Abstract
Background
Cis-regulatory elements (CREs) are crucial for regulating gene expression, and G-quadruplexes (G4s), as prototypal non-canonical DNA structures, may play a role in this regulation. However, the relationship between G4s and CREs, especially with non-promoter-like functional elements, requires further systematic investigation. We aimed to investigate the associations between G4s and human cCREs (candidate CREs) inferred from the Encyclopedia of DNA Elements (ENCODE) data.
Results
We found that G4s are prominently enriched in most types of cCREs, especially those with promoter-like signatures (PLS). The co-occurrence of CTCF signals with H3K4me3 or H3K27ac signals strengthens the association between cCREs and G4s. Genetic variants in G4s, particularly within their G-runs, exhibit higher regulatory potential and deleterious effects compared to cCREs. The G-runs within G4s near transcriptional start sites (TSSs) are more evolutionarily constrained compared to G-runs in cCREs, while those far from the TSS are relatively less conserved. The presence of G4s is often linked to a more favorable local chromatin environment for the activation and execution of regulatory function of cCREs, potentially attributable to the formation of G4 secondary structures. Finally, we discovered that G4-associated cCREs exhibit widespread activation in a variety of cancers.
Conclusions
Our study suggests that G4s are integral components of human cis-regulatory elements, extending beyond their potential role in promoters. The G4 primary sequences are associated with the localization of CREs, while the G4 structures are linked to the activation of these elements. Therefore, we propose defining G4s as pivotal regulatory elements in the human genome.
Similar content being viewed by others
Background
Eukaryotic gene expression is intricately regulated by cis-regulatory elements (CREs) [1, 2]. CREs are sequences located within the non-coding regions of the genome, some of which are positioned near the gene transcription start site (TSS), known as promoters, while others, including enhancers, may be located away from the genes and regulate their expression through spatial and proximal mechanisms facilitated by the three-dimensional chromatin architecture [3]. These cis-regulatory elements can interact with transcription factors, acting like switches to precisely control gene expression. Moreover, there are other types of cis-regulatory elements, including silencers and insulators [3]. Silencers repress gene expression [4], while insulators isolate two adjacent chromosomal regions to prevent mutual interference in gene regulations [5]. The coordinated operation of cis-regulatory elements plays a crucial role in cell differentiation and development, and the utilization of cell-type-specific cis-regulatory elements forms the cornerstone of cellular functional diversity.
G-quadruplexes (G4s) are unusual nucleic acid secondary structures, typically formed in guanine-rich regions [6, 7]. The basic unit of G4s is called a G-tetrad or G-quartet, in which four guanines are linked by Hoogsteen hydrogen bonds to form a stable planar - or near-planar - assembly [6, 7]. Multiple G-tetrads can stack to create a stable G4 structure. G4-prone sequences often match a consensus motif (e.g., GxN1-7GxN1-7GxN1-7Gx, where x ≥ 3 and N can be any base), keeping in mind that other motifs deviating from this consensus may actually form stable G4 structures [6, 7]. Therefore, various G4 prediction tools have been developed, including G4Hunter [8], that do not rely entirely on a specific consensus sequence pattern. Computational studies have shown that G4s are not randomly distributed in the human genome; instead, they are concentrated in functional regions such as promoters [9], telomeres [10], and intron-exon borders [11], suggesting that G4s may have biological functions. Furthermore, in recent years, with the widespread application of single-molecule techniques in G4 research and the development of whole genome methods, including G4 ChIP-seq [12] and G4Access [13], more and more evidence indicates that G4 secondary structures are not genomic “junk” or accidents but possess important biological functions. Extensive studies have revealed that G4s are widely involved in the regulation of a variety of biological processes, including gene transcription [9], protein translation [14], genome stability [15, 16], telomere regulation [17], and genome replication [18]. However, it remains uncertain whether there are any differences in the association between G4s and different groups of CREs, and whether G4s might be the functional component of CREs. Additionally, it is worth exploring whether the activation of CREs is related to the G4 structure itself or the G-rich primary sequence compatible with G4 formation, and how significant the differences in the effects of these two factors on CRE activation would be.
Due to the widespread use of sequencing technologies and the ease of storing and sharing epigenetic data, it is now possible to study and annotate the human genome in depth. An international, large-scale study constructed sets of candidate cis-regulatory elements (cCREs) for the human and mouse genomes by comprehensively analyzing epigenetic data from the ENCODE (Encyclopedia of DNA Elements) project [19]. This provides us with the opportunity to systematically assess the association between G4s and cis-regulatory elements, revealing whether G4s may constitute a key component of cis-regulatory elements in the human genome.
In this study, we focused on exploring the importance of G4s in the candidate cis-regulatory elements of the human genome. We first determined whether G4 sequences were enriched among various groups of cCREs, as the preferential distribution of G4s often hints at their potential biological functions. Subsequently, we assessed whether variants in G4-associated cCREs were more likely to have regulatory effects or be more deleterious compared to cCREs themselves, based on RegulomeDB scores and LINSIGHT scores. Additionally, we interrogated the conservation of these cCRE-G4s from an evolutionary perspective based on 241 mammalian species. Furthermore, we investigated whether the presence of G4s, especially their structures, might correlate with a more favorable local chromatin environment for the activation and utilization of cCREs. Finally, we examined whether the activation of cCREs remains closely associated with G4s across multiple cancers. Our research demonstrates that G4s are indeed fundamental components of CREs, laying the theoretical foundation for subsequent in-depth studies and interpretations of CRE functionality.
Results
Candidate cis-regulatory elements are enriched with G4s
To comprehensively characterize the relationship between G4s and cis-regulatory elements, we employed the G4Hunter software [8] to predict all potential G4 sequences in the human genome (see methods), and these predicted G4s were used for subsequent analysis. In parallel, we retrieved the human candidate cis-regulatory elements (cCREs) from the SCREEN (Search Candidate cis-Regulatory Elements by ENCODE) database, in which human cCREs are primarily categorized into five groups, namely elements (or cCRE) with promoter-like signatures (PLS), proximal enhancer-like signatures (pELS), distal enhancer-like signatures (dELS), high DNase and CTCF signals only (CTCF-only), and high DNase and H3K4me3 signals only (DNase-H3K4me3). These cCREs are all defined based on epigenetic signals, including DNase-seq, H3K4me3, H3K27ac, as well as CTCF, and their distance from transcription start sites (see methods).
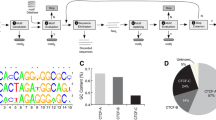
We first examined the proportion of cCREs overlapping with G4 motifs (Fig. 1A). More than half of the PLS elements (57.2%, Fig. 1B) overlap with G4 sequences, followed by pELS elements (35.3%, Fig. 1B). The dELS and DNase-H3K4me3 groups had around 20% of their elements overlapping with G4 sequences (Fig. 1B). The lowest overlap was observed in the CTCF-only group, with only 13.6% of its elements overlapping with G4 sequences (Fig. 1B).
Overview of the enrichment patterns of G4s on candidate cis-regulatory element (cCREs). A Illustration of the definitions of G4-associated cCRE, cCRE-associated G4, etc. We define a cCRE that has an overlap with G4 as a G4-associated cCRE, and similarly, a G4 that has an overlap with a cCRE is defined as a cCRE-associated G4. The proportion of bases on a cCRE that participate in the composition of the G4 sequence is referred to as the coverage of G4 on that cCRE. B Pie charts illustrate the proportion of cCREs (PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 elements) that contain G4s or not. Red represents the proportion of cCREs that contain G4s, while gray represents the opposite. PLS: promoter-like signatures; pELS: proximal enhancer-like signatures; dELS: distal enhancer-like signatures; CTCF-only: high DNase and CTCF signals only. A complete list of abbreviations is provided at the end of the article. C The fold enrichment of G4 in different categories of cCREs compared to the whole genome, with dark and light bars representing the differences in fold enrichment with and without GC correction, respectively
We assessed the fold difference of G4 enrichment across different categories of cCREs compared to the whole genome. We found that the fold difference in G4 enrichment was highest in PLS and pELS elements (Fig. 1C). Given that G4s are, by definition, formed by G-rich sequences, we then corrected for GC content using a method adapted from Guiblet et al [20]. This adjustment considers that both G-rich and C-rich regions (indicating the presence of G4s on the reverse strand) in the human genome have the potential to form G4 structures, while the cCREs defined by the ENCODE project do not include strand information (see methods). After correction, we observed a decrease in fold difference for all cCRE categories (Fig. 1C). However, G4s still exhibited relatively higher fold enrichment compared to the whole genome in most of the cCRE categories, except for the CTCF-only elements (Fig. 1C). Additionally, G4s in different cCRE categories exhibited variations in stability (Kruskal-Wallis test, P = 0), with those in PLS elements showing the highest stability (Additional file 1: Fig. S1A; Wilcoxon test, P < 2.2×10-16 holds true for all comparisons using PLS-G4s as baselines; Additional file 2: Table S1). Moreover, the enrichment pattern of stable G4s in PLS elements showed the most pronounced divergence from that of relatively unstable G4s (Additional file 1: Fig. S1B).
G4s are enriched at CTCF-bound cCREs accompanied by H3K4me3 or H3K27ac signals
We and others previously reported that G4s are tightly associated with the binding of CTCF, with CTCF binding sites significantly enriched around G4s [21,22,23]. However, in this study, our analysis showed a weak correlation between G4s and CTCF-only cCREs, which seems to challenge the existing conclusions and findings. Indeed, other groups of cCREs may also contain CTCF binding (CTCF-bound) signals that, together with enhancer (H3K27ac) or promoter (H3K4me3) signals, forming the basis of PLS and ELS elements. To elucidate the precise association patterns between G4s and CTCF binding elements, we proceeded to perform comparative analysis between CTCF-only cCREs, which exclusively contain CTCF-bound signals, and cCREs containing both CTCF and H3K4me3 or H3K27ac signals (referred to as CTCF-hybrid cCREs or CTCF-hybrid elements for simplicity, Fig. 2A).
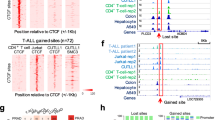
Association mode of G4s with cCREs exhibiting different CTCF occupancy patterns. A Flowchart illustrates our approach to categorize cCREs based on the occupancy patterns of CTCFs. B Z-score values for the enrichment of CTCF-only cCREs compared to CTCF-hybrid cCREs on TAD boundaries of different sizes (5, 10, 20 or 50 kb) in different cells or tissues. Red cell indicates that the enrichment level of CTCF-only cCREs at that TAD boundary is higher than CTCF-hybrid cCREs, while blue cell indicates a lower enrichment level. C Relative density of chromatin loops aound CTCF-hybrid and CTCF-only cCREs, with blue and brown curves signify CTCF-hybrid and CTCF-only cCREs, respectively. D Proportion of CTCF-hybrid and CTCF-free cCREs containing G4s. E, F Relative density of G4 ChIP-seq high-confidence peaks around CTCF-hybrid and CTCF-free cCREs for K562 (E) and HepG2 (F) cell lines. The relative density G4 ChIP-seq high-confidence peaks around CTCF-only cCREs was also put on the same scale for comparison
Considering that G4s are preferentially located at the boundaries of topologically associating domains (TADs) [21], we suspected that the distribution of CTCF-only and CTCF-hybrid elements at TAD boundaries would differ. We compared the enrichment differences of CTCF-only and CTCF-hybrid elements at TAD boundaries across various cell lines and tissues (see methods). As expected, the enrichment level of CTCF-hybrid elements was higher than CTCF-only elements in most cell lines and tissues (Fig. 2B). A similar phenomenon was found when we focused on cohesin-mediated anchor regions of chromatin loops (Fig. 2C). That is, CTCF-hybrid elements are more enriched at the anchor regions compared to CTCF-only elements (Fig. 2C). We speculate that the differential enrichment of CTCF-hybrid elements in specialized regions of higher-order chromatin structure may facilitate a closer association with G4s, as compared to CTCF-only elements.
In addition, we separately calculated the distribution of G4 sequences around CTCF-hybrid and CTCF-only cCREs. CTCF-hybrid cCREs are more enriched in G4 sequences in their vicinity than CTCF-only cCREs (Additional file 1: Fig. S2A), indicating a clear association between G4 and CTCF binding. However, the simultaneous presence of enhancer or promoter signals in CTCF-hybrid cCREs makes it challenging to rule out the possibility that the association of G4s with CTCF-hybrid cCREs might be due to enhancers or promoters alone. Therefore, we compared the differences in the proportions of CTCF-hybrid and CTCF-free cCREs overlapped with G4s. The results showed that a higher proportion of cCREs were associated with G4s when they simultaneously contain CTCF signals, which implies a genuine positive close connection between CTCF binding and G4 localization, but only for the PLS and pELS cCRE groups (Fig. 2D). Of note, the G4 density around CTCF-hybrid cCREs tended to be the highest (Additional file 1: Fig. S2B). To further confirm this association, we constructed logistic regression models to predict the inclusion of G4s in cCREs. For PLS and DNase-H3K4me3 elements, which are predominantly marked by H3K4me3, we modeled using H3K4me3, CTCF, and their interaction term as features; for ELS elements marked by H3K27ac, we modeled using H3K27ac, CTCF and their interaction term as features. As a result, we found that the interaction terms of CTCF with H3K4me3 or H3K27ac both had a positive and significant effect, while CTCF alone did not share this property and might even have an opposite effect (Additional file 1: Fig. S3). This suggests that the coexistence of CTCF and epigenetic signals in cCREs is associated with G4 occupancy, which is consistent with the above observations.
Since the association between CTCF binding and G4 localization can be explained by CTCF binding preference for G4 structures [22, 23], we subsequently conducted cell type-specific analysis to investigate whether there are any differences in the occupancy of G4 structures around cCREs with different CTCF binding states. We obtained cCREs for the K562 and HepG2 cell lines from the SCREEN database and, in addition, retrieved high-confidence G4 ChIP-seq peak data for these two cell lines (see methods). Similarly, we divided the elements in different cCRE groups into CTCF-hybrid and CTCF-free subgroups and calculated the distribution of G4 ChIP-seq peaks around these subgroups. We also calculated the distribution of G4 ChIP-seq peaks around CTCF-only cCREs for comparison. Indeed, we observed an enrichment in G4 structures around the CTCF-only cCREs (Additional file 1: Fig. S4A-B), but this enrichment almost disappeared when we compared the CTCF-only cCREs with other cCREs at the same scale (Fig. 2E-F), indicating that the enrichment of the G4 structures around the CTCF-only cCREs is much less significant than that around the other types of cCREs. Moreover, the average distribution density of the G4 secondary structures around these cCREs tends to be higher when CTCF binding signals are present (Fig. 2E-F), suggesting that the co-occurrence of CTCF and H3K4me3 or H3K27ac signals is associated with a stronger G4 structure occupancy, and G4 structures do not strongly preferentially present in open chromatin regions containing only CTCF signals.
cCRE-G4s are inclined to possess regulatory significance and are under negative selection in the human genome
The preferential distribution of G4s on cCREs raises a question: could variants in cCRE-G4s potentially affect the regulatory functions of the genome? We then conducted a comprehensive investigation into the regulatory potential of all variants mapped to G4 or cCRE regions. For this, we used RegulomeDB [24], which leverages epigenomic data to provide in-depth functional interpretation of variants or regions in the human genome. First, we found that the variants in PLS-G4s exhibited the highest RegulomeDB scores, while the regulatory variant scores for cCRE-G4s were significantly higher than for other G4s, suggesting the potential significant regulatory roles of cCRE-G4s (Fig. 3A; Wilcoxon test, P < 2.2×10-16 holds true for all comparisons using PLS-G4s and other-G4s as baselines, respectively; Additional file 2: Table S2). This observation remains valid when examining the RegulomeDB ranking scores in these G4 regions, where cCRE-G4s showed a higher proportion of high-ranking scores (Additional file 1: Fig. S5A). Furthermore, we inspected whether the regulatory scores of variants in cCREs associated with G4s or not would show differences. As a result, variants in G4-associated cCREs displayed higher regulatory scores and higher proportions of high-ranking scores (Additional file 1: Fig. S5B-C; Wilcoxon test, P = 1.76×10-70, P = 1.04×10-210, P = 0, P = 1.27×10-86, and P = 3.42×10-62 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively), which implies that the presence of G4s may enhance the regulatory potential of their corresponding cCREs.
Overview of RegulomeDB and LINSIGHT scores for G4s and cCREs. A The RegulomeDB probability scores of variants located in different groups of G4s. The y-axis shows the RegulomeDB probability scores for all variants assigned to G4s located in different groups of cCREs. B Boxplot showing the differences in RegulomeDB probability scores for variants located in G4 G-runs versus variants in other nucleotides of G4s (the remaining nucleotides that are not contribute to G-runs) across five groups of cCREs. Wilcoxon test, *: P < 0.05, **: P < 0.01, ****: P < 0.0001. C Similar to (B), with the distinction that we consider the differences in RegulomeDB probability scores for variants within G-runs of G4s and cCREs. Wilcoxon test, ****: P < 0.0001. D The average LINSIGHT scores for different groups of G4s. The mean and the standard deviation error limits are visualized by error plots. E The average LINSIGHT scores for G4 G-runs and the remaining parts of G4s excluding G-runs (other portion) in all five groups of cCREs. The red and blue error plots indicate the mean and the standard deviation error limits of the average LINSIGHT scores for G4 G-runs and the other portion in G4s. Wilcoxon test, *: P < 0.05, ****: P < 0.0001. F Resembling (E), with the comparison conducted between G4 G-runs and cCRE G-runs. Wilcoxon test, ****: P < 0.0001
Given the important role of G-runs for the stability of the G4 sequence, we next compared the regulatory scores of variants harbored in G-runs within G4 sequences with those in the remaining sequences of G4s. Generally, nucleotides within runs of three or more G are more likely to be involved in the formation of a G-tetrad, although this is not absolute since the actual formation of G-quadruplex secondary structures may involve complex sequence patterns. We found that the variants in PLS and ELS G-runs present higher regulatory scores compared to the remaining variants in G4 sequences (Fig. 3B; Wilcoxon test, P = 8.07×10-29, P = 4.55×10-32, P = 1.58×10-51, P = 3.61×10-2, and P = 9.25×10-3 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively), while also having a higher proportion of 1a ranking scores (see methods; 1a corresponding to the category with the highest likelihood being functional; Additional file 1: Fig. S5D). However, the regulatory potential of variants in G4 sequences, especially those in G-runs, may be influenced by the nucleotide composition. To eliminate this effect, we proceeded to analyze the regulatory score differences of variants in G-runs derived from G4s and those originating from cCREs that are not involved in the composition of G4 sequences (see methods). The results revealed that variants within the G-runs of G4s exhibited higher regulatory scores compared to those found in cCRE G-runs (Fig. 3C; Wilcoxon test, P = 1.94×10-130, P = 1.82 ×10-166, P = 0, P = 7.27×10-9, and P = 1.78×10-7 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively), along with a higher proportion of 1a ranking scores (Additional file 1: Fig. S5E). This suggests that variants occurring in G-runs of G4 sequences have a greater potential impact on genome regulation, while the functionality of G4s cannot be simply explained by their high G/C content.
Since G4s are potential functional elements within human cCREs, are they less prone to mutation in the human genome? With this question, we then assessed whether cCRE-G4s in the human genome are under negative selection using the similar approach as mentioned above, but with the assistance of LINSIGHT scores (see methods) [25]. LINSIGHT employs a generalized linear model to infer the probability of negative selection in non-coding regions of the human genome, evaluating the functional impact and potential risk of variants in specific regions, such as enhancers [25]. First, we observed that G4s located in the PLS and pELS regions displayed relatively higher average LINSIGHT scores (Fig. 3D; Wilcoxon test, P < 2.2×10-16 holds true for all comparisons using PLS-G4s and pELS-G4s as baselines, respectively; Additional file 2: Table S3). This suggests that G4s in these regions are under stronger negative selection, making variants in these G4 regions more likely to be deleterious. Intriguingly, we found that PLS and ELS (pELS and dELS) cCREs containing G4 sequences also had higher average LINSIGHT scores (Additional file 1: Fig. S6; Wilcoxon test, P = 0, P = 0, and P = 0 for comparisons in the PLS, pELS, and dELS groups, respectively), implying that the presence of G4 sequences may render these regions more susceptible to negative selection rather than neutral selection, which could be intertwined with the potential functional role of G4s, despite the fact that the average LINSIGHT scores for G4-associated cCREs in both CTCF-only and DNase-H3K4me3 groups were slightly lower compared to non-G4-associated cCREs (Additional file 1: Fig. S6; Wilcoxon test, P = 1.10×10-3 and P = 3.12×10-13 for comparisons in the CTCF-only, and DNase-H3K4me3 groups, respectively). Besides, the average LINSIGHT scores for G4 G-runs were also greater than those of the remaining part of the G4 sequences (Fig. 3E; Wilcoxon test, P = 9.46×10-58, P = 1.64×10-39, P = 8.36×10-18, P = 3.76×10-2, and P = 2.47×10-2 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively). We propose that G4 G-runs play a critical role in stabilizing these secondary structures and are therefore subject to higher constraints in the human genome.
To further confirm that the stronger negative selection on G4 G-runs was not simply caused by high G/C content, we then examined the average LINSIGHT scores of G-runs in cCREs that are not involved in G4 sequence composition. Consequently, we observed that the average LINSIGHT scores of the G4 G-runs were significantly higher than those of the cCRE G-runs in the PLS, ELS, and DNase-H3K4me3 groups (Fig. 3F; Wilcoxon test, P = 0, P = 0, P = 0, P = 1.65×10-231 for comparisons in the PLS, pELS, dELS, and DNase-H3K4me3 groups, respectively). This indicates that the heightened negative selection on G4 G-runs was not merely due to the nucleotide composition of G4 sequences, though we noted that the average LINSIGHT scores for G4 G-runs in the CTCF-only group were lower (Fig. 3F; Wilcoxon test, P = 2.63×10-60).
Finally, we used the human genome-wide mutational constraint map from the gnomAD (Genome Aggregation Database) database [26], which aggregates large-scale human variants data, for further quantification. Due to the resolution limit of the constraint map (1 kb), we were unable to accurately compute and compare the constraints on regions with shorter widths, such as G4 G-runs and cCRE G-runs. Therefore, we no longer discuss the gnomAD constraint differences between them. Overall, the G4s in PLS and pELS presented higher constraint z-score values (Additional file 1: Fig. S7A; Additional file 2: Table S4), which indicates that the observed number of variants in these G4 regions was lower than expected. This is consistent with the results based on the LINSIGHT model, except that the gnomAD constraint z-scores were derived from a substantial amount of human genomic mutation data. Moreover, we found that those cCREs containing G4 sequences showed higher constraint z-score values as well (Additional file 1: Fig. S7B; Wilcoxon test, P = 1.87×10-230, P = 0, P = 0, P = 1.14×10-95, and P = 2.47×10-148 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively).
In general, these findings suggest that the presence of G4s makes the cCRE sequences more constrained, potentially attributable to the significant genomic regulatory functions associated with G4s.
G4 G-runs in cCREs near the transcription start site are highly constrained across 241 mammalian genomes
We obtained the phyloP scores from the Zoonomia Project database, which are derived from the alignment of 241 mammalian genomes. These scores were then used to estimate the evolutionary constraints on G4s, as well as cCREs, and to determine whether G4s in cCREs were under selective pressures during evolution. Similar to our aforementioned findings, G4s in PLS elements are under greater selection pressure, followed by the G4s in pELS. In contrast, the remaining three groups of G4s and those G4s that are not associated with cCREs undergo lower selection pressure (Fig. 4A; Additional file 2: Table S5). Moreover, the average phyloP scores of G4 G-runs tend to be higher than the remaining sequences in G4s (Fig. 4B; Wilcoxon test, P = 0, P = 0, P = 0, P = 1.13×10-187, and P = 0 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively). Nevertheless, when we focus on the comparison of average phyloP scores between G4s and cCREs, we found that although the average phyloP scores of G4s in pELS are higher than those in pELS cCREs, for the other three groups of cCREs (including dELS, CTCF-only, and DNase-H3K4me3 groups), the average phyloP scores of G4s in them are lower (Additional file 1: Fig. S8; Wilcoxon test, P = 3.14×10-1, P = 8.46×10-87, P = 0, P = 0, and P = 5.01×10-268 for comparisons in the PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 groups, respectively). A similar phenomenon was observed when we analyzed the average phyloP scores of the G4 G-runs and the cCRE G-runs that do not comprise G4 sequences (Fig. 4C). Specifically, the average phyloP score of G4 G-runs in PLS and pELS was higher than that of G-runs in cCREs (Fig. 4C; Wilcoxon test, P = 0 and P = 9.05×10-60 for comparisons in the PLS and pELS groups, respectively). However, in the other three groups of cCREs, the situation is reversed (Fig. 4C; Wilcoxon test, P = 0, P = 4.39×10-106, and P = 5.88×10-66 for comparisons in the dELS, CTCF-only, and DNase-H3K4me3 groups, respectively). Since PLS and pELS are elements located relatively close (< 2 kb) to the transcription start site (TSS), we suspect that those G4s further away from the TSS may not be as conserved compared to cCREs during the process of evolution.
Assessing the extent of constraints on cCRE-G4s in mammalian evolution. A The average phyloP scores for different groups of G4s, with dots and ranges indicate the mean and standard deviation errors, respectively. B, C The differences in average phyloP scores between G4 G-runs and other portions in G4s across the five types of cCREs (B). While in (C), the error plots depict the distinctions between G4 G-runs and cCRE G-runs in different cCRE groups. Wilcoxon test, ****: P < 0.0001. D-H The distribution pattern of constraint bases around G4 G-runs and cCRE G-runs in PLS, pELS, dELS, CTCF-only, and DNase-H3K4me3 cCREs, respectively. Red and blue curves denote the constraint base density surrounding G4 G-runs and cCRE G-runs, respectively. (I) Proportion of G-runs in G4s and cCREs covered by constraint bases across all five groups of cCREs. The error bars correspond to the standard deviations
To further confirm this, we first defined bases with phyloP scores ≥ 2.27 as constrained bases, using the threshold proposed by Sullivan et al. [27]. Subsequently, we calculated the distribution of constrained bases upstream and downstream of G4s that overlap with cCREs, as well as the distribution of constrained bases around cCREs. Surprisingly, the constrained bases exhibited a localized depletion around cCRE-G4s, while they displayed an enriched distribution in the vicinity of cCRE centers (Additional file 1: Fig. S9A-E). Considering that the bases within G4 sequences that are not part of G4 G-runs are likely to be less conserved, we proceeded to investigate the distribution of constrained bases centered on both G4 G-runs and cCRE G-runs, respectively. We noticed that G4 G-runs are more constrained than cCREs G-runs in PLS and pELS elements (Fig. 4D-E), while the opposite is true in the remaining three groups of cCREs (Fig. 4F-H), which is consistent with our analysis above. The similar phenomenon was observed when we calculated the proportion of constrained bases in G4 G-runs and cCRE G-runs (Fig. 4I), which strongly suggests that the G-runs in G4s near the transcription start site are highly constrained in mammals, while others may undergo dynamic selection.
The presence of G4s is associated with a favorable local environment for cis-regulatory elements
Previous studies have shown that the presence of G4s is associated with epigenetic marks [28, 29]. We wondered whether the occupancy of G4s in cCREs is also linked to a local environment that is more favorable for the activation or functioning of cCREs. Since the functions of promoters and enhancers are relatively clear, in this section, we only consider PLS and ELS elements.
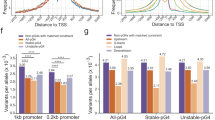
We first divided the cCREs into two subgroups based on whether they overlapped with G4s or not, and then compared the z-score signal values of DNase, H3K4me3, H3K27ac, and CTCF in these two subgroups across all samples utilized in the SCREEN database. We found that G4-associated cCREs exhibited higher DNase signal values in most samples, irrespective of the cCRE type (Fig. 5A). G4-associated PLS elements displayed significantly higher H3K4me3 and H3K27ac z-score signal values in nearly all samples compared to non-G4-associated PLS elements (Fig. 5A). This is also the case in pELS, except that, in a small subset of samples, the H3K27ac signals of G4-associated cCREs were lower than those of non-G4-associated cCREs (Fig. 5A). In most samples, both H3K4me3 and H3K27ac signal values of G4-associated cCREs were significantly greater than those of non-G4-associated cCREs, which was roughly similar to the proportion observed in CTCF signal values (Fig. 5A). Nevertheless, in most samples, we do observe higher values of these epigenetic marks in G4-associated cCREs compared to non-G4-associated cCREs.
Distribution pattern of epigenetic marks surrounding the cCREs associated with G4s or not. A Proportion of the samples used in the SCREEN database that exhibit z-score differences in DNase, H3K4me3, H3K27ac, and CTCF signals between the two subtypes of cCREs associated with G4s or not. Pink bar represents the proportion of samples in which the signal value of G4-associated cCREs on that epigenetic mark is significantly greater than that of non-G4-associated cCREs, while blue bar indicates the opposite. Green represents the proportion of samples where there is no difference between the two subgroups. PLS, pELS and dELS cCREs are considered separately. B Loop anchor distribution around different groups of cCREs (PLS, pELS, dELS). The red curve represents the density of loop anchors around G4-associated cCREs, while the brown curve shows the density of loop anchors around non-G4-associated cCREs. (C) Similar to (B), except that the density of the TF binding motif is calculated. D Heatmap of the unmethylated intensity around G4-associated and non-G4-associated PLS, pELS, dELS cCREs in diverse tissues with red blocks represent higher unmethylated intensity, while blue blocks indicate lower unmethylated intensity
Given that enhancers can interact with promoters through spatial proximity, thereby regulating gene expression, we were intrigued by the potential differences in the distribution of chromatin loop anchor regions between these two cCRE subgroups. For this purpose, we retrieved data on human pan-cell type cohesin-mediated chromatin loops [30] and calculated the distribution densities of loop anchors around them. As expected, G4-associated cCREs tended to be preferentially distributed in anchor regions compared to non-G4-associated cCREs (Fig. 5B). This is particularly pronounced in dELS elements, indicating that the presence of G4s may enhance the regulation of gene expression by cCREs through three-dimensional mechanisms, which is in line with our previous findings [21]. Such difference cannot be simply attributed to the association between G4s and enhancer signals (H3K27ac), as indicated by the logistic regression models (Additional file 1: Fig. S10). Additionally, using a similar approach, we also calculated the distribution density of transcription factor binding sites around these two subgroups of cCREs. We utilized the motif instances derived from the study of Andrews et al. [31] and the transcription factor binding sites sourced from the ReMap2022 database [32] for our calculations. The motif instances of transcription factors exhibited a relatively higher density around G4-associated cCREs (Fig. 5C), and the same trend was also observed when analyzing the transcription factor binding sites data provided by the ReMap2022 database (Additional file 1: Fig. S11A-C), suggesting that the presence of G4s in cCREs may enhance the interaction between transcription factors and cCREs, which coincides with a previous study indicating that G4s serve as hubs for transcription factor binding [33]. Considering that DNA methylation can influence the activity of cCREs [3], we next examined whether the occurrence of G4s might be potentially associated with a lower methylation landscape in cCREs. In this regard, we characterized the intensity of genome-wide unmethylated regions in human cells in the vicinity of cCREs in both subgroups. As a result, hypomethylation patterns were observed around G4-associated cCREs in most cell types (Fig. 5D). These patterns were particularly pronounced in both PLS and pELS elements, suggesting that the presence of G4s may lead to a low methylation state in cCRE regions, making them more susceptible to activation and utilization. Furthermore, by establishing logistic regression models to determine the presence of unmethylated blocks within cCREs, we found that G4s significantly contribute to the hypomethylation tendency of cCREs, suggesting that G4s can act as independent regulatory units to shape the methylation landscape of cCREs (Additional file 1: Fig. S12A-C).
The G4 structure itself, and not simply its associated G-rich sequence, is associated with cCRE activation
To confirm whether it is the G4 secondary structure or the corresponding G-rich sequence that is associated with the activation of cCREs, we parsed the pattern of linkage between cell-type specific G4s and cCREs in the K562 and HepG2 cell lines. We initially evaluated the proportion of G4s supported by G4 ChIP-seq experiments (hereafter referred to as endogenous G4s) that were located in activated cCREs (e.g., PLS elements, see methods). Besides, the background proportion was also estimated without considering whether G4s are supported by ChIP-seq experiments or not (see methods). As a result, endogenous G4s are overwhelmingly located in activated cCREs rather than quiescent ones, especially in PLS elements in both K562 and HepG2 cells (Fig. 6A). Interestingly, endogenous G4s are selectively enriched around activated cCREs, with no apparent enrichment observed around quiescent cCREs (Additional file 1: Fig. S13A-B). In contrast, those G4s that do not form secondary structures exhibit an association with cCREs independent of their activation status (Additional file 1: Fig. S13A-B). For instance, in K562 cells, we observed the enrichment of those G4s around both activated and quiescent PLS regions (Additional file 1: Fig. S13A). This suggests that the activation of cCREs cannot be determined by G4-compatible sequences alone and potentially requires the formation of secondary structures.
Comparing the association between G4 structures and G4 sequences with cCRE activation. A The histogram shows the statistics of the proportion of G4s located in the activated cCRE in each of the 1,000 randomized experiments. Both G4 ChIP-seq supported G4s (orange) and background (all G4s; grey) proportions were estimated separately. Up-panel: K562 cell line; bottom-panel: HepG2 cell line. B Comparison of the average binding coverage of transcription factors on two types of G4s with and without G4 ChIP-seq experimental support. Red dots: the average binding coverage of this transcription factor on G4s with G4 ChIP-seq support is higher than that on other G4s and is higher than 0.5. Orange dots: similar to red dots, except that the average binding coverage is lower than 0.5. Green dots: the average coverage of this transcription factor binding on G4s with G4 ChIP-seq support is lower than that on other G4s. Top panels: K562 cells; bottom panels, HepG2 cells. C Differences in average methylation levels between K562 cell line (top panel) and HepG2 cell line (bottom panel) in regions (center ± 100 bp) where endogenous G4s (with ChIP-seq support, orange) and other G4s (grey) are located. Wilcoxon test, ****: P < 0.0001. D Similar to (C) but calculating the differences in ATAC-seq signals
To further investigate the connection between G4 structures, as opposed to sequences, and the activation of cCREs, we then performed a differential analysis of the transcription factor occupancy levels in the regions where two classes of G4s (with or without ChIP-seq support) are located. We found that almost all transcription factors preferentially bind to endogenous G4 regions (Fig. 6B), with some previously reported to interact with G4s, including SP1 [34, 35] and YY1 [36] (Fig. 6B). In addition, regions containing endogenous G4s in cCREs exhibit lower methylation levels (Fig. 6C; Wilcoxon test, P = 1.12×10-34, P = 1.19×10-52, P = 4.64×10-112 for comparisons of PLS, pELS, and dELS groups in K562 cells, respectively; P = 2.24×10-8, P = 6.02×10-17, P = 4.28×10-41 for comparisons of PLS, pELS, and dELS groups in HepG2 cells, respectively) and more open chromatin state (Fig. 6D; Wilcoxon test, P = 0, P = 0, P = 0 for comparisons of PLS, pELS, and dELS groups in K562 cells, respectively; P = 0, P = 6.71×10-295, P = 1.94×10-179 for comparisons of PLS, pELS, and dELS groups in HepG2 cells, respectively). We speculate that the G4 structures may regulate the activation of cCREs by shaping the local epigenetic environment.
G4-associated cCREs are extensively activated in cancers
We then investigated whether G4-associated cCREs are more inclined to be activated in cancer cells, given the higher levels of G4s in cancer tissues compared to non-malignant tissues [7, 37]. To investigate this, we collected the pan-cancer chromatin accessibility profiles, along with the pan-cancer enhancer data inferred from the H3K27ac signals and the promoter activity estimated from the pan-cancer RNA-seq data (see methods).
We first examined the differences in distribution density and average coverage of ATAC-seq peaks between two distinct subgroups of cCREs: those associated with G4s and those not, across various types of cancers. Our findings revealed that the distribution density and average coverage of ATAC-seq peaks in G4-associated cCREs were higher than that of non-G4-associated cCREs in all cancer types (Fig. 7A-B, Additional file 1: Fig. S14-S17). The proportion of G4-associated cCREs overlapping with ATAC-seq peaks was also higher than that of the remaining cCREs in all types of cancers (Additional file 1: Fig. S18). This suggests that the presence of G4s in cCREs potentially correlates with a more accessible chromatin in cancers, thereby establishing a favorable environment for the activation of cCREs. Next, we evaluated the activation status of enhancer-like cCREs in various types of cancers. We discovered an elevated density of typical enhancers around G4-associated pELS elements in different cancers (Fig. 7C), while the disparity of enrichment is particularly prominent in the vicinity of dELS (Fig. 7D). Since the PLS element is potentially a cis-regulatory element acting as a promoter, we then assigned PLS elements to their nearest promoters and directly substitute the PLS activity with promoter activity in various cancer tissues (see methods). As a result, G4-associated PLS elements exhibited relatively higher promoter activity (Fig. 7E; Wilcoxon test, P < 2.2×10-16 for all cancer types), independent of the type of cancer. Besides, a higher proportion of G4-associated PLS elements were highly activated across different cancer types (Additional file 1: Fig. S19).
Quantification of the differences in pan-cancer chromatin accessibility, enhancer density, and promoter activity between G4-associated and non-G4-associated cCREs. A Heatmap and density plots of the ATAC-seq signals around G4-associated and non-G4-associated PLS elements. Heatmap legend indicating the ATAC-seq signal values around G4-associated PLS elements and non-G4-associated PLS elements in different cancer types. B The average coverage of pan-cancer ATAC-seq peaks in G4-associated cCREs and non-G4-associated cCREs. C The distribution of pan-cancer enhancers upstream and downstream of G4-associated pELS elements and non-G4-associated pELS elements is depicted. The Y-axis represents the magnitude of relative density. D Same as (C), but the distribution of enhancers around dELS was examined. E Boxplot illustrates the disparities in promoter activity between G4-associated PLS elements (in red) and non-G4-associated PLS elements (in yellow) in different cancer types. Wilcoxon test, ****: P < 0.0001
Furthermore, we downloaded cancer-specific gained regulatory sequences from the TSCRE (Tumor-specific Cis-Regulatory Elements) database [38] to examine whether the activity of G4-associated cCREs might be specifically enhanced in cancers. We analyzed cancers that contained both H3K27ac and H3K4me3 marks data. As a result, G4-associated cCREs exhibited a higher enrichment of cancer-specific gained regulatory sequences in most of the cancer types (Additional file 1: Fig. S20). Nonetheless, this pattern did not hold true for all cancer types (Additional file 1: Fig. S20), suggesting that the association between G4-associated cCREs and cancer-specific gained regulatory sequences may vary depending on cancer types.
In conclusion, our analysis suggests that G4-associated cCREs are also susceptible to be activated and used in cancers.
Discussion
Over the past decades, researchers have conducted extensive investigations into the physicochemical properties of G4s. However, in contrast, the study of the biological roles of G4s has only flourished in the last decade, thanks to the explosion of omics data. G4s are highly likely to become widely recognized as regulatory elements in the future. While relatively short, a G4 motif can mediate and regulate various biological functions in a non-coding manner due to its unique secondary structure. Our study addressed a fundamental question: are cis-regulatory elements in non-coding regions of the human genome associated with G4s? In accordance with our research findings, we suggest that G4s are crucial components of some, but not all cis-regulatory elements in the human genome, and G4s may even constitute a core part of them, although not all cCREs are necessarily linked to G4s (Fig. 1B).
We noticed that the association between cCREs and G4s varies among different groups. Specifically, cCREs closer to TSS show a potentially stronger association with G4s, primarily including PLS and pELS elements. This is not unexpected, as G4s are inherently located near transcription start sites (TSSs), especially in upstream regions [39]. Interestingly, we observed that the G4Hunter scores of PLS-G4s tend to be the highest. G4Hunter scores can be used to characterize the stability of G4 structures, at least in vitro, with higher G4Hunter scores often indicating greater stability of G4 secondary structures [8]. Typically, G4s exert their biological functions mainly through the formation of folded secondary structures [40]. This aligns with their enrichment near TSSs, suggesting that G4s in cCREs near TSSs potentially have stronger biological functions. Besides, these G4s are more closely associated with cCREs, possibly serving as their core functional elements, which is consistent with the results we obtained from a conservation perspective in mammalian genomes, namely that G4 G-runs near the TSS regions are more conserved compared to cCRE G-runs in the vicinity of TSSs (Fig. 4D-E). A previous study also found that G-runs in promoter regions are indeed conserved [41]. However, we suggest here that this may also be the case for some enhancers provided they are relatively close to a transcription start site. Besides, we observed differences in evolutionary constraints among different cCRE groups. This holds true for both humans and across species. It should also be noted that the conservation of G-runs is surprisingly reduced when far from the TSS compared to that in cCREs across species (Fig. 4C; Fig. 4F-H). We hypothesize that these G4s may not be strongly selected, thus contributing to the rapid evolution of those elements, which also reflects the diversity of species. Interestingly, this trend is not in agreement with the conclusions drawn from the RegulomeDB regulatory scores (Fig. 3C). We speculate that this discrepancy may arise from the fact that the CREs we studied are regulatory elements within the human genome, whereas the RegulomeDB annotation relies on human epigenome data to infer regulatory scores. Consequently, this reveals the unique characteristics of G4s within the regulatory regions of the human genome. Strikingly, we only observe a weak association between CTCF-only cCREs and G4s, which was unexpected, as both our prior work and that of others had indicated a significant correlation between CTCF binding sites and G4 localization [21, 22]. Upon closer examination, it becomes evident that there are prerequisites for the association between G4s and CTCF-bound cCREs. When only CTCF signals are present, the association between cCREs and G4s is weak; however, when CTCF-bound cCREs also possess H3K4me3 or H3K27ac signals, a stronger association with G4s emerges (Fig. 2D). This strong association cannot be simply explained as a correlation between G4s and H3K4me3 or H3K27ac signals themselves, because when we classified cCREs based on CTCF alone, a higher proportion of cCREs containing CTCF signals were associated with G4s, especially in PLS and pELS elements. This difference becomes even more pronounced when we directly observe cCREs and G4s in the K562 and HepG2 cell lines. We speculate that the interaction between CTCF and G4 in regions such as PLS or ELS may contribute to the formation of chromatin loops and other chromatin conformations, thereby facilitating the function of PLS or ELS elements.
Previous studies have suggested an association between G4s and various epigenetic marks [28, 29, 42]. In this study, we showed that the presence of G4s is often linked to the local chromatin environment that supports the activation or functioning of cCREs, such as increased occupancy of transcription factors and a higher unmethylated state. This prompts us to formulate a hypothesis that some cCREs may become regulatory elements precisely because of the presence of G4s. In other words, the presence of G4s is an important factor ensuring their biological functions. Intriguingly, we found that it is the G4 structures rather than the sequences that is associated with the activation of cCREs (Fig. 6). The G4 structure regions often display higher transcription factor occupancy as well as lower methylation levels, creating a favorable epigenetic environment for the activation of cCREs. Nevertheless, the relationship between the formation of G4 secondary structures and changes in the epigenetic environment is akin to a 'chicken and egg' problem, rendering it difficult for us to make causal inferences via computational methods. Yet, existing research has provided us with some clues. For instance, it has been confirmed that G4 structures can shape the local hypomethylation landscape of the genome [43]. Additionally, G4 structures can interact with transcription factors, serving as hubs for their binding [33]. Investigating how G4 structures influence the local chromatin environment and, consequently, impact cis-regulatory elements will be a very intriguing topic.
Interpreting the consequences of variants in noncoding regions has long been a challenging endeavor [44, 45]. Our study indicates that variants in G4s, especially those within G-runs that are likely to be important for the formation of G4 structures, tend to possess a higher regulatory potential and therefore tend to be conserved, as substitutions often lead to deleterious variants. This indirectly underscores the importance of G4s in cCREs. This aligns with previous studies [20, 46]. However, it is crucial to note that we observed differences in regulatory or evolutionary constraints among different cCRE groups. Using data from the Zoonomia Project indicates that G4s in distinct cis-regulatory regions have experienced varying levels of selection during evolution. Thus, we recommend that G4 should be considered as a feature when developing new models or algorithms to predict the regulatory capacity of non-coding regions.
We noted that, in multiple cancers, G4-associated cCREs are indeed more active, as evidenced by higher levels of chromatin accessibility, promoter activity, as well as enhancer activity. We also observed a clear enrichment tendency of cancer type-specific gained regulatory sequences in G4-associated cCREs. Since cis-regulatory elements also regulate the transcriptome expression of cancer cells or tissues, it would be crucial in the future to further identify those G4-associated cCREs that are aberrantly activated compared to adjacent non-cancerous tissues. We also look forward to the extensive application of G4 ChIP-seq technology in the identification of G4 secondary structures in cancer tissues as well as their paired normal tissues. Identifying G4s in normal cell lines that match the cancer cell line types will be essential to understand which G4 structures are abnormally formed in cancer, whether these abnormally formed G4 structures can affect their associated cis-regulatory elements, and ultimately, whether they can influence the regulation of target genes, potentially leading to tissue carcinogenesis. However, due to the limitations in G4 sequencing technology, we are currently unable to obtain such sample data and analyze the structural alteration of G4s in cancer tissues in conjunction with the activation status of cCREs.
Conclusions
In summary, our study reveals the crucial role of G4s in human cis-regulatory elements, establishing their central position in the genomic regulatory network. G4s are crucial regulatory components of cCREs, and not only promoters; they not only serve as the basic backbone of these elements but also as key enhancers of their regulatory capacity. The formation of G4 structures can provide strong support for the activation of cis-regulatory elements, which may be attributed to their complex interactions with epigenomic marks as well as trans-acting factors. Our findings lay the groundwork for a deeper comprehension of cis-regulatory elements and the exploration of their underlying mechanisms in future research studies.
Methods
G-quadruplex sequence prediction
We employed the G4Hunter software [8] for the prediction and identification of all possible G-quadruplex sequences in both the forward and reverse strands of the human genome, with a window size of 25 (default value) and a score threshold set at 1.5. The threshold default value is generally 1.2, but the chosen value of 1.5 is more stringent: it decreases the number of potential candidates but ensures each candidate sequence actually forms a G4 structure with more than 98% confidence in vitro. In this study, only the 22 autosomes and the X chromosome were considered and studied (i.e., the Y-chromosome was excluded due to the unavailability of partial epigenetic datasets). Unless otherwise specified, the assembly version of the reference genome we used was hg38. As a result, a total of 1,435,201 human G4 sequences were obtained, with no strand asymmetry (717,551 motifs on the forward strand and 717,650 on the reverse strand), and an average width of 33 base pairs (bp, Additional file 1: Fig. S21A). In a particular section, we also used relatively unstable G4s as a reference control set. The prediction of relatively unstable G4s was consistent with the method described above, using a lower of 1.2 and removing G4s with absolute scores greater than 1.5.
The G4 coverage in specific elements or regions is defined as the proportion of bases covered by G4 sequences. Likewise, G4 density is calculated as the count of G4s divided by the total length of the elements or regions. We excluded the gap regions from our calculations when calculating the coverage and density of G4s across the entire genomic. These gap regions were downloaded from the UCSC Genome Browser. We adapted the method from Guiblet et al. [20] to correct for GC content. They directly corrected for G content; however, in this study, we did not correct based on G content because cCREs are not strand-specific. Instead, we corrected for GC content. This approach is preferable because C-rich regions may indicate the presence of G4s on the opposite strand. The corrected G4 coverage and density are obtained by dividing the measurements with the GC content of these elements or regions; see ref [20] for details.
Endogenous G-quadruplex dataset
The endogenous G4 data for the K562 and HepG2 cell lines were obtained from the Gene Expression Omnibus (GEO) with accession number GSE145090. Unlike the human genomic G4 sequences predicted by G4Hunter, these G4s were captured through G4 ChIP-seq experiments, thereby representing the G4 secondary structures that were formed in specific cellular environments. We utilized high-confidence endogenous G4 data (GSE145090_20180108_K562_async_rep1-3.mult.5of8.bed for K562 cells and GSE145090_HepG2_async_rep1-3.mult.6of9.bed for HepG2 cells) in this study and performed genome assembly version conversion using liftOver software. In this study, when referring to G4s supported by G4 ChIP-seq experiments, it specifically denotes endogenous G4s.
Candidate cis-regulatory elements (cCREs) datasets and annotation
The human candidate cis-regulatory elements (cCREs) were retrieved from the SCREEN (Search Candidate cis-Regulatory Elements by ENCODE) database (Registry V3), which encompasses five distinct groups: promoter-like (PLS), proximal enhancer-like (pELS), distal enhancer-like (dELS), CTCF-only, and DNase-H3K4me3 (Additional file 1: Fig. S21B). These various cCREs groups are primarily classified based on epigenomic signals, including DNase, H3K4me3, H3K27ac, and CTCF, as well as genomic context annotations. In brief, PLS cCREs are characterized by high DNase and high H3K4me3 signals and are located within 200 bp of the nearest transcription start site (TSS). The cCREs with high H3K4me3 signals and low H3K27ac signals, positioned more than 200 bp from the TSS, are considered as DNase-H3K4me3 cCREs. Elements with high H3K27ac signals are regarded as candidate enhancer-like (ELS) cCREs, with their classification also dependent on their distance to the TSS. If the distance is less than 2,000 bp, they will be categorized as pELS (proximal ELS, with low H3K4me3 signal as well). Otherwise, they are designated as dELS (distal ELS). CTCF-only cCREs refer to elements with high CTCF signals but low H3K4me3 and H3K27ac signals. Please note that PLS, ELS, etc. may also contain high CTCF signals. Additionally, chromatin accessibility, as indicated by high DNase signals, is a prerequisite for all cCREs elements.
We annotated the presence of G4s in these cCREs (Fig. 1A). For a given cCRE, we will consider it to be a G4-associated cCRE if it overlaps any of the predicted G4 sequences (Fig. 1A). Likewise, this G4 is referred to as a cCRE-associated G4 (or, more briefly in the manuscript, as a cCRE G4, Fig. 1A). Of note, a very small proportion (0.065%) of G4s may be associated with more than one cCRE. Therefore, in this study, we excluded these G4s when comparing the differences between various categories of G4s, such as the disparities in G4Hunter scores between PLS-G4s and pELS-G4s.
In individual analyses, we also used cell type-specific cCREs data, mainly for the K562 and the HepG2 cell lines, which were also obtained from the SCREEN database.
Relative density calculation
In this study, we extensively calculated the relative density of B objects (e.g., G4s) in both the upstream and downstream regions of A objects (e.g., PLS elements). For instance, we examined the distribution of G4 sequences around different groups of cCREs, as well as the distribution of loop anchors upstream and downstream of G4-associated PLS elements and non-G4-associated PLS elements. All these calculations were performed using the EnrichedHeatmap package (V1.28.1) in R. We utilized the coverage mode of EnrichedHeatmap (V1.28.1) for calculation in all scenarios. The lengths of the upstream and downstream extensions, as well as the resolution of the density calculations, were adapted to the specific scenarios we analyzed, as outlined in the results section.
The principle of the calculation is as follows:
The regions upstream and downstream of the A objects are divided into non-overlapping windows with a width (resolution) of \(L\). The average G4 density \({d}_{i}\) was
where \(N\) represents the number of A objects, while \({w}_{i}^{n}\) signifies the width (\(w\)) of B object that intersects with the \(i\)-th window for \(n\)-th A object.
TAD dataset and boundary definition
We extracted topologically associating domain (TAD) coordinates from the 3D Genome Browser database, using the hg38 version, for a variety of human cell lines and tissues. We defined TAD boundaries following the methods of McArthur & Capra [47] we used in our previous study [15], which involved extending a specific width upstream from the TAD start coordinate and downstream from the TAD end coordinate. For instance, if the coordinates of a TAD were (chr1, X, Y), we then generated two boundary regions, with the coordinates of boundary 1 being (chr1, X-N, X) and the coordinates of boundary 2 being (chr1, Y, Y+N), where N is the length (width) of the extension. In this study, we examined the robustness of the results by testing TAD boundaries with widths of 5 kb, 10 kb, 20 kb, and 50 kb, respectively.
Comparative analysis of cCRE enrichment at TAD boundaries
In this study, we compared the enrichment differences between CTCF-hybrid cCREs and CTCF-only cCREs at TAD boundaries. Assuming we have a set of TAD boundaries specific to a particular cell line or tissue, let the number of CTCF-only cCREs be represented as k. We first determined the count of CTCF-only cCREs that are located inside these boundaries and denoted this count as m. Considering that the number of CTCF-only cCREs is substantially smaller than that of CTCF-hybrid cCREs, we performed 1,000 random samplings. In each of these sampling, k samples were randomly selected from the CTCF-hybrid cCREs collection to form a subset. The number of elements in this subset located at TAD boundaries was signified as n. This process allowed us to estimate the background distribution of the number of CTCF-hybrid cCREs enriched at TAD boundaries when the quantity of CTCF-hybrid cCREs equaled that of CTCF-only cCREs, based on the results obtained from 1,000 random samplings.
We applied z-score to evaluate whether CTCF-hybrid cCREs are more enriched or depleted at TAD boundaries compared to CTCF-only cCREs, that is,
where \(m\) is the total number of CTCF-only cCREs located inside TAD boundaries, whereas \({n}_{i}\) represents the total number of CTCF-hybrid cCREs located inside TAD boundaries in \(i\)-th sampling.
When the z-score is less than 0, it means that the enrichment level of CTCF-only cCREs at TAD boundaries is lower than the average level of CTCF-hybrid cCREs. Conversely, when it is greater than 0, it is higher.
Regression model construction
The glm function in R was used to construct the logistic regression models. The dependent variable is binary, for instance, whether a cCRE contains a loop anchor. The selection of independent variables varies according to different analytical tasks. When considering features, the presence or absence of G4 sequences is treated as a binary variable, while the strength of signals such as CTCF and H3K4me3 are treated as continuous variables. We decide whether to introduce interaction terms based on the specific objectives of the research task.
Cohesin-mediated chromatin loop dataset
The human pan-cell cohesin-mediated chromatin loop data was acquired from Grubert et al. [30]. The genomic coordinates of loop anchors were then mapped to the hg38 assembly version using the liftOver software.
In accordance with different task scenarios, we computed and compared the relative density of loop anchors around CTCF-hybrid and CTCF-only cCREs. Additionally, we also examined the relative density of loop anchors around G4-associated cCREs and non-G4-associated cCREs. The calculation method for relative density have been described above.
Comparison of epigenetic z-score values for different cCREs
The z-score matrix files for DNase, H3K4me3, H3K27ac, and CTCF epigenetic marks were gathered from the SCREEN database, which contains the raw signal data for each ENCODE sample used to identify cCREs. For each epigenetic mark (e.g., DNase) and each cCRE group (e.g., PLS), we examined the z-score differences of this epigenetic mark between G4-associated and non-G4-associated cCREs in each sample separately. Subsequently, we computed the proportion of ENCODE samples in which the z-score values for the epigenetic marks of G4-associated cCREs were significantly higher than those of the non-G4-associated cCREs. We also determined the proportion of samples in which these values were lower or showed no statistically significant distinction.
Transcription factor binding sites and motif data
We downloaded non-redundant human transcription factor binding site data from the ReMap2022 database (https://remap.univ-amu.fr). This dataset consists of a total of 68.6 M binding sites, with an average length of 313 bp. These binding sites were identified using various experimental techniques, including ChIP-seq, ChIP-exo, and others.
In addition, we collected human transcription factor motif data from Andrews et al. [31], resulting in an extensive collection of approximately 25.8 M motif instances. On average, these motif instances are around 11 bp in length. We refrained from merging motif instances derived from different transcription factors since this would obscure the binding preferences of multiple transcription factors for the same region.
We employed the EnrichedHeatmap package (V1.28.1) to compute the disparities in transcription factor binding density between G4-associated cCREs and non-G4-associated cCREs.
DNA unmethylation data acquisition and intensity calculation
We obtained genome-wide unmethylated regions (blocks) of normal human tissues from Loyfer et al. [48], where each of these blocks comprises a minimum of four CpGs, with the majority of CpGs within the block being identified as unmethylated. We used the liftOver software to convert the coordinates of unmethylated genomic blocks from the hg19 version to the hg38 version. We also obtained all of the blocks used in that study (regardless of whether they were methylated or not), which we filtered, retaining only the blocks containing at least four CpGs and employed them as background for comparisons.
First, for each type of tissue, we calculated the distribution density D of background blocks surrounding G4-associated cCREs and non-G4-associated cCREs (window width: 10 bp). Using the same approach, we subsequently computed the distribution density d of unmethylated blocks around them. Then the unmethylated intensities I of the window n around the two categories of cCREs in human tissue t can be computed as follows,
where the determination of \({d}_{n}^{t}\) and \({D}_{n}^{t}\) refer to the methods of relative density calculation above.
Identification of G4 and cCRE G-runs
We implemented a customized script to identify G-runs within G4 sequences as well as within non-G4 sequences in cCREs. Typically, G4s with three or more quartets tend to be formed from sequences having at least the same number of consecutive guanines on each G-run, although there are exceptions (e.g., sequences with bulges). Three-quartet G4s tend to be more stable than 2-quartets G4s; therefore, in this study, we only consider G-runs with at least 3 consecutive guanines. While some stable G-quadruplexes may be formed with interrupted G-runs (bulges), incorporating these possibilities into our computational analysis would introduce a significant level of complexity. Therefore, we do not take them into account here, aiming to simplify our analytical model.
We used regular expressions to detect G-runs in G4 sequences (G4s that overlapped with cCREs) on the forward strand. All potential G-runs in the forward strand cCREs were also identified, excluding those that intersected with forward strand G4s. Using the same approach, we also identified G4 G-runs and cCRE G-runs on the reverse strand of the human genome. Consequently, we can assess the differences between G4 G-runs and cCRE G-runs in terms of RegulomeDB regulatory scores, LINSIGHT scores, and other relevant factors.
Acquisition and comparison of RegulomeDB, LINSIGHT, and gnomAD constraint scores
We devised four sets of comparative experiments to assess differences in the regulatory potential of regions associated with cCREs, as well as differences in the negative selection pressures to which they are subjected. These four sets include: (1) Comparing the differences in these metrics across different groups of G4s (e.g., PLS-G4s, pELS-G4s, etc.). (2) Comparing the differences in these metrics across different group of cCREs (e.g., G4-associated PLS versus non-G4-associated PLS, etc.). (3) Evaluating the differences in these metrics between G4 G-runs and the remaining sequences in G4s. Please note that the remaining sequences in G4s were obtained by subtracting G4 G-runs sequences from G4 sequences using the 'subtract' function provide by the bedtoolsr package (V2.30.0-5). (4) Examining the differences in these metrics between G4 G-runs and cCRE G-runs (excluding G4 G-runs).
We obtained the pre-calculated regulatory scores for common SNPs (dbSNP v153) with MAF ≥ 0.01 from the RegulomeDB website (https://regulomedb.org/regulome-search), which contains both probability scores and ranking scores, with the probability scores ranging from 0 to 1, where 1 represents the highest likelihood of being a regulatory variant, while the ranking scores taking the values from 1a to 7, where 1a indicates the one with the most supportive information and, therefore, potentially possess the strongest functional impact. We assigned these variants to different genomic regions for comparative analysis. For example, we evaluated the differences in regulatory probability scores as well as ranking scores among all potential variants within G4 G-runs and cCREs G-runs. If a certain region possesses a more pivotal role in regulatory functions, we can then observe that the regulatory scores in that region are generally higher.
We acquired the pre-calculated LINSIGHT score file (http://compgen.cshl.edu/LINSIGHT/LINSIGHT.bw) and mapped it to the hg38 assembly version (bigwig format). LINSIGHT scores can be used to estimate the extent of negative selection in non-coding regions of the human genome, such as the extent of negative selection differences between G4 G-runs and cCRE G-runs, with higher scores implying stronger negative selection. The bigWigAverageOverBed (V2) software was utilized to calculate the average negative selection strength for genomic regions such as G4 G-runs.
In addition, we acquired the human mutational constraint map from the gnomAD database (V3), which was imputed through the analysis of variations from ~76k human genomes. However, due to the resolution of this constraint map being 1 kb, we were unable to perform precise analysis on short sequences. Consequently, when evaluating disparities in gnomAD constraint scores (gnomAD uses z-scores to represent constraints. For simplicity, we will refer to them as 'scores' below), we only considered the distinctions between G4s located in different groups of cCREs, as well as the differences between G4-associated cCREs and non-G4-associated cCREs. Assuming that a genomic element (e.g., a G4-associated PLS) overlap with m windows of this constraint map, and the widths (base pairs covered by this element) of the overlaps with these windows are (w1, w2, ..., wm), the constraint scores for these m windows are (s1, s2, ..., sk), then the constraint score for this element was calculated as,
PhyloP conservation scores for 241 mammals and the definition of conserved bases
The phyloP conservation scores were obtained from the Zoonomia project (https://zoonomiaproject.org), which includes evolution-based conservation scores calculated for each position in the human genome. These scores were derived through the alignment of sequences from a comprehensive set of 241 mammalian species. We conducted comparisons of the differences in phyloP scores between different groups of G4s and between G4 G-runs and cCRE G-runs, etc. This analysis was carried out using the same methodology as for the assessment of LINSIGHT scores, as described above.
We also adopted the definition presented by Sullivan et al. [27], defining bases with phyloP scores ≥ 2.27 as constrained bases. Using this threshold, we identified a total of 100,651,377 bases across the entire genome that are conserved during evolution.
Distribution of constraint bases around G4s, cCREs, and G-runs
We compared the distribution of constrained bases around a certain group of cCREs (e.g., PLS) and their associated G4s (e.g., PLS-G4s). The calculations were performed using the EnrichedHeatmap package (V1.28.1), as detailed in the methods section above, with a resolution set to 10 bp.
Furthermore, we depicted the distribution of constrained bases in the upstream and downstream regions of G4 G-runs and cCRE G-runs located in distinct groups of cCREs regions. Given that the width of G-runs is usually very short, the resolution of the calculations here was increased to 1 bp. To estimate the distribution of constrained bases around G-runs, we conducted 100 random sampling experiments. In each random sampling, we independently selected 1,000 samples from both the G4 G-runs collection (e.g., PLS G4 G-runs) and the cCRE G-runs collection (e.g., PLS cCRE G-runs). We then calculated the relative density of constrained bases around the sampled samples. Finally, by analyzing the results from 100 random sampling experiments, we estimated the standard deviation errors in the distribution density of constrained bases around G4 G-runs and cCRE G-runs.
Calculation of the proportion of G4s in activated cCREs
We calculated the proportion of endogenous G4s (as supported by G4 ChIP-seq experiments) and all G4s (without considering G4 ChIP-seq experiments) located in various types of activated cCRE elements for K562 and HepG2 cell lines, respectively. Activated cCREs are those that are supported by corresponding epigenetic signals (including DNase, H3K4me3, etc.) in specific cells. In contrast, quiescent cCREs are those lacking corresponding epigenetic data support in specific cells. For example, if a cCRE in the K562 cell line is found to be located in a heterochromatic region, then that cCRE is considered a quiescent cCRE.
As an example, the procedures for calculating the proportion of endogenous G4 located in activated PLS elements are as follows.
Due to the significant disparity in the quantities of the two types of G4s, we employed a sampling method to estimate the distribution of the proportions. The number of random samplings was set to 1,000. For each sampling, we randomly select 1,000 G4s from the set of endogenous G4s and calculated the number of these selected G4s that located in the active PLS elements; the number of selected G4s that located in quiescent PLS elements was also determined. The proportion of endogenous G4s located in activated PLS elements was then calculated as,
where \({n}_{i}^{active}\) represents the number of endogenous G4s located in the activated PLS elements in the \(i\)-th sampling, while \({n}_{i}^{quiescent}\) refers to the number of endogenous G4s located in the quiescent PLS elements.
The proportions of endogenous G4s in other types of activated cCREs, and the proportions of all G4s in activated cCRE elements were calculated with reference to the methods described above.
Cell type-specific analysis
In this study, we compared the differences in transcription factor occupancy, DNA methylation, and chromatin accessibility between G4s supported by G4 ChIP-seq experiments and other G4s in the K562 and HepG2 cell lines.
The cell type-specific transcription factor binding data were sourced from the UCSC Genome Browser database (https://hgdownload.soe.ucsc.edu/goldenPath/hg38/encRegTfbsClustered/). We filtered all transcription factor binding sites for the K562 as well as the HepG2 cell line from this file, and in addition, the definition of transcription factors was strictly referenced to Lambert et al. [49]. G4s predicted by G4Hunter software were divided into two categories based on whether they overlapped with G4 ChIP-seq experiments, and the coverage of various transcription factors on these two categories of G4s was calculated using bedtoolsr package (V2.30.0-5).
DNA methylation data and ATAC-seq data for the K562 and HepG2 cell lines were publicly obtained from the GEO database under accession number GSM2308596, GSM3633977, GSE170378 and GSE170251, respectively. We utilized the bigWigAverageOverBed (V2) software to calculate the average methylation and ATAC-seq signal values for the 200 bp regions centered G4s.
Pan-cancer data for chromatin accessibility, enhancer, and promoter activity
To investigate the activation status of G4-associated cCREs in cancer tissues, we acquired data on pan-cancer chromatin accessibility, enhancer, and promoter activity from publicly available databases, which were assessed using ATAC-seq, H3K27ac, and RNA-seq techniques. In this section of the study, we specifically focused on the cancer types for which all three types of data were available simultaneously.
The chromatin accessibility data were derived from the NIH Genomic Data Commons portal (https://gdc.cancer.gov/about-data/publications/ATACseq-AWG). In this study, cancer type-specific ATAC-seq peak datasets were utilized. We examined the coverage of ATAC-seq peaks for both G4-associated and non-G4-associated cCREs, as well as the proportion of these cCREs located in open chromatin regions. Please note that chromatin accessibility is a necessary requirement for our analysis of pan-cancer enhancer data and promoter activity data. In other words, only the pan-cancer enhancers that overlap with ATAC-seq peaks will be included in subsequent analysis. The same requirement applies to both ELS (including pELS and dELS) elements and PLS elements. This definition is similar to the ENCODE project consortium definition of cis-regulatory elements [19].
We retrieved enhancer data identified through H3K27ac ChIP-seq in cancer tissues or cancer cell lines from the CenhANCER database [50] (https://cenhancer.chenzxlab.cn) with the typical enhancers were being considered for use. We assessed the distribution density of typical enhancers around enhancer-like cCREs (ELS), which encompass pELS and dELS.
The pan-cancer promoter activity was downloaded from the ICGC data portal. We utilized the raw promoter activity data from the PCAWG project, initially filtering out promoter activity data from normal samples within that dataset, retaining only cancer samples relevant to the cancer type used in our analysis. Subsequently, the PLS elements were assigned to the nearest promoters (TSS ± 200 bp). The activity of a specific promoter for a given cancer type was defined as the average activity of that promoter across all samples of that cancer type. If a promoter overlapped with both G4-associated PLS elements and non-G4-associated PLS elements, it was excluded (~12%) to ensure the accuracy of subsequent comparative analysis. We compared the differences in promoter activity between G4-associated PLS elements and non G4-associated PLS elements. Besides, we also evaluated the proportion of highly activated promoters associated with these two PLS element types, considering promoters with an activity exceeding 1.5 as highly activated [51].
The cancer type-specific gained regulatory sequences were obtained from the TSCRE (Tumor-specific Cis-Regulatory Elements; http://tscre.zsqylab.com/home) website. Gained regulatory sequences refer to those that are specifically upregulated in cancer tissues relative to the corresponding non-malignant tissues. To maintain consistency with the standard marks used by the ENCODE project, only the cancer types that contained both H3K4me3 and H3K27ac marks were retained for further analysis.
Availability of data and materials
All data generated or analyzed during this study are included in this published article, its supplementary information files and publicly available repositories. The data and code from this study, including analysis scripts, human G4 sequences predicted by G4Hunter, and the annotation file indicating the presence of G4s in cCREs, are available on the Zenodo repository at https://zenodo.org/doi/https://doi.org/10.5281/zenodo.13147639 [52].
Abbreviations
- G4:
-
G-quadruplex
- cCRE:
-
candidate cis-regulatory element
- PLS:
-
promoter-like signatures
- pELS:
-
proximal enhancer-like signatures
- dELS:
-
distal enhancer-like signatures
- CTCF-only:
-
high DNase and CTCF signals only
- DNase–H3K4me3:
-
high DNase and H3K4me3 signals only
- BLCA:
-
Bladder Urothelial Carcinoma
- BRCA:
-
Breast Invasive Carcinoma
- CESC:
-
Cervical Squamous Cell Carcinoma
- COAD:
-
Colon Adenocarcinoma
- GBM:
-
Glioblastoma Multiforme
- HNSC:
-
Head and Neck Squamous Cell Carcinoma
- KIRC:
-
Kidney Renal Clear Cell Carcinoma
- LGG:
-
Low Grade Glioma
- LIHC:
-
Liver Hepatocellular Carcinoma
- LUAD:
-
Lung Adenocarcinoma
- PAAD:
-
Pancreatic adenocarcinoma
- PRAD:
-
Prostate Adenocarcinoma
- READ:
-
Rectum adenocarcinoma
- SKCM:
-
Skin Cutaneous Melanoma
- STAD:
-
Stomach Adenocarcinoma
- THCA:
-
Thyroid Carcinoma
- UCS:
-
Uterine Carcinosarcoma
- UCEC:
-
Uterine Corpus Endometrial Carcinoma
References
Lewis EB. A gene complex controlling segmentation in Drosophila. Nature. 1978;276(5688):565–70.
Venters BJ, Pugh BF. How eukaryotic genes are transcribed. Crit Rev Biochem Mol Biol. 2009;44(2–3):117–41.
Preissl S, Gaulton KJ, Ren B. Characterizing cis-regulatory elements using single-cell epigenomics. Nat Rev Genet. 2023;24(1):21–43.
Kolovos P, Knoch TA, Grosveld FG, Cook PR, Papantonis A. Enhancers and silencers: an integrated and simple model for their function. Epigenet Chromatin. 2012;5(1):1.
Li Y, Chen CY, Kaye AM, Wasserman WW. The identification of cis-regulatory elements: a review from a machine learning perspective. BioSyst. 2015;138:6–17.
Varshney D, Spiegel J, Zyner K, Tannahill D, Balasubramanian S. The regulation and functions of DNA and RNA G-quadruplexes. Nat Rev Mol Cell Biol. 2020;21(8):459–74.
Spiegel J, Adhikari S, Balasubramanian S. The structure and function of DNA G-Quadruplexes. Trends Chem. 2020;2(2):123–36.
Bedrat A, Lacroix L, Mergny J-L. Re-evaluation of G-quadruplex propensity with G4Hunter. Nucleic Acids Res. 2016;44(4):1746–59.
Lago S, Nadai M, Cernilogar FM, Kazerani M, Domíniguez Moreno H, Schotta G, et al. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat Commun. 2021;12(1):3885.
Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33(9):2908–16.
Georgakopoulos-Soares I, Parada GE, Wong HY, Medhi R, Furlan G, Munita R, et al. Alternative splicing modulation by G-quadruplexes. Nat Commun. 2022;13(1):2404.
Hänsel-Hertsch R, Spiegel J, Marsico G, Tannahill D, Balasubramanian S. Genome-wide mapping of endogenous G-quadruplex DNA structures by chromatin immunoprecipitation and high-throughput sequencing. Nat Protoc. 2018;13(3):551–64.
Esnault C, Magat T, Zine El Aabidine A, Garcia-Oliver E, Cucchiarini A, Bouchouika S, et al. G4access identifies G-quadruplexes and their associations with open chromatin and imprinting control regions. Nat Genet. 2023;55(8):1359–69.
Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat Chem Biol. 2007;3(4):218–21.
Zhang R, Shu H, Wang Y, Tao T, Tu J, Wang C, et al. G-Quadruplex structures are key modulators of somatic structural variants in cancers. Cancer Res. 2023;83(8):1234–48.
Guiblet WM, Cremona MA, Harris RS, Chen D, Eckert KA, Chiaromonte F, et al. Non-B DNA: a major contributor to small- and large-scale variation in nucleotide substitution frequencies across the genome. Nucleic Acids Res. 2021;49(3):1497–516.
Jansson LI, Hentschel J, Parks JW, Chang TR, Lu C, Baral R, et al. Telomere DNA G-quadruplex folding within actively extending human telomerase. Proc Natl Acad Sci. 2019;116(19):9350–9.
Prorok P, Artufel M, Aze A, Coulombe P, Peiffer I, Lacroix L, et al. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat Commun. 2019;10(1):3274.
Abascal F, Acosta R, Addleman NJ, Adrian J, Afzal V, Ai R, et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature. 2020;583(7818):699–710.
Guiblet WM, DeGiorgio M, Cheng X, Chiaromonte F, Eckert KA, Huang Y-F, et al. Selection and thermostability suggest G-quadruplexes are novel functional elements of the human genome. Genome Res. 2021;31(7):1136–49.
Hou Y, Li F, Zhang R, Li S, Liu H, Qin ZS, et al. Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics. 2019;14(9):894–911.
Wulfridge P, Yan Q, Rell N, Doherty J, Jacobson S, Offley S, et al. G-quadruplexes associated with R-loops promote CTCF binding. Mol Cell. 2023;83(17):3064-79.e5.
Tikhonova P, Pavlova I, Isaakova E, Tsvetkov V, Bogomazova A, Vedekhina T, et al. DNA G-Quadruplexes contribute to CTCF recruitment. Int J Mol Sci. 2021;22(13):7090. https://doi.org/10.3390/ijms22137090.
Dong S, Zhao N, Spragins E, Kagda MS, Li M, Assis P, et al. Annotating and prioritizing human non-coding variants with RegulomeDB vol 2. Nat Genet. 2023;55(5):724–6.
Huang Y-F, Gulko B, Siepel A. Fast, scalable prediction of deleterious noncoding variants from functional and population genomic data. Nat Genet. 2017;49(4):618–24.
Chen S, Francioli LC, Goodrich JK, Collins RL, Kanai M, Wang Q, et al. A genomic mutational constraint map using variation in 76,156 human genomes. Nature. 2024;625(7993):92–100.
Sullivan PF, Meadows JRS, Gazal S, Phan BN, Li X, Genereux DP, et al. Leveraging base-pair mammalian constraint to understand genetic variation and human disease. Science. 2023;380(6643):eabn2937.
Fang S, Liu S, Yang D, Yang L, Hu C-D, Wan J. Decoding regulatory associations of G-quadruplex with epigenetic and transcriptomic functional components. Front Genet. 2022;13:957023.
Korsakova A, Phan AT. Prediction of G4 formation in live cells with epigenetic data: a deep learning approach. NAR Genomics Bioinf. 2023;5(3):lqad071.
Grubert F, Srivas R, Spacek DV, Kasowski M, Ruiz-Velasco M, Sinnott-Armstrong N, et al. Landscape of cohesin-mediated chromatin loops in the human genome. Nature. 2020;583(7818):737–43.
Andrews G, Fan K, Pratt HE, Phalke N, Zoonomia Consortium§, Karlsson EK, et al. Mammalian evolution of human cis-regulatory elements and transcription factor binding sites. Science. 2023;380(6643):eabn7930.
Hammal F, de Langen P, Bergon A, Lopez F, Ballester B. ReMap 2022: a database of Human, Mouse, Drosophila and Arabidopsis regulatory regions from an integrative analysis of DNA-binding sequencing experiments. Nucleic Acids Res. 2022;50(D1):D316–25.
Spiegel J, Cuesta SM, Adhikari S, Hänsel-Hertsch R, Tannahill D, Balasubramanian S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021;22(1):117.
Da Ros S, Nicoletto G, Rigo R, Ceschi S, Zorzan E, Dacasto M, et al. G-Quadruplex Modulation of SP1 Functional Binding Sites at the KIT Proximal Promoter. Int J Mol Sci. 2021;22(1):329.
Raiber E-A, Kranaster R, Lam E, Nikan M, Balasubramanian S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2011;40(4):1499–508.
Li L, Williams P, Ren W, Wang MY, Gao Z, Miao W, et al. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat Chem Biol. 2021;17(2):161–8.
Biffi G, Tannahill D, Miller J, Howat WJ, Balasubramanian S. Elevated Levels of G-Quadruplex Formation in Human Stomach and Liver Cancer Tissues. PLoS One. 2014;9(7): e102711.
Peng G, Liu B, Zheng M, Zhang L, Li H, Liu M, et al. TSCRE: a comprehensive database for tumor-specific cis-regulatory elements. NAR Cancer. 2024;6(1):zcad063.
Huppert JL, Bugaut A, Kumari S, Balasubramanian S. G-quadruplexes: the beginning and end of UTRs. Nucleic Acids Res. 2008;36(19):6260–8.
Harkness RW, Hennecker C, Grün JT, Blümler A, Heckel A, Schwalbe H, et al. Parallel reaction pathways accelerate folding of a guanine quadruplex. Nucleic Acids Res. 2021;49(3):1247–62.
Li G, Su G, Wang Y, Wang W, Shi J, Li D, et al. Integrative genomic analyses of promoter G-quadruplexes reveal their selective constraint and association with gene activation. Commun Biol. 2023;6(1):625.
Hänsel-Hertsch R, Beraldi D, Lensing SV, Marsico G, Zyner K, Parry A, et al. G-quadruplex structures mark human regulatory chromatin. Nat Genet. 2016;48(10):1267–72.
Mao S-Q, Ghanbarian AT, Spiegel J, Martínez Cuesta S, Beraldi D, Di Antonio M, et al. DNA G-quadruplex structures mold the DNA methylome. Nat Struct Mol Biol. 2018;25(10):951–7.
Zhu Y, Tazearslan C, Suh Y. Challenges and progress in interpretation of non-coding genetic variants associated with human disease. Exp Biol Med. 2017;242(13):1325–34.
Ellingford JM, Ahn JW, Bagnall RD, Baralle D, Barton S, Campbell C, et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14(1):73.
Gong J-y, Wen C-j, Tang M-l, Duan R-f, Chen J-n, Zhang J-y, et al. G-quadruplex structural variations in human genome associated with single-nucleotide variations and their impact on gene activity. Proceedings of the National Academy of Sciences. 2021;118(21):e2013230118.
McArthur E, Capra JA. Topologically associating domain boundaries that are stable across diverse cell types are evolutionarily constrained and enriched for heritability. Am J Hum Genet. 2021;108(2):269–83.
Loyfer N, Magenheim J, Peretz A, Cann G, Bredno J, Klochendler A, et al. A DNA methylation atlas of normal human cell types. Nature. 2023;613(7943):355–64.
Lambert SA, Jolma A, Campitelli LF, Das PK, Yin Y, Albu M, et al. The Human Transcription Factors. Cell. 2018;172(4):650–65.
Luo Z-H, Shi M-W, Zhang Y, Wang D-Y, Tong Y-B, Pan X-L, et al. CenhANCER: a comprehensive cancer enhancer database for primary tissues and cell lines. Database. 2023;2023:baad022.
Demircioğlu D, Cukuroglu E, Kindermans M, Nandi T, Calabrese C, Fonseca NA, et al. A Pan-cancer Transcriptome Analysis Reveals Pervasive Regulation through Alternative Promoters. Cell. 2019;178(6):1465-77.e17.
Zhang R, Wang Y, Wang C, Sun X, Mergny J-L. G-quadruplexes as pivotal components of cis-regulatory elements in the human genome. Zenodo. https://zenodo.org/doi/10.5281/zenodo.13147639 2024.
Acknowledgements
J.L.M. thanks L. Guittat for helpful discussions, and C. Français and L. Lachapelle for administrative support.
Funding
This work was supported by ANR G4Access (ANR-20-CE12-0023) and INCa G4Access grants to J.L.M, by the Leading Technology Program of Jiangsu Province (BK20222008), the National Natural Science Foundation of China (61972084) and China Scholarship Council (202106090125) to R.Z.
Author information
Authors and Affiliations
Contributions
J.L.M and X.S. supervised this study. J.L.M. and R.Z. conceptualized this study. R.Z. and C.W. designed analysis. R.Z. collected data and performed bioinformatic analysis. R.Z. wrote the initial draft of the manuscript. J.L.M., Y.W., and X.S. revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no conflict of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
12915_2024_1971_MOESM1_ESM.pdf
Additional file 1: Figure S1. Association of G4 stability with cCREs. Figure S2. Distribution of G4s in cCREs with different CTCF occupancy patterns. Figure S3. Coefficient estimates and P-values from logistic regression models. Figure S4. Relative densities of G4 ChIP-seq high confidence peaks around CTCF-only cCREs. Figure S5. Overview of RegulomeDB ranking and probability scores in G4s and cCREs. Figure S6. The differences in average LINSIGHT scores between G4-associated and non-G4-associated cCREs. Figure S7. An overview of gnomAD constraint z-scores for G4 and cCRE regions. Figure S8. Differences in average phyloP scores between cCREs and the G4s overlapping with the corresponding cCREs. Figure S9. Distribution of constrained bases around various groups of cCREs and the G4s overlapping with the corresponding cCRE groups. Figure S10. Coefficient estimates of the G4 and H3K27ac features in the logistic regression model. Figure S11. Transcription factor binding density around G4-associated and non-G4-associated cCREs. Figure S12. Coefficient estimates of features in the logistic regression models predicting the methylation status of CREs. Figure S13. Distribution of activated and quiescent cCREs around G4s supported by G4 ChIP-seq experiments or not. Figure S14. Heatmap shows the pan-cancer ATAC-seq signals around G4-associated pELS elements and non-G4-associated pELS elements. Figure S15. Heatmap shows the pan-cancer ATAC-seq signals around G4-associated dELS elements and non-G4-associated dELS elements. Figure S16. Heatmap shows the pan-cancer ATAC-seq signals around G4-associated CTCF-only elements and non-G4-associated CTCF-only elements. Figure S17. Heatmap shows the pan-cancer ATAC-seq signals around G4-associated DNase-H3K4me3 elements and non-G4-associated DNase-H3K4me3 elements. Figure S18. Bar plot indicates the proportion of G4-associated and non-G4-associated cCREs overlapping with ATAC-seq peaks in distinct cancer types. Figure S19. Proportion of G4-associated and non-G4-associated PLS with activity greater than 1.5in different cancer types. Figure S20. Distribution of cancer-specific gained regulatory sequences upstream and downstream of G4-associated and non-G4-associated cCREs. Figure S21. Overview of the widths of G4s and cCREs.
12915_2024_1971_MOESM2_ESM.xlsx
Additional file 2: Table S1. P-values for the comparison of G4Hunter scoresfor G4s in different genomic regions. Table S2. P-values for the comparison of RegulomeDB scores for G4s in different genomic regions. Table S3. P-values for the comparison of LINSIGHT scores for G4s in different genomic regions. Table S4. P-values for the comparison of gnomAD constraint Z-scores for G4s in different genomic regions. Table S5. P-values for the comparison of phyloP scoresfor G4s in different genomic regions.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, R., Wang, Y., Wang, C. et al. G-quadruplexes as pivotal components of cis-regulatory elements in the human genome. BMC Biol 22, 177 (2024). https://doi.org/10.1186/s12915-024-01971-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12915-024-01971-5