Abstract
Background
Myopia is one of the most common eye diseases in children and adolescents worldwide, and scleral remodeling plays a role in myopia progression. However, the identity of the initiating factors and signaling pathways that induce myopia-associated scleral remodeling is still unclear. This study aimed to identify biomarkers of scleral remodeling to elucidate the pathogenesis of myopia.
Methods
The gene expression omnibus (GEO) and comparative toxicogenomics database (CTD) mining were used to identify the miRNA-mRNA regulatory network related to scleral remodeling in myopia. Real-time quantitative PCR (RT-qPCR), Western blot, immunofluorescence, H&E staining, Masson staining, and flow cytometry were used to detect the changes in the FOXO signaling pathway, fibrosis, apoptosis, cell cycle, and other related factors in scleral remodeling.
Results
miR-15b-5p/miR-379-3p can regulate the FOXO signaling pathway. Confirmatory studies confirmed that the axial length of the eye was significantly increased, the scleral thickness was thinner, the levels of miR-15b-5p, miR-379-3p, PTEN, p-PTEN, FOXO3a, cyclin-dependent kinase (CDK) inhibitor 1B (CDKN1B) were increased, and the levels of IGF1R were decreased in Len-induced myopia (LIM) group. CDK2, cyclin D1 (CCND1), and cell cycle block assessed by flow cytometry indicated G1/S cell cycle arrest in myopic sclera. The increase in BAX level and the decrease in BCL-2 level indicated enhanced apoptosis of the myopic sclera. In addition, we found that the levels of transforming growth factor-β1 (TGF-β1), collagen type 1 (COL-1), and α-smooth muscle actin (α-SMA) were decreased, suggesting scleral remodeling occurred in myopia.
Conclusions
miR-15b-5p/miR-379-3p can regulate the scleral cell cycle and apoptosis through the IGF1R/PTEN/FOXO signaling pathway, thereby promoting scleral remodeling in myopia progression.
Graphical Abstract

Similar content being viewed by others
Introduction
The number of people with myopia is increasing rapidly. One study predicts that by 2050, there will be 4758 million people who will suffer from myopia [1]. When it develops into high myopia, myopia often leads to cataracts, retinal detachment holes, and other irreversible complications [2]. Therefore, to improve eye health in children and adolescents, it is urgent to identify the pathogenesis of myopia and explore precise treatment methods to reduce myopia-related complications and visual impairment.
In the human eye, the sclera experiences growth in infancy, and the scleral enlargement in myopia reflects active scleral extracellular matrix remodeling. A study has shown differences in microRNAs (miRNAs) in the sclera of infants and adults with normal vision in the GEO database [3]. It is reported that some myopia-related mRNAs, including transforming growth factor β (TGF-β), tissue inhibitor of metalloproteinases (TIMP), and increased in matrix metalloproteinases (MMP), are associated with myopic scleral remodeling [4].
miRNAs are non-coding RNAs that regulate the expression of target genes by causing mRNA degradation, cleavage, or translation inhibition [5]. However, the role of miRNA-mRNA interactions in myopia has not yet been investigated. The pathogenesis of myopia is complex, and the specific pathways and mechanisms involved in myopic scleral matrix remodeling are still unknown.
This study aimed to identify additional genes associated with scleral remodeling in myopia. A comprehensive search was conducted in the GEO database to identify miRNAs related to scleral remodeling. Target mRNA prediction was performed using Targetscan. Additionally, myopia-related mRNAs were screened from the CTD database. Following a rigorous screening process, 10 hub mRNAs associated with myopic scleral remodeling were identified. Our findings suggest that the FOXO signaling pathway may play a role in the process of myopia. Subsequently, we conducted molecular experiments to validate these findings. The results showed that miR-15b-5p/miR-379-3p could disrupt the FOXO signaling pathway (IGF1R/PTEN/FOXO3a/CDKN1B) in myopia. The FOXO signaling pathway is involved in regulating the cell cycle, apoptosis, and the reversal of the fibrotic process, participating in scleral remodeling. miRNA-mRNA mining may contribute to identifying new molecular markers of myopia, providing a foundation for further investigation into the pathogenesis of myopia.
Materials and methods
Screening of miRNA-mRNA associated with myopic scleral remodeling
The workflow chart for the bioinformatics analysis related to myopic scleral remodeling is shown in Fig. 1. The differentially expressed miRNAs in the sclera between fetal and adult groups were obtained from the Gene Expression Omnibus database (GSE46435: GPL10850, GPL16770. GEO, https://www.ncbi.nlm.nih.gov/gds/). miRNA differentially expressed analysis was explored using GEO2R (P < 0.001, |log FC|> 4). Targetscan [(http://www.mirdb.org/) (cumulative weighted context + + score < − 0.3)] and miRDB [(http://www.mirdb.org/) (target score ≥ 80)] databases predicted the miRNA-targeted mRNA.
The comparative toxic genomics database (CTD, http://ctdbase.org/) was used to obtain targets for myopia-related diseases using “myopia” as a search term (inference score > 5). Then, the key myopia-related mRNAs are the common mRNAs between the target dataset of differentially expressed miRNAs and the myopia dataset.
CytoHub was used to screen 10 hub mRNAs [6]. GO and KEGG pathway analyses of all target mRNAs, key mRNAs, and hub mRNAs were processed by Metascape (https://metascape.org/) (P < 0.05). Cytoscape was used to integrate hub mRNAs and miRNA information to construct the miRNA-hub mRNA network.
Validation of animal experiments
Animals
The 2-week-old guinea pigs (Cavia porcellus, British Shorthair, tricolor strain) with an average weight between 100 and 120 g (Zhenjiang, China) were used to perform the relevant experiments. Before enrollment, the guinea pigs with cataracts or corneal diseases were excluded. Then the guinea pigs were randomly divided into a normal control (NC) group (n = 16) and a lens-induced myopia (LIM) group (n = 16). No intervention was performed in the NC group, while the right eyes of the animals in the LIM group were covered with − 6.0D glasses to induce myopia. After 4-week myopic induction, the animals were sacrificed, and 4 eyeballs in each group were paraffin-embedded for staining and immunofluorescence assay, meanwhile, another 6 scleral tissues from the eyes in each group were collected for flow cytometry. Finally, 6 scleral tissues from each group were dissected for mRNA and protein expression analysis, respectively.
Biometric measurements
Ophthalmic type A ultrasonography (Cinescan, Quantel Medical, France) was used to measure the ocular axial length. The average of the 10 measurements is the final reading.
Determination of apoptosis and cell cycle arrest
After 4-week myopic induction, the scleral tissues were separated. Subsequently, the scleral tissues were digested using collagenase I for 30 min, followed by filtration through a 70-mesh filter to obtain a single-cell suspension. Further, the cell suspensions were used to determine apoptosis and cell cycle arrest using a flow cytometer (Agilent NovoCyte D2040R). To assess apoptotic levels, the cells in each group were detected using Annexin V-EGFP/PI Apoptosis Detection (40303ES50, Yeasen Biotechnology, China). The cells were fixed with 70% ethanol (− 20 °C precooling) for more than 1 h, and the cell cycle was detected using the cell cycle analysis kit (C1052, Beyotime, China).
Histopathological assessment and Masson staining
Eyeballs were immersed in FAS eyeball fixative (Servicebio, Wuhan, China) (n = 3), then rinsed twice with cold PBS. Histopathological evaluation and Masson staining were performed as per the reports [7, 8]. Scleral thickness was measured at 1000 μm above the optic nerve.
RT-qPCR
RNA was extracted with Trizol (Invitrogen, 15596026, CA, USA), and reverse transcription detection was performed using a reverse transcription kit (Sparkjade, Jinan, China). Each gene was repeated using diluted cDNA. Primer sequences were listed in Table 1 and the gene level was analyzed by a 2−△△Ct method [9].
Western blotting
The sclera of guinea pigs in each group was supplemented with RIPA-buffered lysate containing PMSF (Sparkjade, Jinan, China). The tissue was ground on ice and then centrifuged at 5000 r/min for five minutes in a centrifuger (NEST Biotech., Wuxi, China), and the supernatants were collected. The target proteins are electrophoresis and membrane transfer. Primary antibodies are listed in Table 2. Finally, images were captured using the FUSION-FX7 imaging (Vilber Lourmat, Marne-la-Vall é e, France).
TUNEL assay
After 4-week myopic induction, the eyeballs (n = 4) were immediately isolated and rinsed in sterile saline in 6-well plates (NEST, Wuxi, China). Dehydration and sectioning were then performed. The tissues were covered with a breaking-film working solution and incubated. After sectioning, the TUNEL assay kit (G1502, Servicebio, Wuhan, China) was used to perform the TUNEL assay, and after washing with PBS, DAPI dyeing was used to stain cell nuclei. Slides were closed with an anti-fluorescence quenching sealer. Cell nuclei in tissues stained with FITC-stained positive apoptotic nuclei are green. To determine the number of apoptotic cells, two independent researchers calculated the proportion of positive cell numbers from more than five randomly selected microscopic fields.
Immunofluorescence staining
After completion of antigen repair, the samples were rinsed with cold PBS and incubated with primary antibodies (Table 3). Slides were then rinsed with PBS and incubated with HRP-labeled secondary antibody (1:5000 dilution, Servicebio, Wuhan, China) for 50 min at room temperature. Subsequently, slides were washed with PBS and incubated with a DAPI dye solution. Finally, the slides were then sealed with an anti-fluorescence bursting agent.
Statistical analysis
Results were presented as the mean ± standard deviation (SD), and a statistical software package (SPSS 25.0, SPSS, Chicago, IL) was used to assess the outcome parameters. Student´s t-test for independent samples was used to analyze significant differences between the two groups, and a two-tailed P-value less than 0.05 was considered a significant difference.
Results
MiRNA-mRNA network prediction and enrichment analysis
A total of 1818 miRNAs were harvested in GSE46435 (GPL10850 and GPL16770 both selected for the top 909 miRNAs ranked by P-value) for subsequent data analysis. Two miRNAs were upregulated in 12 fetal samples and 30 in 8 adult samples based on GEO [(|log2 FC)|> 4 and P < 0.001, respectively (Fig. 2A, B).
miRNA-target mRNA network and target mRNA enrichment analysis. A Volcanic maps of differentially expressed miRNAs. B Differentially expressed miRNAs in the sclera between fetuses and adults. The higher the expression of miRNA, the deeper the red color. C Biological process; D molecular function; E cellular component. F KEGG pathway enrichment. GO: Gene Ontology; miRNA: microRNA; mRNA: messenger RNA
A total of 1869 targeted mRNAs were obtained from the intersection of the TargetScan 7.2 and miRDB databases. Subsequently, a miRNA-mRNA network diagram was constructed (Fig. S1).
Furthermore, the targeted mRNAs were enriched for the top 10 significantly enriched GO terms (i.e., biological process (BP) (Fig. 2C), molecular function (MF) (Fig. 2D), cellular component (CC) (Fig. 2E), and enriched KEGG pathways (Fig. 2F) were mapped.
We identified 897 differentially expressed mRNAs based on the CTD database. Among these, 96 mRNAs were duplicated with 32 differentially expressed miRNA targets (from Targetscan and miRDB databases), defined as key mRNAs. To assess the functions of these key mRNAs, we used Medscape for KEGG analysis of the 96 key mRNAs (Fig. 3A). Further, 75 mRNAs were screened from the 96 key mRNAs using the PPI network. (Hide disconnected nodes in the network, highest confidence: 0.4) (Fig. 3B, C). 10 hub mRNAs (i.e., PTEN, SMAD2, FOS, RHOA, MAP2K1, IGF1R, CDKN1B, MAPK8, FGF2, MAP3K1) were analyzed by cytoHub (Fig. 3D).
Key mRNAs enrichment analysis, PPI analysis, and 10 hub mRNAs enrichment analysis. A KEGG pathway enrichment for 96 key mRNAs. miRNA, microRNA. B PPI network constructed by STRING database for the 75 mRNAs out of 96 key mRNAs. C Correlation heat map of 75 mRNAs from 96 key mRNAs. The darker the red, the stronger the correlation. D The subnetwork is reconstructed with the selected hub nodes and their first neighbor mRNA. Red-colored nodes represent 10 hub mRNAs. E 10 hub nodes with significant KEGG pathway enrichment. miRNA: microRNA; PPI: protein–protein interaction; mRNA: messenger RNA
We analyzed the 10 hub mRNAs enriched (Fig. 3E) and searched the PubMed database to confirm the relationship between the 10 hub mRNAs and myopia (Table 4). Furthermore, we observed that the FOXO signaling pathway was the enrichment result of miRNAs targeting 10 hub mRNAs (Fig. 2F) and key mRNAs enrichment annotation (Fig. 3A), following the KEGG pathway analysis.
The results demonstrated a network regulation relationship between 10 hub mRNAs related to sclera remodeling and 17 miRNAs involved in sclera remodeling (Fig. 4A). The results of the prediction in the Target database indicated that hsa-miR-15b-5p, hsa-miR-379-3p, hsa-miR-411-3p, and hsa-miR-494-3p could regulate IGF1R (Fig. 4B). The protein interactions between hub mRNAs were mapped (Fig. 4C, D), and IGF1R plays a pivotal role in the FOXO signaling pathway. Consequently, it was hypothesized that hsa-miR-15b-5p, hsa-miR-379-3p, hsa-miR-411-3p, and hsa-miR-494-3 regulate the FOXO signaling pathway and myopic sclera remodeling. Thus, the follow-up experiment was conducted to validate the findings.
miRNA-mRNA network predicated biomarkers in Sclera remodeling. A miRNA-mRNA regulatory pairs from miRNAs and 10 hub mRNAs. B Schematic diagram of miR-15b-5p/miR-379-3p regulating IGF1R from TargetScan. C Correlation heat map of 10 hub mRNAs. The darker the red, the stronger the correlation. D The PPI network of 10 hub mRNAs. miRNA: microRNA; mRNA: messenger RNA; PPI: protein–protein interaction
Changes in axial length and scleral histopathology
After 4- and 6-week myopic induction, the axial length of the animals in the LIM group increased compared to that of the NC group (all P < 0.05) (Fig. 5A). The mean scleral thickness was significantly lower in the LIM group (all P < 0.05) (Fig. 5B, C). Furthermore, we found that the scleral layer cells of the NC group were neatly arranged and intact, whereas the scleral fibroblasts were disordered and thinned in the arrangements of all layers in the LIM group (Fig. 5B, C).
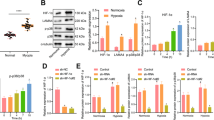
Axial length, scleral thickness analyses, and expression analysis of the FOXO signaling pathway-related molecules. A Measurement of axial length in NC and LIM groups (n = 16). B Measurement of scleral thickness by PAS staining in NC and LIM groups (n = 4). C PAS stained pathological section of the sclera. (n = 4) D–K RT-qPCR analysis. Measurement of miR-15b-5p, miR-379-3p, miR-411-3p, miR-494-3p, IGF1R, PTEN, FOXO3a, and CDKN1B expression at gene level in the sclera of guinea pigs in NC and LIM groups (n = 6). M SDS-PAGE electrophoresis. L, N–Q Measurement of PTEN, p-PTEN, FOXO3a, CDKN1B, and IGF1R expression at the protein level by Western blot in the sclera of guinea pigs in NC and LIM groups. (Western Blot cropped) (n = 6) (*P < 0.05 compared with the NC group). NC: normal control group; LIM: lens-induced myopia group
miR-15b-5p/miR-379-3p regulates the FOXO signaling pathway and participates in myopia development
We verified the miRNA-mRNA results of data mining based on experiments. After 4- and 6-week myopic induction, compared with the NC group, the levels of miR-15b-5p and miR-379-3p of the four miRNAs (miR-15b-5p, miR-494-3p, miR-411-3p, and miR-379-3p) that regulate the FOXO signaling pathway were increased in the LIM group (all P < 0.05) (Fig. 5D–G), whereas the levels of hsa-miR-411-3p and hsa-miR-494-3 showed no significant change. The results of RT-qPCR and WB detection results demonstrated that the levels of PTEN, P-PTEN, FOXO3a, and CDKN1B were increased in the LIM group, whereas the level of IGF1R reduced in comparison to the NC group (all P < 0.05) (Fig. 5H–Q).
The FOXO signaling pathway regulates the scleral cell cycle, and apoptosis, and is associated with the fibrotic process
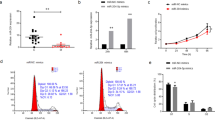
The Foxo signaling pathway affects cell cycle and apoptosis. After 4 and 6 weeks of myopic induction, the results of RT-qPCR, Western blotting, and immunofluorescence demonstrated that the levels of cytoplasmic cycle-related factors CDK2 and CCND1 in sclera tissues of the LIM group were decreased compared with that of the NC group (P < 0.05) (Fig. 6A–F). Furthermore, cell cycle tests revealed myopic scleral cells were arrested in the G1 phase (Fig. 7A, B). Meanwhile, TUNEL and flow cytometry analyses showed increased apoptosis of the LIM scleral cells after 4-week myopic induction. The levels of Bcl-2 decreased, whereas the Bax level increased in the LIM animals (Fig. 7C–F). These results indicate that the FOXO signaling pathway regulates G1 cell cycle arrest and apoptosis.
Scleral cell cycle measurement and apoptotic analysis. A, B Measurement of CDK2 and CCND1 expression at the mRNA level in the sclera of the guinea pigs in NC and LIM groups (n = 4–6). C CDK2 and CCND1 immunoblots. D, E Measurement of CDK2 and CCND1 expression at protein level by Western blot in the sclera of the guinea pigs in NC and LIM groups (n = 6). F Measurement of CDK2 and CCND1 expression at protein level by immunofluorescence in the sclera of the guinea pigs in NC and LIM groups. (Western Blot cropped) (n = 6)
A Determination of cell cycle G1 arrest by flow cytometry (n = 6) and B statistical analysis. C TUNEL assay of scleral tissues in NC and LIM groups. DAPI staining is shown in blue, and TUNEL staining is shown in green (n = 4). D, E Apoptosis measurements of scleral tissues in NC and LIM groups by flow cytometry. F The levels of BAX and BCL-2 proteins detected by immunofluorescence staining (n = 4). (*P < 0.05 vs. the NC group). NC: normal control group; LIM: lens-induced group
Additionally, after 4- and 6-week myopic induction, collagen fibers (blue) and muscle fibers (red) in the LIM group reduced, and muscle fibers became sparse (Fig. 8A), and the expression of TGF-β1, α-SMA, and COL-1 at mRNA and protein levels in LIM sclera tissues was decreased (Fig. 8B–I) (all P < 0.05). These results indicated decreased expression of fibrotic factors and scleral remodeling in myopia.
Analysis of factors associated with scleral fibrosis. A Masson’s staining. Collagen fibers were stained with blue, and muscle fibers were stained with red (n = 4). B–D Measurements of TGF-β1, α-SMA, and COL-1 expression at the mRNA level in the sclera of the guinea pigs in the NC and LIM groups (n = 6). E TGF-β1, α-SMA, and COL-1 immunoblots. F–H Measurement of TGF-β1, α-SMA, and COL-1 expression at protein level by Western blot in the sclera of the guinea pigs in NC and LIM groups (Western Blot cropped) (n = 6). I Measurement of TGF-β1, α-SMA, and COL-1 expression at the protein level by immunofluorescence in the sclera of the guinea pigs in NC and LIM groups (n = 4) (*P < 0.05 vs. the NC group). NC: normal control group; LIM: lens-induced group
Discussion
In this study, we analyzed differentially expressed miRNAs during scleral growth in fetuses and obtained relevant miRNAs associated with scleral remodeling. These miRNAs target mRNAs were intersected with myopia-related mRNAs using CTD databases for enrichment analysis, and we obtained the relevant mRNAs associated with scleral remodeling in the process of myopia. Based on enrichment analysis, we also found that the FOXO signaling pathway was related to myopic scleral remodeling, among which, IGF1R, PTEN, FOXO3a, CDKN1B, and other FOXO signaling pathway-related factors are key nodes. In addition, we confirmed that miR-15b-5p and miR-379-3p regulate IGF1R [10, 11]. Using experimental verification, we found that cell cycle G1 activity and the expression of related factors CDK2 and CCND1 decreased, whereas the apoptotic rate increased; moreover, the levels of TGF-b1, COL1, and α-SMA also reduced, indicating that the FOXO signaling pathway participated in the sclera fibrosis process by regulating the cell cycle, thereby affecting sclera remodeling.
MiR-15b-5p and miR-379 affect the processes of cell proliferation, migration, invasion, and apoptosis. When miR-15b-5p is overexpressed in neuroblastoma, cell proliferation, migration, and invasion are weakened, whereas cell apoptosis is decreased [12]. MiR-379-5p is involved in the proliferation, migration, and invasion of breast cancer [13]. Nevertheless, the role of miR-15b-5p and miR-379-3p in myopia has not been addressed. In this study, we found that the level of miR-15b-5p was up-regulated in the scleral tissue growth stage through bioinformatics analysis, and IGF1R is regulated by miR-15b-5p and miR-379 [10, 11]. So, we speculate that miR-15b-5p and miR-379-3p may play a role in myopic scleral remodeling by negatively regulating the IGF1R expression.
IGF-1 is involved in cell proliferation, differentiation, and apoptosis. One study suggests that IGF1 is associated with the development of myopia [14]. IGF1R inhibited the expression of PTEN [15]. PTEN has phosphatase-dependent and -independent effects, and PTEN affects cell cycle, migration, and growth [16, 17]. PTEN affects scleral fibroblast proliferation, but its specific mechanism has not been studied [18]. FOXO is a downstream factor of PTEN [19]. Activation of FOXO promotes the expression of genes related to cell cycle arrest [20, 21]. Cyclin-dependent kinase (CDK) inhibitor 1B (CDKN1B) is a FOXO3a target gene [22], and CDKN1B binds and inhibits the cyclin E-or cyclin A-associated CDK2 and other CDKs and inhibits G1-G2 cell cycle progression [23]. In the molecular verification of this study, the level of IGF1R in the scleral tissue of myopic decreased, consistent with the above results. In our study, expressions of miR-15b-5p, miR-379-3p, PTEN, p-PTEN, FOXO3a, and CDKN1B were increased, indicating that myopic scleral cell cycle arrest occurred. Our findings suggested that miR-15b-5p and miR-379-3p regulate the IGF1R/PTEN/FOXO3a/CDKN1B signaling pathway and participate in scleral remodeling.
Our study confirmed that apoptosis and G1 phase cell cycle arrest are important mediators in myopic scleral remodeling. Bax, a pro-apoptotic member of the Bcl-2 protein family, plays an essential role in initiating programmed cell death and stress-induced apoptosis [24, 25]. A study has shown that apoptosis in scleral fibroblasts promotes myopia progression [26], meanwhile, promoting fibroblast apoptosis can also reduce pulmonary fibrosis [27]. CDK2 and CCND1 are G1 phase cell cycle markers that drive cell cycle progression [28, 29], and down-regulation of CDK-2 and CCND1 leads to G1 cell cycle arrest [30]. Fibroblasts exhibit reversible plasticity in phenotype and cell fate by easily re-entering the cell cycle. When these characteristics are abnormally activated, they will drive human fibrotic diseases [31]. Lidocaine is a drug used to treat tendon injuries, promotes the expression of CDKN1B, and significantly inhibits the expression of cyclin A, CDK2, and type I collagen, thus decreasing cell proliferation and G1/S transformation, thereby reducing the production of ECM [32]. These studies suggest that apoptosis and G1 phase arrest are important reasons for reversing fibrosis. In this study, apoptosis and G1 cycle arrest occurred in the sclera in myopia. Therefore, we believe that apoptosis and G1 cell cycle arrest lead to the development of myopic sclera in the direction of reverse fibrosis, which is an important reason for myopic sclera remodeling.
We further examined the fibrosis levels only in the sclera. It is known that TGF-β1, COL-1, and α-SMA are typical biomarkers of fibrosis. In this study, TGF-β1, COL-1, and α-SMA expression decreased in the sclera. TGF-β1 regulates the expression of COL-1, the main component of ECM, and their expressions are down-regulated in experimental myopic scleral tissues induced by form deprivation [33, 34]. Changes in collagen state are crucial for myopia progression, and scleral type I collagen is significantly reduced in myopic models [35]. A previous study found that α-SMA expression was increased in the myopic retina, accompanied by fibrosis [36], and this result is consistent with the findings of our study, indicating that the process of reverse fibrosis is an important factor in scleral remodeling. Overall, these findings provide a new basis for the pathogenesis of myopia and precise treatment of myopia.
We constructed a miRNA-mRNA network based on bioinformatics analysis and validated the bioinformatics findings by animal experiments. We found that miR-15b-5p and miR-379-3p can negatively regulate the IGF1R expression via the miR-15b-5p/miR-379-3p-IGF1R/PTEN/FOXO/CDKN1B axis, and thus influence the cell cycle and apoptosis of the sclera, fibrosis process, and scleral remodeling, thereby aggravating the ocular axis elongation and leading to myopia (Fig. 9).
In myopia, miR-15b-5p and miR-379-3p can negatively regulate the IGF1R expression, influence the cell cycle and apoptosis of the sclera through the miR-15b-5p/miR-379-3p-IGF1R/PTEN/FOXO/CDKN1B axis, and thus regulate the fibrosis process and scleral remodeling, thereby promoting the elongation of the ocular axis and leading to myopia. (Created with BioRender.com)
Conclusions
The miR-15b-5p/miR-379-3p/IGF1R/PTEN/FOXO/CDKN1B axis can block the G1 cell cycle, promote apoptosis, and influence the sclera fibrosis process, thereby aggravating the development of myopia. Our findings provide a solid basis for further understanding of the pathogenesis of myopia, facilitating the precise treatment of myopia in clinical practice.
Availability of data and materials
All writers have reviewed and given their approval to the final draft. Publicly accessible datasets were examined in this study. The datasets supporting the conclusions of this article are available in the GEO (https://www.ncbi.nlm.nih.gov), Targetscan (https://www.targetscan.org/), miRDB (http://www.mirdb.org/), CTD (http://ctdbase.org/), Metascape (https://metascape.org/), STRING (http://string-db.org) and Cytoscape (v.3.7.0, https://cytoscape.org/).
Abbreviations
- CDK:
-
Cyclin-dependent kinase
- CDKN1B:
-
CDK inhibitor 1B
- LIM:
-
Len-induced myopia
- NC:
-
Normal control
- CCND1:
-
Cyclin D1
- PTEN:
-
Tensin homolog deleted on chromosome ten
- p-PTEN:
-
Phosphorylation PTEN
- FOXO3a:
-
Forkhead box O3a
- IGF1R:
-
Insulin-like growth factor 1 receptor
- TGF-β1:
-
Transforming growth factor-β1
- COL-1:
-
Collagen type 1
- α-SMA:
-
α-Smooth muscle actin
- miRNAs:
-
MicroRNAs
- TIMP:
-
Tissue inhibitor of metalloproteinases
- MMP:
-
Matrix metalloproteinases
- BP:
-
Biological process
- MF:
-
Molecular function
- CC:
-
Cellular component
- CTD:
-
Comparative toxic genomics database
- GEO:
-
Gene expression omnibus
- RT-qPCR:
-
Reverse transcriptase quantitative PCR
- ECM:
-
Extra cellular matrix
References
Holden BA, Fricke TR, Wilson DA, Jong M, Naidoo KS, Sankaridurg P, Wong TY, Naduvilath TJ, Resnikoff S. Global prevalence of myopia and high myopia and temporal trends from 2000 through 2050. Ophthalmology. 2016;123:1036–42. https://doi.org/10.1016/j.ophtha.2016.01.006.
Bullimore MA, Ritchey ER, Shah S, Leveziel N, Bourne RRA, Flitcroft DI. The risks and benefits of myopia control. Ophthalmology. 2021;128:1561–79. https://doi.org/10.1016/j.ophtha.2021.04.032.
Metlapally R, Gonzalez P, Hawthorne FA, Tran-Viet KN, Wildsoet CF, Young TL. Scleral micro-RNA signatures in adult and fetal eyes. PLoS ONE. 2013;8: e78984. https://doi.org/10.1371/journal.pone.0078984.
Lin MY, Lin IT, Wu YC, Wang IJ. Stepwise candidate drug screening for myopia control by using zebrafish, mouse, and Golden Syrian Hamster myopia models. EBioMedicine. 2021;65: 103263. https://doi.org/10.1016/j.ebiom.2021.103263.
Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–55. https://doi.org/10.1016/j.cell.2009.01.035.
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–504. https://doi.org/10.1101/gr.1239303.
Bao B, Liu J, Li T, Yang Z, Wang G, Xin J, Bi H, Guo D. Elevated retinal fibrosis in experimental myopia is involved in the activation of the PI3K/AKT/ERK signaling pathway. Arch Biochem Biophys. 2023;743:109663. https://doi.org/10.1016/j.abb.2023.109663.
Xu F, Guo D, Jiang Q, Zhang R, Yu T, Yin X, Wu S, Liu D, Wen Y, Wu J, Bi A, Jiang W, Bi H. Association between anti-fibrillin-2 protein induced retinal degeneration via intravitreous delivery and activated TGF-β signalling in mice. Clin Exp Pharmacol Physiol. 2022;49(5):586–95. https://doi.org/10.1111/1440-1681.13631.
Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–8. https://doi.org/10.1006/meth.2001.1262.
Li Z, Cai B, Abdalla BA, Zhu X, Zheng M, Han P, Nie Q, Zhang X. LncIRS1 controls muscle atrophy via sponging miR-15 family to activate IGF1-PI3K/AKT pathway. J Cachexia Sarcopenia Muscle. 2019;10(2):391–410. https://doi.org/10.1002/jcsm.12374.
Cao C, Duan P, Li W, Guo Y, Zhang J, Gui Y, Yuan S. Lack of miR-379/miR-544 cluster resists high-fat diet-induced obesity and prevents hepatic triglyceride accumulation in mice. Front Cell Dev Biol. 2021;9:720900. https://doi.org/10.3389/fcell.2021.720900.
Chava S, Reynolds CP, Pathania AS, Gorantla S, Poluektova LY, Coulter DW, Gupta SC, Pandey MK, Challagundla KB. miR-15a-5p, miR-15b-5p, and miR-16-5p inhibit tumor progression by directly targeting MYCN in neuroblastoma. Mol Oncol. 2020;14(1):180–96. https://doi.org/10.1002/1878-0261.12588.
Yang K, Li D, Jia W, Song Y, Sun N, Wang J, Li H, Yin C. MiR-379-5p inhibits the proliferation, migration, and invasion of breast cancer by targeting KIF4A. Thorac Cancer. 2022;13:1916–24. https://doi.org/10.1111/1759-7714.14437.
Meng B, Wang K, Huang Y, Wang Y. The G allele of the IGF1 rs2162679 SNP is a potential protective factor for any myopia: updated systematic review and meta-analysis. PLoS ONE. 2022;17(7): e0271809. https://doi.org/10.1371/journal.pone.0271809.
Rochester MA, Riedemann J, Hellawell GO, Brewster SF, Macaulay VM. Silencing of the IGF1R gene enhances sensitivity to DNA-damaging agents in both PTEN wild-type and mutant human prostate cancer. Cancer Gene Ther. 2005;12:90–100. https://doi.org/10.1038/sj.cgt.7700775.
Worby CA, Dixon JE. PTEN. Annu Rev Biochem. 2014;83:641–69. https://doi.org/10.1146/annurev-biochem-082411-113907.
Hopkins BD, Hodakoski C, Barrows D, Mense SM, Parsons RE. PTEN function: the long and the short of it. Trends Biochem Sci. 2014;39:183–90. https://doi.org/10.1016/j.tibs.2014.02.006.
Yang Q, Lv S, Zhu H, Zhang L, Li H, Song S. A potential research target for scleral remodeling: effect of MiR-29a on scleral fibroblasts. Ophthalmic Res. 2022;65:566–74. https://doi.org/10.1159/000525189.
Sanese P, Forte G, Disciglio V, Grossi V, Simone C. FOXO3 on the road to longevity: lessons from SNPs and chromatin hubs. Comput Struct Biotechnol J. 2019;17:737–45. https://doi.org/10.1016/j.csbj.2019.06.011.
Liu Y, Ao X, Ding W, Ponnusamy M, Wu W, Hao X, Yu W, Wang Y, Li P, Wang J. Critical role of FOXO3a in carcinogenesis. Mol Cancer. 2018;17(1):104. https://doi.org/10.1186/s12943-018-0856-3.
Furukawa-Hibi Y, Kobayashi Y, Chen C, Motoyama N. FOXO transcription factors in cell-cycle regulation and the response to oxidative stress. Antioxid Redox Signal. 2005;7(5–6):752–60. https://doi.org/10.1089/ars.2005.7.752.
Karger S, Weidinger C, Krause K, Sheu SY, Aigner T, Gimm O, Schmid KW, Dralle H, Fuhrer D. FOXO3a: a novel player in thyroid carcinogenesis? Endocr Relat Cancer. 2009;16(1):189–99. https://doi.org/10.1677/ERC-07-0283.
Dai L, Liu Y, Liu J, Wen X, Xu Z, Wang Z, Sun H, Tang S, Maguire AR, Quan J, et al. A novel cyclinE/cyclinA-CDK inhibitor targets p27(Kip1) degradation, cell cycle progression and cell survival: implications in cancer therapy. Cancer Lett. 2013;333(1):103–12. https://doi.org/10.1016/j.canlet.2013.01.025.
Oltvai ZN, Milliman CL, Korsmeyer SJ. Bcl-2 heterodimerizes in vivo with a conserved homolog, Bax, that accelerates programmed cell death. Cell. 1993;74(4):609–19. https://doi.org/10.1016/0092-8674(93)90509-o.
Plikus MV, Wang X, Sinha S, Forte E, Thompson SM, Herzog EL, Driskell RR, Rosenthal N, Biernaskie J, Horsley V. Fibroblasts: origins, definitions, and functions in health and disease. Cell. 2021;184(15):3852–72. https://doi.org/10.1016/j.cell.2021.06.024.
Wu J, Zhao Y, Fu Y, Li S, Zhang X. Effects of lumican expression on the apoptosis of scleral fibroblasts: in vivo and in vitro experiments. Exp Ther Med. 2021;21(5):495. https://doi.org/10.3892/etm.2021.9926.
Hohmann MS, Habiel DM, Coelho AL, Verri WA Jr, Hogaboam CM. Quercetin enhances ligand-induced apoptosis in senescent idiopathic pulmonary fibrosis fibroblasts and reduces lung fibrosis in vivo. Am J Respir Cell Mol Biol. 2019;60(1):28–40. https://doi.org/10.1165/rcmb.2017-0289OC.
Chen MJ, Cheng AC, Lee MF, Hsu YC. Simvastatin induces G1 arrest by up-regulating GSK3β and down-regulating CDK4/cyclin D1 and CDK2/cyclin E1 in human primary colorectal cancer cells. J Cell Physiol. 2018;233(6):4618–25. https://doi.org/10.1002/jcp.26156.
Zhang JP, Li XL, Li GH, Chen W, Arakaki C, Botimer GD, Baylink D, Zhang L, Wen W, Fu YW, et al. Efficient precise knockin with a double cut HDR donor after CRISPR/Cas9-mediated double-stranded DNA cleavage. Genome Biol. 2017;18(1):35. https://doi.org/10.1186/s13059-017-1164-8.
Xu W, Wang B, Yang M, Zhang Y, Xu Z, Yang Y, Cao H, Tao L. Tebufenozide induces G1/S cell cycle arrest and apoptosis in human cells. Environ Toxicol Pharmacol. 2017;49:89–96. https://doi.org/10.1016/j.etap.2016.12.002.
Song Y, Wei J, Li R, Fu R, Han P, Wang H, Zhang G, Li S, Chen S, Liu Z, et al. Tyrosine kinase receptor B attenuates liver fibrosis by inhibiting TGF-β/SMAD signaling. Hepatology. 2023;78(5):1433–47. https://doi.org/10.1097/HEP.0000000000000319.
Chen YC, Chang HN, Pang JS, Lin LP, Chen JM, Yu TY, Tsai WC. Lidocaine inhibited tendon cell proliferation and extracellular matrix production by down regulation of cyclin A, CDK2, type I and type III collagen expression. Int J Mol Sci. 2022;23(15):8787. https://doi.org/10.3390/ijms23158787.
Jobling AI, Nguyen M, Gentle A, McBrien NA. Isoform-specific changes in scleral transforming growth factor-beta expression and the regulation of collagen synthesis during myopia progression. J Biol Chem. 2004;279(18):18121–6. https://doi.org/10.1074/jbc.M400381200.
Yuan Y, Li M, Chen Q, Me R, Yu Y, Gu Q, Shi G, Ke B. Crosslinking enzyme lysyl oxidase modulates scleral remodeling in form-deprivation myopia. Curr Eye Res. 2018;43(2):200–7. https://doi.org/10.1080/02713683.2017.
Frost MR, Norton TT. Alterations in protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Invest Ophthalmol Vis Sci. 2012;53(1):322–36. https://doi.org/10.1167/iovs.11-8354.
Ouyang X, Han Y, Xie Y, Wu Y, Guo S, Cheng M, Wang G. The collagen metabolism affects the scleral mechanical properties in the different processes of scleral remodeling. Biomed Pharmacother. 2019;118:109294. https://doi.org/10.1016/j.biopha.2019.109294.
Acknowledgements
We acknowledge the GEO, Targetscan, miRDB, CTD, Metascape, STRING, and Cytoscape databases for free use.
Funding
This study was supported by the National Key R&D Program of China (2021YFC2702103, 2021YFC2702100), the Key R&D Program of Shandong Province (2019GSF108252), Science & Technology Project of Medicine and Health of Shandong Province (202307021591) and “Taishan Scholar” Project Special Fund (tsqnz20231252).
Author information
Authors and Affiliations
Contributions
Data curation, Ruixue Zhang, Jinpeng Liu and Miao Zhang; investigation, Ruixue Zhang, Jiawen Hao and Miao Zhang; methodology, Ruixue Zhang and Dadong Guo; resources, Ying Wen, Jinpeng Liu, Yuan Peng and Hongsheng Bi; software, Ying Wen, Yunxiao Xie and Zhaohui Yang; supervision, Hongsheng Bi; validation, Jinpeng Liu, Yuan Peng, Yunxiao Xie and Yongwei Shi; visualization, Dadong Guo; writing—original draft, Ruixue Zhang and Dadong Guo.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The present study was approved by the Experimental Animal Ethics Review Committee of the Affiliated Hospital of Shandong University of Traditional Chinese Medicine (Approval number: AWE-2022-055) and strictly followed the principles of the Statement of Animals Research in Vision and Ophthalmic (ARVO).
Consent for publication
Not Applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Zhang, R., Wen, Y., Liu, J. et al. The miR-15b-5p/miR-379-3p-FOXO axis regulates cell cycle and apoptosis in scleral remodeling during experimental myopia. J Transl Med 22, 710 (2024). https://doi.org/10.1186/s12967-024-05523-x
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12967-024-05523-x