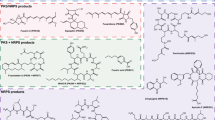
Abstract
Fungal non-ribosomal peptide synthetase (NRPS)-encoding products play a paramount role in new drug discovery. Fusarium, one of the most common filamentous fungi, is well-known for its biosynthetic potential of NRPS-type compounds with diverse structural motifs and various biological properties. With the continuous improvement and extensive application of bioinformatic tools (e.g., anti-SMASH, NCBI, UniProt), more and more biosynthetic gene clusters (BGCs) of secondary metabolites (SMs) have been identified in Fusarium strains. However, the biosynthetic logics of these SMs have not yet been well investigated till now. With the aim to increase our knowledge of the biosynthetic logics of NPRS-encoding products in Fusarium, this review firstly provides an overview of research advances in elucidating their biosynthetic pathways.
Similar content being viewed by others
Introduction
Fungal non-ribosomal peptide synthetases (NRPS) are large modular multifunctional enzymes that generate compounds by sequential condensation of amino acids and hydroxycarboxylic acid units [1]. Fungal NRPS-encoding products are a prolific source of bioactive compounds, some of which have been commercially used as therapeutic agents, such as cyclosporin A, echinocandins and emodepsides [2, 3]. As one of the most common filamentous fungi in nature, Fusarium is well-known for its potential of production of NRPS products with a wide array of biological properties [4,5,6]. With a substantial increase in fungal genome sequences and the incremental optimization of software tools (e.g., anti-SMASH, NCBI, UniProt), bioinformatic analysis of the link between secondary metabolites (SMs) and their biosynthetic gene cluster (BGCs) has become simple and efficient [7,8,9]. A growing number of Fusarium-derived NRPS products and their BGCs have been isolated and characterized [6, 10, 11]. However, the biosynthetic pathways of these SMs have not been well unveiled till now. By extensive literature search and analysis, this review comprehensively summarizes 15 biosynthetic pathways of NRPS-type compounds from Fusarium spp., highlighting the key enzymatic domains involved in their biosynthetic pathways. Additionally, the supporting information summarizes some of the common methods, which can provide valid references for further research.
Canonical NRPS-encoding compounds
One fungal NRPS module usually consists of at least three essential domains including the adenylation (A), the thiolation (T) and the condensation (C) [12,13,14,15]. The other family members also can replace the C domain in the biosynthesis or work together with C domain, including the epimerization (E) domain, the heterocyclization (Cy) domain, the CT domain (a subset of the C domain) etc., which can meet diverse and novel functions [16, 17]. The released products are subsequently further modified by additional enzymes, which are encoded by genes located near the NRPS and thus form the final product [18, 19].
Fusahexin
Fusahexin (1), originally derived from strain F. graminearum PH-1, represents a cyclic hexapeptide consisting of six amino acid residues and containing an uncommon ether bond between the C-δ of proline and the C-β of threonine [20, 21]. Phytopathological investigation showed that this substance plays a key role in hyphal growth, attachment, water–air interface penetration and plant infection through regulation of surface hydrophobicity of conidia and the cell wall as well as hydrophobin rodlet formation in Aspergillus nidulans [22,23,24,25].
Knockout and overexpression experiments revealed that an NRPS4 cluster in F. graminearum was responsible for the production of compound 1 [22, 26]. This cluster contains four genes that respectively encode for glucoside hydrolase, NRPS synthetase (gene NRPS4), ABC transporter and major facilitator superfamily (MFS) transporter (Fig. 1A). The NRPS4 enzyme consists of five modules, in which modules 1–4 are respectively responsible for linking D-alanine, L-leucine, D-allo-threonine, and L-proline, and module 5 is serially reusable in assembly of D-leucine and L-leucine (Fig. 1B) [20]. However, the function of other three enzymes in the NRPS4 cluster had not yet been characterized till now.
Fusaoctaxin
Fusaoctaxins A (2) and B (3), two unusual linear and C-terminally reduced octapeptide with D-amino acid-rich residues, were novel virulence factors during wheat infection and were firstly derived from strain F. graminearum PH-1 [27, 28]. The N-terminal residue of compound 2 is γ-aminobutyric acid (GABA) unit, while it is replaced by guanidoacetic acid (GAA) in compound 3 [28, 29].
Two core NRPS genes nrps5 and nrps9 together with six adjacent genes located in the fg3_54 cluster responsible for the biosynthesis of compounds 2 and 3 (Fig. 2A) were identified by laser microdissection and microarray approach [29, 30]. The essentiality of the fg3_54 gene cluster was unambiguously verified through cluster deletion and individual knockout of several biosynthesis-associated genes including FG-Δnrps9, and FG-Δnrps5, FG-Δfgm4, FG-Δfgm3 and FG-Δfgm1 [28]. The functions of the two key enzymes, NRPS9 and NRPS5, were further characterized by overexpression experiments [31]. The NRPS9 is a M1(A1-T1) di-domain protein that acts as a load module for initiating unit binding, while the NRPS5 harbors seven similar extension modules, M2(A2a-C2-A2b-T2)-M3(C3-A3-T3-E3)-M4(C4-A4-T4-E4)-M5(C5-A5-T5-E5)-M6(C6-A6-T6-E6)-M7(C7-A7-T7-E7)-M8(C8-A8-T8-R) and collaborates with the NRPS9 to biosynthesize octapeptides. These enzymes utilize GABA or GAA as a starting unit and extend the sequence with additional units including L-Ala, L-allo-Ile, L-Ser, L-Val and L-Leu residues (Fig. 2B) [32]. Each residue attached to the module containing the E domain (M3–M7) can undergo epimerization to acquire a D-configuration before transpeptidation. The peptidyl elongation was terminated by L-Leu through binding mediated by module M8, where the release (R) domain catalyzed a four-electron reduction to offload the octapeptide from the assembly line [29, 33].
Overexpression of genes fgm1, fgm2 and fgm3 along with their diverse combinations in Pichia pastoris GS115 showed these genes are responsible for the formation of GAA (Fig. 2C), which is a guanosine residue that serves as the initiating unit for the biosynthesis of compound 3. Fgm1, Fgm2 and Fgm3 respectively encode cytochrome P450, metallo-dependent amidohydrolase, pyridoxal-5′-phosphate (PLP)-dependent lyase. Fgm1 oxidizes L-Arg to 4(R)-hydroxy-L-Arg (4), which selectively enables the activation of inert C4 atom by hydroxylation for subsequent C3-C4 cleavage [34]. Fgm3 catalyzes the cleavage of the Cβ-Cγ bond in 4 to produce 5 and L-Ala [35]. Fgm2 effectively hydrolyzes glycociamidine (6) to produce linearized GAA. The pathway for GAA formation in F. graminearum differs significantly from the well-known pathway that utilizes the L-Arg:L-Gly aminidotransferase (AGAT) to transfer amino group between L-Arg and L-Gly residues. Instead, it relies on L-Arg as a precursor through a series of chemical reactions including inert C−H bond activation, selective C−C bond cleavage, cyclization-based alcohol dehydrogenation, and amidohydrolysis-associated linearization [36].
Gramillin
Gramillins A (8) and B (9) are two host-specific virulence factors initially isolated from several F. graminearum strains [37]. They possess a fused bicyclic structure in which the main peptide ring is cyclized through the carboxylic group of glutamic acid and the side chain of 2-amino adipic acid [38,39,40]. It was the first occurrence of anhydride bond being involved in the cyclization of a cyclic peptide [37, 41].
The functions of the NRPS8 gene cluster were determined through targeted gene disruption [42]. Gene GRA1 encodes a multi-modular NRPS synthase that contains seven A and C domains [43]. GRA2 encodes a transcription factor (TF) and is responsible for the regulation of cyclic peptide production (Fig. 3A) [44, 45]. By combining the Stachelhaus model and analyzing the conservation of the two adjacent A domains, the probable pathway for gramillins biosynthesis was identified. The biosynthetic pathway begins with Glu or 2-amino adipic acid and sequentially connects to Leu, Ser, HO-glutamine (HO-Gln), 2-amino decanoic acid, cysteine B (Cys B), and Cys A via other modules (Fig. 3B) [46, 47]. However, the functions of the other genes still need to be confirmed through additional specific experiments.
Chrysogine
Chrysogine (10) is a natural pigment that was first obtained and studied in Penicillium chrysogenum [48]. Although this substance does not possess remarkablely biological property, its core scaffold, 4(3H)-quinazolinone, is the primary functional group in various first-line antitumor or sedative agents such as idelalisib, raltitrexed, and methaqualone and other marketed drugs (e.g. nolatrexed, albaconazole, and halofuginone) for treatment of malarial, inflammatory, HIV and diabetic diseases [49,50,51,52].
In the past decade great progress had been made in the biosynthetic investigation of 10 in F. tricinctum CGMCC 3.4731, which offers an alternative synthetic pathway for constructing the 4(3H)-quinazolinone scaffold [50, 53]. A highly homologous NRPS gene cluster named ftchy (Fig. 4A) was identified and confirmed to be responsible for the formation of 10 through heterologous expression in Aspergillus nidulans and in vitro incubation experiments in E. coli [50, 54, 55]. The results also indicated that gene ftchyA encodes a fungal two-module NRPS (ftChyA) for the biosynthesis of 11, and the genes ftchyC, ftchyD, ftchyE, ftchyH, and ftchyM respectively encode a dehydrogenase (ftChyC), an amidotransferase (ftChyD), a tripeptide hydrolase (ftChyE), a flavin-dependent oxidase (ftChyH), and α-ketoglutaratedependent dioxygenase (α-KGD; ftChyM) [56, 57]. The enzyme ftChyD catalyses the amidation of 11 to 12 and 13 to 14 by utilizing inorganic ammonium ions or amides of L-Gln and ftChyE transforms 12 to 14 [48]. An unfamiliar α-KGD (ftChyM) catalyses the oxidative cleavage of the C-N bond for the production of 15 from 12. The oxidase ftChyH only catalyses the dehydrogenation reaction and corrects the additional reduction of ftChyC towards 15, ensuring the primary pathway (15 → 16) in the rapid construction of the 4(3H)-quinazolinone scaffold. These additional branching pathways depended on the nonenzymatic cyclization of ftChyM (17 → 10) or promiscuous substrate selectivity (18 → 16 → 10) (Fig. 4B).
Beauvericin
Beauvericin (BEA, 19) is a cyclic hexadepsipeptide that consists of a repetitive linkage between a D-hydroxy-isovaleryl (D-Hiv) and an N-methyl-phenylalanyl residue. It was firstly obtained from Beauveria bassiana and commonly discovered in several pathogenic Fusarium spp. [58, 59]. Bioassay results suggested that this alkaloid displays a wide range of biological activities including cytotoxic, apoptotic, anti-inflammatory, antimicrobial, and nematicidal activities [60,61,62,63,64,65,66].
A deeper understanding of the compound 19 biosynthesis gene cluster (bea cluster) in F. proliferatum LF061 was achieved by knocking out the specified genes using Agrobacterium AGL-1 mediated transformation (ATMT) protocol [67, 68]. A gene of 9413 bp (BEA1) responsible for encoding a hexadepsipeptide synthetases (NRPS22) was revealed, and the kivr gene encodes a novel NADPH-dependent 2-ketoisovalerate reductase (KIVR) responsible for the metabolism of pyruvate to D-Hiv was also unveiled [69]. Sequence analysis of other genes showed that orf1, orf3, orf4, orf5, orf6, and orf10 respectively encode putative thioesterase, triacylglycerol lipase, chitinase, zinc-dependent metalloproteinase, furinase, and multidrug transporter [70, 71].
The small two-gene cluster for BEA biosynthesis in strain LF061 consists of an NRPS gene and a KIVR-encoding gene [72]. D-Hiv is recognized by the A1 domain in module 1 of NRPS22 and attached to the T1 domain as a thioester. L-Phe is specifically activated by the A2 domain and is loaded to the twin T2 domain in module 2. An integrated N-methyltransferase domain is also present in NRPS22, which is responsible for the methylation of the L-Phe residue (Fig. 5) [67, 71]. This serves as a classic example of acting through the core NRPS synthase and provides valuable insights for subsequent studies [60].
Sansalvamide A
Sansalvamide A (20) is a cyclic pentadepsipeptide composed of an α-hydroxyisocaproic acid (α-HICA) unit and four protein amino acids (L-Val, L-Leu, L-Phe, L-Leu). It was originally discovered in the crude extract of an unknown Fusarium strain, which was collected from the surface of the seagrass Halodule wrightii [73,74,75]. Bioassay tests indicated that compound 20 is an effective cytotoxin in the colon cancer cell lines COLO 205 and HCT116 and the melanoma cell line SK-MEL-2 [75, 76].
The BGC NRPS30, which is responsible for the formation of compound 20 in F. solani FGSC 9596, was characterized through a gene knockout experiment using the ATMT approach [77, 78]. This cluster contains at least four genes that encode NRPS30 synthetase (gene NRPS30), oxidoreductase, short-chain dehydrogenase/reductase, and MFS transporter (Fig. 6A). Among the five modules of the NRPS30 enzyme, only the first amino acid of the A3 domain is glycine, while the remaining four are aspartic acid [46, 79]. This suggests that α-HICA is loaded as the third substituent during the biosynthesis of compound 20, as the lack of an acidic residue in the first position is only observed for A domain with non-amino acid substrates [80]. NRPS30 utilizes L-Phe as a starting unit and extends the sequence with additional units, including L-Leu, α-HICA, L-Val, and L-Leu (Fig. 6B).
Apicidin F
Apicidin F (APF, 21) is a cyclic tetrapeptide produced by F. fujikuroi [81]. Structurally, APF consists of N-methoxy-L-tryptophan (25), L-2-aminooctanedioic acid (26), D-pipecolic acid (D-pip; 23) and L-phenylalanine [82, 83]. Biological evaluation showed that this compound has the ability to inhibit histone deacetylase and is a therapeutic agent for antimalarial treatment against Plasmodium falciparum [84, 85].
A highly homologous NRPS gene cluster named APF was uncovered through homologous comparison and genomic sequence analysis (Fig. 7A) [86, 87]. Further exploration of the APF cluster and targeted gene replacement of APF1 revealed that Apf1, a key NRPS enzyme, is responsible for the biosynthesis of compound 21 [88,89,90]. The deletion of other functional genes suggested that the APF gene cluster consists with APF2, APF3, APF4, APF5, APF6, APF7, AFP8, APF9, APF11, and APF12, which respectively encode a transcription factor (Apf2), a putative Δ1-pyrroline-5-carboxylic acid reductase (Apf3), an aminotransferase (Apf4), a fatty acid synthase (Apf5), an O-methyltransferase (Apf6), two cytochrome P450 oxidases (Apf7/Apf8), a FAD-dependent monooxygenase (Apf9), a MFS transporter (Apf11), and a cytochrome b5-like reductase (Apf12).
The comparison of metabolite profile of the knockout mutants revealed that only six genes (APF1, APF3, APF4, APF5, APF6, APF7/AFP8/APF9) directly participate in the biosynthesis of APF [85]. Apf3 reduces L-lysine to L-piperidinic acid (22), which is subsequently converted to 23 by Apf1. L-tryptophan is initially oxidized to N-hydroxyl-L-tryptophan (24) by one of the two P450 enzymes (Apf7/Apf8), followed by conversion to 25 by Apf6. Apf5 is responsible for the condensation of three malonyl-CoA units and an acetyl-CoA into the octanoic acid backbone, which is then oxidized to form 28 by a P450 oxygenase. Apf4 catalyzes the exchange of the keto group of 28 with the amino group to form 27. Apf7/Apf9 may be involved in the conversion of 27 to 26. Ultimately, APF is generated by combining the four precursors in the presence of Apf1 (Fig. 7B). This represents a unique case of NRPS synthase function, where the NRPS enzyme is not fully functional until the final step.
Fusarochromene (NRPS-like)
Fusarochromene (29) firstly isolated from F. sacchari has structural similarities to fusarochromanone (30), which is a lead compound for cancer treatment [91, 92]. Compound 30 demonstrates a wide range of biological activities, such as angiogenesis inhibition, prevention of cell reproduction, and induction of apoptosis in numerous cancer cells, especially COS7 and HEK293 cells [93, 94].
Retro-biosynthetic analysis and 13C-labelled tryptophan experiments suggested that compounds 29 and 30 were actually obtained through oxidative cleavage of tryptophan [91]. The fsc gene cluster was identified by searching the genome of F. equiseti for potential tryptophan dioxygenase (TDO) and dimethylallyl diphosphate transferase (DMAT) genes. Through homologous comparison, the functions of these genes showed that fscA, fscB, fscC, fscD, fscE, fscF, fscG, fscH, fscI, and fscJ respectively encode two oxidoreductases (FscA, FscI), a TF (FscB), an NRPS-like enzyme (FscC), a dioxygenase (FscD), two P450 enzymes (FscE, FscF), a DMAT enzyme (FscG), a kynurenine formamidase-like hydrolase (FscH), and an aromatic peroxidase/chloroperoxidase (FscJ) (Fig. 8A) [95, 96].
A biosynthetic pathway for 29 and 30 is proposed in Fig. 8B. L-tryptophan is converted to D-tryptophan (36) in the presence of FscC, and subsequently hydroxylated by FscE to yield 6-hydroxytryptophan (35) [97]. The pyrrole ring undergoes cleavaged by FscD and is finally converted to 4-hydroxykyrunenine (34). FscA reduces the carboxyl group to primary alcohol (33) and FscG, a DMATS-type prenyltransferase, performs prenylation to 32 with the formation of a chromene ring. 32 is catalyzed by FscJ, leading to the formation of desacetyl-fusarochromene (31). Epoxidation (FscF) and rearrangement reactions of chromene double bonds convert compound 31 to 30. Although specific acetyltransferases were not found near the fsc BGC, several predicted enzymes containing the N-acetyltransferase superfamily domain were discovered in the genome of F. equiseti. These predicted enzymes may have the potential to convert compound 31 to 29 [98].
Hybrid PKS-NRPS products
Polyketide synthase (PKS) and NRPS hybrid systems typically rely on intricate protein–protein interactions to enable the seamless transfer of intermediates between these multimodular enzymes [99,100,101,102]. The PKS in Fusarium strain includes the β-keto synthase (KS) domain, the acyltransferase (AT) domain, the β-keto reductase (KR) domain, dehydrogenase (DH) domain, methyltransferase (MT) domain, enoyl reductase (ER) domain and acyl carrier protein (ACP) domain.
Fusaristatin A
Fusaristatin A (37) is a lipopeptide composed of three amino acid residues (glutamine, dehydroalanine, and β-aminoisobutyric acid) along with their attached polyketide chains. It was originally separated from Fusarium sp. YG-45 and lately detected in Phomopsis longicolla S1B4 and other Fusarium strains including F. graminearum, F. avenaceuma and Fusarium sp. FN080326 [103,104,105,106,107]. Cytotoxic assay indicated that compound 37 displays growth-inhibitory activity against lung cancer cells LU 65 with an IC50 value of 23 μM [103, 108].
As shown in Fig. 9A, the FGSG cluster in F. graminearum consists of at least five genes: PKS6, NRPS7, FGSG-A, FGSG-B, and FGSG-C. Deletion of NRPS7/PKS6 resulted in the absence of 37, confirming that PKS6 and NRPS7 are the two key enzymes jointly responsible for its production. Additionally, FGSG-C is predicted to encode a cytochrome P450 monooxygenase, FGSG-A encodes an aminotransferase, and FGSG-B encodes a putative protein containing a stress response A/B barrel domain [108]. The biosynthetic pathway of product 37 is mainly accomplished by PKS6 and NRPS7. As the FGSG cluster lacks acyltransferases, the polyketide synthesized by PKS6 is directly transferred to NRPS7. Then module 1–3 of NRPS7 sequentially adds Ala, Gln, and β-aminoisobutyric acid, and is finally released through cyclization (Fig. 9B). Although the β-aminoisobutyric acid units are most likely not freely available to the NRPS7, the FGSG cluster harbors cytochrome P450 and aminotransferases, which could potentially obtain it from thymidine.
W493 B
W493 B (38) is a lipopeptide consisting of six amino acid residues [D-allo-Thr, L-Ala, D-Ala, L-Gln, D-Tyr, and L-valine/isoleucine (Val/Ile)], which are linked to a polyketide chain of 3-hydroxy-4-methyltetradecanoic acid. It was initially isolated from Fusarium sp. and displayed inhibitory effect on the growth of Venturia inaequalis, Monilinia mali, and Cochliobolus miyabeanus [109, 110].
The FPSE cluster, consisting of at least four genes (PKS40, NRPS32, FPSE-A, FPSE-B), was identified in F. pseudograminearum through the analysis of the conserved genes [108]. These genes were respectively predicted to encode a PKS enzyme (PKS40), a NRPS enzyme (NRPS32), an acyl-CoA ligase and a thioesterase (Fig. 10A). The biosynthetic pathway of W493 B is primarily catalyzed by PKS40 and NRPS32, which respectively play important roles in the formation of 4-methyltridecanoic acid thioester and a hexapeptide (Fig. 10B). The T1 domain of NRPS32 is responsible for accepting threonine, which is adenylated by the A1 domain and then combined with D-allo-threonine formed by the E1 domain. Five consecutive modules bind Ala, Ala, Gln, Tyr, and Val/Ile to form the final product and release it through the cyclization domain [108]. The biosynthetic pathways of compounds 37 and 38 provide a comprehensive overview of lipopeptide biosynthesis.
Fusaric acid
Fusaric acid (FA, 39), formed by adding a butyl group to the 5-position C of 2-picolinic acid, is a mycotoxin produced by numerous Fusarium species, including F. oxysporum, F. heterosporum, F. verticillioides, and F. fujikuroi [111, 112]. FA is a broad-spectrum plant toxin with high phytotoxicity, and exhibits potent acanthamoebicidal activity and inhibits HIV-1 tat-induced transactivation and apoptosis [113,114,115,116,117].
The FUB cluster in F. fujikuroi was identified through targeted gene deletion, complementation, and overexpression experiments (Fig. 11A) [118,119,120]. These experiments suggest that a total of 12 genes are responsible for FA biosynthesis [121]. As illustrated in Fig. 11A, the functions of these genes showed that FUB1-12 respectively encode a PKS enzyme (FUB1), a putative protein (FUB2), an aspartate kinase (FUB3), a serine hydrolase (FUB4), a homoserine O-acetyltransferase (FUB5), a NAD(P)-dependent dehydrogenase (FUB6), an O-acylhomoserine (thiol) lyase (FUB7), an NRPS-like enzyme (FUB8), a flavin mononucleotide (FMN)-dependent dehydrogenase (FUB9), two fungal-type Zn(II)2Cys6 transcription factors (FUB10 and FUB12), and a MFS transporter (FUB11) [122, 123].
The FA biosynthetic pathway has been proposed in Fig. 11B. With the combined action of FUB3 and FUB5, L-aspartate is converted to O-acetyl-homoserine (42). FUB1 generates the triketide trans-2-hexenal (41), which is potentially released by FUB4 and linked to the NRPS-bound amino acid precursor by Fub6. After further modification by FUB7, the NRPS-bound amino acid precursor is released by FUB8 to form 40, which is finally oxidized by FUB9 to form FA.
Hybrid PKS/NRPS products
The compounds generated by PKS/NRPS hybrid megaenzymes are especially intriguing due to their structural complexity [124, 125]. This hybrid megaenzymes consists of an NRPS module and a PKS module together.The PKS module synthesizes the linear polyketide backbone, which is released after ligating with amino acids through the action of the NRPS module [126,127,128,129]. It is then further converted to more complex metabolites by oxidase or other enzymes.
Fusarin C
Fusarin C (43), a representative of substituted 2-pyrrolidinone metabolites, was firstly isolated in F. moniliforme and is widely present in Fusarium spp., including F. graminearum, F. oxysporum, F. verticillioides and F. fujikuroi [130,131,132,133,134,135]. Biological assays suggested that compound 43 acts as an estrogenic agonist, which stimulates the growth of the breast cancer cell line MCF-7 in concentrations ranging from 0.1 to 20 μM and inhibits its growth in concentrations exceeding 50 μM [136, 137]. Interestingly, 43 was found to induce esophageal and forestomach carcinoma in mouse and rat models, while this effect was not observed by Gelderblom and co-workers [138,139,140,141].
Gene knockout experiment showed that the fus cluster in F. fujikuroi consists of nine coregulated genes, of which fus2-fus9 are adjacent to gene fus1 (the hybrid PKS/NRPS; Fus1) [142,143,144]. Fus2 is related to a putative α/β hydrolase, which is probably involved in the 2-pyrrolidone ring formation. Deduced proteins show similarity to a subunit of elongation factor (Fus3), a peptidase A1 (Fus4), a serine hydrolase family (FSH; Fus5), a major facilitator superfamily transporter (MFS; Fus6), an aldehyde dehydrogenase (Fus7), a cytochrome P450 (Fus8), a characterized methyltransferase (Fus9) (Fig. 12A) [135].
The intermediates of compound 43 were only identified in the Δfus2, Δfus8, Δfus9, and Δfus2-9 mutants, suggesting that the genes fus3, fus4, fus5, fus6, and fus7 are largely uninvolved in the production of fusarin C. The proposed fusarin C biosynthetic pathway is as follows: Fus1 is responsible for the condensation of one acetyl-CoA with six malonyl-CoA and homoserine to form prefusarin (47). Fus8 then oxidizes 47 to form 46, which is an essential reaction until Fus2 catalyzes the formation of 20-hydroxy-prefusarin (45). 45 is further oxidized to produce 44 by Fus8. The final step involves the methylation of the hydroxyl group of C-21 by Fus9, resulting in the production of fusarin C (Fig. 12B). The co-cultivation of different mutants and intermediates analysis further confirms that Fus1, Fus2, Fus8, and Fus9 are sufficient for the biosynthesis (see Additional file 1).
Oxysporidinone
Oxysporidinone (48), a novel antifungal product with 4-hydroxy-2-pyridone backbone and a unique hydroxy-substituted cyclohexane ring, was firstly isolated from F. oxysporum [145, 146]. The oxysporidinone biosynthesis gene cluster (osd cluster) was identified in F. oxysporum ACCC 36465 by regulator activation and gene knockout studies (Fig. 13A) [147]. The osd cluster, containing 21 putative encoding genes (osdA-P and orf1-5), includes a core PKS/NRPS hybrid enzyme (OsdE), a trans-enoyl reductase (OsdF), two short-chain dehydrogenases/reductases (SDR; OsdB and H), four methyltransferases (MT; OsdA, C, D and K), four P450 monooxygenases (OsdG, I, J and M), a fungus-specific transcription factor (OsdL), a flavin oxidoreductase/nicotinamide adenine dinucleotide (NADH) oxidase (OsdN), a flavin adenine dinucleotide (FAD)-conjugated oxidoreductase (OsdO), a cycloheximide lyase (OsdP), an ankyrin (ORF3), a f-box protein (ORF2) and three unknown proteins (ORF1,ORF4,ORF5).
The biosynthetic pathway of 48 was proposed through heterologous expression and in vitro enzyme assays [147,148,149]. In the presence of PKS/NRPS enzyme (OsdE) with OsdF, six malonyls and four SAMs combine to form the backbone structure of tetrameric acid (52). Compound 52 undergoes a classic ring-expansion reaction catalyzed by OsdG to produce 2-pyridone (51). The formation from 51 to 50 is catalyzed by OsdH-J. OsdK is responsible for the N-methylation process, which converts 50 to form 49. Compound 49 is then converted to 53 by OsdM, a TenA-like cytochrome P450 enzyme that oxidizes the phenol ring and forms a [6–5–6] ring system. OsdN carries out two consecutive reduction steps to produce 54. Finally, OsdM adds another hydroxyl group to 54, resulting in the formation of compound 48 (Fig. 13B). Two enzymes (OsdM, OsdN) repeatedly act on the phenol moiety in the substrate. This pathway enhances the current understanding of the mechanism of enzymatic phenol dearomatization.
Fusaridione A
Fusaridione A (55) is an unstable tyrosine-derived 2,4-pyrrolidinedione produced by F. heterosporum [150,151,152,153]. Genomic analysis has revealed a silence gene, fsdS, which consists of a hybrid PKS and NRPS module. The putative biosynthesis pathway of fusaridione A was unveiled by fsdS gene knockout experiments [154]. The polyketide chain is first synthesized by the addition of seven acetyl-CoA units. Each extension requires the involvement of the KS, AT, KR, DH and ACP domain. Then, the tyrosine is activated and attached to the polyketide chain in the presence of the C, A and T domains. Compound 55 is finally released through the Dieckmann cyclase R* domain [16, 155]. The unstable pyrrolidinedione ring is opened by a reverse Dieckmann reaction, resulting in the formation of product 56 (Fig. 14) [156]. Further exploration is required to elucidate the genes that are closely related to gene fsdS.
Equisetin
Equisetin (58) is an HIV-I integrase inhibitor isolated from strain F. equiseti NRRL 5537 [157, 158]. Compound 58 and its N-desmethyl derivative trichosetin (57) represent tetramic acids, which are also widely present in several Fusarium species, including F. heterosporum, F. fujikuroi, and Fusarium sp. FN080326 [150, 159]. These compounds exhibit a broad spectrum of biological activities, including antibacterial, antiviral, antifungal, phytotoxic, and cytotoxic effects [158,159,160,161,162,163]. Gene deletion and overexpression experiments revealed that the trichosetin biosynthesis gene cluster in F. fujikuroi did not contain N-methyltransferase (EqxD), resulting in the isolation of the terminal product 57 [151, 162]. The comparison of gene functions for the biosynthesis of equisetin and its derivatives in F. heterosporum, F. fujikuroi and Fusarium sp. FN080326 is presented in Fig. 15A and Table 1.
The proposed biosynthetic scheme for compound 58 and its derivatives involves the utilization of an acetyl-CoA, seven malonyl-CoA, two S-adenosyl-L-methionine (SAM) and L-serine to form the backbone [164]. The PKS module of EqxS catalyzes with the enoyl reductase (EqxC) to produce a polyketide unit followed by conjugation with a L-serine (in red) through the condensation of the NRPS module. The Dieckmann cyclase domain activity (R*) leads to the release of 57. Compound 57 is then N-methylated by EqxD to form 58, which was further converted to fusarisetin A (59) in Fusarium sp. FN080326 (Fig. 15B).
Conclusions
Fusarium is one of excellent producers of NRPS products with a wide range of biological properties. To the best of our knowledge, over 800 SMs produced by Fusarium strains have been recorded in the Dictionary of Natural Products (DNP) database and nearly 300 chemicals related to NRPS pathway [165]. This review highlights only fifteen biosynthetic pathways that linked NRPS products with their corresponding BGCs identified in Fusarium. Therefore, most of these NRPS compounds linked to their BGCs need to be investigated. More efforts should be made to apply genetic engineering approaches to elucidate the biosynthetic pathways of other Fusarium NRPS-encoding compounds and to characterize their key genes and functions.
Availability of data and materials
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- NRPS:
-
Non-ribosomal peptide synthetases
- BGCs:
-
Biosynthetic gene clusters
- SMs:
-
Secondary metabolites
- NCBI:
-
National Center for Biotechnology Information
- A domain:
-
The adenylation domain
- T domain:
-
The thiolation domain
- C domain:
-
The condensation domain
- CT domain:
-
A subset of the C domain
- E domain:
-
The epimerization domain
- R domain:
-
The release domain
- M:
-
Modules
- MFS:
-
Major facilitator superfamily transporter
- GAA:
-
Guanidoacetic acid
- GABA:
-
γ-Aminobutyric acid
- PLP:
-
Pyridoxal-5′-phosphate
- AGAT:
-
A process of amidino transfer that requires
- L-Arg:
-
L-Gly aminidotransferase activity
- HOGln:
-
HO-Glutamine
- Cys:
-
Cysteine
- LC–MS:
-
Liquid Chromatograph Mass Spectrometer
- NMR:
-
Nuclear Magnetic Resonance Spectroscopy
- α- KGD:
-
α-Ketoglutaratedependent dioxygenase
- ENNs:
-
Enniatins
- KIVR:
-
A novel NADPH-dependent 2-ketoisovalerate reductase
- D-Hiv:
-
D-Hydroxy-isovaleryl
- SAM:
-
S-adenosylmethionine
- BEA:
-
Beauvericin
- HICA:
-
α-Hydroxyisocaproic acid
- ATMT:
-
The Agrobacterium-mediated transformation approach
- APF:
-
Apicidin F
- TFs:
-
The transcription factor
- L-pip:
-
L-Piperidinic acid
- TDO:
-
Tryptophan dioxygenase
- DMAT:
-
Dimethylallyl diphosphate transferase
- PKS:
-
Polyketide synthases
- KS:
-
β-Keto synthase
- AT:
-
Acyltransferase
- KR:
-
β-Keto reductase
- DH:
-
Dehydrogenase
- MT:
-
Methyltransferase
- ER or R:
-
Reductase
- ACP:
-
Acyl carrier protein
- FA:
-
Fusaric acid
- OE:
-
Overexpression
- SDR:
-
Short-chain dehydrogenases/reductases
- FSH:
-
A serine hydrolase family
- FA:
-
Fusaric acid
- Leu:
-
Leucine
- Pro:
-
Proline
- Thr:
-
Threonine
- Ala:
-
Alanine
- Gln:
-
Glutamine
- Tyr:
-
Tyrosine
- Val:
-
Valine
- Ile:
-
Isoleucine
- Phe:
-
Phenylalanine
- Arg:
-
Arginine
- Ser:
-
Serine
- Lys:
-
Lysine
- DNP:
-
The Dictionary of Natural Products
References
Bushley KE, Turgeon BG. Phylogenomics reveals subfamilies of fungal nonribosomal peptide synthetases and their evolutionary relationships. BMC Evol Biol. 2010;10:26.
Oide S, Turgeon BG. Natural roles of nonribosomal peptide metabolites in fungi. Mycoscience. 2020;61(3):101–10.
Krücken J, Holden-Dye L, Keiser J, Prichard RK, Townson S, Makepeace BL, et al. Development of emodepside as a possible adulticidal treatment for human onchocerciasis-the fruit of a successful industrial-academic collaboration. PLoS Pathog. 2021;17(7): e1009682.
Li M, Yu R, Bai X, Wang H, Zhang H. Fusarium: a treasure trove of bioactive secondary metabolites. Nat Prod Rep. 2020;37(12):1568–88.
Xu M, Huang Z, Zhu W, Liu Y, Bai X, Zhang H. Fusarium-derived secondary metabolites with antimicrobial effects. Molecules. 2023;28(8):3424.
Lin C, Feng X, Liu Y, Li Z, Li X, Qi J. Bioinformatic analysis of secondary metabolite biosynthetic potential in pathogenic Fusarium. J Fungi. 2023;9(8):850.
Niehaus E-M, Kim H-K, Münsterkötter M, Janevska S, Arndt B, Kalinina SA, et al. Comparative genomics of geographically distant Fusarium fujikuroi isolates revealed two distinct pathotypes correlating with secondary metabolite profiles. PLoS Pathog. 2017;13(10): e1006670.
Hoogendoorn K, Barra L, Waalwijk C, Dickschat JS, van der Lee TAJ, Medema MH. Evolution and diversity of biosynthetic gene clusters in Fusarium. Front Microbiol. 2018;9:1158.
Villani A, Proctor RH, Kim H-S, Brown DW, Logrieco AF, Amatulli MT, et al. Variation in secondary metabolite production potential in the Fusarium incarnatum-equiseti species complex revealed by comparative analysis of 13 genomes. BMC Genomics. 2019;20(1):314.
Liu Y, Xu M, Tang Y, Shao Y, Wang H, Zhang H. Genome features and AntiSMASH analysis of an endophytic strain Fusarium sp. R1. Metabolites. 2022;12(6):521.
Wiemann P, Sieber CMK, von Bargen KW, Studt L, Niehaus E-M, Espino JJ, et al. Deciphering the cryptic genome: genome-wide analyses of the rice pathogen Fusarium fujikuroi reveal complex regulation of secondary metabolism and novel metabolites. PLoS Pathog. 2013;9(6): e1003475.
Bloudoff K, Schmeing TM. Structural and functional aspects of the nonribosomal peptide synthetase condensation domain superfamily: discovery, dissection and diversity. Biochim Biophys Acta Proteins Proteom. 2017;1865(11PtB):1587–604.
Liu X, Walsh CT. Cyclopiazonic acid biosynthesis in Aspergillus sp.: characterization of a reductase-like R* domain in cyclopiazonate synthetase that forms and releases cyclo-acetoacetyl-L-tryptophan. Biochemistry. 2009;48(36):8746–57.
Süssmuth RD, Mainz A. Nonribosomal peptide synthesis—principles and prospects. Angew Chem Int Ed Engl. 2017;56(14):3770–821.
Hur GH, Vickery CR, Burkart MD. Explorations of catalytic domains in non-ribosomal peptide synthetase enzymology. Nat Prod Rep. 2012;29(10):1074–98.
Bloudoff K, Fage CD, Marahiel MA, Schmeing TM. Structural and mutational analysis of the nonribosomal peptide synthetase heterocyclization domain provides insight into catalysis. Proc Natl Acad Sci USA. 2017;114(1):95–100.
Rui Z, Zhang W. Engineering biosynthesis of non-ribosomal peptides and polyketides by directed evolution. Curr Top Med Chem. 2016;16(15):1755–62.
Stein DB, Linne U, Marahiel MA. Utility of epimerization domains for the redesign of nonribosomal peptide synthetases. FEBS J. 2005;272(17):4506–20.
Bushley KE, Ripoll DR, Turgeon BG. Module evolution and substrate specificity of fungal nonribosomal peptide synthetases involved in siderophore biosynthesis. BMC Evol Biol. 2008;8:328.
Westphal KR, Bachleitner S, Severinsen MM, Brundtø ML, Hansen FT, Sørensen T, et al. Cyclic, hydrophobic hexapeptide fusahexin is the product of a nonribosomal peptide synthetase in Fusarium graminearum. J Nat Prod. 2021;84(8):2070–80.
Turgeon BG, Oide S, Bushley K. Creating and screening Cochliobolus heterostrophus non-ribosomal peptide synthetase mutants. Mycol Res. 2008;112(Pt 2):200–6.
Hansen FT, Droce A, Sørensen JL, Fojan P, Giese H, Sondergaard TE. Overexpression of NRPS4 leads to increased surface hydrophobicity in Fusarium graminearum. Fungal Biol. 2012;116(8):855–62.
Quarantin A, Hadeler B, Kröger C, Schäfer W, Favaron F, Sella L, et al. Different hydrophobins of Fusarium graminearum are involved in hyphal growth, attachment, water-air interface penetration and plant infection. Front Microbiol. 2019;10:751.
Grünbacher A, Throm T, Seidel C, Gutt B, Röhrig J, Strunk T, et al. Six hydrophobins are involved in hydrophobin rodlet formation in Aspergillus nidulans and contribute to hydrophobicity of the spore surface. PLoS ONE. 2014;9(4): e94546.
Dubey MK, Jensen DF, Karlsson M. Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol. 2014;14:18.
Kim KH, Cho Y, Rota LA, Cramer RA, Lawrence CB. Functional analysis of the Alternaria brassicicola non-ribosomal peptide synthetase gene AbNPS2 reveals a role in conidial cell wall construction. Mol Plant Pathol. 2007;8(1):23–39.
Mentges M, Glasenapp A, Boenisch M, Malz S, Henrissat B, Frandsen RJN, et al. Infection cushions of Fusarium graminearum are fungal arsenals for wheat infection. Mol Plant Pathol. 2020;21(8):1070–87.
Tang Z, Tang H, Wang W, Xue Y, Chen D, Tang W, et al. Biosynthesis of a new fusaoctaxin virulence factor in Fusarium graminearum relies on a distinct path to form a guanidinoacetyl starter unit priming nonribosomal octapeptidyl assembly. J Am Chem Soc. 2021;143(47):19719–30.
Jia L, Tang H, Wang W, Yuan T, Wei W, Pang B, et al. A linear nonribosomal octapeptide from Fusarium graminearum facilitates cell-to-cell invasion of wheat. Nat Commun. 2019;10(1):922.
Zhang X-W, Jia L-J, Zhang Y, Jiang G, Li X, Zhang D, et al. In planta stage-specific fungal gene profiling elucidates the molecular strategies of Fusarium graminearum growing inside wheat coleoptiles. Plant Cell. 2012;24(12):5159–76.
Westphal KR, Nielsen KA, Wollenberg RD, Møllehøj MB, Bachleitner S, Studt L, et al. Fusaoctaxin A, an example of a two-step mechanism for non-ribosomal peptide assembly and maturation in fungi. Toxins. 2019;11(5):277.
Moktali V, Park J, Fedorova-Abrams ND, Park B, Choi J, Lee Y-H, et al. Systematic and searchable classification of cytochrome P450 proteins encoded by fungal and oomycete genomes. BMC Genomics. 2012;13(1):525.
Mihara H, Esaki N. Bacterial cysteine desulfurases: their function and mechanisms. Appl Microbiol Biotechnol. 2002;60(1):12–23.
Sharer JD, Bodamer O, Longo N, Tortorelli S, Wamelink MM, Young S. Laboratory diagnosis of creatine deficiency syndromes: a technical standard and guideline of the American College of Medical Genetics and Genomics. Genet Med. 2017;19(2):256–63.
Sarasa SB, Mahendran R, Muthusamy G, Thankappan B, Selta DRF, Angayarkanni J. A brief review on the non-protein amino acid, gamma-amino butyric acid (GABA): its production and role in microbes. Curr Microbiol. 2020;77(4):534–44.
Humm A, Fritsche E, Steinbacher S, Huber R. Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis. EMBO J. 1997;16(12):3373–85.
Bahadoor A, Brauer EK, Bosnich W, Schneiderman D, Johnston A, Aubin Y, et al. Gramillin A and B: cyclic lipopeptides identified as the nonribosomal biosynthetic products of Fusarium graminearum. J Am Chem Soc. 2018;140(48):16783–91.
Harris LJ, Desjardins AE, Plattner RD, Nicholson P, Butler G, Young JC, et al. Possible role of trichothecene mycotoxins in virulence of Fusarium graminearum on maize. Plant Dis. 1999;83(10):954–60.
Gardiner DM, Kazan K, Manners JM. Novel genes of Fusarium graminearum that negatively regulate deoxynivalenol production and virulence. Mol Plant Microbe Interact. 2009;22(12):1588–600.
Jonkers W, Dong Y, Broz K, Kistler HC. The Wor1-like protein Fgp1 regulates pathogenicity, toxin synthesis and reproduction in the phytopathogenic fungus Fusarium graminearum. PLoS Pathog. 2012;8(5): e1002724.
Harris LJ, Balcerzak M, Johnston A, Schneiderman D, Ouellet T. Host-preferential Fusarium graminearum gene expression during infection of wheat, barley, and maize. Fungal Biol. 2016;120(1):111–23.
Sieber CM, Lee W, Wong P, Münsterkötter M, Mewes HW, Schmeitzl C, et al. The Fusarium graminearum genome reveals more secondary metabolite gene clusters and hints of horizontal gene transfer. PLoS ONE. 2014;9(10): e110311.
Nielsen MR, Wollenberg RD, Westphal KR, Sondergaard TE, Wimmer R, Gardiner DM, et al. Heterologous expression of intact biosynthetic gene clusters in Fusarium graminearum. Fungal Genet Biol. 2019;132: 103248.
Shostak K, González-Peña Fundora D, Blackman C, Witte T, Sproule A, Overy D, et al. Epistatic relationship between MGV1 and TRI6 in the regulation of biosynthetic gene clusters in Fusarium graminearum. J Fungi. 2023;9(8):816.
Shostak K, Bonner C, Sproule A, Thapa I, Shields SWJ, Blackwell B, et al. Activation of biosynthetic gene clusters by the global transcriptional regulator TRI6 in Fusarium graminearum. Mol Microbiol. 2020;114(4):664–80.
Stachelhaus T, Mootz HD, Marahiel MA. The specificity-conferring code of adenylation domains in nonribosomal peptide synthetases. Chem Biol. 1999;6(8):493–505.
Kalb D, Lackner G, Hoffmeister D. Fungal peptide synthetases: an update on functions and specificity signatures. Fungal Biol Rev. 2013;27(2):43–50.
Viggiano A, Salo O, Ali H, Szymanski W, Lankhorst PP, Nygård Y, et al. Pathway for the biosynthesis of the pigment chrysogine by penicillium chrysogenum. Appl Environ Microbiol. 2018;84(4):e02246-e2317.
Khan I, Zaib S, Batool S, Abbas N, Ashraf Z, Iqbal J, et al. Quinazolines and quinazolinones as ubiquitous structural fragments in medicinal chemistry: an update on the development of synthetic methods and pharmacological diversification. Bioorg Med Chem. 2016;24(11):2361–81.
Chen X, Rao L, Chen J, Zou Y. Unexpected assembly machinery for 4(3H)-quinazolinone scaffold synthesis. Nat Commun. 2022;13(1):6522.
Auti PS, George G, Paul AT. Recent advances in the pharmacological diversification of quinazoline/quinazolinone hybrids. RSC Adv. 2020;10(68):41353–92.
Kshirsagar UA. Recent developments in the chemistry of quinazolinone alkaloids. Org Biomol Chem. 2015;13(36):9336–52.
Varga J, Kocsubé S, Tóth B, Mesterházy A. Nonribosomal peptide synthetase genes in the genome of Fusarium graminearum, causative agent of wheat head blight. Acta Biol Hung. 2005;56(3–4):375–88.
Wollenberg RD, Saei W, Westphal KR, Klitgaard CS, Nielsen KL, Lysøe E, et al. Chrysogine biosynthesis is mediated by a two-module nonribosomal peptide synthetase. J Nat Prod. 2017;80(7):2131–5.
Hai Y, Huang A, Tang Y. Biosynthesis of amino acid derived α-pyrones by an NRPS-NRPKS hybrid megasynthetase in fungi. J Nat Prod. 2020;83(3):593–600.
Haynes SW, Gao X, Tang Y, Walsh CT. Assembly of asperlicin peptidyl alkaloids from anthranilate and tryptophan: a two-enzyme pathway generates heptacyclic scaffold complexity in asperlicin E. J Am Chem Soc. 2012;134(42):17444–7.
Garcia-Cela E, Kiaitsi E, Medina A, Sulyok M, Krska R, Magan N. Interacting environmental stress factors affects targeted metabolomic profiles in stored natural wheat and that inoculated with F. graminearum. Toxins. 2018;10(2):56.
Hamill RL, Higgens CE, Boaz HE, Gorman M. Structure of beauvericin, a new depsipeptide antibiotic toxic to Artemia salina. Tetrahedron Lett. 1969;49(10):4255–8.
Logrieco A, Moretti A, Castella G, Kostecki M, Golinski P, Ritieni A, et al. Beauvericin production by Fusarium species. Appl Environ Microbiol. 1998;64(8):3084–8.
Wu Q, Patocka J, Nepovimova E, Kuca K. A review on the synthesis and bioactivity aspects of beauvericin, a Fusarium mycotoxin. Front Pharmacol. 2018;20(9):1338.
Fernández-Blanco C, Frizzell C, Shannon M, Ruiz MJ, Connolly L. An in vitro investigation on the cytotoxic and nuclear receptor transcriptional activity of the mycotoxins fumonisin B1 and beauvericin. Toxicol Lett. 2016;257:1–10.
Wätjen W, Debbab A, Hohlfeld A, Chovolou Y, Proksch P. The mycotoxin beauvericin induces apoptotic cell death in H4IIE hepatoma cells accompanied by an inhibition of NF-κB-activity and modulation of MAP-kinases. Toxicol Lett. 2014;231(1):9–16.
Yoo S, Kim MY, Cho JY. Beauvericin, a cyclic peptide, inhibits inflammatory responses in macrophages by inhibiting the NF-κB pathway. Korean J Physiol Pharmacol. 2017;21(4):449–56.
Zhang H, Ruan C, Bai X, Zhang M, Zhu S, Jiang Y. Isolation and identification of the antimicrobial agent beauvericin from the endophytic Fusarium oxysporum 5–19 with NMR and ESI-MS/MS. Biomed Res Int. 2016;2016:1084670.
Shimada A, Fujioka S, Koshino H, Kimura Y. Nematicidal activity of beauvericin produced by the fungus Fusarium bulbicola. Z Naturforsch C J Biosci. 2010;65(3–4):207–10.
Tao YW, Lin YC, She ZG, Lin MT, Chen PX, Zhang JY. Anticancer activity and mechanism investigation of beauvericin isolated from secondary metabolites of the mangrove endophytic fungi. Anticancer Agents Med Chem. 2015;15(2):258–66.
Zhang T, Zhuo Y, Jia X, Liu J, Gao H, Song F, et al. Cloning and characterization of the gene cluster required for beauvericin biosynthesis in Fusarium proliferatum. Sci China Life Sci. 2013;56(7):628–37.
Frandsen RJ, Andersson JA, Kristensen MB, Giese H. Efficient four fragment cloning for the construction of vectors for targeted gene replacement in filamentous fungi. BMC Mol Biol. 2008;9(1):70.
Zhang T, Jia X, Zhuo Y, Liu M, Gao H, Liu J, et al. Cloning and characterization of a novel 2-ketoisovalerate reductase from the beauvericin producer Fusarium proliferatum LF061. BMC Biotechnol. 2012;12(1):55.
Sharom FJ. ABC multidrug transporters: structure, function and role in chemoresistance. Pharmacogenomics. 2008;9(1):105–27.
Niehaus EM, Studt L, von Bargen KW, Kummer W, Humpf HU, Reuter G, et al. Sound of silence: the beauvericin cluster in Fusarium fujikuroi is controlled by cluster-specific and global regulators mediated by H3K27 modification. Environ Microbiol. 2016;18(11):4282–302.
Zobel S, Boecker S, Kulke D, Heimbach D, Meyer V, Süssmuth RD. Reprogramming the biosynthesis of cyclodepsipeptide synthetases to obtain new enniatins and beauvericins. ChemBioChem. 2016;17(4):283–7.
Belofsky GN, Jensen PR, Fenical W. Sansalvamide: a new cytotoxic cyclic depsipeptide produced by a marine fungus of the genus Fusarium. Tetrahedron Lett. 1999;40(15):2913–6.
Kunicki JB, Petersen MN, Alexander LD, Ardi VC, McConnell JR, McAlpine SR. Synthesis and evaluation of biotinylated sansalvamide A analogs and their modulation of Hsp90. Bioorg Med Chem Lett. 2011;21(16):4716–9.
Hwang Y, Rowley D, Rhodes D, Gertsch J, Fenical W, Bushman F. Mechanism of inhibition of a poxvirus topoisomerase by the marine natural product sansalvamide A. Mol Pharmacol. 1999;55(6):1049–53.
Lee H, Lee C. Structural analysis of a new cytotoxic demethylated analogue of neo-N-methylsansalvamide with a different peptide sequence produced by Fusarium solani isolated from potato. J Agric Food Chem. 2012;60(17):4342–7.
Malz S, Grell MN, Thrane C, Maier FJ, Rosager P, et al. Identification of a gene cluster responsible for the biosynthesis of aurofusarin in the Fusarium graminearum species complex. Fungal Genet Biol. 2005;42(5):420–33.
Romans-Fuertes P, Sondergaard TE, Sandmann MIH, Wollenberg RD, Nielsen KF, Hansen FT, et al. Identification of the non-ribosomal peptide synthetase responsible for biosynthesis of the potential anti-cancer drug sansalvamide in Fusarium solani. Curr Genet. 2016;62(4):799–807.
Rodriguez RA, Pan P-S, Pan C-M, Ravula S, Lapera S, Singh EK, et al. Synthesis of second-generation sansalvamide A derivatives: novel templates as potential antitumor agents. J Org Chem. 2007;72(6):1980–2002.
Khayatt BI, Overmars L, Siezen RJ, Francke C. Classification of the adenylation and acyl-transferase activity of NRPS and PKS systems using ensembles of substrate specific hidden Markov models. PLoS ONE. 2013;8(4): e62136.
von Bargen KW, Niehaus EM, Bergander K, Brun R, Tudzynski B, Humpf HU. Structure elucidation and antimalarial activity of apicidin F: an apicidin-like compound produced by Fusarium fujikuroi. J Nat Prod. 2013;76(11):2136–40.
Darkin-Rattray SJ, Gurnett AM, Myers RW, Dulski PM, Crumley TM, Allocco JJ, et al. Apicidin: a novel antiprotozoal agent that inhibits parasite histone deacetylase. Proc Natl Acad Sci USA. 1996;93(23):13143–7.
Niehaus EM, Janevska S, von Bargen KW, Sieber CMK, Harrer H, Humpf HU, et al. Apicidin F: characterization and genetic manipulation of a new secondary metabolite gene cluster in the rice pathogen Fusarium fujikuroi. PLoS ONE. 2014;9(7): e103336.
Kouraklis G, Theocharis S. Histone deacetylase inhibitors and anticancer therapy. Curr Med Chem Anticancer Agents. 2002;2(4):477–84.
Matore BW, Banjare P, Guria T, Roy PP, Singh J. Oxadiazole derivatives: histone deacetylase inhibitors in anticancer therapy and drug discovery. Eur J Med Chem. 2022;5: 100058.
Jin JM, Lee S, Lee J, Baek SR, Kim JC, Yun SH, et al. Functional characterization and manipulation of the apicidin biosynthetic pathway in Fusarium semitectum. Mol Microbiol. 2010;76(2):456–66.
Cheng Y, Ahn JH, Walton JD. A putative branched-chain-amino-acid transaminase gene required for HC-toxin biosynthesis and pathogenicity in Cochliobolus carbonum. Microbiology (Reading). 1999;145(Pt12):3539–46.
Suciati, Garson MJ. Isolation of the tetrapeptide apicidins G, H and I from the fungus Fusarium semitectum. Nat Prod Commun. 2014;9(2):233–6.
Olsen CA, Ghadiri MR. Discovery of potent and selective histone deacetylase inhibitors via focused combinatorial libraries of cyclic alpha3beta-tetrapeptides. J Med Chem. 2009;52(23):7836–46.
Janevska S, Tudzynski B. Secondary metabolism in Fusarium fujikuroi: strategies to unravel the function of biosynthetic pathways. Appl Microbiol Biotechnol. 2018;102(2):615–30.
Marshall JW, de Mattos-Shipley KMJ, Ghannam IAY, Munawar A, Killen JC, Lazarus CM, et al. Fusarochromene, a novel tryptophan-derived metabolite from Fusarium sacchari. Org Biomol Chem. 2021;19(1):182–7.
Xie W, Mirocha CJ, Wen Y. Isolation and structure identification of two new derivatives of the mycotoxin fusarochromenone produced by Fusarium equiseti. J Nat Prod. 1995;58(1):124–7.
Gu Y, Barzegar M, Chen X, Wu Y, Shang C, Mahdavian E, et al. Fusarochromanone-induced reactive oxygen species results in activation of JNK cascade and cell death by inhibiting protein phosphatases 2A and 5. Oncotarget. 2015;6(39):42322–33.
Badal S, Williams SA, Huang G, Francis S, Vendantam P, Dunbar O, et al. Cytochrome P450 1 enzyme inhibition and anticancer potential of chromene amides from Amyris plumieri. Fitoterapia. 2011;82(2):230–6.
Wogulis M, Chew ER, Donohoue PD, Wilson DK. Identification of formyl kynurenine formamidase and kynurenine aminotransferase from Saccharomyces cerevisiae using crystallographic, bioinformatic and biochemical evidence. Biochemistry. 2008;47(6):1608–21.
Li W, Fan A, Wang L, Zhang P, Liu Z, An Z, Yin WB. Asperphenamate biosynthesis reveals a novel two-module NRPS system to synthesize amino acid esters in fungi. Chem Sci. 2018;9(9):2589–94.
Ishihara A, Sugai N, Bito T, Ube N, Ueno K, Okuda Y, et al. Isolation of 6-hydroxy-L-tryptophan from the fruiting body of Lyophyllum decastes for use as a tyrosinase inhibitor. Biosci Biotechnol Biochem. 2019;83(10):1800–6.
Yow GY, Uo T, Yoshimura T, Esaki N. D-amino acid N-acetyltransferase of Saccharomyces cerevisiae: a close homologue of histone acetyltransferase Hpa2p acting exclusively on free D-amino acids. Arch Microbiol. 2004;182(5):396–403.
Zhang J, Wang H, Liu X, Hu C, Zou Y. Heterologous and engineered biosynthesis of nematocidal polyketide-nonribosomal peptide hybrid macrolactone from extreme thermophilic fungi. J Am Chem Soc. 2020;142(4):1957–65.
Miyanaga A, Kudo F, Eguchi T. Protein-protein interactions in polyketide synthase-nonribosomal peptide synthetase hybrid assembly lines. Nat Prod Rep. 2018;35(11):1185–209.
Minami A, Ugai T, Ozaki T, Oikawa H. Predicting the chemical space of fungal polyketides by phylogeny-based bioinformatics analysis of polyketide synthase-nonribosomal peptide synthetase and its modification enzymes. Sci Rep. 2020;10(1):13556.
Keller NP, Hohn TM. Metabolic pathway gene clusters in filamentous fungi. Fungal Genet Biol. 1997;21(1):17–29.
Shiono Y, Tsuchinari M, Shimanuki K, Miyajima T, Murayama T, Koseki T, et al. Fusaristatins A and B, two new cyclic lipopeptides from an endophytic Fusarium sp. J Antibiot (Tokyo). 2007;60(5):309–16.
Lim C, Kim J, Choi JN, Ponnusamy K, Jeon Y, Kim SU, et al. Identification, fermentation, and bioactivity against Xanthomonas oryzae of antimicrobial metabolites isolated from Phomopsis longicolla S1B4. J Microbiol Biotechnol. 2010;20(3):494–500.
Jang JH, Asami Y, Jang JP, Kim SO, Moon DO, Shin KS, et al. Fusarisetin A, an acinar morphogenesis inhibitor from a soil fungus, Fusarium sp. FN080326. J Am Chem Soc. 2011;133(18):6865–7.
Sørensen LQ, Lysøe E, Larsen JE, Khorsand-Jamal P, Nielsen KF, Frandsen RJ. Genetic transformation of Fusarium avenaceum by Agrobacterium tumefaciens mediated transformation and the development of a USER-Brick vector construction system. BMC Mol Biol. 2014;15:15.
Hegge A, Lønborg R, Nielsen DM, Sørensen JL. Factors influencing production of fusaristatin A in Fusarium graminearum. Metabolites. 2015;5(2):184–91.
Sørensen JL, Sondergaard TE, Covarelli L, Fuertes PR, Hansen FT, Frandsen RJN, et al. Identification of the biosynthetic gene clusters for the lipopeptides fusaristatin A and W493 B in Fusarium graminearum and F. pseudograminearum. J Nat Prod. 2014;77(12):2619–25.
Nihei K, Itoh H, Hashimoto K, Miyairi K, Okuno T. Antifungal cyclodepsipeptides, W493 A and B, from Fusarium sp.: isolation and structural determination. Biosci Biotechnol Biochem. 1998;62(5):858–63.
Burmeister HR, Vesonder RF, Peterson RE, Costello CE. Production and purification of a peptide of Fusarium tricinctum that causes conidia of Penicillium to swell. Mycopathologia. 1985;91(1):53–6.
Bacon CW, Porter JK, Norred WP, Leslie JF. Production of fusaric acid by Fusarium species. Appl Environ Microbiol. 1996;62(11):4039–43.
Ernst G, Stephi N-R, Hans K. Fusaric acid, a second toxin of wilt produced by Fusarium lycopersici. Compt rend. 1952;234:173–4.
Wang H, Ng TB. Pharmacological activities of fusaric acid (5-butylpicolinic acid). Life Sci. 1999;65(9):849–56.
D’Mello JPF, Placinta CM, Macdonald AMC. Fusarium mycotoxins: a review of global implications for animal health, welfare and productivity. Anim Feed Sci Tech. 1999;80(3):183–205.
Porter JK, Bacon CW, Wray EM, Hagler WM Jr. Fusaric acid in Fusarium moniliforme cultures, corn, and feeds toxic to livestock and the neurochemical effects in the brain and pineal gland of rats. Nat Toxins. 1995;3(2):91–100.
Ramautar A, Mabandla M, Blackburn J, Daniels WM. Inhibition of HIV-1 tat-induced transactivation and apoptosis by the divalent metal chelators, fusaric acid and picolinic acid-implications for HIV-1 dementia. Neurosci Res. 2012;74(1):59–63.
Boonman N, Prachya S, Boonmee A, Kittakoop P, Wiyakrutta S, Sriubolmas N, et al. In vitro acanthamoebicidal activity of fusaric acid and dehydrofusaric acid from an endophytic fungus Fusarium sp. Tlau3. Planta Med. 2012;78(14):1562–7.
Brown DW, Butchko RA, Busman M, Proctor RH. Identification of gene clusters associated with fusaric acid, fusarin, and perithecial pigment production in Fusarium verticillioides. Fungal Genet Biol. 2012;49(7):521–32.
Niehaus EM, von Bargen KW, Espino JJ, Pfannmüller A, Humpf HU, Tudzynski B. Characterization of the fusaric acid gene cluster in Fusarium fujikuroi. Appl Microbiol Biotechnol. 2014;98(4):1749–62.
Zeng T, Zeng H, Fu M, Huang K, Guo J, Hu X. Kynurenine pathway as alternative biosynthetic pathway for fusaric acid in Fusarium oxysporum f. sp. cubense. Australas Plant Path. 2021;50(4):415–26.
Michielse CB, Studt L, Janevska S, Sieber CMK, Arndt B, Espino JJ, et al. The global regulator FfSge1 is required for expression of secondary metabolite gene clusters but not for pathogenicity in Fusarium fujikuroi. Environ Microbiol. 2015;17(8):2690–708.
Brown DW, Lee SH, Kim LH, Ryu JG, Lee S, Seo Y, et al. Identification of a 12-gene fusaric acid biosynthetic gene cluster in Fusarium species through comparative and functional genomics. Mol Plant Microbe Interact. 2015;28(3):319–32.
Studt L, Janevska S, Niehaus EM, Burkhardt I, Arndt B, Sieber CMK, et al. Two separate key enzymes and two pathway-specific transcription factors are involved in fusaric acid biosynthesis in Fusarium fujikuroi. Environ Microbiol. 2016;18(3):936–56.
Chooi YH, Tang Y. Navigating the fungal polyketide chemical space: from genes to molecules. J Org Chem. 2012;77(22):9933–53.
Maiya S, Grundmann A, Li X, Li S, Turner G. Identification of a hybrid PKS/NRPS required for pseurotin A biosynthesis in the human pathogen Aspergillus fumigatus. ChemBioChem. 2007;8(14):1736–43.
Fisch KM, Bakeer W, Yakasai AA, Song Z, Pedrick J, Wasil Z, et al. Rational domain swaps decipher programming in fungal highly reducing polyketide synthases and resurrect an extinct metabolite. J Am Chem Soc. 2011;133(41):16635–41.
Ames BD, Nguyen C, Bruegger J, Smith P, Xu W, Ma S, et al. Crystal structure and biochemical studies of the trans-acting polyketide enoyl reductase LovC from lovastatin biosynthesis. Proc Natl Acad Sci USA. 2012;109(28):11144–9.
Sims JW, Schmidt EW. Thioesterase-like role for fungal PKS-NRPS hybrid reductive domains. J Am Chem Soc. 2008;130(33):11149–55.
Boettger D, Hertweck C. Molecular diversity sculpted by fungal PKS-NRPS hybrids. ChemBioChem. 2013;14(1):28–42.
Wiebe LA, Bjeldanes LF. Fusarin C, a mutagen from Fusarium Moniliforme grown on corn. J Food Sci. 1981;46(5):1424–6.
Thrane U, Adler A, Clasen PE, Galvano F, Langseth W, et al. Diversity in metabolite production by Fusarium langsethiae, Fusarium poae, and Fusarium sporotrichioides. Int J Food Microbiol. 2004;95(3):257–66.
Gelderblom WCA, Marasas WFO, Steyn PS, Thiel PG, Merwe KJ, Rooyen PH, et al. Structure elucidation of fusarin C, a mutagen produced by Fusarium moniliforme. J Chem Soc Chem Commun. 1984;2:122–4.
Cantalejo MJ, Torondel P, Amate L, Carrasco JM, Hernández E. Detection of fusarin C and trichothecenes in Fusarium strains from Spain. J Basic Microbiol. 1999;39(3):143–53.
Díaz-Sánchez V, Avalos J, Limón MC. Identification and regulation of fusA, the polyketide synthase gene responsible for fusarin production in Fusarium fujikuroi. Appl Environ Microbiol. 2012;78(20):7258–66.
Kleigrewe K, Aydin F, Hogrefe K, Piecuch P, Bergander K, Würthwein E-U, et al. Structure elucidation of new fusarins revealing insights in the rearrangement mechanisms of the Fusarium mycotoxin fusarin C. J Agric Food Chem. 2012;60(21):5497–505.
Maragos CM, Busman M, Plattner RD. Development of monoclonal antibodies for the fusarin mycotoxins. Food Addit Contam Part A Chem Anal Control Expo Risk Assess. 2008;25(1):105–14.
Sondergaard TE, Hansen FT, Purup S, Nielsen AK, Bonefeld-Jørgensen EC, Giese H, et al. Fusarin C acts like an estrogenic agonist and stimulates breast cancer cells in vitro. Toxicol Lett. 2011;205(2):116–21.
Li M, Jiang Y, Bjeldanes LF. Carcinogenicity of fusarin C isolated from Fusarium moniliforme. Chinese J Cancer Res. 1990;2(3):1–5.
Lu F, Li M, Cheng S. In vitro transformation of rat esophageal epithelial cells by fusarin C. Sci China B. 1991;34(12):1469–77.
Bever RJ Jr, Couch LH, Sutherland JB, Williams AJ, Beger RD, Churchwell MI, et al. DNA adduct formation by Fusarium culture extracts: lack of role of fusarin C. Chem Biol Interact. 2000;128(2):141–57.
Gelderblom WC, Thiel PG, Jaskiewicz K, Marasas WF. Investigations on the carcinogenicity of fusarin C: a mutagenic metabolite of Fusarium moniliforme. Carcinogenesis. 1986;7(11):1899–901.
Song Z, Cox RJ, Lazarus CM, Simpson TJTJ. Fusarin C biosynthesis in Fusarium moniliforme and Fusarium venenatum. ChemBioChem. 2004;5(9):1196–203.
Niehaus EM, Kleigrewe K, Wiemann P, Studt L, Sieber CM, Connolly LR, et al. Genetic manipulation of the Fusarium fujikuroi fusarin gene cluster yields insight into the complex regulation and fusarin biosynthetic pathway. Chem Biol. 2013;20(8):1055–66.
Boecker S, Zobel S, Meyer V, Süssmuth RD. Rational biosynthetic approaches for the production of new-to-nature compounds in fungi. Fungal Genet Biol. 2016;89:89–101.
Jayasinghe L, Abbas HK, Jacob MR, Herath WH, Nanayakkara NP. N-Methyl-4-hydroxy-2-pyridinone analogues from Fusarium oxysporum. J Nat Prod. 2006;69(3):439–42.
Breinhold J, Ludvigsen S, Rassing BR, Rosendahl CN, Nielsen SE, Olsen CE. Oxysporidinone: a novel, antifungal N-methyl-4-hydroxy-2-pyridone from Fusarium oxysporum. J Nat Prod. 1997;60(1):33–5.
Li D, Wang W, Xu K, Li J, Long B, Li Z, et al. Elucidation of a dearomatization route in the biosynthesis of oxysporidinone involving a TenA-like cytochrome P450 enzyme. Angew Chem Int Ed Engl. 2023;62(25): e202301976.
Chiang YM, Oakley CE, Ahuja M, Entwistle R, Schultz A, Chang SL, et al. An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J Am Chem Soc. 2013;135(20):7720–31.
Yin WB, Chooi YH, Smith AR, Cacho RA, Hu Y, et al. Discovery of cryptic polyketide metabolites from dermatophytes using heterologous expression in Aspergillus nidulans. ACS Synth Biol. 2013;2(11):629–34.
Kakule TB, Sardar D, Lin Z, Schmidt EW. Two related pyrrolidinedione synthetase loci in Fusarium heterosporum ATCC 74349 produce divergent metabolites. ACS Chem Biol. 2013;8(7):1549–57.
Janevska S, Arndt B, Baumann L, Apken LH, Mauriz Marques LM, Humpf HU, et al. Establishment of the inducible Tet-On system for the activation of the silent trichosetin gene cluster in Fusarium fujikuroi. Toxins. 2017;9(4):126.
Kato N, Nogawa T, Hirota H, Jang JH, Takahashi S, Ahn JS, et al. A new enzyme involved in the control of the stereochemistry in the decalin formation during equisetin biosynthesis. Biochem Biophys Res Commun. 2015;460(2):210–5.
Heneghan MN, Yakasai AA, Williams K, Kadir KA, Wasil Z, Bakeer W, et al. The programming role of trans-acting enoyl reductases during the biosynthesis of highly reduced fungal polyketides. Chem Sci. 2011;2(5):972–9.
Schmidt K, Riese U, Li Z, Hamburger M. Novel tetramic acids and pyridone alkaloids, militarinones B, C, and D, from the insect pathogenic fungus Paecilomyces militaris. J Nat Prod. 2003;66(3):378–83.
Gui C, Li Q, Mo X, Qin X, Ma J, Ju J. Discovery of a new family of Dieckmann cyclases essential to tetramic acid and pyridone-based natural products biosynthesis. Org Lett. 2015;17(3):628–31.
Diec KW. Ueber cyklische β-Ketoncarbonsäureester. Liebigs Ann. 1901;317(1):27–109.
Vesonder RF, Tjarks LW, Rohwedder WK, Burmeister HR, Laugal JA. Equisetin, an antibiotic from Fusarium equiseti NRRL 5537, identified as a derivative of N-methyl-2,4-pyrollidone. J Antibiot (Tokyo). 1979;32(7):759–61.
Burmeister HR, Bennett GA, Vesonder RF, Hesseltine CW. Antibiotic produced by Fusarium equiseti NRRL 5537. Antimicrob Agents Chemother. 1974;5(6):634–9.
Zhao H, Cui Z, Gu Y, Liu Y, Wang Q. The phytotoxicity of natural tetramic acid derivatives. Pest Manag Sci. 2011;67(9):1059–61.
Mo X, Li Q, Ju J. Naturally occurring tetramic acid products: isolation, structure elucidation and biological activity. RSC Adv. 2014;4(92):50566–93.
Burkhardt I, Siemon T, Henrot M, Studt L, Rösler S, Tudzynski B, et al. Mechanistic characterisation of two sesquiterpene cyclases from the plant pathogenic fungus Fusarium fujikuroi. Angew Chem Int Ed Engl. 2016;55(30):8748–51.
Marfori EC, Kajiyama S, Fukusaki E, Kobayashi A. Trichosetin, a novel tetramic acid antibiotic produced in dual culture of trichoderma harzianum and catharanthus roseus callus. Z Naturforsch C J Biosci. 2002;57(5–6):465–70.
Desjardins AE, Proctor RH. Molecular biology of Fusarium mycotoxins. Int J Food Microbiol. 2007;119(1–2):47–50.
Marfori EC, Bamba T, Kajiyama Si, Fukusaki EI, Kobayashi A. Biosynthetic studies of the tetramic acid antibiotic trichosetin. Tetrahedron. 2002;58(33):6655–8.
Web links and URLs
The Dictionary of Natural Products. https://dnp.chemnetbase.com. Accessed 15 Dec 2023.
Acknowledgements
This work was co-financially supported by the National Key Research and Development Program of China (2022YFC2804203 and 2018YFC0311004) and the National Natural Science Foundation of China (41776139).
Author information
Authors and Affiliations
Contributions
HW, ZJ, BF: writing—original draft preparation, writing—figures of this review; BL, ZW: writing—review and editing. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1. Table S1
. Detail information for NRPS-type secondary metabolites in Fusarium strains and their research methods.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Huang, Z., Zhu, W., Bai, Y. et al. Non-ribosomal peptide synthetase (NRPS)-encoding products and their biosynthetic logics in Fusarium. Microb Cell Fact 23, 93 (2024). https://doi.org/10.1186/s12934-024-02378-1
Received:
Accepted:
Published:
Version of record:
DOI: https://doi.org/10.1186/s12934-024-02378-1