value of ribonucleotide reductase regulatory subunit M1 and excision repair cross-complementation group 1 in advanced urothelial carcinoma (UC) treated with first-line gemcitabine plus platinum combination chemotherapy. PLoS One. 2015;10:e0133371. https://doi.org/10.1371/journal.pone.0133371. <PMID: 26200905>.
Hoffmann AC, Wild P, Leicht C, Bertz S, Danenberg KD, Danenberg PV, Stöhr R, Stöckle M, Lehmann J, Schuler M, Hartmann A. MDR1 and ERCC1 expression predict outcome of patients with locally advanced bladder cancer receiving adjuvant chemotherapy. Neoplasia. 2010;12:628–36. https://doi.org/10.1593/neo.10402. <PMID: 20689757>.
Sun JM, Sung JY, Park SH, Kwon GY, Jeong BC, Seo SI, Jeon SS, Lee HM, Jo J, Choi HY, Lim HY. ERCC1 as a biomarker for bladder cancer patients likely to benefit from adjuvant chemotherapy. BMC Cancer. 2012;12:187. https://doi.org/10.1186/1471-2407-12-187. <PMID: 22616552>.
Klatte T, Seitz C, Rink M, Rouprêt M, Xylinas E, Karakiewicz P, Susani M, Shariat SF. ERCC1 as a prognostic and predictive biomarker for urothelial carcinoma of the bladder following radical cystectomy. J Urol. 2015;194:1456–62. https://doi.org/10.1016/j.juro.2015.06.099. <PMID: 26162296>.
Kawashima A, Takayama H, Kawamura N, Doi N, Sato M, Hatano K, Nagahara A, Uemura M, Nakai Y, Nishimura K, Miyoshi S, Kawano K, Nishimura K, Nonomura N, Tsujimura A. Co-expression of ERCC1 and Snail is a prognostic but not predictive factor of cisplatin-based neoadjuvant chemotherapy for bladder cancer. Oncol Lett. 2012;4:15–21. https://doi.org/10.3892/ol.2012.689. <PMID: 22807952>.
Kawashima A, Nakayama M, Kakuta Y, Abe T, Hatano K, Mukai M, Nagahara A, Nakai Y, Oka D, Takayama H, Yoshioka T, Hoshida Y, Itatani H, Nishimura K, Nonomura N. Excision repair cross-complementing group 1 may predict the efficacy of chemoradiation therapy for muscle-invasive bladder cancer. Clin Cancer Res. 2011;17:2561–9. https://doi.org/10.1158/1078-0432.CCR-10-1963. <PMID: 21177407>.
Sakano S, Ogawa S, Yamamoto Y, Nishijima J, Miyachika Y, Matsumoto H, Hara T, Matsuyama H. ERCC1 and XRCC1 expression predicts survival in bladder cancer patients receiving combined trimodality therapy. Mol Clin Oncol. 2013;1:403–10. https://doi.org/10.3892/mco.2013.85. <PMID: 24649183>.
Givechian KB, Garner C, Garban H, Rabizadeh S, Soon-Shiong P. CAD/POLD2 gene expression is associated with poor overall survival and chemoresistance in bladder urothelial carcinoma. Oncotarget. 2018;9:29743–52. https://doi.org/10.18632/oncotarget.25701. <PMID: 30038717>.
Del Caño-Ochoa F, Ramón-Maiques S. Deciphering CAD. Structure and function of a mega-enzymatic pyrimidine factory in health and disease. Protein Sci. 2021;30:1995–2008. https://doi.org/10.1002/pro.4158. <PMID: 34288185>.
Brown KK, Spinelli JB, Asara JM, Toker A. Adaptive Reprogramming of De Novo Pyrimidine Synthesis Is a Metabolic Vulnerability in Triple-Negative Breast Cancer. Cancer Discov. 2017;7:391–9. https://doi.org/10.1158/2159-8290.CD-16-0611. <PMID: 28255083>.
Lee YS, Gregory MT, Yang W. Human Pol ζ purified with accessory subunits is active in translesion DNA synthesis and complements Pol η in cisplatin bypass. Proc Natl Acad Sci U S A. 2014;111:2954–9. https://doi.org/10.1073/pnas.1324001111. <PMID: 24449906>.
Elgaaen BV, Haug KB, Wang J, Olstad OK, Fortunati D, Onsrud M, Staff AC, Sauer T, Gautvik KM. POLD2 and KSP37 (FGFBP2) correlate strongly with histology, stage and outcome in ovarian carcinomas. PLoS One. 2010;5:e13837. https://https://doi.org/10.1371/journal.pone.0013837. <PMID: 21079801>.
Faltas BM, Prandi D, Tagawa ST, Molina AM, Nanus DM, Sternberg C, Rosenberg J, Mosquera JM, Robinson B, Elemento O, Sboner A, Beltran H, Demichelis F, Rubin MA. Clonal evolution of chemotherapy-resistant urothelial carcinoma. Nat Genet. 2016;48:1490–9. https://doi.org/10.1038/ng.3692. <PMID: 27749842>.
Barakat KH, Jordheim LP, Perez-Pineiro R, Wishart D, Dumontet C, Tuszynski JA. Virtual screening and biological evaluation of inhibitors targeting the XPA-ERCC1 interaction. PLoS One. 2012;7:e51329 <PMID: 23272099>.
Barakat KH, Torin Huzil J, Luchko T, Jordheim L, Dumontet C, Tuszynski J. Characterization of an inhibitory dynamic pharmacophore for the ERCC1-XPA interaction using a combined molecular dynamics and virtual screening approach. J Mol Graph Model. 2009;28:113–30. https://doi.org/10.1016/j.jmgm.2009.04.009. <PMID: 19473860>.
Gentile F, Tuszynski JA, Barakat KH. New design of nucleotide excision repair (NER) inhibitors for combination cancer therapy. J Mol Graph Model. 2016;65:71–82. https://doi.org/10.1016/j.jmgm.2016.02.010. <PMID: 26939044>.
Saijo M, Matsuda T, Kuraoka I, Tanaka K. Inhibition of nucleotide excision repair by anti-XPA monoclonal antibodies which interfere with binding to RPA, ERCCl, and TFIIH. Biochem Biophys Res Commun. 2004;321:815–22. https://doi.org/10.1016/j.bbrc.2004.07.030. <PMID: 15358100>.
Wojtaszek JL, Chatterjee N, Najeeb J, Ramos A, Lee M, Bian K, Xue JY, Fenton BA, Park H, Li D, Hemann MT, Hong J, Walker GC, Zhou P. A small molecule targeting mutagenic translesion synthesis improves chemotherapy. Cell. 2019;178:152–9. https://doi.org/10.1016/j.cell.2019.05.028. <PMID: 31178121>.
Chang DW, Gu J, Wu X. Germline prognostic markers for urinary bladder cancer: obstacles and opportunities. Urol Oncol. 2012;30(4):524–32. https://doi.org/10.1016/j.urolonc.2012.04.003. <PMID: 22742565>.
Garcia-Grossman IR, Kim PH, Przybylo JA, Alanee S, Riches JC, Regazzi AM, Milowsky MI, Offit K, Bajorin DF. Germline single nucleotide polymorphisms associated with response of urothelial carcinoma to platinum-based therapy: the role of the host. Ann Oncol. 2013;24(9):2414–21. https://doi.org/10.1093/annonc/mdt225. <PMID: 23897706>.
Sakano S, Wada T, Matsumoto H, Sugiyama S, Inoue R, Eguchi S, Ito H, Ohmi C, Matsuyama H, Naito K. Single nucleotide polymorphisms in DNA repair genes might be prognostic factors in muscle-invasive bladder cancer patients treated with chemoradiotherapy. Br J Cancer. 2006;95(5):561–70. https://doi.org/10.1038/sj.bjc.6603290. <PMID: 16880786>.
Gu J, Zhao H, Dinney CP, Zhu Y, Leibovici D, Bermejo CE, Grossman HB, Wu X. Nucleotide excision repair gene polymorphisms and recurrence after treatment for superficial bladder cancer. Clin Cancer Res. 2005;11(4):1408–15. https://doi.org/10.1158/1078-0432.CCR-04-1101. <PMID: 15746040>.
Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non-small-cell lung cancer. J Natl Cancer Inst. 2002;94(14):1091–9. https://doi.org/10.1093/jnci/94.14.1091. <PMID: 12122100>.
Vodicka P, Kumar R, Stetina R, Sanyal S, Soucek P, Haufroid V, Dusinska M, Kuricova M, Zamecnikova M, Musak L, Buchancova J, Norppa H, Hirvonen A, Vodickova L, Naccarati A, Matousu Z, Hemminki K. Genetic polymorphisms in DNA repair genes and possible links with DNA repair rates, chromosomal aberrations and single-strand breaks in DNA. Carcinogenesis. 2004;25(5):757–63. https://doi.org/10.1093/carcin/bgh064. <PMID: 14729591>.
Matullo G, Peluso M, Polidoro S, Guarrera S, Munnia A, Krogh V, Masala G, Berrino F, Panico S, Tumino R, Vineis P, Palli D. Combination of DNA repair gene single nucleotide polymorphisms and increased levels of DNA adducts in a population-based study. Cancer Epidemiol Biomarkers Prev. 2003;12(7):674–7.
Gallagher DJ, Vijai J, Hamilton RJ, Ostrovnaya I, Iyer G, Garcia-Grossman IR, Kim PH, Przybylo JA, Alanee S, Riches JC, Regazzi AM, Milowsky MI, Offit K, Bajorin DF. Germline single nucleotide polymorphisms associated with response of urothelial carcinoma to platinum-based therapy: the role of the host. Ann Oncol. 2013;24(9):2414–21. https://doi.org/10.1093/annonc/mdt225. <PMID: 23897706>.
Acknowledgements
Our gratitude goes to Michaela Ďuratná from the Translation Research Unit, 2nd Department of Oncology, Comenius University, Faculty of Medicine in Bratislava, Slovakia for the technical support. The authors thank Dr. Barbora Baďurová for fruitful discussion and for critical reading of the manuscript.
Funding
This work was supported by the VEGA Agency of the Slovak Republic (grant number 2/0075/23); the Integrated infrastructure operational program for the projects 'Systemic public research infrastructure - Biobank for cancer and rare diseases' (ITMS grant number 313011AFG5) co-financed by the European Regional Development Fund; OncoReSearch, Slovakia (grant number NA).
Author information
Authors and Affiliations
Contributions
PP, AH and MC conceived and designed the study; PP, AH, PM, EO, KK, MV and CB performed the experiments, and collected, assembled, analyzed and interpretated data; PP, AH, KK and MC wrote the manuscript; PP and JR performed statistical analysis; PP, KK and JR prepared the figured and tables; PP, JR, JM, JO, TAW and MC revised the manuscript; PP and MC secured finding, and administrated and supervised the project. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The study was conducted in accordance with the Declaration of Helsinki and was approved by the Institutional Review Board of the Ethics Review Committee of the SECI (approval number 6-2022/EK, date of approval February 3, 2022) and NCI (approval number UC-SK002/2023, date of approval April 24, 2023). Informed consent was obtained from all study participants.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Palacka, P., Holíčková, A., Roška, J. et al. Prognostic value of nucleotide excision repair and translesion DNA synthesis proteins in muscle-infiltrating bladder carcinoma. BMC Cancer 24, 1103 (2024). https://doi.org/10.1186/s12885-024-12865-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12865-8