Abstract
Neonatal sepsis is a severe medical condition that contributes significantly to neonatal mortality. However, early diagnosis and treatment can help manage the condition effectively. Predictive analytics can assist neonatal sepsis diagnosis and treatment and offer a reassuring solution. This study presents a systematic literature review of various predictive analytics methods for neonatal sepsis diagnosis and treatment. It thoroughly reviews 16 studies between 2014 and 2024, including retrospective and prospective data and utilizing various predictive modeling techniques, such as Machine Learning (ML) and Deep Learning (DL). The review unveils that the Predictive analytics models can rapidly detect early and late-onset neonatal sepsis. This detection aids clinicians in decision-making and improves healthcare management for neonates, especially in low-resource settings. This study lays the groundwork for utilizing advanced analytics technologies to address challenges in this domain. It inspires further research in innovative and unexplored methods for neonatal sepsis diagnosis.
Similar content being viewed by others
Explore related subjects
Discover the latest articles, news and stories from top researchers in related subjects.Avoid common mistakes on your manuscript.
1 Introduction
Neonatal infections are responsible for 25% of infant deaths globally and 10% of deaths in children under 5 years old. In developing countries, they account for 99% of the world’s mortality due to bacterial infections [1]. Due to the uneven distribution of health services between urban and rural areas, developing countries such as India face a high Neonatal Mortality Rate (refer to Table 1 for abbreviations used in this paper) of 22 [2] and the highest number of neonatal deaths within the first 24 h of life [3]. Specifically, the rural NMR is twice the urban NMR [4]. India contributes to one-fifth of global live births and over a quarter of neonatal deaths [4]. In 2013 alone, nearly 0.75 million neonates died in India, which is the highest in the world [4]. Significantly, 70% of all infant deaths and over 50% of deaths in children under five occur in the neonatal period. Deaths within the first week of the neonatal period alone account for 45% of infant mortality [4]. Neonatal sepsis, along with antibiotic resistance, kills more than 0.6 million babies annually in India [5, 6]. The United Nations SDG for NMR is 12 [7]. However, India faces significant challenges due to limited infrastructure and resources, which result in delays in diagnosing and treating neonates. This difficulty is further intensified because one or two clinical parameters cannot diagnose neonatal sepsis.
Neonatal sepsis is the presence of symptoms of sepsis in the neonatal period combined with bacteriological isolation of an infectious agent from blood or CSF [8] and the radiographic evidence of pneumonia [9]. About one million newborn babies across the world die due to neonatal infections during the neonatal period (from 0 to 28 days of birth) [1]. Neonatal sepsis includes serious infections, such as bacteremia, pneumonia, and meningitis in newborn babies. It is manifested in various forms, including very early-onset sepsis, the sepsis that occurs within the first three days of life; EOS the sepsis occurring within the first week; and finally, LOS occurring after the first week of life till the end of the neonatal period (28 days of life) [10]. The neonatal period carries the highest daily risk of mortality in the first four weeks than any other period and is 30-fold higher than the post-neonatal period from month one of birth to 59 months of age [4].
Neonatal sepsis can be managed by early diagnosis and treatment [8]. The study established that 90% of the babies sampled had more than one-factor causing sepsis. The critical challenge therefore in the diagnosis of neonatal sepsis arise from the nonspecific nature of symptoms, making both diagnosis and treatment difficult. A blood culture test, the current gold standard of diagnosis, takes more than 2 days to generate results [11, 12]. Blood culture tests are, at times, inadequate, tend to throw up false negatives, and have not always demonstrated complete accuracy [13]. Moreover, in the absence of specific laboratory tests for neonatal sepsis, and the inherent protean nature of the non-specific symptoms, accurate clinical diagnosis of neonatal sepsis continues to remain a challenge [9, 14] with delays in diagnosis and treatment as common issues [15] across the globe. Blood culture test continues to be a gold standard even though its sensitivity is challenged by 18% false negative rates [15, 16]. Additionally, the volume of blood obtained from a neonate is more often less than 0.5 ml while the recommended range to detect bacteria is between 0.5 and 1.0 ml [13]. To determine the volume of blood necessary for conducting culture tests among babies, it is essential to know the detection systems sensitivity, and the probability of locating at least one microorganism in the collected blood culture sample. Sampling small volumes of blood to spot low-density bacteraemia or fungemia could run into a risk of not finding an organism in the culture bottle [13]. Also, in critically ill babies or very low birth weight babies, it is very hard to draw 1 ml blood sample and the low volume of the sample is usually inadequate to diagnose neonatal sepsis [16, 17]. Especially in preterm babies, it is hard to make a suspicion of infection, as a result of which antibiotic usage is common and even leads to higher rates of mortality and morbidity when administered during the first seven days among new-borns [18]. Failure to quickly diagnose neonatal sepsis, primarily due to its indefinite sign and symptoms, makes the disease more lethal and destructive. Blood culture test, the only method for confirmatory diagnosis of neonatal sepsis, has however not been effectively addressing issues around neonatal morbidity and mortality, leading to a practice of rampant of empirical antibiotics [15] which in turn is contributing to yet another global health crisis, AMR.
According to [19], antibiotic resistance arises because of mutations in microbes and resulting selection pressure due to antibiotic use which offers a competitive advantage for mutated strains. Especially in LMICs, antibiotic use has gone up due to rising incomes, growing rates of hospitalization, and increasing prevalence of hospital infections [19]. NICUs in LMICs report between 15.2 and 62.0 infections per 1000 patient days and are up to nine times more than that in the USA which averages at about 6.9 infections per 1000 patient-days [16, 19]. In [16] state that for every culture-positive sepsis result, on average, 11–23 infants in developed countries receive antibiotic treatment leading to antibiotic resistance and a rise in healthcare costs. On many occasions, despite negative blood cultures for 98% of patients, 47% of very low birth weight infants continued to receive antibiotics [16]. A review titled Antimicrobial Resistance: Tackling a crisis for the health and wealth of nations commissioned by the government of the United Kingdom and published in 2014 reported that an estimated 10 million people could die because of AMR by 2050 [20, 21]. A study published in the Lancet [21], which performed a comprehensive review of the burden of AMR across 204 countries, 23 bacterial pathogens, and 88 drug combinations, reports that the magnitude of AMR, especially that caused by bacterial pathogens surpasses that of malaria or HIV. According to the Lancet study, one critical way to address this growing problem is to increase the quality of data collection, especially in low-income settings where surveillance is weak and data is sparse which could pave the way for addressing this global health crisis and guide policymaking. Research has also demonstrated that continuous use of sepsis screens is indeed effective in reducing antibiotic usage among neonates [22].
In developing countries like India, resistance to WHO recommended treatment regimens of antibiotics like ampicillin and gentamicin among pathogens causing neonatal infections in the first 28 days of their life is highly prevalent. A high proportion of these isolates, especially 71% Klebsiella spp and 50% E-coli are found to be resistant to gentamicin, which is the first line of treatment for neonatal infections [19]. Other harmful pathogens like Enterobacteriaceae and Acinetobacter spp causing neonatal infections are also associated with high antibiotic resistance and high mortality in neonatal nurseries [19]. In high-income countries, the culture-positive to culture-negative sepsis ratio ranges from 1:6 to 1:16 [23]. This ratio is likely skewed towards culture-negative sepsis in South Asia. Additionally, the pathogen profile in South Asia is observed to be different from that in high-income countries. In South Asia, Gram-negative pathogens (>60%) are dominant, while group B streptococci are low, which typically have a high incidence in high-income countries. The predominance of Gram-negative pathogens in South Asia indicates that the transition from Gram-Negative to Gram-positive organisms such as group B streptococci, that occurred about five to six decades ago in developed countries, is largely attributed to improved aseptic routines, including hand hygiene in neonatal intensive care units.
This disparity in pathogen profile across geographies requires localized algorithms in developing countries’ context rather than deploying the ones built in the developed country’s context since they will only render themselves ineffective in aiding health professionals, clearly demonstrating that ‘one size fits all’ is not the perfect solution in healthcare. Researchers have therefore indicated towards criticality in the requirement of the development of novel technology-based approaches for fast prediction of neonatal sepsis [11] that could perform diagnosis as effectively or more.
Predictive analytics methods have attracted researchers’ attention for their ability to diagnose neonatal sepsis early. They used statistical methods [8], Machine learning (ML) [15], and Artificial Neural Networks (ANNs) [24] to aid in predicting the onset of sepsis in neonates. Predictive analytics has generated significant interest in healthcare because of its potential to transform economies, especially in primary health services [25]. Acknowledged as a non-medical intervention, predictive analytics promises to address health crises, build preparedness for next-generation epidemics, and aid in resilient recoveries during global health emergencies or calamities [26]. EHR alerts and CDSS all under the “classic” predictive analytics category [25], which, in recent times, has seen a significant rise in budgetary allocations.
Predictive analytics use data to build algorithms capable of deriving insights, mimicking intelligent human decision-making processes, thereby enhancing the effectiveness of decision-making processes [27]. Predictive analytics in healthcare broadly refers to ML, a subset of AI, and DL based on ANNs is a subset of ML. An example of a widespread use case of predictive analytics in healthcare is predicting patients at risk of developing certain conditions, estimating the likelihood of unexpected transfers to ICUs or emergency departments, and forecasting unplanned hospitalization expenses, among other scenarios [25].
While the health sector has pioneered predictive analytics [28], its adoption in health management focusing on neonatal sepsis detection has been uneven globally [27]. As a consequence, the predictive analytics adoption curve traverses a non-linear trajectory owing to diverse stages of innovation in several countries, resulting in varied success rates amidst the unaddressed challenges around their respective legal, regulatory, ethical, and cultural factors [29]. Thus, the current study fills the gap in predictive analytics literature in early predicting neonatal sepsis by providing a pathway to improve its early diagnosis and screening and address related antibiotic resistance. It also builds upon theory to foster world-class research in predictive analytics applications.
Therefore, following a rigorous scientific research design, this study endeavors to holistically assimilate knowledge concerning prediction analytics models for early diagnosis of Neonatal Sepsis in the healthcare sector in the context of developing countries. A systematic literature review is considered a reliable method to synthesize the literature and is deemed appropriate to assimilate the fragmented literature to determine the current state of the art [30] literature.
The objectives of the current review are as follows:
-
1.
To systematically review predictive analytics methods used for the early diagnosis of neonatal sepsis (see Sect. 2 and Sect. 3).
-
2.
To identify gaps in the current research efforts toward building advanced predictive analytics methods for neonatal sepsis diagnosis (see Sect. 4). The review is up-to-date and explores different dimensions of neonatal sepsis diagnosis in the context of the nature of the disease (EOS/LOS), country of study, performance analysis of different predictive analytics methods, and reviews of prominent studies on the implementation of predictive analytics methods in neonatal sepsis diagnosis by different researchers. This review will benefit researchers in the domain of neonatal sepsis diagnosis and predictive analytics, providing an in-depth understanding of the different predictive analytics methods used for early diagnosis of neonatal sepsis.
The rest of the paper is organized as follows: Sect. 2 presents the method used for systematic literature review. Later, Sect. 3 presents the results obtained and their discussion of key findings, strengths, and limitations. Section 4 presents the gaps in the literature and future directions for further research. Finally, the paper is concluded with acknowledgements.
2 Methods
The four-step process outlined in the PRISMA guidelines [31] was used to create a rigorous and repeatable review, as recommended by [32]. The search protocol is classified into three phases:
2.1 Phase1: Planning the review
Articles were searched from Scopus, PubMed, Google Scholar, etc. An exhaustive search using boolean operators with a combination of “OR” and “AND” was used along with the following keywords- “Neonatal Sepsis” AND “Neonatal Sepsis Diagnosis” AND, “Predictive Analytics” OR “Artificial Intelligence” OR “Machine Learning.” Research papers were searched from the year 2014 to 2024.
All the articles were screened independently by all the researchers; however, only those articles with the main keywords in the abstract that were relevant were considered for review.
Inclusion Criteria: Further, the articles were selected based on the inclusion criteria mentioned below:
-
1.
Neonates with positive blood culture before 72 h of life were considered Early-onset Neonatal Sepsis cases. Neonates with positive blood culture reports after 72 h of life were considered late neonatal sepsis cases.
-
2.
Both prospective and retrospective studies that used electronic health records and vital signal measurement devices to try to figure out how to predict neonatal sepsis.
-
3.
Clinical Studies using various statistical methods for predicting Neonatal Sepsis were considered.
Exclusion Criteria: The following are excluded from our study
-
1.
Non-English articles, conference papers, and editorial communication
-
2.
Systematic reviews of previously published studies
-
3.
Meta-analysis of studies
2.2 Phase II: Conducting the review
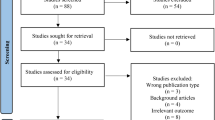
Based on the inclusion and exclusion criteria, 1690 articles were screened for document type and subjects, 72 from Scopus, 4 from PubMed, and 1612 from Google Scholar were considered for further evaluation. 994 articles were exported based on accessibility, out of which 924 articles were excluded and which met the inclusion and exclusion criteria defined for the study. A final set of 48 full-text articles were identified for the literature review, and 16 articles with predictive models developed and validated were analyzed.
2.3 Phase III: Reporting
Table 1 presents the review’s results and abbreviations used in the paper. Figure 1 depicts the PRISMA approach for choosing the final set of 16 relevant studies. Figure 2 presents the distribution of studies year wise, journal wise and country wise respectively. These studies are briefly described as follows:
Mani et al. [16] proposed machine learning algorithms for early diagnosis of late-onset neonatal sepsis, which include SVM, NB, Variants such as TAN, AODE, a sample-based classifier (KNN), decision tree classifiers, CART, RF, LR and LBR. Authors picked variables from a subset of NICU variables deemed relevant, aided by clinical expertise and literature evidence. They included 781 (71 X 11) temporal variables (laboratory) and 30 nontemporal (demographics). Data missing values were addressed using the LOCF approach [33]. The Gaussian single imputation method is considered superior over simple mean, median, and mode value imputation because it introduces less bias and reduces the missing values from 91 to 64%. Variables with more than 90% missing data were removed. The final dataset consisted of 93 variables and 299 instances. An ensemble of machine learning methods used included classifier algorithms such as SVM; decision support algorithms such as NB and CART, LR; KNN, TAN, AODE, RF, and finally LBR. Feature selection was done using a 5-fold nested cross-validation procedure in MATLAB. Performance was measured using the AUC since it works well independent of class size and classification threshold [34]. Since the sample size is smaller relative to the number of variables, authors used SVM algorithms such that they could select a subset of features that are highly predictive of the outcome variable. SVM algorithms have qualitative and quantitative advantages and can pick informative features quite well [35]. The treatment specificity of ML algorithms was higher than the physicians criterion measured as a prescription of an antibiotic within 12 h of phlebotomy. The authors concluded that most ML algorithms treatment sensitivity and specificity exceeded that of the physicians.
Thakur et al. [36] proposed two different prediction models for low-resource and developing countries using non-invasive and invasive parameters extracted from the MIMIC III database. The authors applied binary logistic regression to develop prediction models and used Chi-square test and Fishers exact test as goodness-of-fit tests. They extracted a total of seven parameters within a time window of 12 h, including blood culture reports, temperature, CSF culture result, HR, systolic and diastolic pressure, white blood cell count, and bands. In the derivation and validation phase, 1472 neonates were divided into two datasets in the ratio of 70/30. Binary logistic regression was used to calculate the prediction power of invasive parameters (platelets, bands, and WBC) and non-invasive parameters such as temperature, blood pressure, and heart rate. The authors concluded that both prediction models derived from invasive and non-invasive parameters performed equally well, with an AUROC of 0.777 and 0.824 in the derivation dataset and an AUROC of 0.830 and 0.824 in the validation dataset.
Lopez-Martinez et al. [24] proposed a deep learning approach to predict early neonatal sepsis. They constructed a sequence of ANNs with increasing hidden layers and nodes, trained the networks on a set of hyper parameters, and normalized the data using Gaussian normalization. The network model results show a sensitivity of 80.3%, a specificity of 90.4%, a precision value of 83.1%, and an AUC of 92.5%. The authors concluded that the neural network model could be used as an inference system to aid in detecting neonatal sepsis.
Gomez et al. [37] proposed a noninvasive machine learning algorithm for early detection of neonatal sepsis using HRV monitoring features. The study included 79 newborn babies with less than 48 h of life and a gestational age between 36 and 41 weeks. Electrocardiogram signal data was recorded, and HRV parameters were calculated. The authors developed supervised machine learning algorithms, and critical metrics such as sensitivity, specificity, positive predictive value, negative predictive value, and area under the ROC curve were calculated. The study results indicated an AUC of 0.94 using the adaptive boosting machine learning model, an AUC of 0.88 for the bagged trees model, and an AUC of 0.84 for random forests. The authors concluded that neonatal sepsis can be identified using HRV parameters with the use of the adaptive boosting algorithm, which promises better results
Hu et al. [38] proposed an Artificial Neural Network model for detecting late onset of neonatal sepsis. The authors placed data collectors in NICUs to gather real-time vital sign data, including HR, RR, and blood oxygen saturation (SpO2). They transformed the vital signs data into images and labeled the data as normal or sepsis category. After extracting features, they developed a 14-layer CNN. The authors concluded that they successfully demonstrated the feasibility of using a CNN for sepsis detection using vital signs. However, they suggested further work using more powerful methods such as LSTM.
Thakur et al. [39] proposed a noninvasive prediction model to detect sepsis in neonates in developing countries and compared its performance with an invasive prediction model. The study used data extracted from the MIMIC III database, with 1447 samples filtered from a sample of 7875 EHR records. Two logistic regression models were developed for invasive and noninvasive parameters separately. The results showed that the area under the receiver operating characteristic curve for predicting sepsis in neonates using noninvasive parameters was 0.879 (95% CI), with a sensitivity of 74.81% (95% CI), specificity of 88.75% (95% CI), and PPV of 49.75% (95% CI). The authors concluded that both invasive and noninvasive models performed well and suggested that further improvements to these models could help frontline health workers and semi-skilled workers in the early prediction of neonatal sepsis, leading to a decrease in mortality rates in developing countries.
Masino et al. [15] developed ML models for early recognition of sepsis in the NICU using electronic health records at least 4 h before clinical manifestation. The authors conducted a retrospective control study considering 36 features derived from electronic health records and developed 8 machine learning models using 10-fold nested cross-validation. The results demonstrate that 6 out of 8 models showed a mean area under the ROC of 0.80–0.82, and by including both culture and clinically positive neonatal sepsis-identified cases, the same models achieved a mean ROC of 0.85–0.87. The authors concluded that machine learning predictive models could identify neonates with sepsis before their clinical recognition.
Goldberg et al. [40] introduced a validated prediction model for early diagnosis of late-onset neonatal sepsis. The model incorporates the clinical evaluation of the NLR and CRP. The authors conducted a retrospective case–control study, extracting data from medical files, and utilized univariate and multivariate logistic regression methods to identify the risk of sepsis. The authors concluded that the combination of clinical evaluation with NLR and CRP values can be utilized to predict LOS in neonates.
Song et al. [12] conducted a study to develop a prediction model for LONS using noninvasive vital sign data and machine learning technology. The model was designed to detect clinical sepsis 48 h before it occurred. The model’s performance was comparable to those using invasive data, with key markers being blood pressure, oxygen saturation, and body temperature. The authors confirmed that a machine learning-based LONS prediction model could be developed using vital sign data regularly measured in clinical settings.
Alvi et al. [41] developed an ANN model for early detection of neonatal sepsis. The authors utilized under-sampling and oversampling techniques to balance the dataset. They fed a set of hyperparameters to the ANN. They concluded that the model achieved a true positive rate of 98.4%, a true negative rate of 98.1%, a precision of 96.8%, an AUC of 99.8%, and an accuracy of 98.2%. The better performance of the ANN model was attributed to selecting the correct set of features based on the strength score of the dependent variable. It was also concluded that the developed ANN model helps detect positive and negative sepsis cases and can be used as a decision-support tool.
Helguera Repetto et al. [42] introduced a predictive model by training and validating an ANN algorithm using a balanced dataset that included preterm and term neonates. The authors found that the ANN model’s performance was better than physician diagnosis based on traditional scoring systems. Additionally, the model performed well compared to other advanced methods that used maternal and neonatal variables.
Alvi et al. [43] proposed a deep-learning model for predicting the early onset of neonatal sepsis using non-invasive data. The deep learning models are designed to work with time series, sequential, or image data. The authors built two neural networks, CNN and LSTM-RNN, to classify subjects and determine whether they are affected by sepsis. The study’s results indicated that with identical precision and recall percentages, CNN can predict both positive and negative cases well. The recall rate of LSTM-RNN is better, with a result of 1.0. LSTM RNN is considered good at predicting the early onset of neonatal sepsis, with an F-measure of 98.04% and an accuracy of 99.40%. The authors concluded that although CNN’s performance is lower than that of ANN and RNN, it performed better than traditional ML algorithms. LSTM RNN outperforms CNN and ANN.
Honore et al. [44] conducted a study investigating the predictive value of machine learning-assisted analysis of non-invasive, high-frequency monitoring data and demographic factors to detect neonatal sepsis. The study involved 325 infants with a total of 2866 hospitalization days. Personalized event timelines were created to monitor interventions and clinical findings. Time-domain features from heart rate, respiratory rate, and oxygen saturation values, as well as demographic factors, were analyzed. The Naive Bayes algorithm within a maximum a posteriori framework was used to predict sepsis up to 24 h before clinical suspicion. The study identified twenty cases of sepsis and found that combining multiple vital signs improved the algorithm’s performance compared to using only heart rate characteristics. This allowed for sepsis prediction with an area under the receiver operating characteristics curve of 0.82 up to 24 h before clinical suspicion. Additionally, the risk of sepsis increased 150-fold 10 h before clinical suspicion. The non-invasive patient data used in the algorithm provides valuable predictive value for the early detection of neonatal sepsis. The authors concluded that the non-invasive patient data algorithm provides applicable predictive value for neonatal sepsis detection.
Van den Berg et al. [45] conducted a study to develop a predictive algorithm to assist doctors in the early detection of LOS in very preterm infants. In a meticulously designed retrospective cohort study, all consecutively admitted preterm infants (GA of 32 weeks) from 2008 until 2019 were included. The subjects were classified as LOS or control according to blood culture results, the gold standard in the field. Routine and continuously measured oxygen saturation and heart rate data with a minute-by-minute sampling rate were extracted from electronic medical records to generate features. Machine learning techniques such as generalized additive models, logistic regression, and XGBoost) were then employed to build a robust classification algorithm. Upon evaluation, the LR algorithm demonstrated a promising AUC of 0.73 (p< 0.05) at the moment of clinical suspicion (t = 0). In the longitudinal simulation, the algorithms detected LOS in at least 47% of the patients before clinical suspicion without exceeding the alarm fatigue threshold of 3 alarms per day. This suggests that the algorithm, based on routinely collected data, has the potential to significantly accelerate clinical decisions in the early detection of LOS, even with limited inputs, thereby revolutionizing the way LOS is managed in clinical practice.
Robi et al. [46] have applied a classification stacking model for neonatal diseases such as sepsis, a leading cause of neonatal deaths. The developed stacking model was rigorously compared to three related machine-learning models: XGB, RF, and SVM. The results were precise, and the proposed stacking model outperformed the other models, boasting an impressive accuracy of 97.04%. This robust performance underscores the potential of the developed predictive models in contributing to the early detection and accurate diagnosis of neonatal diseases, particularly in resource-limited health facilities.
Kallonen et al. [47] conducted a study aimed at predicting late-onset sepsis (LOS) in newborns by analyzing noninvasive biosignal measurements from NICU monitors. The authors developed a machine learning algorithm based on a CNN structure, which achieved 83% sensitivity and 73% specificity for detecting LOS 44 h before clinical suspicion. The electrocardiogram and respiratory impedance signals were crucial in improving predictive accuracy. The authors concluded that this approach offers state-of-the-art prediction performance for LOS without requiring artifact removal.
Apart from studies done using Machine Learning and Artificial Neural Networks, there are few other studies that have used traditional statistical methods by researchers to predict neonatal sepsis clinically which were summarized as below.
Selimovic et al. [48] developed a predictive score to diagnose early onset of sepsis using CBC and CRP levels. The authors concluded that the predictive score for EONS is useful in the diagnostic evaluation of neonates suspected of Early onset of neonatal sepsis.
Okascharoen et al. [49] proposed a practical, clinical prediction scoring model for the diagnosis of Late onset of Sepsis using weighted coefficients from Coxs proportional hazards model ROC analysis. The authors also validated the model which indicated a good performance.
Kuzniewicz et al. [50] proposed multivariable prediction models to estimate the risk of Early onset of sepsis in preterm as well as term neonates. The study was conducted on a cohort of infants born at Kaiser Permanente Northern California Hospital during the period 2010 to 2015 to test the use of antibiotic administration, blood culture tests, clinical outcomes, and re admissions at the hospital for early onset of sepsis. Segmented regression models were used to predict the onset of sepsis. The authors concluded that usage of the Early onset of Sepsis calculator resulted in a decrease in blood culture test from 14.5 to 4.9%, while empirical administration of antibiotic use in the first 24 h reduced from 5.0 to 2.6% and a reduction of antibiotic administration from 0.5 to 0.4% was observed between 24 and 72 h of birth. Their study clearly indicated the importance of the clinical care multi variable risk prediction model in reducing the proportion of neonates undergoing extensive laboratory testing and empirical antibiotic treatment without any adverse effects.
Goldberg et al. [40] proposed a validated prediction model for early diagnosis of late-onset neonatal sepsis with clinical evaluation of NLR and CRP. The authors conducted a retrospective case–control study with data extracted from medical files and applied univariate and multivariate logistic regression methods to identify the risk of sepsis. The authors concluded that a clinical evaluation in conjunction with NLR and CRP values could be used to predict the late onset of sepsis in Neonates.
Huang et al. [51] proposed using nomograms to predict the Late onset of sepsis in preterm infants. A development cohort of 1256 preterm infants and a validation cohort of 452 preterm infants were included, and the nomograms were built with and without thyroid function. The authors concluded that thyroid hypofunction in preterm infants may increase the incidence of sepsis.
3 Results and discussion
Of the 16 studies selected (as per Table 1), 15 were retrospective, and one was conducted on prospective data. Most studies (70%) are based in developed countries such as the USA, Finland, Netherlands, Australia, Israel, Spain, etc., and few studies on neonatal sepsis diagnosis were conducted in developing countries. It was also observed that late-onset neonatal sepsis diagnosis is of significant interest in studies conducted in developed countries. Most models (62.5%) were based on non-invasive parameters without involving any blood tests. ML models such as SVM, NB Classifiers, AODE Estimators, Sample-based classifiers, Decision tree classifier, CART, RF, LR, LBR, AB, BT, GB, GP, KNN, GAM, XGBoost etc were used in majority of the studies. ANNs, CNNs, and DL techniques were used to predict sepsis in 6 studies analyzed.
3.1 Key findings
Sepsis is a severe medical condition that requires prompt diagnosis and treatment. However, traditional diagnostic methods have inherent limitations that can cause delays in recognizing the onset of sepsis. Fortunately, machine learning algorithms have proven and consistently demonstrated their high effectiveness in predicting sepsis hours before it is clinically recognized [15]. The systematic review of various studies reaffirmed these impressive results, showing better sensitivity and specificity than clinicians’ diagnosis of sepsis [42].
Both invasive and non-invasive models were equally effective in predicting sepsis, with the latter providing applicable predictive value and supporting clinical decision-making in the early diagnosis of late-onset neonatal sepsis [12, 36]. Additionally, NLR and C-reactive protein prediction can rapidly identify neonatal sepsis’s late onset [40]. HRV is identified as one of the critical parameters for sepsis diagnosis. Among machine learning algorithms, Adaboost ML was an effective tool for predicting sepsis, and several other machine learning algorithms were used in the studies [37].
As illustrated in a few articles, ANNs, and CNN models were effectively used as an intelligent systems inference engine to enhance the detection of neonatal sepsis [24]. These models demonstrated high accuracy in predicting neonatal sepsis, with the CNN model capable of predicting its onset 24 h ahead [38]. Furthermore, ANN can accurately predict neonatal sepsis and outperform physicians’ diagnoses [41]. The LSTM RNN is the best-performing prediction model, with an accuracy of 99.40% [43]. When combined with PSD transformation, biosignals from NICU monitors provide excellent prediction performance for late-onset neonatal sepsis [47]. With its potential to support early diagnosis of neonatal sepsis, machine learning holds promise for healthcare, particularly in resource-limited facilities [46]. The performance of predictive models using non-invasive features is comparable to invasive models, offering a ray of hope for more accessible and efficient healthcare [45]. Machine learning algorithms can improve patient outcomes by predicting sepsis hours before clinical recognition [16].
3.2 Strengths and limitations
3.2.1 Strengths
The systematic review rigorously analyzed an equal proportion of studies investigating early and late-onset sepsis. It thoroughly evaluated the use of both invasive and non-invasive parameters for developing machine learning models. The study was up to date with the articles published till March 2024.
3.2.2 Limitations
While comprehensive, it is crucial to note that this review was limited to studies conducted in English, excluding a significant number of articles. Moreover, most of these studies were conducted in developed countries, leaving a need for more research in developing countries. The potential impact of predictive models in diagnosing neonatal sepsis, especially in low-resource settings, is immense. To underline the urgency and importance of this issue, it is clear that more evidence is needed. Urgent and extensive research is required in the field of neonatal sepsis detection, particularly in the context of developing countries.
4 Literature review gaps and future directions
4.1 Gaps identified
Most existing ML studies on the early prediction of neonatal sepsis are focused on the LOS from developed countries and are retrospective using EHR data ([15, 16, 18]). According to these studies, the essential causative organisms for EOS are Streptococcus agalactiae and Escherichia coli (in Western countries data). Studies indicate that due to a sporadic rise in AMR, neonatal mortality is also increasing, and hence, non-culture-based diagnosis needs to be encouraged. Moreover, in high-income countries, the culture-positive to culture-negative sepsis ratio ranges from 1:6 to 1:16 ([23]), and this ratio is likely skewed towards culture-negative sepsis in South Asia. Additionally, variation in pathogen profile in South Asia compared to that from high-income countries calls for localized algorithms to address the issue.
Most research in Predictive Analytics is empirical in nature; therefore, a more significant number of theoretical studies are also needed to develop the body of literature and advance academic research in the context of developing countries.
4.2 Future research directions
Research from developed countries states that early diagnosis and treatment of such critical medical conditions using predictive analytics methods reduce antibiotic use by at least 50% ([52]) and infections by 84% upon continuous use [23]. Additionally, due to low resource constraints in LMICs, there is an imperative need for applications powered by predictive analytics methods to address issues of low access to healthcare facilities
A wider academic view is, however the need of the hour to bring to the fore a better perspective of how new technologies perform or are accepted in radically different cultural contexts from the oft-tried and tested spaces of the American or European environments and encompass the sociological milieu of gender, ethnicity, and other factors that influence the adoption of novel ways and means of addressing global health, especially among healthcare providers, facilitators, and their local and global partners. The field of AI thus entails increased studies to enhance the body of literature further and necessitates independent studies to address region-specific issues with global impact.
5 Conclusions
Predictive analytics-based models can predict the early manifestation of Neonatal Sepsis. They can be used as a rapid detection tool for the early onset and late onset of Neonatal Sepsis. These methods can be valuable tools for clinicians to aid in effective decision-making and improved healthcare management of neonates, particularly in low-resource settings. Most of the prediction models were developed on clinical data sets from countries in North America, Europe, or Asian countries such as China, and their applicability to low-resource regions has seen subpar success rates. Therefore, it is imperative to develop a prediction model with localized data sets and validate their adoption with healthcare practitioners.
Data availability
There is no data used in this paper.
Abbreviations
- AI:
-
Artificial intelligence
- AB:
-
Adaptive boosting
- AMR:
-
Anti-microbial resistance
- ANN:
-
Artificial neural network
- AODE:
-
Averaged one dependence estimator
- AUC:
-
Area under curve
- AUROC:
-
Area under receiver operating curve
- BT:
-
Bagged trees
- CART:
-
Classification and regression tree
- CBC:
-
Complete blood count
- CDSS:
-
Clinical decision support system
- CI:
-
Confidence interval
- CNN:
-
Convolutional neural network
- CRP:
-
C-Reactive protein
- CSF:
-
Cerebro spinal fluid
- DL:
-
Deep learning
- EHR:
-
Electronic health record
- EONS:
-
Early onset of neonatal sepsis
- EOS:
-
Early onset of sepsis
- GA:
-
Gestational age
- GAM:
-
Generalized additive model
- GB:
-
Gradient boosting
- GBS:
-
Group B strep (streptococcus)
- GP:
-
Gaussian process
- HIV:
-
Human immunodeficiency virus
- HR:
-
Heart rate
- HRV:
-
Heart rate variability
- ICU:
-
Intensive care unit
- KNN:
-
K nearest neighbor
- LBR:
-
Lazy bayesian rules
- LMIC:
-
Low and middle income countries
- LOCF:
-
Last observation carried forward
- LOS:
-
Late onset of sepsis
- LONS:
-
Late onset of neonatal sepsis
- LR:
-
Logistic regression
- LSTM:
-
Long short term memory
- ML:
-
Machine learning
- NB:
-
Naive bayes classifier
- NICU:
-
Neonatal intensive care unit
- NLR:
-
Meta-analysis
- PPV:
-
Positive prediction value
- PRISMA:
-
Preferred reporting items for systematic reviews and meta analysis
- PROM:
-
Premature rupture of membrane
- RF:
-
Random forest
- RR:
-
Respiratory;Rate
- RNN:
-
Recurrent neural network
- ROC:
-
Receiver operating characteristic curve
- SDG:
-
Sustainable development goal
- SVM:
-
Support vector machine
- TAN:
-
Tree augmented naive bayes
- USA:
-
United States of America
- WBC:
-
White blood count
- WHO:
-
World Health Organization
- XGB:
-
EXtreme gradient boosting
References
Waters D, Jawad I, Ahmad A, Luksic I, Nair H, Zgaga L, Theodoratou E, Rudan I, Zaidi A, Campbell H. Aetiology of community-acquired neonatal sepsis in low and middle income countries. J Glob Health. 2011;1(2):154–70231981163484776.
NHM: Child Health
Roy M, Gupta R, Sehgal R. Neonatal transport in India: from public health perspective. Med J Dr D Y Patil Univ. 2016;9(5):566. https://doi.org/10.4103/0975-2870.192156.
Sankar M, Neogi S, Sharma J, Chauhan M, Srivastava R, Prabhakar P, Khera A, Kumar R, Zodpey S, Paul V. State of newborn health in India. J Perinatol. 2016;36(3):3–8.
Laxminarayan R, Bhutta ZA. Antimicrobial resistance—a threat to neonate survival. Lancet Glob Health. 2016;4(10):676–7. https://doi.org/10.1016/s2214-109x(16)30221-2.
Panigrahi P, Chandel DS, Hansen NI, Sharma N, Kandefer S, Parida S, Satpathy R, Pradhan L, Mohapatra A, Mohapatra SS, Misra PR, Banaji N, Johnson JA, Morris JG, Gewolb IH, Chaudhry R. Neonatal sepsis in rural India: timing, microbiology and antibiotic resistance in a population-based prospective study in the community setting. J Perinatol. 2017;37(8):911–21. https://doi.org/10.1038/jp.2017.67.
Mfateneza E, Rutayisire PC, Biracyaza E, Musafiri S, Mpabuka WG. Application of machine learning methods for predicting infant mortality in rwanda: analysis of rwanda demographic health survey 2014–15 dataset. BMC Pregnancy Childbirth. 2022. https://doi.org/10.1186/s12884-022-04699-8.
Singh M, Narang A, Bhakoo ON. Predictive perinatal score in the diagnosis of neonatal sepsis. J Trop Pediatr. 1994;40(6):365–8. https://doi.org/10.1093/tropej/40.6.365.
Gerdes JS, Polin RA. Sepsis screen in neonates with evaluation of plasma fibronectin. Pediatr Infect Dis J. 1987;6(5):443–6. https://doi.org/10.1097/00006454-198705000-00005.
Zaidi AKM, Thaver D, Ali SA, Khan TA. Pathogens associated with sepsis in newborns and young infants in developing countries. Pediatr Infect Dis J. 2009;28(1):10–8. https://doi.org/10.1097/inf.0b013e3181958769.
Sahu P, Raj Stanly EA, Simon Lewis LE, Prabhu K, Rao M, Kunhikatta V. Prediction modelling in the early detection of neonatal sepsis. World J Pediatr. 2022;18(3):160–75. https://doi.org/10.1007/s12519-021-00505-1.
Song W, Jung SY, Baek H, Choi CW, Jung YH, Yoo S. A predictive model based on machine learning for the early detection of late-onset neonatal sepsis: development and observational study. JMIR Med Inform. 2020;8(7):15965. https://doi.org/10.2196/15965.
Schelonka RL, Chai MK, Yoder BA, Hensley D, Brockett RM, Ascher DP. Volume of blood required to detect common neonatal pathogens. J Pediatr. 1996;129(2):275–8. https://doi.org/10.1016/s0022-3476(96)70254-8.
Hammerschlag MR, Klein JO, Herschel M, Chen FCJ, Fermin R. Patterns of use of antibiotics in two newborn nurseries. N Engl J Med. 1977;296(22):1268–9. https://doi.org/10.1056/nejm197706022962206.
Masino AJ, Harris MC, Forsyth D, Ostapenko S, Srinivasan L, Bonafide CP, Balamuth F, Schmatz M, Grundmeier RW. Machine learning models for early sepsis recognition in the neonatal intensive care unit using readily available electronic health record data. PLoS One. 2019;14(2):0212665. https://doi.org/10.1371/journal.pone.0212665.
Mani S, Ozdas A, Aliferis C, Varol HA, Chen Q, Carnevale R, Chen Y, Romano-Keeler J, Nian H, Weitkamp J-H. Medical decision support using machine learning for early detection of late-onset neonatal sepsis. J Am Med Inform Assoc. 2014;21(2):326–36. https://doi.org/10.1136/amiajnl-2013-001854.
Neal PR, Kleiman MB, Reynolds JK, Allen SD, Lemons JA, Yu PL. Volume of blood submitted for culture from neonates. J Clin Microbiol. 1986;24(3):353–6. https://doi.org/10.1128/jcm.24.3.353-356.1986.
Ting JY, Synnes A, Roberts A, Deshpandey A, Dow K, Yoon EW, Lee K-S, Dobson S, Lee SK, Shah PS. Association between antibiotic use and neonatal mortality and morbidities in very low-birth-weight infants without culture-proven sepsis or necrotizing enterocolitis. JAMA Pediatr. 2016;170(12):1181. https://doi.org/10.1001/jamapediatrics.2016.2132.
Laxminarayan R, Duse A, Wattal C, Zaidi AKM, Wertheim HFL, Sumpradit N, Vlieghe E, Hara GL, Gould IM, Goossens H, Greko C, So AD, Bigdeli M, Tomson G, Woodhouse W, Ombaka E, Peralta AQ, Qamar FN, Mir F, Kariuki S, Bhutta ZA, Coates A, Bergstrom R, Wright GD, Brown ED, Cars O. Antibiotic resistance—the need for global solutions. Lancet Infect Dis. 2013;13(12):1057–98. https://doi.org/10.1016/s1473-3099(13)70318-9.
Kraker MEA, Stewardson AJ, Harbarth S. Will 10 million people die a year due to antimicrobial resistance by 2050? PLoS Med. 2016;13(11):1002184. https://doi.org/10.1371/journal.pmed.1002184.
Murray CJL, Ikuta KS, Sharara F, Swetschinski L, Robles Aguilar G, Gray A, Han C, Bisignano C, Rao P, Wool E, Johnson SC, Browne AJ, Chipeta MG, Fell F, Hackett S, Haines-Woodhouse G, Kashef Hamadani BH, Kumaran EAP, McManigal B, Achalapong S, Agarwal R, Akech S, Albertson S, Amuasi J, Andrews J, Aravkin A, Ashley E, Babin F-X, Bailey F, Baker S, Basnyat B, Bekker A, Bender R, Berkley JA, Bethou A, Bielicki J, Boonkasidecha S, Bukosia J, Carvalheiro C, Castañeda-Orjuela C, Chansamouth V, Chaurasia S, Chiurchiù S, Chowdhury F, Clotaire Donatien R, Cook AJ, Cooper B, Cressey TR, Criollo-Mora E, Cunningham M, Darboe S, Day NPJ, De Luca M, Dokova K, Dramowski A, Dunachie SJ, Duong Bich T, Eckmanns T, Eibach D, Emami A, Feasey N, Fisher-Pearson N, Forrest K, Garcia C, Garrett D, Gastmeier P, Giref AZ, Greer RC, Gupta V, Haller S, Haselbeck A, Hay SI, Holm M, Hopkins S, Hsia Y, Iregbu KC, Jacobs J, Jarovsky D, Javanmardi F, Jenney AWJ, Khorana M, Khusuwan S, Kissoon N, Kobeissi E, Kostyanev T, Krapp F, Krumkamp R, Kumar A, Kyu HH, Lim C, Lim K, Limmathurotsakul D, Loftus MJ, Lunn M, Ma J, Manoharan A, Marks F, May J, Mayxay M, Mturi N, Munera-Huertas T, Musicha P, Musila LA, Mussi-Pinhata MM, Naidu RN, Nakamura T, Nanavati R, Nangia S, Newton P, Ngoun C, Novotney A, Nwakanma D, Obiero CW, Ochoa TJ, Olivas-Martinez A, Olliaro P, Ooko E, Ortiz-Brizuela E, Ounchanum P, Pak GD, Paredes JL, Peleg AY, Perrone C, Phe T, Phommasone K, Plakkal N, Ponce-de-Leon A, Raad M, Ramdin T, Rattanavong S, Riddell A, Roberts T, Robotham JV, Roca A, Rosenthal VD, Rudd KE, Russell N, Sader HS, Saengchan W, Schnall J, Scott JAG, Seekaew S, Sharland M, Shivamallappa M, Sifuentes-Osornio J, Simpson AJ, Steenkeste N, Stewardson AJ, Stoeva T, Tasak N, Thaiprakong A, Thwaites G, Tigoi C, Turner C, Turner P, Doorn HR, Velaphi S, Vongpradith A, Vongsouvath M, Vu H, Walsh T, Walson JL, Waner S, Wangrangsimakul T, Wannapinij P, Wozniak T, Young Sharma TEMW, Yu KC, Zheng P, Sartorius B, Lopez AD, Stergachis A, Moore C, Dolecek C, Naghavi M. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. The Lancet 2022;399(10325), 629–655. https://doi.org/10.1016/s0140-6736(21)02724-0
Philip AG. Decreased use of antibiotics using a neonatal sepsis screening technique. J Pediatr. 1981;98(5):795–9.
Chaurasia S, Sivanandan S, Agarwal R, Ellis S, Sharland M, Sankar MJ. Neonatal sepsis in south Asia: huge burden and spiralling antimicrobial resistance. BMJ. 2019. https://doi.org/10.1136/bmj.k5314.
López-Martínez F, Núñez-Valdez ER, Lorduy Gomez J, García-Díaz V. A neural network approach to predict early neonatal sepsis. Comput Electric Eng. 2019;76:379–88. https://doi.org/10.1016/j.compeleceng.2019.04.015.
Lin S. A clinician’s guide to artificial intelligence (ai): Why and how primary care should lead the health care AI revolution. J Am Board Family Med. 2022;35(1):175–84. https://doi.org/10.3122/jabfm.2022.01.210226.
BMJ: Artificial Intelligence and covid-19
Yu M, Tang A, Brown K, Bouchakri R, St-Onge P, Wu S, Reeder J, Mullie L, Chassé M. Integrating artificial intelligence in bedside care for Covid-19 and future pandemics. BMJ. 2021. https://doi.org/10.1136/bmj-2021-068197.
Laï M-C, Brian M, Mamzer M-F. Perceptions of artificial intelligence in healthcare: findings from a qualitative survey study among actors in France. J Transl Med. 2020. https://doi.org/10.1186/s12967-019-02204-y.
BMJ: Get ready for AI in pandemic response and healthcare
Ganeshkumar P, Gopalakrishnan S. Systematic reviews and meta-analysis: understanding the best evidence in primary healthcare. J Family Med Primary Care. 2013;2(1):9. https://doi.org/10.4103/2249-4863.109934.
Liberati A, Altman DG, Tetzlaff J, Mulrow C, Gotzsche PC, Ioannidis JPA, Clarke M, Devereaux PJ, Kleijnen J, Moher D. The prisma statement for reporting systematic reviews and meta-analyses of studies that evaluate healthcare interventions: explanation and elaboration. BMJ. 2009;339(jul 211):2700–2700. https://doi.org/10.1136/bmj.b2700.
Tranfield D, Denyer D, Smart P. Towards a methodology for developing evidence-informed management knowledge by means of systematic review. Br J Manag. 2003;14(3):207–22. https://doi.org/10.1111/1467-8551.00375.
Overall JE, Tonidandel S, Starbuck RR. Last-observation-carried-forward (locf) and tests for difference in mean rates of change in controlled repeated measurements designs with dropouts. Soc Sci Res. 2009;38(2):492–503.
Fawcett T. An introduction to roc analysis. Pattern Recogn Lett. 2006;27(8):861–74.
Guyon I, Weston J, Barnhill S, Vapnik V. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422.
Thakur J, Pahuja SK, Pahuja R. Neonatal sepsis prediction model for resource-poor developing countries. In: 2018 2nd International Conference on Electronics, Materials Engineering & Nano-Technology (IEMENTech). IEEE, 2018. https://doi.org/10.1109/iementech.2018.8465268
Gomez R, Garcia N, Collantes G, Ponce F, Redon P. Development of a non-invasive procedure to early detect neonatal sepsis using hrv monitoring and machine learning algorithms. In: 2019 IEEE 32nd International Symposium on Computer-Based Medical Systems (CBMS). IEEE, 2019. https://doi.org/10.1109/cbms.2019.00037
Hu Y, Lee VCS, Tan K. An application of convolutional neural networks for the early detection of late-onset neonatal sepsis. In: 2019 International Joint Conference on Neural Networks (IJCNN). IEEE, 2019. https://doi.org/10.1109/ijcnn.2019.8851683
Thakur J, Pahuja S, Pahuja R. Non-invasive prediction model for developing countries to predict sepsis in neonates. Biomed Eng. 2019;31(01):1950001.
Goldberg O, Amitai N, Chodick G, Bromiker R, Scheuerman O, Ben-Zvi H, Klinger G. Can we improve early identification of neonatal late-onset sepsis? A validated prediction model. J Perinatol. 2020;40(9):1315–22. https://doi.org/10.1038/s41372-020-0649-6.
Alvi RH, Habibur Rahman M, Al Shaeed Khan A, Rahman RM. Predicting early neonatal sepsis using neural networks and other classifiers. In: 2020 IEEE 10th International Conference on Intelligent Systems (IS). IEEE, (2020). https://doi.org/10.1109/is48319.2020.9199937
Helguera-Repetto AC, Soto-Ramírez MD, Villavicencio-Carrisoza O, Yong-Mendoza S, Yong-Mendoza A, León-Juárez M, González-y-Merchand JA, Zaga-Clavellina V, Irles C. Neonatal sepsis diagnosis decision-making based on artificial neural networks. Front Pediatr. 2020. https://doi.org/10.3389/fped.2020.00525.
Alvi RH, Rahman MH, Khan AAS, Rahman RM. Deep learning approach on tabular data to predict early-onset neonatal sepsis. J Inf Telecommun. 2020;5(2):226–46. https://doi.org/10.1080/24751839.2020.1843121.
Honoré A, Forsberg D, Adolphson K, Chatterjee S, Jost K, Herlenius E. Vital sign-based detection of sepsis in neonates using machine learning. Acta Paediatr. 2023;112(4):686–96.
Berg MAM, Medina OOAG, Loohuis IIP, Flier MM, Dudink JJ, Benders MMJNL, Bartels RRT, Vijlbrief DDC. Development and clinical impact assessment of a machine-learning model for early prediction of late-onset sepsis. Computers in Biology and Medicine 2023;163, 107156 https://doi.org/10.1016/j.compbiomed.2023.107156
Robi YG, Sitote TM. Neonatal disease prediction using machine learning techniques. J Healthcare Eng. 2023;2023:1–16. https://doi.org/10.1155/2023/3567194.
Kallonen A, Juutinen M, Värri A, Carrault G, Pladys P, Beuchée A. Early detection of late-onset neonatal sepsis from noninvasive biosignals using deep learning: a multicenter prospective development and validation study. Int J Med Inform. 2024;184: 105366. https://doi.org/10.1016/j.ijmedinf.2024.105366.
Selimovic A, Skokic F, Bazardzanovic M, Selimovic Z, et al. The predictive score for early-onset neonatal sepsis. Turk J Pediatr. 2010;52(2):139–44.
Okascharoen C, Sirinavin S, Thakkinstian A, Kitayaporn D, Supapanachart S. A bedside prediction-scoring model for late-onset neonatal sepsis. J Perinatol. 2005;25(12):778–83.
Kuzniewicz MW, Mukhopadhyay S, Li S, Walsh EM, Puopolo KM. Time to positivity of neonatal blood cultures for early-onset sepsis. Pediatr Infect Dis J. 2020;39(7):634–40.
Huang Y, Yu X, Li W, Li Y, Yang J, Hu Z, Wang Y, Chen P, Li W, Chen Y. Development and validation of a nomogram for predicting late-onset sepsis in preterm infants on the basis of thyroid function and other risk factors: Mixed retrospective and prospective cohort study. J Adv Res. 2020;24:43–51.
Divall P, Camosso-Stefinovic J, Baker R. The use of personal digital assistants in clinical decision making by health care professionals: a systematic review. Health Inform J. 2013;19(1):16–28. https://doi.org/10.1177/1460458212446761.
Acknowledgements
This study received no funding from any international or domestic agencies. Authors confirm that there is no financial or non-financial benefits have been received or will be received from any party related directly or indirectly to the subject of this article. None of the authors were involved in the journals review of, or decisions related to this manuscript. Ethics approval was not required for systematic review.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License, which permits any non-commercial use, sharing, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if you modified the licensed material. You do not have permission under this licence to share adapted material derived from this article or parts of it. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by-nc-nd/4.0/.
About this article
Cite this article
Narasimha Rao, K.V.K.L., Dadabada, P.K. & Jaipuria, S. A systematic literature review of predictive analytics methods for early diagnosis of neonatal sepsis. Discov Public Health 21, 96 (2024). https://doi.org/10.1186/s12982-024-00219-5
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12982-024-00219-5