Abstract
Background
Colorectal cancer ranks among the most prevalent malignancies globally. Accurate prediction of metachronous liver metastasis is crucial for optimizing postoperative management. Tripartite motif-containing protein 27 (TRIM27), an E3 ubiquitin ligase, is implicated in diverse cellular functions and tumorigenesis.
Methods
This study aimed to develop and validate a TRIM27-based nomogram for prognostication in colorectal cancer patients. Transcriptome sequencing of five paired tumor and normal tissue samples identified TRIM27 as a potential prognostic biomarker. Immunohistochemistry was employed to assess TRIM27 expression in colorectal cancer cohorts from two institutions.
Results
TRIM27 expression correlated significantly with both the prognosis of colorectal cancer patients and the occurrence of metachronous liver metastasis. A nomogram incorporating TRIM27 and clinical factors was constructed and demonstrated robust predictive accuracy in an independent validation cohort.
Conclusion
The TRIM27-based nomogram is a valuable prognostic tool for predicting prognosis and metachronous liver metastasis in colorectal cancer patients, aiding in personalized treatment decisions.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Colorectal cancer (CRC) is a major global health burden, ranking as the third most common malignancy and the second leading cause of cancer-related mortality [1]. Liver metastasis is the primary determinant of prognosis in CRC patients, with a significant survival disparity between metastatic and non-metastatic disease [2]. While synchronous liver metastases are diagnosed concurrently with the primary tumor, metachronous metastases occur more than six months later after post-radical resection, affecting approximately 15–20% of CRC patients [3]. Despite advancements, current prognostic factors inadequately stratify patients for the development of metachronous liver metastases [4, 5], underscoring the pressing need for specific molecular biomarkers to refine prognostic assessments and tailor therapeutic approaches.
The Tripartite motif-containing (TRIM) family proteins are implicated in diverse biological processes, including tumorigenesis and metastasis [6,7,8,9]. TRIM27, a member of this family, is characterized by its RING finger domain, two B-boxes, and a coiled-coil domain. Initially identified as a fusion protein with the RET proto-oncogene, TRIM27 has been associated with promoting cancer cell proliferation in multiple malignancies, such as esophageal, lung, and breast cancers. Nevertheless, its role in CRC remains elusive [10].
To address this knowledge gap, we conducted a multicenter, retrospective study to investigate the association between TRIM27 expression and metachronous liver metastasis in CRC patients. Our aim was to develop and validate a TRIM27-based nomogram to improve prognostication and inform treatment decisions.
Materials and methods
Patients and tissue samples
A total of 35 paired frozen tissue samples, including adjacent non-cancerous tissue, CRC tissue, and liver metastasis tissue, were collected from Sun Yat-Sen Memorial Hospital. Of these, five pairs were allocated for sequencing, while the remaining thirty pairs were designated for qPCR validation and immunohistochemistry.
Additionally, 155 and 145 paraffin-embedded tissue samples, obtained from from Sun Yat-Sen Memorial Hospital (n = 155) and the First People’s Hospital of Foshan (n = 145) between 2015 and 2018, were used for subsequent nomogram construction and validation. Patient selection adhered to strict inclusion and exclusion criteria.
Inclusion Criteria:
-
(1)
Age > 18 years
-
(2)
Histological confirmation of colorectal cancer (preoperative colonoscopy or postoperative pathology)
-
(3)
Underwent radical resection of colorectal cancer with primary anastomosis
-
(4)
Absence of liver metastasis on preoperative imaging
-
(5)
Minimum follow-up of three years with annual CT scans
-
(6)
No neoadjuvant chemotherapy
-
(7)
Availability of complete clinical, pathological, and follow-up data
Exclusion Criteria:
-
(1)
History of liver surgery
-
(2)
Primary liver cancer
-
(3)
Severe liver disease (viral hepatitis, liver abscess, cirrhosis)
Quantitative real-time PCR (qRT-PCR)
Total RNA was isolated from the samples using TRIZOL reagent (Invitrogen, Carlsbad, USA). The synthesis of cDNA was performed utilizing a cDNA synthesis kit (TaKaRa Bio, Inc., Shiga, Japan). Quantitative real-time PCR (qRT-PCR) was conducted on a Bio-Rad CFX Connect real-time PCR system (Hercules, CA, USA) using the following cycling conditions: initial denaturation at 95 °C for 10 min, followed by 40 cycles of 95 °C for 30 s, annealing at 55 °C for 30 s, and extension at 72 °C for 30 s. Gene expression levels were quantified using the 2−△△CT method and normalized to β-actin expression. The primer sequences used were: β-actin forward primer: ACCGGGCATAGTGGTTGGA, β-actin reverse primer: ATGGTACACGGTTCTCAACATC, TRIM27 forward primer: AGCCCATGATGCTCGACTG, and TRIM27 reverse primer GGGCACGACACGTTAGTCT. All experiments were performed in triplicate.
Immunohistochemistry (IHC)
Formalin-fixed, paraffin-embedded colorectal cancer tissue sections were subjected to deparaffinization, rehydration, and antigen retrieval using 3% hydrogen peroxide in methanol. Nonspecific binding was blocked with normal goat serum. The slides were then incubated with rabbit anti-TRIM27 (PA5-27,619, Invitrogen, Carlsbad, CA, USA, 1:100 dilution) overnight at 4 °C, followed by detection with HRP-conjugated anti-rabbit IgG and visualization with diaminobenzidine (DAB). Stained slides were examined microscopically using a Nikon Ni-U upright microscope (Nikon Corporation, Tokyo, Japan) and photographed. IHC staining intensity and percentage of positive cells were semi-quantitatively, scored 0–3 and 0–4, respectively [11]. Staining intensity was assessed using a 4-point ordinal scale: 0 (no staining), 1 (weak staining), 2 (moderate staining), and 3 (strong staining). Percentage of positive cells was graded on a 5-point scale: 0 (no positive cells), 1 (1–10% positive cells), 2 (11–50% positive cells), 3 (51–80% positive cells), and 4 (> 80% positive cells). The product of these two scores was calculated as the protein expression level. Protein expression levels were categorized as negative (0-1), weak positive (2–3), positive (4–8), or strong positive (9–12) based on the product of these scores. The IHC scoring was independently performed by two senior pathologists to ensure accuracy and consistency.
Bioinformatics
Gene expression data and clinical data for 270 patients, including 270 CRC tissue samples and 43 normal tissue samples, were retrieved from The Cancer Genome Atlas (TCGA) database (https://www.cancer.gov/ccg/research/genome-sequencing/tcga). This data was then used to investigate the association between the expression of specific proteins, like TRIM27, and overall survival in CRC patients..
Statistical analysis
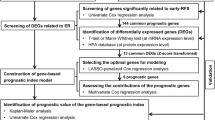
Data were presented as mean ± standard deviation (SD). Statistical analyses were performed using GraphPad Prism (version 8.0) and R software (version 3.6.1). One-way ANOVA and t-tests were applied for group comparisons, with statistical significance defined as a P-value < 0.05. To assess survival outcomes, univariable and multivariable Cox regression analysis was conducted and subsequently used to construct survival nomograms. Logistic regression models were employed to predict the risk of metachronous liver metastasis, followed by the development of corresponding nomograms. The discriminative ability of the nomograms was evaluated using concordance indices (C-indices). Calibration of the nomograms was assessed by comparing predicted survival probabilities with Kaplan–Meier estimates. The “rms” package in R was used for nomogram generation, and the “timeROC” package was utilized for time-dependent receiver operating characteristic (ROC) curve analysis.
Results
Clinicopathological features of patients
A total of 155 samples were collected from Sun Yat-Sen Memorial Hospital to establish a training cohort, while 145 samples were obtained from the First People’s Hospital of Foshan to form an external validation cohort (Fig. 1). Comparative analysis revealed no significant differences between the two cohorts in terms of clinicopathological characteristics (Table 1). All patients underwent radical resection of colorectal cancer with primary anastomosis and were followed up for a minimum of 36 months. Three-year overall survival (OS) rates were comparable between the training and validation cohorts (72.3% vs. 71.0%, P = 0.814). Similarly, disease-free survival (DFS) rates (65.2% vs. 64.8%, P = 0.952) and the incidence of metachronous liver metastasis (24.52% vs. 22.07%, P = 0.617) were not significantly different between the two cohorts.
Screening and validation of target genes
Identifying target genes in high-throughput transcriptome sequencing
High-throughput transcriptome sequencing of five paired samples yielded expression levels for 17,084 mRNAs. Differential expression analysis identified 273 differentially expressed genes (DEGs) with significant variance (P < 0.005) between liver metastasis, tumor tissue, and normal tissues (Fig. 2A). To further prioritize candidate genes, we focused on DEGs exhibiting a consistent decrease in expression from liver metastasis to tumor to normal tissues. This analysis identified 12 genes, including NFRKB, LPCAT2, ZKSCAN1, TFB1M, FAAH2, UBE3D, RCL1, TRIM27, SYNJ2, UHRF2, U2SURP, and BCL11A, as potential biomarkers (Fig. 2B).
Screening and Validation of Target Genes. A Heatmaps of gene expression of 5 paired samples. M1-M5: liver metastasis; C1-C5: colorectal tumor tissues; N1-N5: normal tissues. B Venn diagram analysis of differentially expressed genes meeting consistent expression trend criteria. C Relative expression of LPCAT2, RCL1, TRIM27 and UHRF2 by qRT-PCR in 30 paired samples
Validation of candidate biomarkers by qRT-PCR
The relative expression of all 12 candidate mRNAs were quantified using qRT-PCR in an independent cohort of 30 paired samples. Among the 12 initially screened candidate genes, the result showed that only LPCAT2, RCL1, TRIM27, and UHRF2 met the consistent expression trend criteria (Fig. 2C and Supplementary Figure S1).
Evaluatiion of Candidate Genes in TCGA
We utilized TCGA data to explore the expression of four genes. Firstly, we compared the expression levels of the four mRNAs in 43 paired colorectal cancer and adjacent normal tissues. Our analysis revealed that all four mRNAs were significantly upregulated in colorectal tumor tissues (P < 0.05) (Fig. 3A and Supplementary Figure S2A-C). Subsequent analysis differentiated stage IV from stages I-III colorectal tumor tissues, showing exclusive overexpression of TRIM27 in stage IV (P = 0.006), with no significant differences for LPCAT2, RCL1, and UHRF2 (Fig. 3B and Supplementary Figure S2D-F). Prognostic correlations were assessed by dividing patients into high and low expression groups based on the median expression level. TRIM27 expression was significantly associated with both OS (P = 0.041) and DFS (P = 0.021) (Fig. 3D-E). In contrast, LPCAT2 and UHRF2 correlated only with OS, and RCL1 showed no significant prognostic impact (Supplementary Figure S3).
Marker validation in mRNA and protein level. A Expression levels of TRIM27 in colorectal tumor tissues and normal tissues from TCGA database. B Expression levels of TRIM27 in Stage IV and Stage I-III colorectal tumor tissues from TCGA database. C Expressive level of TRIM27 by IHC in 30 paired samples. D-E OS and DFS curve between high expression group and low expression group of TRIM27
Immunohistochemical validation of protein expression
Immunohistochemistry (IHC) was performed on 30 paired samples to assess protein expression levels of the four candidate mRNAs. Semi-quantitative analysis revealed that TRIM27 was significantly overexpressed in liver metastasis compared to normal and tumor tissues (P = 0.037) (Fig. 3C). These findings, combined with bioinformatics data from the TCGA database, identified TRIM27 as a promising molecular marker for further investigation in the context of metachronous liver metastasis and colorectal cancer prognosis.
TRIM27 as a prognostic biomarker for metachronous liver metastasis in colorectal cancer
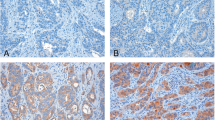
Immunohistochemical (IHC) staining was performed to assess TRIM27 expression, which was categorized into four levels based on established criteria (Fig. 4A) and then samples were then divided into two groups based on expression levels. Negative (0–1) and weak positive (2–3) are defined as low expression, while positive (4–8) and strong positive (9–12) are defined as high expression. We conducted a prognostic analysis by combining the samples from the training cohort and the test cohort. Among patients who developed metachronous liver metastasis, 19 had strong positive and 20 had positive TRIM27 expression, compared to 11 strong positive and 27 positive patients without metastasis (P < 0.001) (Fig. 4D). These findings indicate a significant association between high TRIM27 expression and the development of metachronous liver metastasis in colorectal cancer patients.
TRIM27 is correlated to metachronous liver metastasis of colorectal cancer. A Expression levels of TRIM27 by IHC in CRC tissues. B Kaplan–Meier curves of OS based on the expression of TRIM27 in CRC patients. C Kaplan–Meier curves of DFS based on the expression of TRIM27 in CRC patients. D Proportion of metachronous liver metastasis in different TRIM27 expression levels
Kaplan–Meier survival analysis revealed significantly improved OS and DFS in patients with low TRIM27 expression compared to those with high expression. The 1-year OS rates were 95.07% and 79.22%, respectively (P < 0.001), while the corresponding 3-year OS rates were 80.27% and 46.75% (P < 0.001) (Fig. 4B). Similarly, patients with low TRIM27 expression exhibited significantly better 1-year DFS (92.83% vs. 68.83%, P < 0.001) and 3-year DFS (75.78% vs. 31.17%, P < 0.001) compared to those with high expression (Fig. 4C). These findings strongly support the association between high TRIM27 expression and poor prognosis in colorectal cancer.
Nomogram construction and validation
Constructing prognostic nomograms
To further investigate the prognostic value of TRIM27, univariate and multivariate Cox regression analyses were conducted. Variables achieving statistical significance in the univariate analysis (P ≤ 0.1) were included in the multivariate model. T stage, N stage, TRIM27 expression, preoperative CEA levels, and adjuvant chemotherapy emerged as independent prognostic factors for OS in the multivariate analysis (Supplementary Table S1-S2). A prognostic nomogram incorporating these factors was developed to predict OS in colorectal cancer patients (Fig. 5A). Similarly, multivariate Cox regression analysis identified N stage, TRIM27 expression, and adjuvant chemotherapy as independent predictors of DFS (Supplementary Table S3-S4). A nomogram was constructed to predict DFS based on these factors (Fig. 5B).
To identify potential predictors of metachronous liver metastasis, univariate and multivariate logistic regression analyses were performed. TRIM27 expression, preoperative CEA levels, and adjuvant chemotherapy were retained as significant predictors in the final multivariate model (Supplementary Tables S5-S6). A nomogram was constructed to predict the risk of metachronous liver metastasis based on these factors (Fig. 5C).
Validating the nomograms
The nomogram for predicting OS demonstrated robust performance with C-indices of 0.783 and 0.803 in the training and test cohorts, respectively. Calibration curves confirmed excellent agreement between predicted and observed OS probabilities (Supplementary Figure S4). Similarly, the nomogram for predicting DFS exhibited strong discriminative ability, with C-indices of 0.768 and 0.851 in the training and test cohorts, and the calibration curve also indicated a good calibration effect (Supplementary Figure S5). The 1-year and 3-year time-dependent ROC curve analyses demonstrated that the nomograms significantly outperformed traditional risk factors in predicting both OS and DFS (Supplementary Figure S6). In the training cohort, the AUC for 3-year OS and 3-year DFS were 0.784 (95% CI, 0.694–0.873) and 0.794 (95% CI, 0.715–0.873), respectively, while in the external validation cohort, these values increased to 0.856 (95% CI, 0.780–0.932) and 0.944 (95% CI, 0.906–0.983), respectively (Fig. 6A-B). These robust findings underscore the clinical utility of the proposed nomograms for improving patient management.
A 3-year time-dependent ROC curve to predict OS in the training cohort and external validation cohort. B 3-year time-dependent ROC curve to predict DFS in the training cohort and external validation cohort. C ROC curve to predict metachronous liver metastasis in the training cohort and external validation cohort. AUC: area under curve. D Decision curve analysis of the nomogram and TNM stage to predict metachronous liver metastasis of CRC
The nomogram for predicting metachronous liver metastasis demonstrated strong discriminative ability, with C-indices of 0.824 and 0.818 in the training and validation cohorts, respectively. Calibration curves confirmed the model's accurate prediction of the risk of metachronous liver metastasis (Supplementary Figure S7). ROC curve analysis yielded AUC values of 0.824 (95% CI, 0.747–0.901) and 0.818 (95% CI, 0.736–0.901) in the training and test cohorts, respectively, further supporting the nomogram's predictive performance (Fig. 6C).
These findings underscore the clinical utility of these nomograms in improving risk stratification and guiding treatment decisions for colorectal cancer patients..
Clinical application
The clinical utility of the nomogram was evaluated using decision curve analysis (DCA). As depicted in Fig. 6D, the nomogram demonstrated superior net benefit compared to the traditional TNM staging system in predicting the risk of metachronous liver metastasis, suggesting its potential to improve clinical decision-making.
Discussion
Prognosis for colorectal cancer patients with liver metastasis remains poor following surgery. While synchronous liver metastases are often detected preoperatively, early micrometastases frequently evade diagnosis due to limitations in imaging techniques [12]. This highlights the imperative for specific molecular biomarkers to refine prognostic stratification and tailor therapeutic interventions.
This study introduces TRIM27 as a novel prognostic biomarker for colorectal cancer, demonstrating its ability to stratify patients according to their risk of developing metachronous liver metastasis. Our findings highlight TRIM27 as a valuable adjunct to traditional clinicopathological factors in predicting patient outcomes.
Although TNM staging is a cornerstone for prognostic assessment in cancer patients [13], it may not accurately stratify risk for patients with metachronous liver metastasis. Our model did not include the TNM staging system because patients with synchronous liver metastasis were excluded during the initial modeling phase. Therefore, our model can be considered a supplement to the TNM system. Different from other existing metachronous liver metastasis prediction models [14, 15], our model incorporates the novel biomarker TRIM27, which provides additional insights beyond traditional pathological features, potentially leading to more personalized and effective therapeutic strategies. While further validation studies are warranted, these preliminary findings suggest that TRIM27 may serve as a valuable adjunct to current prognostic models, potentially improving risk stratification and informing personalized treatment strategies.
CEA have been previously recognized as valuable predictors of survival outcomes in colorectal cancer [16]. Consistent with this, our study corroborates the association between elevated preoperative CEA levels and an increased risk of metachronous liver metastasis.
The TRIM protein family has emerged as a critical regulator of tumorigenesis and metastasis. Recent studies have highlighted the overexpression of TRIM29 in multiple malignancies [17, 18], identified TRIM47 as an oncogene driving colorectal cancer progression [19], and linked TRIM11 overexpression to enhanced proliferation and metastasis in lung cancer [20]. Additionally, TRIM14 and TRIM52 have been implicated in promoting metastasis and tumorigenesis through diverse mechanisms [21, 22].
TRIM27 has been implicated as a key player in tumorigenesis across multiple cancer types [23, 24]. However, TRIM27's role in colorectal cancer metastasis remains less understood. We hypothesize that TRIM27 may contribute to CRC metachronous liver metastasis through multiple mechanisms. TRIM27 can enhance various tumor proliferation signaling pathways, such as PI3K/AKT, Wnt/β-catenin, and NF-κB pathways, thereby increasing the proliferation and invasion capabilities of tumor cells [25,26,27]. Additionally, studies have also indicated that TRIM27 promotes tumor chemoresistance and modulates immune function [28,29,30]. Since metachronous liver metastasis occurs after the primary tumor has been resected, overexpression of TRIM27 may lead to CRC chemoresistance and immune escape, allowing residual cancer cells to re-establish and grow in the liver.
Although this retrospective study provides valuable insights into prognosis and metachronous liver metastasis in colorectal cancer, it is subject to inherent limitations. The potential for unobserved confounding variables and the relatively small sample size restrict the generalizability of our findings and the ability to inform immediate treatment decisions based on the nomogram. Nevertheless, this study offers a quantitative foundation for future prospective investigations to validate these predictive factors and optimize clinical management.
Conclusion
This study highlights the significant role of TRIM27 in predicting prognosis and metachronous liver metastasis in colorectal cancer. The developed nomograms, incorporating TRIM27 expression, enhance personalized postoperative prognostic assessments and guide individualized treatment strategies for patients.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Data availability
No datasets were generated or analysed during the current study.
References
Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. 2021;71(3):209–49.
Engstrand J, Nilsson H, Str Mberg C, Jonas E, Freedman J. Colorectal cancer liver metastases – a population-based study on incidence, management and survival. BMC Cancer. 2018;18(1):78.
Tsai M-S, Su Y-H, Ho M-C, Liang J-T, Chen T-P, Lai H-S, et al. Clinicopathological Features and Prognosis in Resectable Synchronous and Metachronous Colorectal Liver Metastasis. Ann Surg Oncol. 2007;14(2):786–94.
Kim HJ, Lee SS, Byun JH, Kim JC, Yu CS, Park SH, et al. Incremental Value of Liver MR Imaging in Patients with Potentially Curable Colorectal Hepatic Metastasis Detected at CT: A Prospective Comparison of Diffusion-weighted Imaging, Gadoxetic Acid-enhanced MR Imaging, and a Combination of Both MR Techniques. Radiology. 2015;274(3):712–22.
Wiering B, Ruers TJM, Krabbe PFM, Dekker HM, Oyen WJG. Comparison of multiphase CT, FDG-PET and intra-operative ultrasound in patients with colorectal liver metastases selected for surgery. Ann Surg Oncol. 2007;14(2):818–26.
Chen D, Li Y, Zhang X, Wu H, Wang Q, Cai J, et al. Ubiquitin ligase TRIM65 promotes colorectal cancer metastasis by targeting ARHGAP35 for protein degradation. Oncogene. 2019;38(37):6429–44.
Brown NJM, Ramalho M, Pedersen EW, Moravcsik E, Grimwade D. PML nuclear bodies in the pathogenesis of acute promyelocytic leukemia: Active players or innocent bystanders? Front Biosci. 2009;14(14):1684–707.
Tn Pathiraja, Kn Thakkar, S Jiang, S Stratton, Z Liu, M Gagea, et al. TRIM24 links glucose metabolism with transformation of human mammary epithelial cells. Oncogene. 2015;34(22):2836–45.
Sho T, Tsukiyama T, Sato T, Kondo T, Cheng J, Saku T, et al. TRIM29 negatively regulates p53 via inhibition of Tip60. Biochimica et Biophysica Acta (BBA) - Mol Cell Res. 2011;1813(6):1245–53.
Yu C, Rao D, Wang T, Song J, Zhang L, Huang W. Emerging roles of TRIM27 in cancer and other human diseases. Front Cell Dev Biol. 2022;10:1004429.
Xu H, Li W, Zhu C, Cheng N, Li X, Hao F, et al. Proteomic profiling identifies novel diagnostic biomarkers and molecular subtypes for mucinous tubular and spindle cell carcinoma of the kidney. J Pathol. 2022;257(1):53–67.
Li S, Yuan L, Yue M, Xu Y, Liu S, Wang F, et al. Early evaluation of liver metastasis using spectral CT to predict outcome in patients with colorectal cancer treated with FOLFOXIRI and bevacizumab. Cancer Imaging. 2023;23(1):30.
Gospodarowicz MK, Benedet L, Hutter RV, Fleming I, Sobin LH. History and international developments in cancer staging. Prév Contrle En Cancérologie Pcc. 1998;2(6):262–8.
Hao M, Li H, Wang K, Liu Y, Liang X, Ding L. Predicting metachronous liver metastasis in patients with colorectal cancer: development and assessment of a new nomogram. World J Surg Oncol. 2022;20(1):80.
Xiao C, Zhou M, Yang X, Wang H, Tang Z, Zhou Z, et al. Accurate Prediction of Metachronous Liver Metastasis in Stage I-III Colorectal Cancer Patients Using Deep Learning With Digital Pathological Images. Front Oncol. 2022;12: 844067.
Arru M, Aldrighetti L, Castoldi R, Di Palo S, Orsenigo E, Stella M, et al. Analysis of Prognostic Factors Influencing Long-term Survival After Hepatic Resection for Metastatic Colorectal Cancer. World J Surg. 2008;32(1):69.
Chen Y, Ma J, Zhang M. TRIM29 promotes the progression of colorectal cancer by suppressing EZH2 degradation. Exp Biol Med (Maywood, NJ). 2023;248(18):1527–36.
Jiang T, Xia Y, Li Y, Lu C, Lin J, Shen Y, et al. TRIM29 promotes antitumor immunity through enhancing IGF2BP1 ubiquitination and subsequent PD-L1 downregulation in gastric cancer. Cancer Lett. 2024;581:216510.
Liu F, Xie B, Ye R, Xie Y, Zhong B, Zhu J, et al. Overexpression of tripartite motif-containing 47 (TRIM47) confers sensitivity to PARP inhibition via ubiquitylation of BRCA1 in triple negative breast cancer cells. Oncogenesis. 2023;12(1):13.
Zhang R, Li S-W, Liu L, Yang J, Huang G, Sang Y. TRIM11 facilitates chemoresistance in nasopharyngeal carcinoma by activating the β-catenin/ABCC9 axis via p62-selective autophagic degradation of Daple. Oncogenesis. 2020;9(5):45.
Nenasheva VV, Kovaleva GV, Uryvaev LV, Ionova KS, Dedova AV, Vorkunova GK, et al. Enhanced expression of trim14 gene suppressed Sindbis virus reproduction and modulated the transcription of a large number of genes of innate immunity. Immunologic Research. 2015;62(3):255–62.
Gómez-Martín D, Galindo-Feria AS, Barrera-Vargas A, Merayo-Chalico J, Juárez-Vega G, Torres-Ruiz J, et al. Ro52/TRIM21-deficient expression and function in different subsets of peripheral blood mononuclear cells is associated with a proinflammatory cytokine response in patients with idiopathic inflammatory myopathies. Clin Exp Immunol. 2017;188(1):154–62.
J Wang, Jl Teng, D Zhao, P Ge, B Li, Pc Woo, et al. The ubiquitin ligase TRIM27 functions as a host restriction factor antagonized by Mycobacterium tuberculosis PtpA during mycobacterial infection. Sci Rep. 2016;6:34827.
Xing L, Tang X, Wu K, Huang X, Yi Y, Huan J. TRIM27 Functions as a Novel Oncogene in Non-Triple-Negative Breast Cancer by Blocking Cellular Senescence through p21 Ubiquitination. Mol Ther Nucleic Acids. 2020;22:910–23.
Lee JT, Shan J, Zhong J, Li M, Zhou B, Zhou A, et al. RFP-mediated ubiquitination of PTEN modulates its effect on AKT activation. Cell Res. 2013;23(4):552–64.
Liu S, Tian Y, Zheng Y, Cheng Y, Zhang D, Jiang J, et al. TRIM27 acts as an oncogene and regulates cell proliferation and metastasis in non-small cell lung cancer through SIX3-β-catenin signaling. Aging (Albany NY). 2020;12(24):25564–80.
Xiao C, Zhang W, Hua M, Chen H, Yang B, Wang Y, et al. TRIM27 interacts with Iκbα to promote the growth of human renal cancer cells through regulating the NF-κB pathway. BMC Cancer. 2021;21(1):841.
Zhang HX, Xu ZS, Lin H, Li M, Xia T, Cui K, et al. TRIM27 mediates STAT3 activation at retromer-positive structures to promote colitis and colitis-associated carcinogenesis. Nat Commun. 2018;9(1):3441.
Yao Y, Liu Z, Cao Y, Guo H, Jiang B, Deng J, et al. Downregulation of TRIM27 suppresses gastric cancer cell proliferation via inhibition of the Hippo-BIRC5 pathway. Pathol Res Pract. 2020;216(9): 153048.
Wang J, Teng JL, Zhao D, Ge P, Li B, Woo PC, et al. The ubiquitin ligase TRIM27 functions as a host restriction factor antagonized by Mycobacterium tuberculosis PtpA during mycobacterial infection. Sci Rep. 2016;6:34827.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Health Commission of Foshan, Guangdong Province, China (grant number 20240718A010025).
Author information
Authors and Affiliations
Contributions
Jiankun Zhu and Shilin Zhi conceptualized the study and were responsible for the drafting and writing of the manuscript. Shengning Zhou analyzed the data. Jintao Zeng collected the data. Yong Ji supervised the experiments and provided leadership for the research team. Fanghai Han managed the project and coordinated the research activities. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
This study was conducted in accordance with the Declaration of Helsinki and was approved by the Ethics Committee of Sun Yat-sen Memorial Hospital, Sun Yat-sen University. The approval number for this study is SYSKY-2024–254-01. Informed consent was obtained from all individual participants included in the study. All participants were assured of confidentiality and the right to withdraw from the study at any time without any consequences.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhu, J., Zhi, S., Zeng, J. et al. Development and validation of a TRIM27-based nomogram for predicting metachronous liver metastasis and prognosis in postoperative colorectal cancer patients. BMC Cancer 24, 1142 (2024). https://doi.org/10.1186/s12885-024-12890-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-024-12890-7