Abstract
Background
Nasopharyngeal carcinoma (NPC) is diagnosed relatively late and has a poor prognosis, requiring early detection to reduce the disease burden. This diagnostic test accuracy meta-analysis evaluated the serological diagnostic value of nine EBV-related IgA antibody panels (EBNA1-IgA, VCA-IgA, EA-IgA, Zta-IgA, EBNA1-IgA + VCA-IgA, VCA-IgA + EA-IgA, VCA-IgA + Rta-IgG, EBNA1-IgA + VCA-IgA + Zta-IgA and VCA-IgA + EA-IgA + Rta-IgG), aiming to identify suitable serological detection biomarkers for NPC screening.
Methods
PubMed, Embase, China National Knowledge Infrastructure and Chinese BioMedical Literature Database were searched from January 1st, 2000 to September 30th, 2023, with keywords nasopharyngeal carcinoma, IgA, screening, early detection, early diagnosis, sensitivity and specificity. Articles on the diagnostic value of serum EBV-related IgA antibody panels for NPC were included. Study selection, data extraction, and quality assessment were performed independently by two researchers, and a third researcher was consulted in the case of disagreement. Bivariate models were used for statistical analysis. The quality of included studies was evaluated through Quality Assessment of Diagnostic Accuracy Studies tool (QUADAS-2).
Results
A total of 70 articles were included, involving 11 863 NPC cases and 34 995 controls. Among the nine EBV-related IgA antibody panels, EBNA1-IgA + VCA-IgA [0.928 (0.898, 0.950)], VCA-IgA + Rta-IgG [0.925 (0.890, 0.949)], EBNA1-IgA + VCA-IgA + Zta-IgA [0.962 (0.909, 0.985)] and VCA-IgA + EA-IgA + Rta-IgG [0.945 (0.918, 0.964)] demonstrated higher pooled sensitivity (95%CI). In terms of diagnostic odds ratio (DOR) (95%CI), EBNA1-IgA + VCA-IgA [107.647 (61.173, 189.430)], VCA-IgA + Rta-IgG [105.988 (60.118, 186.857)] and EBNA1-IgA + VCA-IgA + Zta-IgA [344.450 (136.351, 870.153)] showed superior performance. Additionally, the SROC curves for EBNA1-IgA + VCA-IgA and VCA-IgA + Rta-IgG were more favorable. However, publication bias was detected for VCA-IgA (P = 0.005) and EBNA1-IgA + VCA-IgA (P = 0.042).
Conclusions
In general, parallel detection of serum EBNA1-IgA, VCA-IgA and Zta-IgA antibodies using ELISA demonstrates better pooled sensitivity and DOR among the studied panels. In the cases where fewer indicators are used, serum VCA-IgA and EBNA1-IgA/Rta-IgG antibody panel exhibits a comparable performance.
Trial registration
The International Prospective Register of Systematic Reviews registration number: CRD42023426984, registered on May 28, 2023.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
Nasopharyngeal carcinoma (NPC) is an epithelial malignancy that occurs in the nasopharynx. According to the World Health Organization (WHO), it was estimated to be 120 434 new NPC cases and 73 482 deaths in the world in 2022, mainly distributed in East Asia, Southeast Asia and North Africa [1]. Approximately 70% of NPC patients are already in advanced stages when they are diagnosed. Through comprehensive treatment, such as surgery, chemotherapy and radiotherapy, 5-year survival rate for advanced stages ranges from 51.4 to 76.0%, compared to 82.6–86.6% for early stages, so early detection is needed to reduce the disease burden [2,3,4,5].
The pathogenesis of NPC is closely related to Epstein-Barr virus (EBV) infection, and detection of EBV-related biomarkers is crucial for early detection of NPC. EBV serum antibody (antibodies are labeled as ‘anti-EBV’, or more precisely, they are antibodies against EBV-encoded antigens) detection has the characteristics of rapidity, simplicity and low cost. Enzyme-linked immunosorbent assay (ELISA) has standardized operation procedure and the interpretation of results is not subject to subjective influence of researchers.
So far, there have been a number of studies on the detection of serum EBV-related IgA antibodies or antibody panels associated with IgA by ELISA, but the results were not consistent. Although several meta-analyses have investigated the diagnostic value of relevant antibody tests for NPC, they involved only antibodies alone or in combination, and did not analyze and compare all possible IgA antibody panels simultaneously [6,7,8,9].
Regarding the above issues and preliminary search results, we used a diagnostic test accuracy meta-analysis to evaluate the serological diagnostic value of nine antibody panels of EBV-related IgA antibodies by ELISA, in order to find suitable detection biomarkers/panels and provide more evidence for NPC screening and early detection.
Methods
This meta-analysis was conducted according to the Preferred Reporting Items for Systematic reviews and Meta-Analyses (PRISMA) statement and registered on the International Prospective Register of Systematic Reviews (PROSPERO) with registration number CRD42023426984 [10, 11].
Search strategy
1) Databases: PubMed, Embase, China National Knowledge Infrastructure (CNKI) and Chinese BioMedical Literature Database (CBM). The first two were used for searching English literature, whereas the latter two were specifically for Chinese literature, given that a large number of related studies have been published in Chinese journals. 2) Search period: from January 1st, 2000 to September 30th, 2023. 3) Key words: nasopharyngeal carcinoma, IgA, screening, early detection, early diagnosis, sensitivity and specificity. 4) Languages: Chinese and English. 5) References for other system reviews or meta-analyses.
Inclusion criteria
1) The case group of the study were comprised of newly diagnosed or untreated NPC patients confirmed by histopathology, and the control group were healthy individuals or general population. At least 20 cases and 20 controls were included in each study. 2) The study applied ELISA to detect serum EBV-IgA antibodies alone or in parallel, within nine antibody panels including EBNA1-IgA, VCA-IgA, EA-IgA, Zta-IgA, EBNA1-IgA + VCA-IgA, VCA-IgA + EA-IgA, VCA-IgA + Rta-IgG, EBNA1-IgA + VCA-IgA + Zta-IgA, VCA-IgA + EA-IgA + Rta-IgG. 3) If similar studies were found originated from the same population, only the most complete one was selected. If the same antibody panel in one study was tested multiple times with ELISA kits from different manufacturers, the mean value was calculated and included. 4) Only studies that provide sufficient data were included.
Exclusion criteria
1) Studies in which NPC patients in the case group also had other tumors or the control group were patients from hospital with other head and neck diseases. 2) Studies for which full texts were not available.
Study selection, data extraction and quality assessment
Study selection, data extraction, and quality assessment were performed independently by two researchers, and a third researcher was consulted in the case of disagreement. According to the search strategy and inclusion/exclusion criteria, literature search, duplicate removal, title abstract screening, and full text screening were carried out via EndNote 20. The extracted data included first author, year of publication, journal name, funding sources, study area, number of cases, number of controls, antibody panels, and the numbers for true positive, false positive, false negative and true negative. The Quality Assessment of Diagnostic Accuracy Studies tool (QUADAS-2) was used to evaluate the risk of bias and applicability of the included literature. The tool is consisted of four key domains: patient selection, index test, reference standard and flow and timing. Each was assessed in terms of risk of bias based on signalling questions and the first three in terms of concerns regarding applicability. The process was conducted in RevMan5.4 [12].
Data analysis
Bivariate models were used for statistical analysis. Firstly, the Bivariate I2 value was calculated to detect heterogeneity among the included studies; if I2 > 0.5, heterogeneity was considered present. Secondly, the threshold effect was assessed using the Spearman correlation coefficient between the logarithm of sensitivity and the logarithm of (1-specificity) for each study; if P < 0.05, the threshold effect was present, only a summary receiver operating characteristic (SROC) curve was plotted; otherwise, pooled sensitivity, specificity, diagnostic odds ratio (DOR) and the corresponding 95% confidence intervals (95%CIs) were also obtained. Subsequently, the included studies were classified by study area (whether high-risk) or number of cases (whether ≤ 100). Based on the number of studies in the two categories, subgroup analysis and sensitivity analysis were performed. Additionally, meta-regression analysis was also conducted on these two factors. The difference was statistically significant when P < 0.05. All these analyses were carried out using Meta-DiSc and Meta-DiSc 2.0 [13, 14]. Finally, Deek’s asymmetry test was conducted using the midas package in Stata 12.0 to evaluate publication bias in each antibody panel; a P-value of < 0.10 indicated the presence of publication bias.
Results
Study selection
Based on the search strategy, a total of 762 articles were found. Then, 217 duplicates were removed, leaving 545 articles. According to the inclusion and exclusion criteria, 380 articles were removed due to title and abstract screening, and then 95 articles were removed by full text screening. Finally, 70 articles were included in this meta-analysis [15,16,17,18,19,20,21,22,23,24,25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46,47,48,49,50,51,52,53,54,55,56,57,58,59,60,61,62,63,64,65,66,67,68,69,70,71,72,73,74,75,76,77,78,79,80,81,82,83,84]. The literature screening process was shown in Fig. 1.
Study characteristics
A total of 70 articles were included, published from 2002 to 2023, involving 11 863 NPC cases and 34 995 controls. All studies were conducted in China, and 50 of them were from high-risk areas (Guangdong, Guangxi, Hainan, and Hong Kong). The main characteristics of the included literature and antibody panels were shown in Tables 1 and 2.
Quality appraisal
QUADAS-2 was used to evaluate the quality of the included articles, and the results indicated that they were of high quality. The risk of bias was primarily attributed to flow and timing issues, followed by patient selection concerns. This was because the intervals between the index test and the reference standard were unclear in some studies, and it remained unclear whether subject selection in some studies involved continuous inclusion and random sampling. The quality assessment results were shown in Fig. S10.1 and Fig. S10.2 in Supplementary Materials.
Heterogeneity, threshold effect and publication bias
The heterogeneity, threshold effect and publication bias of the included studies for nine antibody panels were calculated separately. The results indicated that heterogeneity was present among all panels, whereas no significant threshold effect was observed, suggesting that the heterogeneity was primarily attributed to non-threshold factors. With the exception of VCA-IgA (P = 0.005) and EBNA1-IgA + VCA-IgA (P = 0.042), no publication bias was detected in the other panels (see Table 2).
Meta-analysis, subgroup analysis and sensitivity analysis
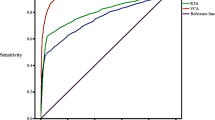
The pooled sensitivity, specificity and DOR of the nine antibody panels were presented in Table 2. Specifically, the sensitivity (95%CI) of EBNA1-IgA + VCA-IgA [0.928 (0.898, 0.950)], VCA-IgA + Rta-IgG [0.925 (0.890, 0.949)], EBNA1-IgA + VCA-IgA + Zta-IgA [0.962 (0.909, 0.985)] and VCA-IgA + EA-IgA + Rta-IgG [0.945 (0.918, 0.964)] was higher. Additionally, EBNA1-IgA + VCA-IgA [107.647 (61.173, 189.430)], VCA-IgA + Rta-IgG [105.988 (60.118, 186.857)], EBNA1-IgA + VCA-IgA + Zta-IgA [344.450 (136.351, 870.153)] had higher DOR (95%CI). The SROC curves for each panel were depicted in Fig. 2. Observing the positions of each summary point and corresponding confidence ellipses, the results for EBNA1-IgA + VCA-IgA and VCA-IgA + Rta-IgG were better. The results of the subgroup analysis classified by study area or number of cases were shown in Fig. S11.1 and Fig. S11.2 in the Supplementary Materials. Moreover, the findings from each meta-regression analysis were not statistically significant. As demonstrated in the sensitivity analysis results presented in Table S10 of the Supplementary Materials, there was no significant change in the DOR values. Therefore, parallel detection of serum VCA-IgA and EBNA1-IgA/Rta-IgG antibodies using ELISA may constitute suitable serological detection strategies for NPC screening and early detection. However, EBNA1-IgA + VCA-IgA + Zta-IgA may potentially represent an even better panel, simply because the small number of studies or individuals included in the analysis limited its evaluation and interpretation. And the characteristics and forest plots for each panel were provided in the Supplementary Materials.
SROC curves for each panel. AUC (95%CI): EBNA1-IgA=0.94 (0.91, 0.96); VCA-IgA=0.93 (0.90, 0.95); EA-IgA=0.92 (0.89, 0.94); Zta-IgA=0.93 (0.90, 0.95); EBNA1-IgA+VCA-IgA=0.97 (0.95, 0.98); VCA-IgA+EA-IgA=0.94 (0.91, 0.95); VCA-IgA+Rta-IgG=0.96 (0.94, 0.98); EBNA1-IgA+VCA-IgA+Zta-IgA=0.99 (0.97, 0.99); VCA-IgA+EA-IgA+Rta-IgG=0.96 (0.94, 0.98)
Discussion
In China, NPC is mainly distributed in Guangdong, Guangxi, Hainan and Hong Kong [85]. Given that all 70 articles included in our study were from China, we designated these regions as high-risk areas for subsequent analysis. The results of this study revealed heterogeneity among the included studies for nine antibody panels. All panels did not have threshold effects, allowing us to calculate their pooled sensitivity, specificity, and DOR. Publication bias was detected for VCA-IgA and EBNA1-IgA + VCA-IgA. In conclusion, parallel detection of serum EBNA1-IgA, VCA-IgA and Zta-IgA antibodies by ELISA showed better pooled sensitivity and DOR among these nine panels. Under the condition that fewer indicators need to be detected, serum VCA-IgA and EBNA1-IgA/Rta-IgG antibody panel has comparable performance.
In addition to EBV-related antibodies serving as diagnostic markers for NPC, EBV-DNA load is also one of the common diagnostic methods. Other biomarkers, including heat shock protein 70 (HSP70), sialic acid (SA) and microRNA (miRNA), are closely associated with the development of NPC. A prospective study has shown that screening with two positive plasma EBV-DNA tests approximately four weeks apart could help in the early detection of NPC in asymptomatic individuals [86]. Another study has found and validated that P85 Ab, a novel serological biomarker for NPC screening, had good screening performance [87]. Although the results in this article are generally more accurate than those of several previous meta-analyses, the gap is not particularly significant [6,7,8].
This study is the first meta-analysis to determine the usefulness of EBNA1-IgA + VCA-IgA + Zta-IgA and comprehensively evaluates the performance of nine serum EBV-related IgA antibody panels detected by ELISA. The overall quality of the included studies was relatively high, and the inclusion and exclusion criteria of the article were considered comprehensively, aiming to align with the screening scenarios of a large population.
However, it also has some limitations. Since all research sources were from China, the conclusions may not be universally applicable to other regions or populations. Most of the included diagnostic trials were single-center studies, and the sample sizes of many studies were not particularly large. Due to the different designs of the included studies and the differences in sex, age, and tumor stage among the study population, the source of heterogeneity cannot be clearly explained. Although the design of this study is as close to the population screening situation as possible, the conditions still need to be verified in large-scale populations. In addition, this article focused solely on analyzing serum IgA antibodies or antibody panels detected by ELISA, and there are numerous other biomarkers and panels with promising diagnostic potential that could be explored in future research based on the current findings.
Conclusion
In summary, parallel detection of serum EBNA1-IgA, VCA-IgA and Zta-IgA antibodies using ELISA demonstrates better pooled sensitivity and DOR among these nine panels generally. When using a reduced number of indicators, serum VCA-IgA and EBNA1-IgA/Rta-IgG antibody panel exhibits comparable diagnostic performance.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2024;4:1–35.
Wei X, Yu S, Wang J, Xiang Z, Liu L, Min Y. Association between time from diagnosis to treatment and survival of patients with nasopharyngeal carcinoma: a population-based cohort study. Curr Probl Cancer. 2024;48:101060.
Koide Y, Kodaira T, Kitayama M, Kawakita D, Kirita T, Yoshimoto S, et al. Definitive radiotherapy for nasopharyngeal carcinoma in Japan: analysis of cases in the National Head and Neck Cancer Registry from 2011 to 2014. Jpn J Clin Oncol. 2024;54(1):54–61.
Huang J, Yang Z-Y, Wu B, Ding Q, Qin Y, Zhang Z-J, et al. Long-term therapeutic outcome and prognostic factors of patients with nasopharyngeal carcinoma receiving intensity-modulated radiotherapy: an analysis of 608 patients from low-endemic regions of China. Curr Med Sci. 2021;41(4):737–45.
Xu M, Zang J, Luo S, Wang J, Li X. Long-term survival outcomes and adverse effects of nasopharyngeal carcinoma patients treated with IMRT in a non-endemic region: a population-based retrospective study. BMJ Open. 2021;11(8):e045417.
Lian M. Combining Epstein–Barr virus antibodies for early detection of nasopharyngeal carcinoma: a meta-analysis. Auris Nasus Larynx. 2023;50(3):430–9.
Liu W, Chen G, Gong X, Wang Y, Zheng Y, Liao X, et al. The diagnostic value of EBV-DNA and EBV-related antibodies detection for nasopharyngeal carcinoma: a meta-analysis. Cancer Cell Int. 2021;21(1):164.
Feng Y, Xia W, He G, Ke R, Liu L, Xie M, et al. Accuracy evaluation and comparison of 14 diagnostic markers for nasopharyngeal carcinoma: a meta-analysis. Front Oncol. 2020;10:1779.
Chen Y, Xin X, Cui Z, Zheng Y, Guo J, Chen Y, et al. Diagnostic value of serum Epstein-Barr virus capsid antigen-IgA for nasopharyngeal carcinoma: a meta-analysis based on 21 studies. Clin Lab. 2016;62(6):1155–66.
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. PLoS Med. 2021;18(3):e1003583.
Page MJ, Moher D, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD, et al. PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews. BMJ (Clinical Res ed). 2021;372:n160.
Review Manager (RevMan). Computer program. Version 5.4. The Cochrane Collaboration.
Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A. Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol. 2006;6:31.
Plana MN, Arevalo-Rodriguez I, Fernández-García S, Soto J, Fabregate M, Pérez T, et al. Meta-DiSc 2.0: a web application for meta-analysis of diagnostic test accuracy data. BMC Med Res Methodol. 2022;22(1):306.
Zhang CQ, Zong YS, Huang BZ, Sun Y, Ye YZ, Feng KT, et al. Enhancing the efficiency of Epstein-Barr viral serologic test in the diagnosis of nasopharyngeal carcinoma. Chin J Oncol. 2002;24(4):356–9.
Cheng WM, Ji MF, Li XL, Yang JL, Zong YS. Screening out nasopharyngeal carcinoma by two-stage ELISA for EB virus. Chin J Immunol. 2003;19(12):834–6.
Gu YL, Zhang CQ, Wu ZB, Zong YS, Liang YJ, Chen YL. Study on sero-diagnosis of nasopharyngeal carcinoma using a dual antibody test against recombinant Epstein-Barr virus antigens. Chin J Cancer. 2003;22(9):903–6.
Huang WG, Lin J, Chen RC. Detection of diagnosis about nasopharyngeal carcinorma combined with EBVCA-IgA and EBVA-IgA in serum. J Pract Med. 2003;10(11):1235–6.
Hu WW, Zong YS, Li FP, Li GM, Zhong BL, Zhang M, et al. Comparison of 6 antibody assays detecting Epstein-Barr virus for serodiagnosis of nasopharyngeal carcinoma. Chin J Clin Oncol. 2006;33(14):795–8.
Li XL, Chen Y, Xu F. Comparison of enzyme-linked immunosorbent assay and immunoenzymatic assay for detection of serum EB-VCA-IgA. Exp Lab Med. 2006;24(2):127–8.
Wu XM, Wei YF. Analysis of EBV antibody detection by ELISA and immunoenzymatic method. Med Forum. 2006;10(7):631–2.
Tang JW, Rohwäder E, Chu IM, Tsang RK, Steinhagen K, Yeung AC, et al. Evaluation of Epstein-Barr virus antigen-based immunoassays for serological diagnosis of nasopharyngeal carcinoma. J Clin Virol. 2007;40(4):284–8.
Liang YJ, Zong YS, Gu YL, Zhang Y, Feng YF, Liu YD, et al. Application of enzyme-linked inununosorbent assay to the serological diagnosis of nasopharyngeal carcinoma. J Pract Med. 2008;24(17):3055–8.
Gu AD, Mo HY, Xie YB, Peng RJ, Bei JX, Peng J, et al. Evaluation of a multianalyte profiling assay and an enzyme-linked immunosorbent assay for serological examination of Epstein-Barr virus-specific antibody responses in diagnosis of nasopharyngeal carcinoma. Clin Vaccine Immunol. 2008;15(11):1684–8.
Cheng JR, Cai YL, Zheng YM, Li J, Mo YK. Detections of immunoglobulin A antibodies against different Epstein-Barr virus antigens in diagnosis of nasopharyn-geal carcinoma diagnosis. Med Innov China. 2009;6(27):101–3.
Deng ZX. Value of detection of serum EBNAI-IGA and EBVCA-IgA by ELISA in diagnosis of nasopharyngeal carcinoma. Chin Trop Med. 2009;9(9):1718–97.
Jiang SQ, Liu Q. Application of logistic regression in combination with multiple diagnostic tests for auxiliary diagnosis of nasopharyngeal carcinoma. Chin J Cancer. 2009;28(2):213–6.
Pan SX, Shi ZS. Evaluate the two ELISA methods and the EIA method in detecting Epstein-Barr virus. Chin J Med Guide. 2009;11(11):1918–9.
Yang MC, Lu AY, Li S, Li RL, Xu MH, Li TJ et al. Comparison of IEMA and ELISA techniques for measurement of EB viral VCA-IgA. Labeled Immunoassays & Clin Med. 2009(2):93–5.
Chen WS, Lu JX, Ye SX. Application of combined detection of EBV VCA-IgA and EA-IgA antibodies in the diagnosis of NPC. J Trop Med. 2010;10(4):434–6.
Hu B, Li ZX, Sheng P, Li L. Expression of VCA genes of Epstein-Barr virus in Pichia pastoris and clinical application. Chin J Microbiol Immunol. 2010;4:365–70.
Tan YJ, SU XK, Cui JH, Qu WH, Xue XY. Comparison of detection of serum VCA-IgA, EA-IgA and EBV-DNA in nasopharygeal carcinoma patients. Chongqing Med J. 2010;39(6):703–6.
Ma DJ, He XF, Li YN. Clinical value of immunoserological and biological detection of Epstein-Barr virus infection in nasopharyngeal carcinoma. Hainan Med J. 2011;22(15):105–7.
Wu YH. Analysis of clinical effect of two different methods for detecting Epstein-Barr virus. Lab Med Clin. 2011;8(6):694–5.
Zhang SL, Li W, Zeng LL, Yang YQ, Zhang WY. Combined determination of Epstein-Barr virus-related antibodies and antigens for diagnosis of nasopharyngeal carcinoma. Mil Med Jnt Log. 2011;25(3):262–3.
Zhou T, Deng YG. Correlation between three serologic tests of Epstein-Barr virus and diagnosis of nasopharyngeal carcinoma. Chin J Clin (Electronic Edition). 2011(11):3308–9.
Zhu HH, Zhang Z, Xie Y, Han L, M.Middeldorp J, Tang CX, et al. The value of double peptide-coated ELISA test in serological diagnosis of nasopharyngeal carcinoma. Chin J Oncol Prev Treat. 2011;3(1):52–5.
Deng WW. Clinical value of triple test(vca-iga,ea-igg、na1-iga)in serological diagnosis of nasopharyngeal carcinoma. Mod Hosp. 2012;12(4):62–4.
Lei DS, Yu J, Tong XL, Wang MW, Wang K, Chen H. Diagnostic value of cytokeratin 19 fragment in nasopharyngeal carcinoma. Chin J Pathol. 2012;41(7):461–5.
Wang ZJ, Cao WJ, Chen F, Shen J, Tang L, Wang SZ. Analysis of serum anti-epstein-barr virus antibody in patients with nasopharyngeal carcinoma in Shanghai. Chin J Ophthalmol Otorhinolaryngol. 2012;12(1):40–6.
Liu Y, Huang Q, Liu W, Liu Q, Jia W, Chang E, et al. Establishment of VCA and EBNA1 IgA-based combination by enzyme-linked immunosorbent assay as preferred screening method for nasopharyngeal carcinoma: a two-stage design with a preliminary performance study and a mass screening in southern China. Int J Cancer. 2012;131(2):406–16.
He J. Study on the labeling and preliminary application of quantum dot technology for nasopharyngeal carcinoma marker EBNA1. Cent South Univ. 2013.
Li XL, Chen Y, Zheng RQ, Peng W, Gao YN, Ye Q. Application value of Epstein-Barr virus rta protein antibody IgG combined with two antibodies in the diagnosis and screening of nasopharyngeal carcinoma. Lab Med Clin. 2013;10(3):321–3.
Yan CE, Wang HJ, Jia DQ, Han XH, Qi J. Diagnostic value of combined measurement of serum SA 、 EA-IgA 、 VCA-IgA and CgA levels in patients with nasopharyngeal carcinoma. Chin J Med. 2013(10):27–8.
Shi DW, Wang PP, Ji MF, Li YB, Zhou HW, Shen S, et al. Evaluation of commercial kits of detecting EBV antibodies for nasopharyngeal carcinoma diagnosis. Labeled Immunoassays & Clin Med. 2014;21(3):310–4.
Xu Q, Zheng SH, Lin SX. To evaluate the clinical value of Epstein-Barr virus (EBV)’s Rta/IgG in the diagnosis of nasopharyngeal carcinoma (NPC) patients in Zhuhai. J Med Infrom. 2014(19):97–8.
Peng YH, Xu YW, Qiu SQ, Hong CQ, Zhai TT, Li EM, et al. Combination of autoantibodies against NY-ESO-1 and viral capsid antigen immunoglobulin A for improved detection of nasopharyngeal carcinoma. Oncol Lett. 2014;8(3):1096–102.
Du MX, Wang WJ. Diagnostic value of combined detection of Rta-IgG,EBNA1-IgA and VCA-IgA antibodies in nasopharyngeal carcinoma in Zhongshan area. Int J Lab Med. 2015;36(13):1856–60.
Peng YH, Xie CL, Zhuang SS, Zhang LQ, Xu YW. Combined analysis of autoantibodies against p53 and Epstein-BarrvirusVCA-IgA for improved diagnosis of nasopharyngeal carcinoma. Cancer Prev Res (Phila). 2015;27(1):26–34.
Wu WQ, Liao SP, Lin XL, Huang QF. Application on the EB virus detection in the physical examination of nasopharyngeal carcinoma screening. All Health. 2015;9(14):18–9.
Xu Q, Zheng SH, Lin SX. Clinical value of Rta protein antibody IgG of EB virus in diagnosis for nasopharynx cancer. Int J Lab Med. 2015;36(17):2500–1.
Yi XH. The detections of EBV-VCA-IgA, EBV-EA-Ig A and EBV-Rta-IgG in diagnosis of nasopharyngeal carcinoma. Fujian Medical Univ. 2015.
Zhang XL, Zhou JL, Cao YP. The value of detection of three anti-epstein-barr viral antibodies for nasopharyngeal carcinoma diagnosis. Chin J Lab Med. 2015(2):111–4.
Gu XM, Li YY, Lv R, Huang JS, Zhou XJ, Du GY. The correlation between serum homocysteine and EB virus three antibodies in nasopharyngeal carcinoma and the evaluation of its diagnosis performance. Chin J Health Lab Tec. 2016(2):231–3.
Pan JZ, Huang HS, Zhu YX, Zhang ZD. Clinical value of three kinds of EB virus antibody detection in screening of nasopharyngeal carcinoma. China Med Pharm. 2016;6(19):225–8.
Wu CY, Qiu MH, Zeng XL, Du MY, Yao M, Chen YX. VCA-IgA and Rta-IgG joint detection diagnosis and effectiveness of nasopharyngeal carcinoma. Chin J Lab Med. 2016;8:609–12.
Yu X, Ji MF, Cheng WM, Huang YL, Li FG. Assessment of EBV antibodies and EBV-DNA in the diagnosis and stages of nasopha-ryngeal carcinoma. Chin J Clin Oncol. 2016;43(15):650–4.
Zheng FJ, Fu YH, Chen WS, Liu N. Expression of Rta recombinant antigen of Epstein-Barr virus and establishment of ELISA method for nasopharyngeal carcinoma screening. J Prev Med Chin PLA. 2016;34(4):464–7.
Lin X, Chen S, Xue X, Lu L, Zhu S, Li W, et al. Chimerically fused antigen rich of overlapped epitopes from latent membrane protein 2 (LMP2) of Epstein-Barr virus as a potential vaccine and diagnostic agent. Cell Mol Immunol. 2016;13(4):492–501.
Chen SC, Peng L, Hu JY, Zhang XJ, Long LN. Application value of three EB virus antibody combined tests in screening of nasopharyngeal carcinoma. Int J Lab Med. 2017;38(14):1949–50.
Yan CE, Chen F, Wang QP, Liu QY, Li J, Gao J, et al. Correlation study on serum EA-IgA、VCA-IgA、Rta-IgG、EBV-DNA levels of nasopharyngeal carcinoma. Chin J Med. 2017;52(8):98–100.
Gao R, Wang L, Liu Q, Zhang LF, Ye YF, Xie SH, et al. Evaluation of seven recombinant VCA-IgA ELISA kits for the diagnosis of nasopharyngeal carcinoma in China: a case-control trial. BMJ Open. 2017;7(6):e013211.
Lin LH, Xu YW, Huang LS, Hong CQ, Zhai TT, Liao LD, et al. Serum proteomic-based analysis identifying autoantibodies against PRDX2 and PRDX3 as potential diagnostic biomarkers in nasopharyngeal carcinoma. Clin Proteom. 2017;14(1):1–10.
Gao J, Peng YP, Zhang CM, Liu LZ, Shen JJ. Diagnosis value of granulysin in patients with nasopharyngeal carcinoma. Zhejiang Med J. 2018;40(14):1599–605.
Li G, Xu Y. The value of EB virus IgA antibody levels in nasopharyngeal carcinoma patients during radiotherapy. Mod Oncol. 2018;26(17):2686–9.
Zhang YN. Changes in serum levels of Rta-IgG, VCA-IgA and EA-IgA antibodies in patients with nasopharyngeal carcinoma before and after radiotherapy. University of South China. 2018.
Li HT, Li ZH, Cui MT, Yang JF, Yang ZZ. Application and clinical significance of three EB virus antibody binding tests in screening nasopharyngeal carcinoma. Int J Lab Med. 2019;40(A01):126–9.
Tang HN, Li YR, Tang LL, Ren YP, Chen YL, Qin LX, et al. Study on diagnostic value of Rta-IgG combined with VCA-IgA and EA-IgA EB viral antibodies detection in nasopharyngeal carcinoma in Hunan area. Lab Med Clin. 2019;16(23):3396–400.
Zhang HW, Chen JH. Correlation analysis between serum NRSN2 with epstein-Barr virus antibody and DNA in nasopharyngeal carcinoma. J Chin Physician. 2019;21(11):1688–92.
Cai XP, Cai WP. Comparison of Epstein-Barr virus serum antibody kits from different manufacturers in the detection of nasopharyngeal carcinoma. Front Med. 2020;10(20):242–3.
Fang QM, Cai WP, Wang Y, Zhong SY, Luo XH. Application value of combined detection of 4 EB Virus antibodies in diagnosis and treatment of nasopharyngeal carcinoma. Lingnan J Emerg Med. 2020;25(3):279–82.
Guo J, Cui Z, Zheng Y, Li X, Chen Y. Comparison of Epstein-Barr virus serological tools for the screening and risk assessment of nasopharyngeal carcinoma: a large population-based study. Pathol Oncol Res. 2020;26(4):2185–90.
Liu W, Li H, Sheng H, Liu X, Chi P, Wang X, et al. A randomized controlled trial on evaluation of plasma Epstein-Barr virus biomarker for early diagnosis in patients with nasopharyngeal carcinoma. Adv Ther. 2020;37(10):4280–90.
Rao D, Fu M, Chen Y, Liu Q, Xiao L, Zhang X, et al. A combination of two ELISA tests for nasopharyngeal carcinoma screening in endemic areas based on a case-control study. PeerJ. 2020;8:e10254.
Yu X, Li F, Cheng W, Wu B, Fang H, Xia F, et al. Efficacy of chemiluminescence immunoassays on VCA-IgA and EBNA1-IgA antibodies of Epstein-Barr virus in diagnosing nasopharyngeal carcinoma. J Cancer. 2020;11(24):7176–83.
Gong YF, Tang BH, Song XM, Tang L, Wang ZJ, Cao WJ. Application of serological detection of EB virus immunoglobin A in nasopharyngeal carcinoma screening. Chin J Ophthalmol Otorhinolaryngol. 2021;21(6):431–4.
Lu SJ, Wu XH, Yang XY, Guo ZH. Study on the diagnostic value of serum EBNA1-IgA,VCA-IgA and VEA-IgA in nasopharyngeal carcinoma. Exp Lab Med. 2021;39(1):142–4.
Wang CY, Zhang SC, Xie ZJ, An L, Zhao CP, Wang CH. Value study of EB virus antibody and scca combined examination on the early diagnosis and prognosis prediction for nasopharyngeal carcinoma patients. Smart Healthc. 2021;7(20):10–15.
Wang J, Xue SW. Detection and analysis of three antibodies of Eb virus NA,ZTA and VCA. Smart Healthc. 2021;7(24):1–3.
Zhang J, Gu XM, Deng HY, Shen HM, Ding JX, Tang FQ, et al. Diagnostic significance of combined detection of Epstein-Barr virus antibodies,EA-IgA,VCA-IgA and Rta-IgG in serum for nasopharyngeal carcinoma. Pract Prev Med. 2021;28(1):44–7.
Deng RH, Liu LY, Xie WK, Liu ML, Zhang GR. Screening value of combined detection of four EB virus antibodies in nasopharyngeal carcinoma. Chin J Otorhinolaryngol Integ Med. 2022;30(2):88–91.
Qu HL, Ye PJ, Liu JH, Lv WB, Yao J. The prognostic value of epstein-barr virus antibodies in nasopharyngeal carcinoma. J Trop Med. 2022;22(1):41–4.
Qin N, Wang S, Liu W. Value of combined measurement of serum epstein-barr virus antibodies and epstein-barr virus DNA in the diagnosis of nasopharyngeal carcinoma. J Qingdao Univ Med Sci. 2023;59(1):64–7.
Xu FX. Clinical value of Epstein-Barr virus antibody detection in the diagnosis of nasopharyngeal carcinoma. Chin Med Digest(Otorhinolaryngology). 2023;38(2):49–52.
Zheng RS, Chen R, Han BF, Wang SM, Li L, Sun KX, et al. Cancer incidence and mortality in China, 2022. Chin J Cancer. 2024;00:221–31.
Chan KCA, Woo JKS, King A, Zee BCY, Lam WKJ, Chan SL, et al. Analysis of plasma Epstein-Barr virus DNA to screen for nasopharyngeal cancer. N Engl J Med. 2017;377(6):513–22.
Li T, Li F, Guo X, Hong C, Yu X, Wu B, et al. Anti-epstein-barr virus BNLF2b for mass screening for nasopharyngeal cancer. N Engl J Med. 2023;389(9):808–19.
Funding
This work was supported by the National Key R&D Program of China (2023YFC2507000); National Science & Technology Fundamental Resources Investigation Program of China (2019FY101105); Sanming Project of Medicine in Shenzhen (SZSM 201911015); and Shenzhen Science and Technology Innovation Program (KCXFZ20211020172542002).
Author information
Authors and Affiliations
Contributions
Concept and design: All authors. Acquisition, analysis, or interpretation of data: All authors. Drafting of the manuscript: All authors. Statistical analysis: Han Liu, Song Song, Jiansong Ren. Administrative, technical, or material support: Lin Lei, Ji Peng, Jiansong Ren. Supervision: Jiansong Ren. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Liu, H., Lei, L., Song, S. et al. The serological diagnostic value of EBV-related IgA antibody panels for nasopharyngeal carcinoma: a diagnostic test accuracy meta-analysis. BMC Cancer 24, 1115 (2024). https://doi.org/10.1186/s12885-024-12878-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12878-3