Abstract
Background
Corynebacterium glutamicum is an attractive host for secretory production of recombinant proteins, including high-value industrial enzymes and therapeutic proteins. The choice of an appropriate signaling peptide is crucial for efficient protein secretion. However, due to the limited availability of signal peptides in C. glutamicum, establishing an optimal secretion system is challenging.
Result
We constructed a signal peptide library for the isolation of target-specific signal peptides and developed a highly efficient secretory production system in C. glutamicum. Based on the sequence information of the signal peptides of the general secretion-dependent pathway in C. glutamicum, a synthetic signal peptide library was designed, and validated with three protein models. First, we examined endoxylanase (XynA) and one potential signal peptide (C1) was successfully isolated by library screening on xylan-containing agar plates. With this C1 signal peptide, secretory production of XynA as high as 3.2 g/L could be achieved with high purity (> 80%). Next, the signal peptide for ⍺-amylase (AmyA) was screened on a starch-containing agar plate. The production titer of the isolated signal peptide (HS06) reached 1.48 g/L which was 2-fold higher than that of the well-known Cg1514 signal peptide. Finally, we isolated the signal peptide for the M18 single-chain variable fragment (scFv). As an enzyme-independent screening tool, we developed a fluorescence-dependent screening tool using Fluorescence-Activating and Absorption-Shifting Tag (FAST) fusion, and successfully isolated the optimal signal peptide (18F11) for M18 scFv. With 18F11, secretory production as high as 228 mg/L was achieved, which was 3.4-fold higher than previous results.
Conclusions
By screening a fully synthetic signal peptide library, we achieved improved production of target proteins compared to previous results using well-known signal peptides. Our synthetic library provides a useful resource for the development of an optimal secretion system for various recombinant proteins in C. glutamicum, and we believe this bacterium to be a more promising workhorse for the bioindustry.
Similar content being viewed by others
Introduction
Corynebacterium glutamicum, a gram-positive, non-spore-forming soil bacterium, has traditionally been used as a potent cell factory for the industrial production of amino acids, fine chemicals, food additives, and biopolymers [1,2,3]. Additionally, C. glutamicum has recently been recognized as an attractive host for the production of recombinant proteins, including high-value industrial enzymes and therapeutic proteins [4,5,6]. As a host for protein production, C. glutamicum has several advantageous properties: (i) much less production of endogenous proteins in the culture medium; (ii) much less proteolytic activity; (iii) endotoxin-free generally recognized as safe host; (iv) rapid growth in defined media; and (v) easy scale-up to industrial-scale cultivation [6,7,8,9]. Compared to the cytoplasmic production of recombinant proteins, secretory production in culture medium precludes the need for cell disruption, which facilitates downstream processing (easy recovery of protein from culture medium) and consequently reduces the overall production costs [10].
C. glutamicum, as with other bacterial hosts, mainly exports proteins across the cytoplasmic membrane via the general secretion (Sec) pathways [8, 11]. This is not a simple process but requires the systematic assistance of multiple components such as signal peptides, Sec-translocons, chaperones, and foldases. Thus, to increase the secretion yield of proteins, much effort is required to optimize a multitude of biological and bioprocess parameters [12, 13]. Particularly, signal peptide, a short amino acid sequence located at the N-terminus of the secreted protein, plays a pivotal role in protein secretion through translocon in the membrane, and the choice of proper signal peptide substantially affects the overall yield of protein secretion [14, 15]. Previously, many efforts have been made to isolate new/potential signal peptides from C. glutamicum, and it is known that several signal peptides, including CspA, CspB PorB, Cg1514 (for the Sec-dependent pathway), TorA, and CgR0949 (for the TAT-dependent pathway), are efficient and have often been used for the secretory production of various recombinant proteins [16,17,18,19]. However, the efficiency of secretion varies among different target proteins, and finding the optimal signal peptide driving the efficient secretion of target proteins generally requires many trial-and-error steps with various signal peptides [20, 21]. These studies are highly time-consuming and laborious, and because of the limited availability of signal peptides, secretory production systems have not been developed for many proteins in C. glutamicum.
Because the secretion efficiency of signal peptides and target proteins is not yet predictable [15], the systematic screening of a large variety of signal peptides (i.e., libraries) has been suggested as the most promising approach for constructing an optimal production system for a target protein. For example, Zhang et al. used a 114 signal peptide library from Bacillus subtilis to isolate an optimal signal peptide for xylanase production in B. subtilis, and the extracellular xylanase activity was improved using the isolated signal peptide [22]. In addition, a bigger signal peptide library (173 B. subtilis signal peptides) was screened to identify the optimal signal peptide for natto phytase expression [23], B. stearothermophilus α-amylase [24], and α-amylase [25] in B. subtilis. The B. subtilis signal peptide library has also been successfully used to optimize secretory production in C. glutamicum. Hemmerich et al. investigated B. subtilis signal peptide library to optimize cutinase secretion from Fusarium solani pisi in C. glutamicum, and some signal peptides from the B. subtilis library showed comparable secretion performance in C. glutamicum [26]. Despite several successful screening results, the size of the employed signal peptide library was relatively small (less than 200 signal peptides), and all were derived from natural sources (particularly Bacillus sp.), which is also a limitation in finding the optimal signal peptide for many diverse target proteins. A large library must be screened, and the source of the signal peptide must be beyond natural sources. Thus, identifying an appropriate signal peptide for each target protein remains a major hurdle in the development of an efficient secretory system.
Here, to address this issue, we report the construction of a fully synthetic signal peptide library for Sec-dependent secretory production in C. glutamicum, and the development of optimal secretion systems for heterologous proteins by screening a signal peptide library. Based on the signal peptide information collected from C. glutamicum, we designed a large synthetic signal peptide library which consisted of 27 amino acids in length. To validate this synthetic library, we isolated optimal signal peptides for three protein models: endoxylanase, ⍺-amylase, and the M18 single-chain variable fragment (scFv). For endoxylanase and ⍺-amylase, the optimal signal peptides were isolated by an enzyme activity-dependent screening strategy on substrate-containing agar plates, and the cells with the isolated signal peptides showed a remarkable secretion yield in fed-batch cultivation. For M18 scFv, we developed a Fluorescence-Activating and Absorption-Shifting Tag (FAST)-derived fluorescence detection system and the optimal signal peptide was successfully isolated by screening FAST-fused M18 scFv with synthetic signal peptides. We believe that this fully synthetic signal peptide library is a useful resource for the development of efficient protein production systems in C. glutamicum.
Materials and methods
Bacterial strains and flask culture conditions
The bacterial strains and plasmids used in this study are listed in Table S1. Escherichia coli XL1-Blue was used for gene cloning and C. glutamicum strains were used for secretory production. E. coli was cultivated in Luria-Bertani media (BD Biosciences, Franklin Lakes, NJ, USA) at 37℃ with 200 rpm shaking. C. glutamicum was cultivated in brain heart infusion (BHI) medium (BD Biosciences) or modified CGXII medium (3 g/L K2HPO4, 1 g/L KH2PO4, 2 g/L urea, 10 g/L (NH4)2SO4, 2 g/L MgSO4, 200 µg/L biotin, 5 mg/L thiamine, 10 mg/L calcium pantothenate, 10 mg/L FeSO4, 1 mg/L MnSO4, 1 mg/L ZnSO4, 200 µg/L CuSO4, 10 mg/L CaCl2, 20 g/L glucose, 7 g/L casamino acid, and 15 g/L yeast extract) at 30℃ with 200 rpm shaking. Kanamycin (50 µg/mL for E. coli strains and 25 µg/mL for C. glutamicum strains) was added to all culture media as a sole antibiotic. For the comparison of secretory production in flasks, all C. glutamicum cells were cultivated in BHI medium at 30℃ for 24 h. Fully grown cells were transferred to a 250 mL baffled flask containing 50 mL of fresh BHI medium or modified CGXII medium and cultivated for 24 h.
Plasmid construction
Polymerase chain reaction (PCR) was performed using a C1000TM Thermal Cycler (Bio-Rad, Hercules, CA, USA) with Prime STAR HS Polymerase (Takara Bio Inc., Shiga, Japan). All oligonucleotides used for PCR are listed in Table S2. To produce endoxylanase (XynA), xynA was amplified from pHCP-S-XynA [27] using PCR with primers XbaI_SapI_XynA_F and EcoRI_NotI_SfiI_R. PCR products were digested with XbaI and NotI restriction enzymes and cloned into pHCMS [27], yielding pHCP-XynA. XynA secretion systems containing signal peptides were constructed using the pHCP-XynA plasmid. XynA and each signal peptide (Cg2052, Cg1514, and PorB) were amplified using PCR, digested with NdeI and NotI restriction enzymes, and cloned into pHCMS to yield pHCP-Cg2052-XynA, pHCP-Cg1514-XynA, and pHCP-PorB-XynA, respectively. To produce ⍺-amylase (AmyA), AmyA was amplified from pCG-S-AmyA [16] using PCR with XbaI_SapI_Amy_F/ EcoRI_NotI_SfiI_Amy_R primers and cloned into pHCMS to yield pHCP-AmyA. The ⍺-amylase secretion system with Cg1514 signal peptide was constructed on pHCP-AmyA, yielding pHCP-Cg1514-AmyA.
To fuse FAST, the sequence information was obtained from the Twinkle Factory (https://www.the-twinkle-factory.com/fluorogens-for-fast-and-splitfast/; France), and synthesized with optimized sequences for codon preferences of C. glutamicum ATCC13032 from GENEER (Daejeon, Republic of Korea). To construct FAST-fused XynA secretion systems with signal peptides (Cg1514, Cg2052, PorB, and C1), each signal peptide fused with XynA and FAST fragments was amplified and overlapped using PCR. The final PCR products were digested with NdeI and NotI restriction enzymes and cloned into pHCMS to yield pHCP-Cg1514-XynA-FAST, pHCP-Cg2052-XynA-FAST, pHCP-PorB-XynA-FAST, and pHCP-C1-XynA-FAST. To produce M18 scFv, FAST was fused to the C-terminus of M18 scFv using Gibson assembly to yield pHCP-PorB-M18-FAST. Specifically, M18 scFv, with a PorB signal peptide attached to the N-terminus of the gene, was amplified from pH36M2 [28] and the FAST fragment was amplified from the synthesized gene using PCR as an assembly insert. All plasmid constructions were carried out in E. coli XL1-Blue or E. coli NEB 10-beta, and each plasmid was transformed into C. glutamicum via electroporation.
Synthetic signal peptide library construction
For the construction of a synthetic signal peptide library, known signal peptide sequences of C. glutamicum R and C. glutamicum ATCC13032 were assembled using Clustal Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/) [29, 30]. The known signal peptides were predicted with SignalP 5.0 (http://www.cbs.dtu.dk/services/SignalP/) for analysis of each N-, H-, C-part, and cleavage site [30, 31]. After aligning the predicted signal peptide sequences, the synthetic signal peptide was designed to 27 amino acids in length. Synthetic signal peptides were amplified by PCR using the degenerate primers SPL_1_F, SPL_2_F, SPL_3_F, SPL_SapI_1_R, SPL_SapI_2_R, and SPL_SapI_3_R, without a template. These primers had short assembled sequences (five amino acids, ALALA). PCR was performed again on the amplified synthetic signal peptides with primers XbaI_RBS_NdeI_F and NotI_SfiI_SapI_R. After amplification, the synthetic signal peptides were digested with XbaI and NotI and cloned into pHCMS with the same restriction enzyme sites, yielding pHCP-SP-L. Endoxylanase secretion (pHCP-SP-XynA) was constructed by cloning the synthetic signal peptide sequences from pHCP-SP-L into pHCP-XynA with KpnI and SapI restriction enzyme sites. Libraries of ⍺-amylase and M18 (pHCP-SP-AmyA and pHCP-SP-M18-FAST, respectively) were constructed with same procedures as above.
Screening of synthetic signal peptide library on agar plate
To screen the endoxylanase library, it was spread on BHI agar plates containing 1% xylan from beechwood (Sigma-Aldrich, St. Louis, MO, USA). The cells were cultivated in a static incubator at 30℃ until a halo was formed on the agar plates.
For screening the ⍺-amylase library, the library was first spread on BHI agar plates and cultivated in a static incubator at 30℃. The colonies on the agar plate were dotted on BHI agar containing 1% starch (Junsei Chemical Co., Ltd., Tokyo, Japan), and cells were cultivated in a static incubator at 30℃ until a halo formed on the agar plates. Next, Lugol’s solution (Sigma-Aldrich) was spread on the starch-containing BHI agar plate to determine the diameter of each halo. For the individual selection of clones, the diameter of the halo and the size of the colony in the middle of the halo were considered simultaneously.
Analysis of FAST-tagged proteins
To test the activity of the FAST-tagged secreted proteins, the signal peptide library system was first transformed into C. glutamicum SP002 and spread on BHI agar plates. Then, each single colony was inoculated into deep 96-well plates with each well containing 900 µL BHI media, and cultured for 24 h at 30℃ using a 96-well shaking incubator (DWMax MBR-034P, Taitec Co., Saitama, Japan). After cultivation and centrifugation, 10 µL of cell culture supernatant was loaded onto a 96-well black plate with 1.25 µM of HBRAA-3E (TFAmber-NP; Twinkle Factory). Fluorescence was measured at an excitation wavelength of 505 nm with a 559 nm filter using a TECAN Infinite M Plex (Tecan Group Ltd., Männedorf, Switzerland).
Protein preparation and analysis
For preparation of extracellular proteins, culture supernatant was mixed with the same volume of cold acetone and incubated at -20℃ for 2 h. Then, the mixture was centrifuged (13,000 rpm, 10 min at 4℃), and the pellet was resuspended with protein sampling buffer (50 mM Tris-HCl, pH 10, 10% glycerol, 4% SDS, 8 M urea, 2% β-mercaptoethanol, and 0.02% bromophenol blue) to approximately a 10 to 30-fold concentration. For the fermentation samples, only the culture supernatant was mixed with the protein sampling buffer without acetone precipitation. For SDS-PAGE analysis, the samples were loaded onto a polyacrylamide gel, stained with Coomassie brilliant blue, and destained with destaining buffer. Densitometric analysis was performed using a GelAnalyzer 2010a [32]. For N-terminal amino acid sequencing, the polyacrylamide gel loaded with the culture sample supernatant was transferred onto a PVDF membrane, which was then stained with Coomassie brilliant blue and destained with destaining buffer. The major secreted protein band was extracted from the membrane and analyzed by EMASS Co. (Seoul, Republic of Korea).
Activity assay
The activity of XynA was determined by the XylX6 kit (Megazyme Co., Ireland) [33]. Briefly, supernatant from the fed-batch culture was dialyzed with 10% phosphate buffered saline (0.16 M NaCl, 3 mM Na2HPO4, and 1.1 mM KH2PO4) and incubated at 40℃ for 3 min. Then, samples were mixed with pre-incubated XylX6 reagent solution containing XylX6 substrate and β-xylosidase and incubated at 40℃. After 10 min, 1.5 mL of stopping reagent (2% Tris, pH 10.0) was treated and the absorbance of the reaction solution was measured at 400 nm by spectrophotometer. As a negative control (N.C.), 10% PBS was used. One unit (U) of XynA activity is defined as the amount of enzyme required to release one micromole of 4-nitrophenol from the XylX6 substrate in one minute.
The activity of ⍺-amylase was measured by 3,5-dinitrosalicylic acid (DNS) methods [34]. Culture supernatant of fed-batch sample was 25-fold diluted with distilled water, and then, sample was mixed with 50 µL of starch solution (1%(w/v) starch, 20 mM sodium phosphate). After reaction for 3 min at room temperature, DNS reagent was added, and sample was incubated at 100℃ for 5 min. After cooling, 1 mL of distilled water was added and the absorbance was measured at 540 nm by spectrophotometer. For the negative control, distilled water was used instead of starch solution. One unit (U) of ⍺-amylase activity is defined as the amount of enzyme required to release one micromole (µmol) of reducing sugar in one minute.
The binding activity of the M18 scFv was determined by ELISA as described previously [18]. Briefly, 50 µL of 5 µg/mL antigen (domain 4 of anthrax toxin PA) dissolved in carbonate-bicarbonate buffer was added to each well of a 96-well plate, and incubated at 37℃ for 2 h. After washing four times with PBS-T (0.5% Tween-20 in PBS), the coated antigens were blocked with 200 µL/well of 5% (w/v) bovine serum albumin (BSA) solution in PBS for 1 h at room temperature. Then, the serially diluted samples were treated to the coated wells and incubated for 1 h at room temperature. Each well was washed with PBS-T, and 50 µL of HRP-conjugated anti-FLAG IgG antibody dissolved in 5% (w/v) BSA solution in PBS (dilution factor 1:5000) was inoculated to the wells and incubated for 1 h at room temperature. After washing, 50 µL of TMB liquid peroxidase substrate system was added until the deep blue color appears, and 50 µL of 2 M H2SO4 was added to stop the reaction. The absorbance at 450 nm of each well was measured.
Fed-batch cultivation
For 2-L scale fed-batch cultivation, a seed culture was conducted with 5 mL of BHI media at 30℃ for 24 h with 200 rpm shaking. The cells were inoculated into a baffled flask containing modified CGXII medium (200 mL). After cultivation, cells were transferred into 2 L of modified CGXII media in a 5 L bioreactor (BioCNS, Daejeon, Korea). The culture condition was maintained at 30℃, pH 7.0 by adding a 20% ammonia solution. The dissolved oxygen (DO) concentration was maintained at 30% (v/v) by online monitoring and automatically increasing the agitation speed to 1200 rpm. Cell growth was determined by measuring optical density at 600 nm (OD600) using a spectrophotometer (Mecasys, Daejeon, Korea). For the production of ⍺-amylase and M18 scFv, fed-batch cultivations were conducted in a mini-bioreactor. The overall fermentation procedures were the same as those for 2 L-scale fed-batch cultivation, except that the volume of the cultivation medium was 200 mL in a 500 mL bioreactor (Applikon Biotechnology, Delft, Netherlands).
Results
Construction of Sec-type synthetic signal peptide library
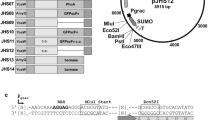
A synthetic signal peptide library was created by analyzing native Sec-type signal peptide sequences from C. glutamicum ATCC13032 and C. glutamicum R using SignalP 5.0 and aligned with Clustal Omega. Sequence analysis was performed to design a library based on the minimal structural components of native signal peptide sequences (Fig. 1A and Fig. S1). This led to the design of a synthetic signal peptide library comprising 27 amino acid sequences (Fig. 1B). These sequences included five amino acids with a positive charge at the N-terminus, a hydrophobic section with sixteen amino acids in the middle, and a polar section with six amino acids at the C-terminus. Based on the amino acid frequency, we designed A-L-A-L-A amino acid sequences from the 10th to 14th positions as a core hydrophobic region, which was also beneficial for library construction by PCR (Fig. 1B). The 9th and 22nd positions were designed using NTT and VCM, respectively, to generate the preferred amino acid sequences at each position: isoleucine (I), phenylalanine (F), leucine (L), or valine (V) at the 9th position and alanine (A), proline (P), or threonine (T) at the 22nd position. Furthermore, amino acid residue at 25th position was designed with the degenerate nucleotide sequence BYN to generate signal peptidase I (SPase I), encompassing A, serine (S), L, P, F, or V. The entire synthetic signal peptide library was generated by PCR using degenerate primers without a template, and was subsequently cloned into pHCMS to produce pHCP-SP-L. Notably, the terminal amino acid residues of the synthetic signal peptide library were cleaved using the SapI site, enabling fusion with a protein of interest without additional amino acids downstream (Fig. S2). By utilizing A, a commonly employed cleavage site amino acid, at the end of the synthetic signal peptide library, the developed construct allowed for the cleavage and secretion of the target protein without introducing extra amino acids. The overall scheme of the library design is depicted in Fig. 1B, and the construction of pHCP-SP-L was accomplished using a library of 1.6 × 107 cells in the E. coli NEB 10-beta strain. To check the sequence construction and diversity of the library (pHCP-SP-L), 18 colonies were randomly selected. We found that they had an accurate signal peptide construction, including the core hydrophobic section (A-L-A-L-A ) and cleavage site, and none of them shared identical signal peptide sequences (Fig. S3).
Sequence analysis of signal peptides and library construction (A) WebLogo depicting frequency of amino acid changes within the signal peptides of C. glutamicum. (B) Overall scheme of the synthetic signal peptide library construction. Nucleotide symbols: V represents G, C or A; M represents A or C; B represents G, T, or C; Y represents C or T; and N represents A, T, G or C. The entire synthetic signal peptide library was generated via PCR using degenerate primers without a template and subsequently cloned into pHCMS, yielding pHCP-SP-L
Enzyme activity-dependent screening of effective signal peptide for endoxylanase
To assess the functionality of the synthetic peptide library, we first used XynA from Streptomyces coelicolor A3(2) as a model protein. By cloning signal peptide sequences from the library into pHCP-XynA containing xynA expression cassette, we constructed the XynA library (pHCP-SP-XynA) in E. coli with a library size of approximately 2.4 × 105 cells. After transformation into C. glutamicum, the cells were spread onto agar plates containing xylan for enzyme activity-dependent screening of the library. On this xylan agar plate, cells containing an effective signal peptide for the secretory production of XynA in the medium can degrade xylan, resulting in a clear halo around the colonies. Among approximately 2 × 104 colonies on xylan-agar plates, 244 colonies displayed halos around colonies (Fig. 2A), and for a comparative assessment of degradation activity under identical conditions, the halo-forming colonies were transferred to fresh xylan-agar plates. From the second screening, three colonies showing larger halo sizes (C1, G1, and G8) were selected, and after cultivation in liquid medium, the secreted XynA into the culture medium in each clone was analyzed by SDS-PAGE. As shown in Fig. 2B, all three clones successfully produced XynA in the culture medium and their sizes were consistent with the predicted XynA size (48 kDa). When we compared the secretion yields with those of other well-known C. glutamicum-derived signal peptides, including Cg1514, Cg2052, and PorB, two clones (C1 and G1) showed superior secretion yields (Fig. 2B). To evaluate the possible effects of the host cell, each plasmid was re-transformed into new C. glutamicum cells, and the secretion ability was determined again. As expected, newly transformed cells showed successful secretion of XynA, with yields almost identical to those of isolated clones (data not shown), indicating that no other unknown host factors affect the secretion of XynA beyond the signal peptide itself. From the isolated clones, the sequences of the signal peptides were determined, and we found that the sequences of these three signal peptides (C1, G1, and G8) were distinct from one another, but all had the same length (27 amino acids) with an Ala residue at the C-terminus, as designed (Fig. 2C). To confirm that cleavage occurred at the correct position, N-terminal amino acid sequences of secreted XynA were determined, and we found that all contain A-E-S-T-L sequences that perfectly match the first five amino acids of mature XynA and indicate the precise cleavage of synthetic signal peptides.
Isolation of signal peptide for endoxylanase (XynA). (A) Screening of pHCP-SP-XynA library on xylan-agar plate. The red arrows indicate halo-forming colonies. (B) SDS-PAGE analysis of culture supernatant from C. glutamicum ATCC13032 in BHI medium. Lane M, protein size marker; lane 1, negative control (pHCP-XynA); lane 2, pHCP-C1-XynA; lane 3, pHCP-G1-XynA; lane 4, pHCP-G8-XynA; lane 5, pHCP-Cg1514-XynA; lane 6, pHCP-Cg2052-XynA; lane 7, pHCP-PorB-XynA. (C) Amino acid sequences of synthetic signal peptides (C1, G1, and G8). N-terminal amino acid sequences of XynA are indicated in grey shading. (D) SDS-PAGE analysis of the culture supernatant from C. glutamicum SP002 in modified CGXII medium. Lane M; protein size marker; lane 1, negative control (pHCP-XynA); lanes 2 and 3, pHCP-C1-XynA; lanes 4 and 5, pHCP-G1-XynA. All culture supernatants are concentrated thirty times and 10 µL was loaded on each well
Next, instead of C. glutamicum ATCC 13032, the secretion efficiencies of two synthetic signal peptides (C1 and G1) were also examined with the C. glutamicum SP002 strain, in which the major secretory proteins, specifically Cg2052 and Cg1514, were removed from C. glutamicum ATCC 13032, resulting in an increase of purity and secretion yield of target proteins in culture medium [35]. After transformation, the cells were cultivated in modified CGXII medium, which is generally used for fed-batch cultivation in bioreactors. Compared to earlier cultivation (Fig. 2B), both signal peptides (C1 and G1) in C. glutamicum SP002 also showed similar levels of secretion yields. Notably, we confirmed that the C1 synthetic signal peptide demonstrated higher secretion efficiency than G1 (Fig. 2D). Based on these results, we selected the C1 signal peptide for subsequent fed-batch cultivation.
To further validate the applicability of the XynA secretion system employing the C1 synthetic signal peptide, we conducted fed-batch cultivation using C. glutamicum SP002 harboring pHCP-C1-XynA. In this culture, the cell-specific growth rate was 0.125 h-1, and the final OD600 reached 226 after 32 h (Fig. 3A). SDS-PAGE analysis confirmed a much lower contamination of native proteins, which was observed in the cultivation of C. glutamicum ATCC 13032. The amount of XynA in the total secreted proteins was then quantified, revealing that the max XynA content at 48 h was 83.1% of the total extracellular protein content (Fig. 3B). Furthermore, the final concentration of XynA reached 3.2 g/L, with a productivity of 77 mg/L/h, which are much higher than our previous result with Cg1514 (1.5 g/L) [16]. By the activity assay, we confirmed that XynA secreted in culture medium were fully functional (209.5±1.3 U/mL) (Fig. S4).
Fed-batch cultivation of C. glutamicum SP002 harboring pHCP-C1-XynA. (A) Time profiles of cell density (OD600), dry cell weight (DCW, g/L), glucose conc. (g/L) and XynA conc. in culture medium (g/L). Symbols: ◆, cell density; ◼, glucose conc; ⬤, dry cell weight; ▲, XynA conc. (B) SDS-PAGE analysis of culture supernatants. Lane M represents protein size makers, and each lane represent protein fraction in each sampling time. All culture supernatants are not concentrated and 1 uL loaded on each well. Arrowhead indicates the band of XynA
Isolation of optimal signal peptide for secretory production of AmyA
Next, to validate the efficacy of the synthetic signal peptide library, we examined AmyA as a second model. We introduced a synthetic signal peptide library into the N-terminus of AmyA and transformed it into C. glutamicum cells for screening on agar plates. Similar to the screening of the XynA library, we harnessed the starch-degrading properties of AmyA for library screening. Secreted AmyA enzymatically degrades starch, creating a halo on starch-agar plates, which can be visualized by staining with Lugol’s solution (Fig. 4A). More than 4,000 individual colonies were screened on starch agar plates. To ensure a fair comparison of the degradation activity under uniform conditions, both colony and halo sizes were considered alongside the control strain that produced AmyA with the Cg1514 signal peptide. Among the screened clones, seven candidates (HS01, HS02, HS03, HS04, HS05, HS06, and HS07) displaying bigger halo sizes than the control strain were selected, and after re-transformation into the C. glutamicum SP002, their production levels were analyzed by flask cultivation. The HS06 cells showed the highest secretory efficiency (Fig. 4B). From DNA sequencing, the amino acid sequence of the signal peptide in HS06 was determined, and it was confirmed that the signal peptide was precisely cleaved after the last Ala residue of the signal peptide (Fig. 4C).
Isolation of signal peptide for ⍺-amylase (AmyA). (A) Screening of pHCP-SP-AmyA library on starch-agar plate. (B) SDS-PAGE analysis of culture supernatant of isolated colonies. Lane M, protein size marker; lanes 1 to 7, HS01, HS02, HS03, HS04, HS05, HS06 and HS07. All culture supernatants are concentrated thirty times and 10 µL was loaded on each well. Arrowhead indicates the band of AmyA. (C) Amino acid sequences of synthetic signal peptide of HS06. N-terminal three amino acid sequences of AmyA are indicated in grey shading
To assess whether the HS06 strain is suitable for large-scale production, we conducted fed-batch cultivation in a bioreactor. After inoculation, the cells grew well with a cell-specific growth rate of 0.096 h-1, and the final OD600 reached 138.5 after 40 h (Fig. 5A). SDS-PAGE analysis of the culture medium revealed that AmyA was successfully produced in the culture medium with high purity (78% of the total extracellular protein). The maximum titer reached 1.48 g/L after 40 h (Fig. 5A and B), and the activity of secreted AmyA was determined as 4,003.4±98.7 U/mL (Fig. S5). Notably, this production titer was almost 2-fold higher than that reported previously using the Cg1514 signal peptide (782.6 mg/L) [16].
Fed-batch cultivation of C. glutamicum SP002 harboring HS06. (A) Time profiles of cell density (OD600), glucose conc. (g/L) and AmyA conc. in culture medium. (g/L). Symbols: ◆, cell density; ◼, glucose conc.; ▲, AmyA conc. (B) SDS-PAGE analysis of culture supernatants. Lane M represents protein size makers, and each lane represent protein fraction in each sampling time. On each well, 2 µL of culture supernatants were loaded. Arrowhead indicates the band of AmyA
FAST-based screening methods for signal peptide library. (A) Overall scheme of FAST-based screening methods using 96-well plates. (B) Validation of FAST-based screening methods with various XynA expression systems. SDS-PAGE analysis of XynA expression systems with FAST. Lane M, protein size marker; lane 1, negative control (pHCP-XynA); lane 2, pHCP-C1-XynA-FAST; lane 3, pHCP-Cg2052-XynA-FAST; lane 4, pHCP-Cg1514-XynA-FAST; lane 5, pHCP-PorB-XynA-FAST. All culture supernatants were concentrated ten times and 10 µL was loaded on each well. Arrowhead indicates the band of XynA (C) Analysis of fluorescent intensity of XynA secretion systems with FAST
FAST-based activity-independent screening strategy for isolation of signal peptide for scFv
As shown earlier with two examples, enzyme activity-based screening methods on agar plates offers a straightforward and visually intuitive way to identify the optimal secretory system. However, many proteins cannot be assessed using enzymatic assays, and the aforementioned halo-based screening strategy is not suitable for these proteins. To address this issue and enable the screening of recombinant proteins that are unsuitable for enzymatic methods, we developed an activity-independent screening system using FAST-based fluorescence. FAST is a small monomeric protein tag (14 kDa) that becomes fluorescent when it binds to a non-permeable fluorogen, such as HBRAA-3E [36] (Fig. 6A). If cells produce FAST-fused proteins in the culture medium, they can interact with the fluorogen, resulting in strong fluorescent signals. This fluorogen is non-permeable; therefore, FAST-fused proteins in the cytoplasm (no secretion) cannot interact with the fluorogen, and consequently, cells with low secretion efficiency give very low fluorescent signals (Fig. 6A). Based on this difference in fluorescence, we isolated a high producer from the library. To demonstrate this FAST-based screening approach, we examined the production of FAST-fused XynA with various signal peptides (C1, Cg1514, Cg2052, and PorB) showing different XynA secretion efficiencies. SDS-PAGE analysis confirmed that FAST-fused XynA was successfully produced with each signal peptide, and the secretion yield of FAST-fused XynA was similar to that of the non-fused forms (C1 > Cg1514 > PorB > Cg2052) (Fig. 6B), which also indicated that the fusion of FAST with the target protein did not interfere with the secretion efficiency of the target protein. To detect the fluorescence of the FAST-fused protein, samples from the same cultures were mixed with fluorogen, and we confirmed that the fluorescence intensity of each clone was highly correlated with the secretion yields (Fig. 6C). Based on these results, we conclude that the FAST-based activity-independent strategy is a useful tool for screening signal peptide libraries.
Isolation of signal peptide from the library for M18 secretion. (A) SDS-PAGE analysis of isolated candidates with FAST. M; protein size marker, 1; pHCP-PorB-M18-FAST, 2; pHCP-18F11-M18-FAST, 3; pHCP-20G1-M18-FAST, 4; pHCP-24G1-M18-FAST. (B) SDS-PAGE analysis of isolated candidate (18F11) without FAST. M; protein size marker, 1; pHCP-PorB-M18, 2; pHCP-18F11-M18. (C) Western blot analysis of isolated candidate (18F11) without FAST. M; protein size marker, 1; pHCP-PorB-M18, 2; pHCP-18F11-M18. All culture supernatants are concentrated thirty times and 10 µL was loaded on each well. (D) Amino acid sequences of synthetic signal peptide of 18F11. N-terminal amino acid sequences of M18 scFv are indicated in grey shading
Using this FAST-based screening strategy, we isolated a signal peptide for M18 scFv that targets anthrax toxin [37]. We constructed a library of FAST-fused M18 scFv (pHCP-SP-M18-FAST) by fusing synthetic signal peptides at the N-terminus and FAST at the C-terminus of M18 scFv. After transformation into C. glutamicum SP002, the individual colonies were inoculated into a 96-well plate for screening. After cultivation, we inoculated 1.25 µM of fluorogen into each well and assessed the fluorescent intensity. We screened 3,072 colonies, isolated individual clones displaying high fluorescence intensity, and analyzed their secretion yields in flask cultivation. All clones successfully produced FAST-fused M18 scFv (41.5 kDa) and demonstrated secretion yields similar to those of the PorB signal peptide (Fig. 7A). Among the isolated candidates, 18F11 exhibited a 1.2-fold increase in fluorescence intensity compared to the control with the PorB signal peptide. After removing the FAST tag, the secretory production of M18 scFv with the isolated signal peptide (18F11) was analyzed, and M18 scFv (27.9 kDa) maintained its secretion yield in the absence of the FAST tag, which was notably higher than that of the PorB signal peptide (Fig. 7B and C). Using DNA sequencing, the amino acid sequence of 18F11 was determined, and the signal peptide was cleaved after the last Ala residue of signal peptide (Fig. 7D).
Fed-batch cultivation of C. glutamicum SP002 harboring 18F11. (A) Time profiles of cell density (OD600), glucose conc. (g/L) and M18 scFv conc. in culture medium (g/L). Symbols: ◆, cell density (OD600); ◼, glucose conc.; ▲, M18 scFv conc. (B) SDS-PAGE analysis of culture supernatants. Lane M represents protein size makers, and each lane represent protein fraction in each sampling time. On each well, 10 µL of culture supernatants were loaded. Arrowhead indicates the band of M18 scFv
Finally, fed-batch cultivation of cells producing M18 scFv with the 18F11 signal peptide was conducted. After inoculation, cells harboring pHCP-18F11-M18 grew well with a cell-specific growth rate of 0.130 h-1, and the maximum cell density reached an OD600 of 264.5 after 25 h, and then gradually decreased (Fig. 8A). As the cells grew, the M18 scFv levels in the culture medium also increased gradually, and a maximum titer of 228 mg/L was achieved after 32 h (Fig. 8B), which was 3.4-fold higher than that of our previous result with the PorB signal peptide (68 mg/L) in fed-batch cultivation [28]. The binding activity of secreted M18 scFv against antigen (anthrax toxin PA) was also determined by ELISA, and it was clearly confirmed that the fully functional M18 scFv was produced in fed-batch cultivation (Fig. S6).
Discussion
As a robust workhorse for the secretory production of recombinant proteins, C. glutamicum has been widely used and many successful results have been achieved [13, 16,17,18]. However, limited availability of signal peptides prevents C. glutamicum from becoming a potential host for recombinant protein production [5]. In this study, we designed a fully synthetic signal peptide library and attempted to develop an optimal secretion system for target proteins through library screening in C. glutamicum. Using this library, the limitations of signal peptides were overcome, and we successfully demonstrated the isolation of signal peptides from three different target proteins (XynA, AmyA, and M18 scFv). As shown by fed-batch cultivation, all optimized systems exhibited significantly improved secretion performance compared to those using well-known signal peptides. Particularly, in the case of XynA, cells producing XynA with isolated C1 signal peptide showed much improved production titer (3.2 g/L) compared to those of Cg1514 (1.5 g/L) or CspB2 Signal peptides (1.77 g/L) [16, 17]. Notably, in this culture, a much higher cell density (OD600 of 256) was achieved and the purity of XynA in the culture medium was very high (> 80%) (Fig. 3A and B). This improved cell growth and high purity indicated that protein secretion with the C1 signal peptide did not cause any issues (i.e., cell lysis) during the translocation of XynA via the Sec-channel in the cytoplasmic membrane, indicating that the C1 signal peptide is ideal for the secretion of XynA in C. glutamicum. Similarly, in the cases of AmyA and M18 scFv, cells also showed improved growth and higher production titers than those with well-known signal peptides (Figs. 5 and 7). Our production titers were also compared with other bacterial hosts such as E. coli, Bacillus sp, and yeast strains (Pichia pastoris, Saccharomyces cerevisiae, etc.), and we confirmed that our production titers were similar or higher compared with previous results (Tables S3, S4 and S5), which indicates our synthetic signal peptide library can be a useful resource for the development of a target-specific secretion system in C. glutamicum. Of course, only the isolation of optimal signal peptide does not guarantee the successful production of any challenging proteins. In general, the secretory production of recombinant proteins can be affected by various factors including protein folding, solubility, proteolytic degradation and expression levels [4, 8], and for the successful production and further increase, we also need to optimize those factors together with signal peptides.
For all examined proteins, the isolated signal peptides exhibited improved secretion performance, but we believe there is a greater chance of isolating more effective signal peptides from the library because we screened only small populations in the synthetic library. In this study, we used two approaches to screen the synthetic library: (i) enzyme activity-dependent selection on agar plates and (ii) fluorescent signal-based selection using FAST fusion. The first approach is simple and fast, but is limited to enzymes capable of hydrolyzing the substrate to produce a clear halo around the colony, and the selection of the best colony is dependent on direct observation of the halo size on the plate. The second approach using FAST fusion is a protein activity-independent method, so it can be applied to a much wider range of proteins. However, the screening speed is relatively slow and work-intensive because it requires the cultivation of individual clones in 96-well plates. Using this approach for FAST-fused M18 scFv screening, only 103–104 cells were screened, which was much smaller than the library size. We need to implement a more feasible approach to facilitate a screening process capable of covering a much larger population of libraries. Fluorescence-activated cell sorting (FACS) has been widely used for high-throughput screening, and positive clones exhibiting higher fluorescence can be sorted from large libraries with extremely high accuracy and speed [38, 39]. For FACS-assisted screening, the secretion event must be converted to fluorescence with high correlation. For this purpose, Bakkes et al. developed a secretion-related stress-response-based biosensor that can quantify protein secretion as fluorescence intensity using split GFP, and the best clone exhibiting the highest fluorescence intensity can be isolated using FACS [40, 41]. A split GFP-based biosensor for protein secretion has also been employed for high-throughput screening of B. subtilis hosts [42, 43]. Alternatively, Abatemarco et al. developed RNA-aptamers-in-droplets, in which the secreted proteins interacted with them, resulting in a high fluorescent signal [44]. Using this approach, the extracellular product titer was transduced into fluorescence, allowing for the high-throughput screening of millions of variants using FACS. FACS-assisted high-throughput screening is independent of protein activity, allowing its application across a wide range of proteins. Therefore, integration of a large synthetic library with this high-throughput screening method is a powerful approach for developing an efficient secretion system in C. glutamicum [45, 46].
In the present study, we only examined C. glutamicum as a production host, but our synthetic library can possibly be applied to the development of a secretion system in other bacterial hosts and can be used for artificial intelligence (AI)-assisted signal peptide generation. To date, the use of signal peptides derived from C. glutamicum for protein secretion in other bacteria has not been reported; however, it has been previously shown that the signal peptide library from low-GC B. subtilis is functional in high-GC C. glutamicum although they are distantly related [26, 47]. Recent advancements in AI technology have brought rapid and substantial changes to biotechnology [48,49,50]. In 2020, Wu et al. reported a machine translation model for generating SP sequences in which the Swiss-Prot protein database from all available organisms was used to train a transformer model and confirmed that 48% of the generated SPs led to secreted enzyme activity in B. subtilis [51]. In general, to obtain meaningful results, machine learning requires training with a database that includes not only information from the database, but also large amounts of experimental data. Through library screening with each target protein, the performance of signal peptides for secretory production can be ranked. In this study, we showed the sequences for successful secretion results, but we can also collect other sequencing data responsible for moderate or poor secretion results. We believe that this massive and qualified experimental dataset can be a useful resource for AI training, which will permit the design of a more efficient secretion system to maximize protein secretion yields.
Conclusions
In this study, we designed a synthetic signal peptide library and successfully developed an optimal secretion system in C. glutamicum for three target proteins (XynA, AmyA, and M18 scFv). All systems exhibited significantly improved production titers compared to those of well-known signal peptides. To the best of our knowledge, this is the first report of the construction of a fully synthetic signal peptide and its use in the development of a target protein specific secretion system in C. glutamicum. By implementing a feasible high-throughput screening strategy, our synthetic library provides one of the useful resources for the development of an optimal secretion system for various high-value recombinant proteins in C. glutamicum. In addition, based on the massive experimental data obtained from library screening and optimization of other factors (promoter/BRS strengths, culture conditions etc.), we can develop a more efficient secretion system and believe C. glutamicum can be a more promising workhorse in the bioindustry.
Data availability
Data is provided within the manuscript or supplementary information files.
References
Becker J, Rohles CM, Wittmann C. Metabolically engineered Corynebacterium glutamicum for bio-based production of chemicals, fuels, materials, and healthcare products. Metab Eng. 2018;50:122–41.
Zha J, Zhao Z, Xiao Z, Eng T, Mukhopadhyay A, Koffas MAG, Tang YJ. Biosystem design of Corynebacterium glutamicum for bioproduction. Curr Opin Biotech. 2023;79:102870.
Choi JW, Jeon EJ, Jeong KJ. Recent advances in engineering Corynebacterium glutamicum for utilization of hemicellulosic biomass. Curr Opin Biotech. 2019;57:17–24.
Lee SM, Jeong KJ. Advances in synthetic biology tools and engineering of Corynebacterium glutamicum as a platform host for recombinant protein production. Biotechnol Bioproc E. 2023;28(6):962–76.
Freudl R. Beyond amino acids: Use of the Corynebacterium glutamicum cell factory for the secretion of heterologous proteins. J Biotechnol. 2017;258:101–9.
Liu X, Zhang W, Zhao Z, Dai X, Yang Y, Bai Z. Protein secretion in Corynebacterium glutamicum. Crit Rev Biotechnol. 2017;37(4):541–51.
Liu X, Yang Y, Zhang W, Sun Y, Peng F, Jeffrey L, Harvey L, McNeil B, Bai Z. Expression of recombinant protein using Corynebacterium Glutamicum: progress, challenges and applications. Crit Rev Biotechnol. 2016;36(4):652–64.
Lee MJ, Kim P. Recombinant protein expression system in Corynebacterium glutamicum and its application. Front Microbiol. 2018;9:2523.
Billman-Jacobe H, Wang L, Kortt A, Stewart D, Radford A. Expression and secretion of heterologous proteases by Corynebacterium glutamicum. Appl Environ Microbiol. 1995;61(4):1610–3.
Anné J, Economou A, Bernaerts K. Protein secretion in gram-positive bacteria: From multiple pathways to biotechnology, in protein and sugar export and assembly in gram-positive bacteria, Bagnoli F and Rappuoli R, Editors. Springer International Publishing. Cham; 2017. pp. 267–308.
Liu L, Yang H, Shin HD, Li J, Du G, Chen J, Chen. Recent advances in recombinant protein expression by Corynebacterium, Brevibacterium, and Streptomyces: from transcription and translation regulation to secretion pathway selection. Appl Microbiol Biotechnol. 2013;97(22):9597–608.
Tsirigotaki A, De Geyter J, Šoštaric´ N, Economou A, Karamanou S. Protein export through the bacterial sec pathway. Nat Rev Microbiol. 2017;15(1):21–36.
Jin Q, Pan F, Hu C-F, Lee SY, Xia X-X, Qian Z-G, Qian. Secretory production of spider silk proteins in metabolically engineered Corynebacterium glutamicum for spinning into tough fibers. Metab Eng. 2022;70:102–14.
Peng C, Shi C, Cao X, Li Y, Liu F, Lu F. Factors influencing recombinant protein secretion efficiency in gram-positive bacteria: Signal peptide and beyond. Front Bioeng Biotechnol. 2019;7.
Freudl R. Signal peptides for recombinant protein secretion in bacterial expression systems. Microb Cell Fact. 2018;17(1):52.
Yim SS, Choi JW, Lee RJ, Lee YJ, Lee SH, Kim SY, Jeong KJ. Development of a new platform for secretory production of recombinant proteins in Corynebacterium glutamicum. Biotechnol Bioeng. 2016;113(1):163–72.
Zhang W, Yang Y, Liu X, Liu C, Bai Z. Development of a secretory expression system with high compatibility between expression elements and an optimized host for endoxylanase production in Corynebacterium glutamicum. Microb Cell Fact. 2019;18(1):72.
An SJ, Yim SS, Jeong KJ. Development of a secretion system for the production of heterologous proteins in Corynebacterium glutamicum using the porin B signal peptide. Protein Expr Purif. 2013;89(2):251–7.
Watanabe K, Tsuchida Y, Okibe N, Teramoto H, Suzuki N, Inui M, Yukawa H. Scanning the Corynebacterium glutamicum R genome for high-efficiency secretion signal sequences. Microbiology. 2009;155(3):741–50.
Low KO, Muhammad Mahadi N, Md. Illias R. Optimisation of signal peptide for recombinant protein secretion in bacterial hosts. Appl Microbiol Biotechnol. 2013;97(9):3811–26.
Kang X-M, Cai X, Huang Z-H, Liu Z-Q, Zheng Y-G. Construction of a highly active secretory expression system in Bacillus subtilis of a recombinant amidase by promoter and signal peptide engineering. Int J Biol Macromol. 2020;143:833–41.
Zhang W, Yang M, Yang Y, Zhan J, Zhou Y, Zhao X. Optimal secretion of alkali-tolerant xylanase in Bacillus subtilis by signal peptide screening. Appl Microbiol Biotechnol. 2016;100(20):8745–56.
Tsuji S, Tanaka K, Takenaka S, Yoshida K. Enhanced secretion of natto phytase by Bacillus subtilis. Biosci Biotechnol Biochem. 2015;79(11):906–14.
Yao D, Su L, Li N, Wu J. Enhanced extracellular expression of Bacillus stearothermophilus α-amylase in Bacillus subtilis through signal peptide optimization, chaperone overexpression and α-amylase mutant selection. Microb Cell Fact. 2019;18(1):69.
Fu G, Liu J, Li J, Zhu B, Zhang D, Zhang. Systematic screening of optimal signal peptides for secretory production of heterologous proteins in Bacillus subtilis. J Agric Food Chem. 2018;66(50):13141–51.
Hemmerich J, Rohe P, Kleine B, Jurischka S, Wiechert W, Freudl R, Oldiges M. Use of a Sec signal peptide library from Bacillus subtilis for the optimization of cutinase secretion in Corynebacterium glutamicum. Microb Cell Fact. 2016;15(1):208.
Choi JW, Yim SS, Jeong KJ. Development of a high-copy-number plasmid via adaptive laboratory evolution of Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2018;102(2):873–83.
Yim SS, An SJ, Choi JW, Ryu AJ, Jeong KJ. High-level secretory production of recombinant single-chain variable fragment (scFv) in Corynebacterium glutamicum. Appl Microbiol Biotechnol. 2014;98(1):273–84.
Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23(21):2947–8.
Sievers F, Higgins DG. Clustal Omega for making accurate alignments of many protein sequences. Protein Sci. 2018;27(1):135–45.
Nielsen H. Predicting secretory proteins with SignalP. Methods Mol Biol. 2017;1611:59–73.
Istvan Lazar ILS Jr. GelAnalyzer 23.1. 2010. http://www.gelanalyzer.com
Mangan D, Cornaggia C, Liadova A, McCormack N, Ivory R, McKie VA, Ormerod A, McCleary BV. Novel substrates for the automated and manual assay of endo-1,4-β-xylanase. Carbohydr Res. 2017;445:14–22.
Msarah MJ, Ibrahim I, Hamid AA, Aqma WS. Optimisation and production of alpha amylase from thermophilic Bacillus spp. and its application in food waste biodegradation. Heliyon. 2020;6(6):e04183.
Jeon EJ, Choi JW, Cho MS, Jeong KJ. Enhanced production of neoagarobiose from agar with Corynebacterium glutamicum producing exo-type and endo-type β-agarases. Microb Biotechnol. 2021;14(5):2164–75.
Plamont M-A, Billon-Denis E, Maurin S, Gauron C, Pimenta FM, Specht CG, Shi J, Quérard J, Pan B, Rossignol J, et al. Small fluorescence-activating and absorption-shifting tag for tunable protein imaging in vivo. PNAS. 2016;113(3):497–502.
Harvey BR, Georgiou G, Hayhurst A, Jeong KJ, Iverson BL, Rogers GK. Anchored periplasmic expression, a versatile technology for the isolation of high-affinity antibodies from Escherichia coli-expressed libraries. PNAS. 2004;101(25):9193–8.
Robinson JP, Ostafe R, Iyengar SN, Rajwa B, Fischer R. Flow cytometry: the next revolution. Cells. 2023;12:14.
Kim T, Ko M, Rha E, Kim H, Lee H. Microfluidics-driven high-throughput phenotyping and screening in synthetic biology: from single cells to cell-free systems. Biotechnol Bioproc E. 2024;29:25–33.
Bakkes PJ, Lenz P, Müller C, Bida A, Dohmen-Olma D, Knapp A, Oldiges M, Jaeger KE, Freudl R. Biosensor-based optimization of cutinase secretion by Corynebacterium glutamicum. Front Microbiol. 2021;12:750150.
Daugherty PS, Iverson BL, Georgiou G. Flow cytometric screening of cell-based libraries. J Immunol Methods. 2000;243:211–27.
Knapp A, Ripphahn M, Volkenborn K, Skoczinski P, Jaeger K-E. Activity-independent screening of secreted proteins using split GFP. J Biotechnol. 2017;258:110–6.
Lenz P, Bakkes PJ, Müller C, Malek M, Freudl R, Oldiges M, Drepper T, Jaeger K-E, Knapp A. Analysis of protein secretion in Bacillus subtilis by combining a secretion stress biosensor strain with an in vivo split GFP assay. Microb Cell Fact. 2023;22:203.
Abatemarco J, Sarhan MF, Wagner JM, Lin J-L, Liu L, Hassouneh W, Yuan S-F, Alper HS, Abate AR. RNA-aptamers-in-droplets (RAPID) high-throughput screening for secretory phenotypes. Nat Commun. 2017;8:332.
Müller C, Bakkes PJ, Lenz P, Waffenschmidt V, Helleckes LM, Jaeger K-E, Wiechert W, Knapp A, Freudl R, Oldiges M. Accelerated strain construction and characterization of C. glutamicum protein secretion by laboratory automation. Appl Microbiol Biotechnol. 2022;106(12):4481–97.
Yim SS, Choi JW, Lee YJ, Jeong KJ. Rapid combinatorial rewiring of metabolic networks for enhanced poly(3-hydroxybutyrate) production in Corynebacterium glutamicum. Microb Cell Fact. 2023;22:29.
Rohe P, Venkanna D, Kleine B, Freudl R, Oldiges M. An automated workflow for enhancing microbial bioprocess optimization on a novel microbioreactor platform. Microb Cell Fact. 2012;11:144.
Beardall WAV, Stan G-B, Dunlop MJ. Deep learning concepts and applications for Synthetic Biology. GEN Biotechnol. 2022;1(4):360–71.
Goshisht MK. Machine learning and deep learning in synthetic biology: key achitectures, applications, and challenges. ACS Omega. 2024;9(9):9921–45.
Radivojević T, Costello Z, Workman K, Garcia Martin H. A machine learning automated recommendation tool for synthetic biology. Nat Commun. 2020;11:4879.
Wu Z, Yang KK, Liszka MJ, Lee A, Batzilla A, Wernick D, Weiner DP, Arnold FH. Signal peptides generated by attention-based neural networks. ACS Synth Biol. 2020;9(8):2154–61.
Funding
This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Ministry of Science and ICT (MSIT) [No. NRF-2020R1A2C2012537], and by the Korea Institute of Planning and Evaluation for Technology in Food, Agriculture and Forestry (IPET) through Crop Viruses and Pests Response Industry Technology Development Program, funded by Ministry of Agriculture, Food and Rural Affairs (MAFRA)(Grant no. 321109-04-1-HD020), Republic of Korea.
Author information
Authors and Affiliations
Contributions
EJJ, SML, and KJJ designed the experiments. EJJ performed experiments for library design, and production of endoxylanase. SML performed experiments for production of M18 scFv. HSH performed experiment for production of amylase. EJJ wrote the draft. SML and KJJ revised manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
12934_2024_2516_MOESM1_ESM.docx
Supplementary Material 1. Additional file 1: Figure S1. Multiple sequence alignment of known sec-type signal peptides from Corynebacterium using CLUSTAL W. Figure S2. Schematic illustration of signal peptide library construction for POI (Protein of Interest). Figure S3. Confirmation of amino acids sequences of signal peptides in 18 random clones of signal peptide library. Figure S4. Activity assay of XynA by XylX6 kit. Figure S5. Activity assay of AmyA by DNS method. Figure S6. Binding assay of M18 scFv by ELISA. Table S1. List of bacterial strains and plasmids used in this work. Table S2. List of oligonucleotides used in this work. Table S3. List of production of endoxylanases and amylases in bacterial hosts. Table S4. List of antibody production in bacterial hosts. Table S5. List of antibody production in yeasts.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Jeon, E.J., Lee, S.M., Hong, H.S. et al. Design of fully synthetic signal peptide library and its use for enhanced secretory production of recombinant proteins in Corynebacterium glutamicum. Microb Cell Fact 23, 252 (2024). https://doi.org/10.1186/s12934-024-02516-9
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12934-024-02516-9